Abstract
The high regenerative capacity of adult skeletal muscle relies on a self-renewing depot of adult stem cells, termed muscle satellite cells (MSCs). Androgens, known mediators of overall body composition and specifically skeletal muscle mass, have been shown to regulate MSCs. The possible overlapping function of androgen regulation of muscle growth and MSC activation has not been carefully investigated with regards to muscle regeneration.Therefore, the aim of this study was to examine coinciding androgen-mediated genetic changes in an in vitro MSC model and clinically relevant in vivo models. A gene signature was established via microarray analysis for androgen-mediated MSC engagement and highlighted several markers including follistatin (FST), IGF-1, C-X-C chemokine receptor 4 (CXCR4), hepatocyte growth factor (HGF) and glucocorticoid receptor (GR). In an in vivo muscle atrophy model, androgen re-supplementation significantly increased muscle size and expression of IGF-1, FST, and HGF, while significantly decreasing expression of GR. Biphasic gene expression profiles over the 7-day re-supplementation period identifed temporal androgen regulation of molecular targets involved in satellite cell engagement into myogenesis. In a muscle injury model, removal of androgens resulted in delayed muscle recovery and regeneration. Modifications in the androgen signaling gene signature, along with reduced Pax7 and MyoD expression, suggested that limited MSC activation and increased inflammation contributed to the delayed regeneration. However, enhanced MSC activation in the androgen-deplete mouse injury model was driven by an androgen receptor (AR) agonist. These results provide novel in vitro and in vivo evidence describing molecular targets of androgen signaling, while also increasing support for translational use of AR agonists in skeletal muscle recovery and regeneration.
Key Words: Androgen, Skeletal Muscle, Regeneration, Satellite Cells, Levator Ani
Introduction
Androgens belong to a class of steroid hormones which serve as ligands for the androgen receptor (AR). Ligand-bound AR modulates target gene regulation via recognition of and binding to androgen response elements (AREs). Androgens and the AR play an essential role in sexual development [Hughes et al., 2001], and can influence the regulation of overall body composition [Mooradian et al., 1987]. As an established anabolic treatment, androgen supplementation has been shown to positively influence bone [Kawano et al., 2003; Turner et al., 1990; Vanderschueren et al., 1993; Venken et al., 2006] and skeletal muscle [Celotti and Negri Cesi, 1992; Herbst and Bhasin, 2004; Tingus and Carlsen, 1993; Yin et al., 2003]. However, therapeutic use of androgens has been limited, due to adverse side effects including significant cardiovascular events [Basaria et al., 2010; Finkle et al., 2014; Vigen et al., 2013] and potential risk for prostate hyperplasia [Acosta et al., 2004]. The recent development of androgen-like molecules (e.g., non-steroidal AR modulators) aims to retain the beneficial influence of androgen on skeletal muscle, while circumventing undesirable cardiovascular and prostate-related side-effects. These modulators have been implicated as possible therapies in instances of skeletal muscle wasting, degeneration, and/or injury, where skeletal muscle mass is in a state of dysregulation [Corona et al., 2011; Li et al., 2007; Manfredi et al., 2007; O'Connell and Wu, 2014; Zhang and Sui, 2013]. While AR modulator efficacy has been widely studied, investigations of the molecular targets downstream of AR-mediated muscle mass regulation remain limited.
Regulation of skeletal muscle mass is a balance between protein synthesis and degradation [Saini et al., 2006]. Catabolic stimuli (e.g., mechanical unloading, starvation, disease, hormone deprivation therapy) induce muscle atrophy via protein degradation and autophagy pathways. Anabolic stimuli (e.g., mechanical loading, nutrient supplementation, hormone therapy) initiate muscle hypertrophy by increasing overall net protein balance. The remarkable regenerative capacity of skeletal muscle following injury is a clear example of coordinated systems involving both catabolic and anabolic pathways at work. Following an insult (injury) to muscle, a distinct process of tissue recovery commences involving degeneration, inflammation, remodeling and fibrosis [Tidball, 2005, 2011; Turner and Badylak, 2012]. A pool of muscle-specific stem cells, termed satellite cells, is activated early and rapidly following muscle injury to directly aid in the recovery process. Muscle satellite cells (MSCs), originally defined on the basis of their location (situated between the sarcolemma and basal lamina of myofibers), are responsible for early growth during development and subsequently act as committed stem cells for adult skeletal muscle undergoing recovery from injury and trauma [Mauro, 1961]. Adult MSCs serve as a self-renewing depot of cells aiding in the repair and growth of skeletal muscle [Campion, 1984; Grounds et al., 2002]. MSCs are typically single, unfused and undifferentiated cells that have withdrawn from the cell cycle and are considered to be quiescent [Allbrook et al., 1971; Schultz et al., 1978]. These cells uniformly express the transcription factor Pax7, and can be stimulated to enter the cell cycle through a range of catabolic and anabolic stimuli resulting in MSC activation. The activation event leads to MSC proliferation and eventual differentiation into myotubes, which fuse to existing myofibers, or each other, directly generating new muscle fibers. In the process of skeletal muscle regeneration, Pax7+ MSCs have been confirmed to be absolutely essential for normal regenerative myogenesis [Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011].
While the molecular mechanisms by which androgen promotes skeletal muscle regeneration are not completely understood, there is evidence that androgen-induced activation of MSCs is a critical mediator of this process [Chen et al., 2005; Joubert and Tobin, 1995; Nnodim, 2001]. The current manuscript identifies key molecular targets that are regulated by androgen using a novel levator ani satellite cell (LASC) in vitro system. In order to identify AR-specific changes in gene expression mediated in the LASC in vitro culture, a non-steroidal AR agonist (NARA) was utilized. This in vitro system identified notable target genes for AR that included HGF, IGF-1, CXCR4, FST, and GR. The changes observed in vitro were then validated in the levator ani (LA) perineal muscle tissue, which is highly responsive to androgen levels. Finally, these changes in gene expression were queried in a muscle injury model wherein endogenous satellite cells mediate the repair and recovery, and were found to provide strong evidence suggesting androgen-mediated effects on satellite cell activity. Collectively, these results provide novel in vitro and in vivo evidence describing the effect of androgen on skeletal muscle, while at the same time enhancing support for use of AR agonists in skeletal muscle diseases where atrophy and injury are present.
Materials and Methods
Materials
Reagents and primers used for quantitative real-time PCR were from Life Technologies (Grand Island, NY). Primer catalog numbers for genes can be found in Supplementary Material Table 1. The L6 rat skeletal muscle line (CRL-1458) was purchased from ATCC (Manassas, VA). For LASC (rat cell line) immunostaining, the anti-Pax7 antibody (MAB1675; R&D systems) was conjugated together with FITC (FITC Conjugation kit; Abcam ab102884). For mouse muscle IHC, the anti-Pax7 primary antibody was purchased from Abcam (Ab34360). The non-steroidal AR agonist (NARA) was synthesized at Eli Lilly & Co (Indianapolis, IN). All other materials were from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Animals
Animal experiments were conducted in normal male ICR (CD-1) mice (20–30 g; Harlan, Indianapolis, IN), or male Wistar rats (3-4 months old; Harlan, Indianapolis, IN), performed with the approval of Institutional Animal Care and Use Committee and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animals were housed in a room with controlled temperature (22 ± 2°C), and a 12:12 h light-dark cycle with ad libitum access to food (TD 5001 with 0.95% calcium and 0.67% phosphorus; Teklad, Madison, WI) and water. For gonadectomy studies, animals (mice or rats) were divided into sham operated and un-castrated (Sham/Intact) or gonadectomized (GDX) groups. Animals were gonadectomized under anesthesia and the testes were bilaterally removed by scrotal incision. Animals were allowed to recover from operation and were put on their respective study at 6 weeks post surgery. Body composition was measured using Quantitative Nuclear Magnetic Resonance analysis (ECHO MRI, 3-1 Composition Analyzer; (Echo Medical Systems, Houston, TX).
NARA-treated animals
Rats: Six weeks post GDX surgery, rats were grouped into separate treatment cohorts; Sham surgery (Veh), GDX (Veh), GDX + Androgen (NARA). The GDX+NARA animals were dosed daily with subcutaneous injections of NARA (1 mg/kg). Animal groups (n=7-8) were sacrificed at specified time points (3, 24, 48, 72 h, and 7 d) post initiation of NARA treatment. The LA muscles were isolated, weighed and frozen for future analysis.
Mice: Six weeks post GDX surgery, mice were grouped into separate treatment cohorts; Intact-Sham (non-gonadectomized, non-injury) with vehicle or NARA treatment, Intact-CTX (cardiotoxin injury) with vehicle or NARA treatment, GDX-Sham (gonadectomized, non-injury) with vehicle or NARA treatment, GDX-CTX with vehicle or NARA treatment. The NARA animals were dosed daily for 7 days with subcutaneous injections of NARA (1 mg/kg) prior to muscle injury. Animals continued daily vehicle or NARA treatment following injury and were sacrificed 4 days post injury in order to harvest gastrocnemius muscles.
Cardiotoxin injury
Six weeks post GDX surgery, mice were subjected to muscle injury by injecting 100 μL of 10 μM cardiotoxin (CTX; from naja nigricollis; Calbiochem, La Jolla, CA) solution (diluted in physiologic saline) into the right gastrocnemius muscle with a three-point injection technique as previously described [Yaden et al., 2014a; Yaden et al., 2014b]. The injured gastrocnemius muscles were subsequently harvested at specified time points for muscle mass, immunohistochemical and gene expression analysis.
Generation of levator ani satellite cell (LASC) line
The levator ani (LA) muscle tissue was isolated from rats (age 6 weeks) and minced with sterile scissors. The muscle homogenate was washed 3 times with Hank’s Balanced Salt Solution (HBSS), and then digested by 100 U/mL collagenase (Invitrogen cat# 17101—015) at 37oC and 5% CO2 for 18 h. The LA cell suspension was filtered through a sterile nylon mesh to separate the dispersed cells and tissue fragments. The suspension was washed 3 times by centrifugation in HBSS with the pellet being resuspended in DMEM/F12 (3:1) with 10% FBS culture medium. The LA cells were immortalized as previously described [Kogan et al., 2006; Stadler et al., 2011]. In brief, cells were transfected using the supernatant from Phoenix/hTERT (amphotropic packaging cell line transfected with human telomerase gene) with 10 µg/mL polybrene at 37oC and 5% CO2 for 3 h. Fresh culture medium was then placed on the cells, which were allowed to remain in culture for 24 h. The LA cells were then screened with 10 µg/mL blasticidin for 10 to 14 days. Single clones were isolated and remained in culture for 24 h. Individual clones were verified for expression of transcripts reflecting muscle satellite cells and AR expression. A single clone was selected that displayed satellite cell markers and was termed the levator ani satellite cell (LASC). A subset of cells from this clone were fixed (in 3.7% formaldehyde), permeabilized (0.1% Triton X-100). Immunofluorescence experiments were performed with the Pax7 primary antibody (10 µg/mL; R&D Systems) conjugated with a fluorescent label via the Alexa Fluor 488 protein labeling kit (Invitrogen). Nuclei were counterstained with DAPI and cells were visualized using confocal microscopy (40X magnification). All experiments were performed on LASCs with a passage number of 20 or less.
Electrical cell impedance sensing (ECIS) assay
Cell impedance measurements were captured using the xCELLigence RTCA System (Roche Applied Science) instrument using the E-Plates 96-well device as previously described [Rakhilin et al., 2011]. LASCs were seeded on a 96-well plate at a density of 5,000 cells per well (in 200 μL). Cells were allowed to attach for 24 h. The respective treatment (NARA or IGF-1) was applied in a volume of 30 μL without a media change to avoid sharp transient changes in the impedance readout. The xCELLigence RTCA System software (Roche Applied Science) measured the cell index value (which corresponds to electrical impedance) after 48 h of treatment. The data were normalized to the impedance value for each well prior to initiation of respective treatment.
Microarray analysis
LASCs were treated with vehicle (DMSO; 0.2%) or NARA (10 nM) for 4 or 48 h. RNA was extracted using the PerfectPure RNA isolation kit (5 Prime, Gaithersburg, MD). Five μg of total RNA from each sample was labeled and hybridized to Affymetrix Rat Genome 230_2.0 microarrays according to Affymetrix protocols. Each microarray was washed and stained using an Affymetrix Fluidics Station 400 and scanned in an Affymetrix confocal GeneArray scanner. Affymetrix MAS5.0 software was used to scale data to a target intensity of 1500 and calculate transcript abundance (signal). Microarray signal intensities were generated by Affymetrix Expression Console using the MAS5 algorithm with targeted trimmed mean signal set to 1500. The gene chips were checked for quality control by their detection calls, chip pairwise correlation, and principal components analysis (PCA). Three chips were identified as outliers and excluded from analysis for obvious low correlation with and deviating on PCA plots from the rest of the chips. ANOVA t-test was carried out to test the statistical significance between groups of interest for each probe set using statistical software SAS (SAS Institute). The resultant p-values were then adjusted for multiple testing using the false discovery rate (FDR) approach by Benjamini and Hochberg [Benjamini, 1995]. The microarray data can be accessed at the GEO repository, accession number GSE63489. Qiagen Ingenuity Pathway Analysis (IPA) software was applied to further examine the data using an FDR cutoff of 0.2 and fold change cutoff of 1.4.
Gene expression analysis
RNA was extracted from cells using the PerfectPure RNA isolation kit and from skeletal muscles using TRIzol reagent (Life Technologies, Grand Island, NY). Total RNAs were reverse transcribed using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). All cDNAs were assayed for genes of interest using TaqMan Gene Expression Analysis (Applied Biosystems, Carlsbad, CA) and the Assay-On-Demand primer/probe set (Life Technologies). Gene of interest mRNA levels were quantitated by determining the cycle number at which amplification detection threshold was achieved. Real-time polymerase chain reactions (rt-PCR) were performed in 20 µl reactions according to manufacturer guidelines. Samples were subjected to quantitative rt-PCR using the ABI 7900HT real-time PCR system (Life Technologies). Gene of interest values were normalized to a housekeeping gene prior to interpretation of results.
Luciferase reporter assay measuring promoter induction
LASCs were seeded (10,000 cells/well) and allowed to attach for 24 h, then transfected with a luciferase reporter plasmid (PGL3 Vector: Promega Cat. # E1761) containing the appropriate promoter construct. The FST luciferase construct contained the ~1.0 kb 5’ upstream region of the FST promoter (reverse primer sequence, 5’-CCCTCGAGGGGCTGCGGTCTTCCATGAATT-3’; forward primer sequence 5’-GGGTACCCCTGCAGGCTGTGGGGGAGTGG-3’). The HGF luciferase construct contained the ~1.4 kb 5’ upstream region of the HGF promoter (reverse primer sequence, 5’-GGCTAGCCGATGCCGGGCTGAAAGAA-3’; forward primer sequence 5’-GGGTACCCTGCCTTTGCTGGTGGAGGTG-3’). Following 36 h, cells were treated with 10 nM of Metribolone (R1881) or 0.1% DMSO (Vehicle). Cells were subjected to lysis and luminescence was measured using a GeniosPRO instrument with substrate injection (Luciferase Reporter Gene Assay Kit; Roche, Basel, Switzerland). Values are representative of transfection experiments performed in triplicate. Relative luciferase units were measured. Putative androgen response elements (AREs) and corresponding predicted scores were determined by JASPAR database modeling [Mathelier et al., 2014].
Muscle histology
Muscle tissue samples were fixed in 10% neutral-buffered formalin, paraffin-embedded, and evaluated using H&E staining of gastrocnemius muscle sections using Aperio ImageScope software (Vista, CA). Images were acquired using digital slide scanning (ScanScope XT; Aperio). Immunofluorescence experiments were performed with anti-PAX7 (1.0 mg/mL: Abcam), followed by secondary anti-mouse Alexa Fluor488 Antibody (1:250). DAPI staining was performed using Fluoro-gel II with DAPI (Electron Microscopy Sciences, Hatfield, PA) during the coverslip process. All experiments were repeated a minimum of three times with at least five animals per group. All quantifications of Pax7 were determined using ImageScope software version 11.2.0.780 (Aperio).
Statistical analysis
Data were expressed as means ± SEM. Differences between two groups were determined by the Student t test. One-way ANOVA was used to evaluate the effects of multiple groups. A three-way ANOVA was used to assess data in Figure 5, main effects of androgen status (Intact vs GDX), injury status (sham vs. CTX), and treatment (vehicle vs. NARA), as well as interactions between androgen status, injury status, and treatment for all variables. Dunnett’s or Bonferonni’s post hoc t test was applied to determine the source of significant variance. A P value of ≤0.05 was considered statistically significant.
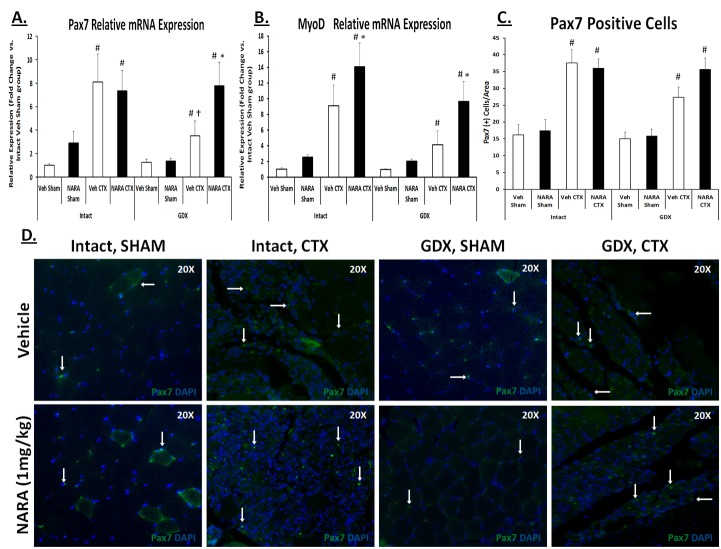
Figure 5. NARA treatment augments satellite cell activation in gonadectomized mouse model following muscle injury.
Gastrocnemius muscle from treated (NARA; 1 mg/kg/day) or untreated (Vehicle), intact or gonadectomized (GDX) mice were removed 4 days post sham or cardiotoxin (CTX) injection. A-B. Pax7 and MyoD relative mRNA expression levels. C-D. Pax7-positive cell quantifications and their representative images of immunofluorescence in gastrocnemius sections muscles 4 days after CTX injury, using an antibody specific for Pax7 (green) with the addition of DAPI (blue) staining for nuclei. Original magnification, x20. Data are expressed as means ± SEM of five mice per group. A three-way ANOVA was used to assess main effects of androgen status (Intact vs GDX), injury status (sham vs. CTX), and treatment (vehicle vs. NARA), as well as interactions between androgen status, injury status, and treatment for all variables; significance was set at P ≤0.05. *Significantly different from corresponding vehicle group. # Significantly different from corresponding sham group within androgen status model (Intact or GDX). † Significantly different between GDX and corresponding intact group.
Results
Rat levator ani (LA) muscle cells defined as novel satellite cell line distinct from mature myoblasts
Using a collagenase-based separation protocol (see Methods), a novel cell line was isolated from the LA muscle. These cells displayed morphology consistent with previously published work describing muscle satellite cells [Montarras et al., 2005; Motohashi et al., 2014]. The predominant phenotype consisted of small, single nucleated cells with an elongated shape expressing Pax7 (representative cell image Figure 1A). These cells were confirmed to be a novel line of muscle satellite cells, which were subsequently termed levator ani satellite cells (LASCs). RT-qPCR analysis indicated that LASCs expressed established satellite cell genes (Pax3, Pax7, Desmin, Myf5), while they lacked the expression of mature myoblast/muscle genes (myogenin, myoD and myostatin) compared to rat myoblast L6 cell line (Table 1). Collectively, these phenotypic and genotypic assessments provide a compelling view of the muscle satellite cell line.
Figure 1. LASC response to androgen receptor agonists.
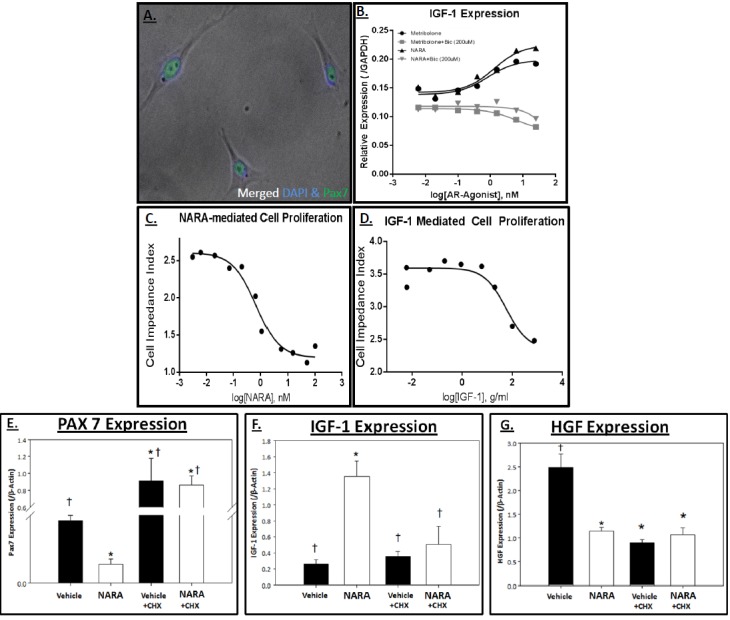
A. Representative images of levator ani satellite cell (LASC) line. B. LASCs were treated with increasing doses of Metribolone (Met; dark circles), Met + Bicalutamide (Bic; 200 μM; gray squares), NARA (black triangles), or NARA + Bic (200 μM; grey inverted triangles). IGF-1 expression (normalized to GAPDH) was used as an indicator of an androgen response. C-D. LASCs seeded in 96-well Acea plates for 24 h in appropriate media were then treated with increasing doses of C. NARA or D. IGF-1, and incubated 48 h. Cell proliferation was measured over a 48 h time period and the cell impedance index was plotted. E-G. LASCs were seeded in 6-well plates for 24 h. Cells were then incubated with or without cycloheximide (CHX; 10 μM) for 1 h followed by 48 h treatment with Vehicle or NARA (10 nM). Relative mRNA expression of E. Pax7, F. IGF-1, G. HGF (normalized to β-actin) was measured via real-time PCR. Data are means ± SEM. * p>0.05 vs Vehicle and † p<0.05 vs NARA, as determined by ANOVA and Dunnet’s post hoc analysis.
Table 1. Rat levator ani (LA) muscle cells defined as novel satellite cell line distinct from mature myoblasts.
| Genes | LASC (Relative Expression/GAPDH) | L6 (Relative Expression/GAPDH) |
|---|---|---|
| Pax3 | ++ (0.0557) | + (0.0146 |
| Pax7 | ++ (0.0085) | -- (Not Detected) |
| Myogenin | -- (Not Detected) | ++ (.3319) |
| MyoD | -- (Not Detected) | -- (Not Detected) |
| Desmin | ++ (0.0052) | + (0.0003) |
| Notch1 | + (0.0192) | + (0.0140) |
| Notch3 | + (0.0133) | ++ (0.0212) |
| HGF | + (0.223) | ++ (0.2734) |
| IGF1 | + (0.0262) | ++ (0.5441) |
| Myf5 | ++ (1.300) | + (0.0003) |
| CXCR4 | + (0.0275) | -- (Not Detected) |
| Follistatin | ++ (0.3881) | + (0.1096) |
| Myostatin | -- (Not Detected) | ++ (0.8634) |
| AR | ++ (0.3669) | -- (Not Detected) |
The Levator Ani Satellite cell (LASC) line was compared to the L6 myoblast cell line. Cells of each line were allowed to attach for 24 hours and maintained for 24 hours before being lysed for gene expression analysis. Relative mRNA expression (measured via real-time PCR) of a specific gene is represented by a plus sign (+), while no expression is indicated by minus signs (--). A 3-fold or greater gene expression difference between cell lines is indicated by double plus signs (++). Relative mRNA expression values (normalized to GAPDH) are displayed in parentheses.
LASCs display robust response to AR agonist
Steroidal AR agonists (e.g., testosterone, metribolone) often display adverse effects due to cross-reactivity with other steroid hormone receptors [Hamann et al., 1998]. Accordingly, exploration of nonsteroidal AR agonists has grown with the increasing interest in AR-mediated therapeutics. To confirm effectiveness between a steroidal AR-agonist (metribolone) versus a nonsteroidal AR-agonist (NARA) in the LASC line, we measured gene regulation of IGF-1, a well-established AR target gene. Both AR agonists displayed a dose-response relationship with comparable effectiveness at inducing IGF-1 expression (Figure 1B). Inability of the non-steroidal AR agonists to be aromatized is a distinguishing and attractive trait for this class of molecule. To investigate whether our NARA demonstrated AR specificity (suggesting a nonaromatizable compound) bicalutamide was utilized as an AR antagonist. Bicalutamide (200 μM) completely inhibited NARA-mediated upregulation of IGF-1 at all doses (Figure 1B), confirming AR specificity for the NARA.
Electric cell-substrate impedance sensing (ECIS) is commonly used to monitor changes in cell attachment, size, migration and/or proliferation. In order to further characterize the response of LASC to NARA, the ECIS assay was applied to measure LASC proliferation in response to increasing NARA doses. A surprising, yet consistent, decrease in LASC proliferation was observed with increasing doses of NARA, as reflected by the decreased cell impedance index (Figure 1C). Morphological changes were apparent as a result of androgen stimulation, suggesting these cells were being driven to differentiate. In order to determine if the activity could be explained by increased NARA-mediated IGF-1 signaling, the ECIS assay was applied to measure LASC proliferation in response to increasing IGF-1 doses (Figure 1D). Similar to NARA treatment, there was a dose-dependent decrease in the cell impendence index, consistent with a NARA-induced, IGF-1-mediated decrease in proliferation. Accordingly, the expression of IGF-1 may contribute in large part to the activation and myogenic differentiation of LASCs.
To confirm LASC myogenic differentiation capacity, LASCs were plated and treated with or without NARA in differentiation media (DM) for up to 96 h. Morphological and gene expression analysis (Supplementary Material Figure 1) was performed at 6, 24, 48, 72, and 96 h post DM introduction. LASCs displayed significantly decreased levels of the classic MSC marker, Pax7, over the differentiation time course. Concurrently, LASCs presented significantly increased levels of MyoD, MyoG and Myh2. Morphologically, myotubes were visible after 72 h in differentiation media (+NARA). The NARA treatment was observed to augment the differentiation process when defined by gene expression. In NARA-treated LASCs, Pax7 was significantly decreased by 24 h (vs. 72 for Veh), while MyoD and MyoG expression increased earlier in the differentiation time course, as well (vs. Veh). Together, differentiation of the novel LASC line into myotubes was established, with NARA treatment augmenting the maturation process of these satellite cells.
Cycloheximide co-treatment was used to determine if androgen-mediated gene regulation required protein synthesis. LASCs were treated with or without NARA (10 nM) in combination with or without cycloheximide (10 μM) for 48 h. Overall, cycloheximide (CHX) inhibited androgen-mediated gene expression. The decrease in Pax7 expression with NARA treatment alone confirmed the loss of satellite cell phenotype, as LASC proliferation decreased, signifying engagement in differentiation (Figure 1E). CHX did not alter IGF-1 expression in vehicle-treated LASCs, though complete inhibition of NARA-mediated IGF-1 expression was observed in CHX-treated LASCs. Of note, CHX by itself altered the regulation of PAX7 and HGF expression (vehicle-treated), suggesting that the stem cell-like feature of proliferating satellite cells, in part, required protein synthesis.
Microarray analysis of LASCs confirms androgen sensitivity and provides novel biomarkers
The LASC line proved to be a unique tool that displayed a robust response to androgen stimulation. To better understand the steps involved in active myogenesis engagement after cessation of cell proliferation, we subjected the NARA-treated (vs. vehicle; 4 or 48 h treatment) LASC mRNA to analysis with Affymetrix microarray chips. The ten genes displaying the greatest up- and down-regulation due to NARA treatment are listed in Table 2. Functional pathways associated with skeletal muscle growth are highlighted in Supplementary Material Table 2, where 48 h of NARA treatment (vs. vehicle) significantly regulated genes involved in pathways driving skeletal muscle cell proliferation (p<0.001; predicted inhibition), differentiation (p<0.001; predicted activation) and hypertrophy (p<0.001; predicted activation). To note, 4 h NARA treatment did not significantly alter these functional pathways, though did have significant effects on various other pathways (e.g., cell cycle genes upregulated). The microarray analysis confirmed activation and differentiation of LASCs within 48 h of NARA treatment, while providing a set of genes that were highly regulated; FST (+4.96 fold change), IGF1 (+4.71), NR3C1 (GR; -4.52) and CXCR4 (-9.28).
Table 2. Top 10 up- and down-regulated genes by androgen.
| 4 hr (vs. Vehicle) | 48 hr (vs. Vehicle) | |
|---|---|---|
| Up-regulated (Fold) | RASSF9 (4.84) | BMPER (12.33) |
| AFMID (4.49) | TFCP2L1 (10.42) | |
| RICTOR (4.09) | SV2B (10.31) | |
| SQSTN1 (2.92) | CRISPLD2 (9.95) | |
| NOG (2.81) | SVIL (9.66) | |
| ERBB4 (2.80) | CES1 (9.05) | |
| CEBPD (2.73) | TUBB4B (8.69) | |
| MAP4K4 (2.66) | FGL2 (8.43) | |
| PLEKHF2 (2.55) | ENPP1 (8.21) | |
| NF2 (2.51) | SULT1A1 (6.74) | |
| Down-Regulated (Fold) | NPPA (-5.72) | RGD1560633 (-11.18) |
| BMP15 (-4.58) | CXCR4 (-9.28) | |
| APOC2 (-3.47) | PTGER3 (-6.76) | |
| LPAR2 (-3.08) | TMEM26 (-5.81) | |
| NKX2-3 (-2.97) | KCNA6 (-5.58) | |
| OXT (-2.93) | CPXM2 (-5.57) | |
| NR1H2 (-2.87) | AP3D1 (-5.39) | |
| FBXO32 (-2.77) | GTPBP1 (-5.36) | |
| PTAFR (-2.64) | PTGS2 (-5.21) | |
| TAC1 (-2.61) | UPK2 (-4.86) |
LASCs were seeded, allowed to attach 24 hours, treated with vehicle (DMSO) or NARA (10 nM) and incubated for 4 or 48 hours. Isolated cell mRNA was reverse transcribed into cDNA, labeled, and analyzed using DNA microarray. Data analysis was performed using Qiagen IPA software with cutoffs set at an FDR ≥ 0.2 and fold change ≥ 1.4. Fold change in parentheses.
Liganded AR regulates FST, CXCR4 and GR expression
Microarray results provided a valuable signature for androgen-mediated gene regulation from which several unique genes were highlighted as possible markers and/or mediators of muscle regeneration. To confirm LASC microarray findings, expression of follistatin (FST), C-X-C chemokine receptor 4 (CXCR4) and glucocorticoid receptor (GR) was measured (via RT-qPCR) in LASCs (Figure 2). A dose-dependent increase in FST (logEC50 = 0.60 nM ± 0.17) expression, and dose-dependent decreases in CXCR4 (logEC50 = 0.35 nM ± 0.07) and GR (logEC50 = 0.31 nM ± 0.08) expression were observed. The striking response in the real time PCR results across a dose range of NARA treatment to androgen treatment reinforced the microarray findings, while at the same time providing a novel set of gene changes mediated by androgen.
Figure 2. Liganded androgen receptor regulates FST, GR, and CXCR4 expression.
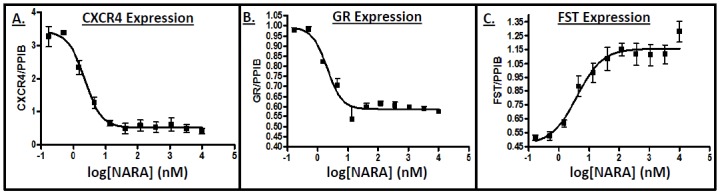
LASCs were seeded and allowed to attach 24 h, then treated with increasing doses of NARA and incubated 48 h. Relative mRNA expression of A. CXCR4, B. GR, and C. FST (normalized to PPIB; measured via real-time PCR) displayed a dose-dependent response. Data are means ± SEM.
In order to further investigate whether androgen directly regulated our target genes, we analyzed the promoter regions for putative androgen response elements (AREs) using the JASPAR transcription factor DNA-binding model [Mathelier et al., 2014]. In addition, in vitro promoter induction analysis was performed using a luciferase reporter system transfected into LASCs. The ~1.0 kb upstream region of the FST promoter was found to have three predicted AREs. Robust induction of the FST promoter by 10 nM metribolone was observed (Supplementary Material Figure 2A). Likewise, the ~1.4 kb upstream region of the HGF promoter was found to have predicted AREs. Induction of the HGF promoter by 10 nM metribolone was observed after 29 h of treatment (Supplementary Material Figure 2B). Vehicle (0.1% DMSO) treamtent did not result in significant induction of either promoter. These results show that the upstream promoter is sufficient to drive the induction seen with AR agonism, and suggests a direct regulation of HGF and FST by AR.
In vivo androgen-mediated gene regulation in LA muscle
The in vitro results from the LASC line provided a valuable gene signature of molecular targets. We aimed to pursue in vivo analysis that could confirm that the androgen-mediated MSC activation and accompanying gene expression changes measured in vitro were physiologically relevant. A clinically-relevant model of muscle atrophy was applied to study the effect of androgen on muscle recovery. Gonadectomized (GDX) male rats were utilized to overcome the variability of circulating endogenous androgens. Daily dosing with NARA (1 mg/kg) was then performed with animals being taken for analysis at 3, 24, 48, 72 h and 7 d. The GDX treatment resulted in significantly decreased total body and LA muscle weight versus sham as a result of the androgen depletion. Supplementing with NARA did not significantly increase body weight over the 7-day treatment period, however LA tissue weight did increase significantly by day 7 (Figure 3A&B).
Figure 3. In vivo androgen-mediated gene regulation in rat LA muscle.
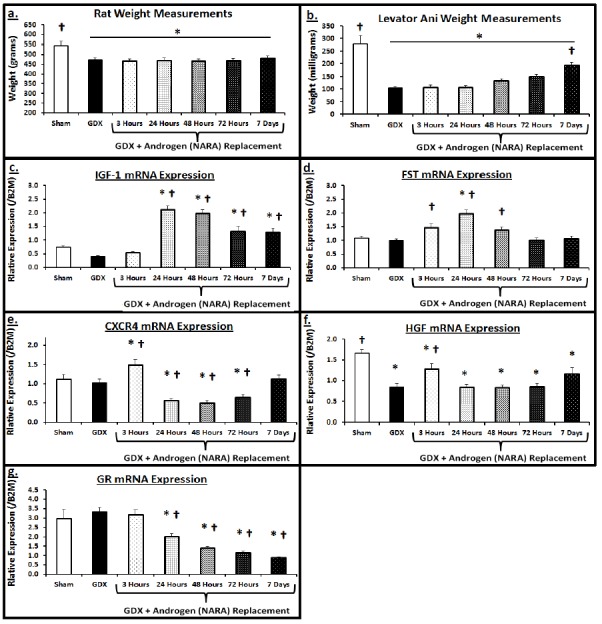
Male rats (8 weeks old) underwent gonadectomy or sham surgery. Six weeks after surgery, GDX animals were dosed daily with NARA (1 mg/kg). Cohorts (n=7-8) of GDX animals were sacrificed at hour 3, 24, 48, 72 and 7 days. A. Body weight at time of sacrifice (grams), B. Levator Ani wet weight (mg), C-G. LA muscle relative mRNA expression of IGF-1, FST, CXCR4, HGF, and GR (normalized to β-2 microglobulin; B2M) was measured via real-time PCR. Data are means ± SEM. ANOVA indicated significant differences among groups. Dunnet’s post hoc analysis set either Sham muscles or GDX (No NARA) as control and significance versus the respective group is indicated; *p<0.05 vs. Sham, † p<0.05 vs GDX.
NARA-treated GDX animals provided novel in vivo time course results, as characterized by the gene response signature. Augmenting the LASC results, NARA treatment initially led to significant increases in expression of IGF-1 (Figure 3C), FST (Figure 3D), and HGF (Figure 3F), while significantly decreasing expression of GR (Figure 3G). Interestingly, several genes displayed a biphasic-like expression profile; daily NARA dosing induced significantly higher CXCR4 levels at the 3 h time point, however, CXCR4 levels reversed by 24 h and remained significantly decreased to the 72 h time point. Similarly, HGF levels displayed an early, yet transient increase at the 3 h point, returning to basal-like levels by 24 h. FST expression followed a similar pattern, displaying a more sustained increase at 3, 24 and 48 h, yet returning to basal-like levels by 72 h. Although GR expression did not decrease initially at 3 h, there was a sustained decrease in its expression for the duration of the time course. These in vivo results demonstrated that NARA-induced, AR-mediated gene expression exhibits unique profiles over time. The correlation between gene response changes in vitro using LASC and those seen in vivo suggest that satellite cells are a primary target of androgen signaling in this tissue, and that additional insight can be gained by studying the role of AR in muscle satellite cells.
Removal of androgens results in delayed recovery and altered gene expression in muscle injury model, suggesting androgen-mediated activation of satellite cells
To extend these findings from the perineal LA muscle to a typical skeletal muscle, we used a mouse model of androgen depletion (gonadectomy; GDX) combined with cardiotoxin-induced gastrocnemius muscle injury. It has been reported that recovery from cardiotoxin-mediated injury requires MSCs and involves specific steps including inflammation, tissue necrosis, macrophage infiltration, and satellite cell activation [Garry et al., 2000; Yaden et al., 2014a; Yaden et al., 2014b]. The mode of injury resulted in an initial increase in tissue weight, explained by acute bleeding and infiltration of cellular components driving the inflammatory response (phagocytes, monocytes, lymphocytes, etc. [Czerwinska et al., 2012]). A sharp decrease in muscle weight was then observed, as severe atrophy presents during the necrosis phase of muscle repair. In androgen-normal (intact) mice, muscle recovery (based on return to sham control muscle weight) was achieved by 21 days, while androgen-null (GDX) mice required 35 days for full muscle recovery. In the androgen-normal (intact) mice, recovery following CTX (vs. control) was characterized by 1) a transient increase followed by a decrease in HGF (Figure 4C) and CXCR4 (Figure 4F), 2) a progressive increase in IGF-1 (Figure 4D) and FST (Figure 4E) and 3) an acute increase followed by an immediate normalization in TNF-α (Figure 4G).
Figure 4. Removal of androgens results in delayed mouse muscle recovery following insult.
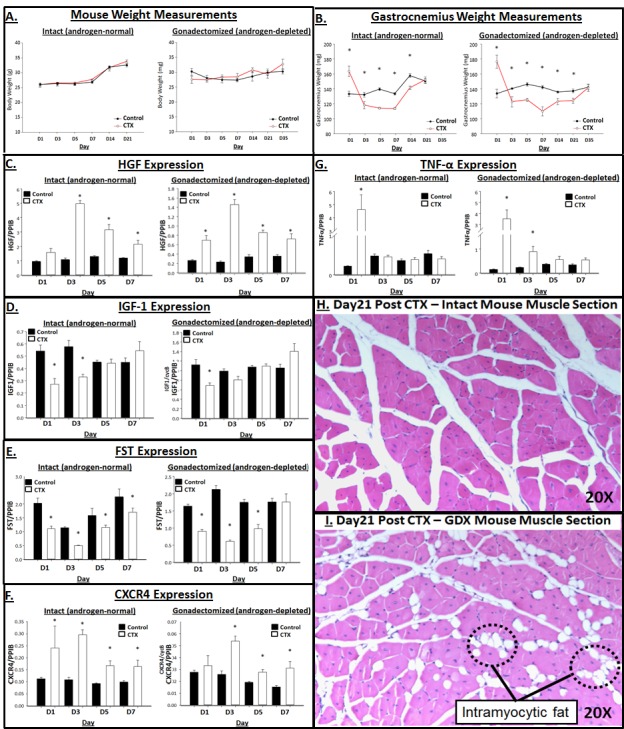
Male mice (8 weeks old) underwent gonadectomy or sham surgery. Six weeks after surgery, a cohort of mice were subjected to skeletal muscle injury via intramuscular injection of cardiotoxin (CTX; 10 µM) in the gastrocnemius muscle. Cohorts (n=7-8) of animals were sacrificed at post CTX injury at day 1, 3, 5, 7, 14, 21, 28, 35. A. Body weight at time of sacrifice (grams), B. Gastrocnemius wet weight (mg), C-H. Gastroc muscle relative mRNA expression of HGF, IGF-1, FST, CXCR4, and TNF-α (normalized to PPIB) was measured via real-time PCR. Data are means ± SEM. Significance (*p<0.05) of CTX vs. Control (within Intact or GDX) was analyzed by ANOVA and Bonferonni’s posthoc analysis. H-I. Representative H&E cross-sectional images (20X) of myofibers and fatty infiltration in the H. Intact (androgen-normal) and I. GDX (androgen-depleted) gastrocnemius muscle at the 21 days post cardiotoxin time point.
Removal of endogenous androgen (GDX mice) resulted in delayed muscle recovery following CTX-induced muscle injury (Figure 4B), as observed by muscle weight. By day 21, the injured muscles from intact (androgen-normal) mice had regained mass comparable to the sham control, suggesting complete recovery. Conversely, the injured muscles from GDX (androgen-depleted) mice did not fully recover muscle mass (vs. uninjured control) until day 35. Histological analysis (H&E) performed on muscles at 21 days post CTX injury reflected interstitial fatty infiltration in the gastrocnemius muscle from GDX mice, while the intact muscles displayed pathology consistent with healthy regenerating muscle (Figure 4H). Injury in the androgen-depleted (GDX) model highlighted genes possibly regulated by androgen. Though subtle, androgen-depletion resulted in delayed upregulation of CXCR4, with androgen-normal muscle displaying a 116% increase in CTX vs. Ctrl muscle, while GDX only had a 19% (CTX vs. Ctrl) increase one day post CTX. Likewise, androgen-depletion resulted in significantly increased TNF-α levels following CTX injury, with a 304% increase (vs. Ctrl) in the GDX group and 4% decrease (vs. Ctrl) in the intact group at day 3 post CTX (see Table 3 for further description of the percent changes between CTX versus control muscles from the intact and GDX mice). While IGF-1 and FST levels displayed little variation between intact vs. GDX groups, differential expression of HGF, CXCR4 and TNF-α provided results suggesting that gene regulation was being mediated in an androgen-specific manner in response to injury.
Table 3. Percent change between control vs. CTX animals.
| Measurement | Androgen Status | Day 1 | Day 3 | Day 5 | Day 7 |
|---|---|---|---|---|---|
| Muscle Weight | Normal (Intact) |
+20%* (CTX > Ctrl) |
-12%* (CTX < Ctrl) |
-20.4%* | -16.7%* |
| Depleted (GDX) | +24%* | -12.9%* | -14.3%* | -22.8%* | |
| HGF | Normal (Intact) | +67% | +364%* | +194%* | +107%* |
| Depleted (GDX) | +163%* | +527%* | +143%* | +105%* | |
| IGF-1 | Normal (Intact) | -50%* | -43%* | +4% | +34% |
| Depleted (GDX) | -38%* | -18% | +5% | +22% | |
| FST | Normal (Intact) | -45%* | -56%* | -28%* | -20%* |
| Depleted (GDX) | -44%* | -71%* | -44%* | +3% | |
| CXCR4 | Normal (Intact) | +116%* | +176%* | +80%* | +76%* |
| Depleted (GDX) | +19% | +71%* | +47%* | +106%* | |
| TNF-α | Normal (Intact) | +1294%* | -4% | +8% | -18% |
| Depleted (GDX) | +1752%* | +304%* | +54% | +52% |
Weight (body and muscle) and gene changes were compared between the Control versus CTX muscles in androgen-normal (intact) and androgen-depleted (GDX) animals, as displayed in Figure 4 (e.g., Normal Muscle Weight on Day 1 of +20% indicates CTX muscles were 20% heavier than Control muscles). *p>0.05; differences calculated to be significant, as shown graphically in Figure 4.
NARA treatment is associated with markers of satellite cell activation following muscle injury
Using the mouse model of androgen depletion (gonadectomy; GDX) combined with cardiotoxin (CTX)-induced gastrocnemius muscle injury, we specifically queried whether there was differential MSC activation as a result of androgen depletion in the injury model, perhaps explaining the delayed regenerative capacity. Moreover, we aimed to understand if a 7-day preventative NARA (1/mg/kg/day) treatment followed by injury (in both intact and GDX mice) could augment MSC activation. The brief 7-day NARA treatment regimen did not significantly alter gastrocnemius muscle size (data not shown). Four days post CTX injury, there was a main effect (p<0.05) of injury (CTX vs. sham) for Pax7 and MyoD gene expression, along with Pax7-positive cells (measured via IHC) in the gastrocnemius muscle (Figure 5). Furthermore, there was a significant main effect (p<0.05) of androgen status (GDX vs. intact) for MyoD gene expression and a significant main effect (p<0.05) of NARA treatment for MyoD gene expression and strong trend for Pax7 gene expression (p=0.083). The GDX-CTX muscle has significantly lower levels of Pax7 compared to intact-CTX, suggesting decreased MSC activation upon injury. NARA treatment in the GDX– CTX mouse resulted in muscle expression of Pax7 similar to that of the NARA-treated intact-CTX mouse, suggesting comparable MSC activation between the groups. A similar pattern of gene expression was observed for MyoD levels; in both intact and GDX mice, NARA treatment significantly increased MyoD levels 4-days post CTX injection. Though not significant across groups, a strong trend for increased Pax7-postive cells in NARA-treated vs. vehicle-treated muscles was observedin the GDX-CTX model (Figure 5C&D). Together, these data provide evidence that a lack of androgen and AR activation limits MSC activation upon muscle injury. Furthermore, AR activation, whether through an endogenous or exogenous AR agonist, augments MSC activation.
Discussion
The primary goal of this study was to establish a gene response signature defining the action of androgen on satellite cell activation in order to support interpretation of clinically relevant muscle recovery and regeneration models. A novel muscle satellite cell line (levator ani satellite cells; LASCs) treated with androgen responded with dose-dependent alterations in gene expression, ultimately presenting with myogenic differentiation. Focusing on this myogenic event in LASCs, microarray analysis established a unique gene signature highlighting markers of muscle satellite cell (MSC) activation (HGF and IGF-1), migration (CXCR4), and growth (FST and GR). Biomarker expression patterns defined by the gene signature predicted androgen-mediated muscle growth in a rat model of muscle atrophy, suggesting MSC engagement in response to liganded AR. A temporal response was observed for CXCR4, FST and HGF expression during the time course of androgen-mediated muscle growth, providing further depth to the gene signature. Finally, a muscle injury model comparing animals of normal versus depleted androgen status emphasized the requirement of androgen during skeletal muscle repair, as androgen-depleted animals displayed delayed regenerative capacity. Evidence for the role of androgen in muscle regeneration was further defined by delayed CXCR4 and HGF expression in the androgen-depleted animals post injury, suggesting hampered satellite cell activation. Furthermore, a direct effect of androgen-deprivation on the inflammatory response was observed in sustained TNF-α expression along with increased fat infiltration post injury. Finally, a connection between AR agonism and Pax7 and MyoD expression in injured muscle provided evidence for androgen-mediated MSC activation. These results are the first to actively connect androgen regulation of satellite cells to muscle recovery and regeneration through a defined set of response genes (summarized in Supplementary Material Figure 3).
There is strong evidence describing androgen-mediated skeletal muscle hypertrophy [Kadi et al., 2000; Sinha-Hikim et al., 2006; Sinha-Hikim et al., 2003]. It has been suggested that this muscle hypertrophy is largely mediated via androgen actions on MSCs [Doumit et al., 1996; Joubert and Tobin, 1995; Sinha-Hikim et al., 2003]. AR is expressed in MSCs and recent evidence has established these cells as androgen targets via gene expression [Doumit et al., 1996; Swift-Gallant and Monks, 2013] and cell growth analysis [Braga et al., 2012; Kamanga-Sollo et al., 2004; Kamanga-Sollo et al., 2011; Sinha-Hikim et al., 2004]. Previous studies have utilized satellite cells isolated from the androgen-responsive levator ani (LA) muscle [Clark et al., 1975; Singh et al., 2009] as a powerful tool to understand androgen action [Braga et al., 2012; Swift-Gallant and Monks, 2013]. Analysis of NARA-treated satellite cells from the LA (LASCs) in this study confirmed a robust androgen response while delivering a novel gene signature defining the effect of an AR agonist in the myogenic activation of satellite cells. Furthermore, changes in gene expression in in NARA-treated (48 h) LASC showing increased IGF-1 and FST, and decreased CXCR4, HGF, and GR, demonstrated that androgen ultimately facilitates entry of the muscle-derived satellite cells into myogenesis.
Up to this point, the role of androgen in MSC proliferation and differentiation remains unclear and is dependent on species and the level of stem cell commitment [Chen et al., 2005; Motohashi and Asakura, 2014]. Characterization of the LASC line using the cell electrical impedance assay showed that NARA treatment decreased cell impedance. This indicated androgen ultimately augmented LASC differentiation, expanding on previous work showing androgen may signal through IGF-1 [Dayton and White, 2008; Machida and Booth, 2004]. We further demonstrated that LASCs treated with IGF-1 displayed decreased proliferation (Figure 1C), akin to the NARA response. These results suggest that LASCs are ultimately being driven to differentiate, perhaps through androgen-mediated IGF-1 signaling. Likewise, the decrease in LASC Pax7 levels after 48 h of NARA treatment confirmed the loss of satellite cell genotype, endorsing activation into myogenesis. We can infer that LASC activation and proliferation occurred prior to the differentiation event based on, 1) microarray results showing NARA-mediated activation of cell cycle at the 4 h time point (vs. vehicle) and 2) previous work showing MyoD and myf5 regulation by androgen in a time-dependent manner [Singh et al., 2009]. However, in the LASC+CHX study, the necessity of protein synthesis in NARA-mediated IGF-1 regulation suggests a cascade of both direct and indirect mechanisms ultimately results in satellite cells differentiating. Our results suggest that this MSC differentiation process is initiated by decreased proliferation, via down regulation in HGF, and driven to differentiate, in part, via upregulation of IGF-1 and FST.
Previous work studying muscle atrophy using androgen-depleted animals re-supplemented with an AR agonist have confirmed a rescuing of muscle mass. We believe the muscle recovery and growth observed by re-supplementation is a result of the direct action of androgen on satellite cells, specifically the activation and differentiation of MSCs that replenish the GDX-induced atrophic tissue. Utilizing the gene response markers established using LASCs to characterize in vivo androgen gene regulation provided further insight into the possible mechanism behind the muscle mass rescuing effect of androgen re-supplementation. CXCR4, a protein known to be important in stem cell migration [Peled et al., 2012], was downregulated in LASCs after 48 h. However, in the rat androgen supplementation model, CXCR4 levels displayed a biphasic response with early, transient upregulation, followed by decreased expression over time. These time-dependent changes in CXCR4 may signal cycles of trafficking and differentiation of MSCs which are required to replenish the muscle tissue upon injury or atrophy-inducing conditions. Together, these results suggests that migratory machinery action in MSCs precedes differentiation, presumably to facilitate migration of activated and proliferating MSCs to the site of required growth or repair [Miller et al., 2008].
Interestingly, a biphasic-like response was observed in the rat NARA time course for not only CXCR4, but also HGF. This biphasic gene response has been investigated previously in models of prostate cancer [Wang et al., 2005] and reproduction [Hazra et al., 2013; Ivanga et al., 2009]. However, to our knowledge, this is the first in vivo evidence showing temporal control of AR-mediated gene regulation in skeletal muscle. Along with the early and transient induction of CXCR4, an early increase in HGF was observed which likely facilitates satellite cell activation and proliferation [Allen et al., 1995; Gal-Levi et al., 1998]. The latter time-dependent decrease observed for HGF and CXCR4 may support a coordinated process of cell proliferation, migration and engagement in myogenic differentiation. Follistatin also displayed a unique response, with peak expression observed at 24 h and a return to basal levels thereafter. These changes may reflect an effect of androgen on the TGF-β pathway in muscle, perhaps acting to inhibit the catabolic effects of myostatin [Yaden et al., 2014a]. Growing evidence suggests that FST may mediate the effects of androgen-induced myogenic differentiation of satellite cells [Braga et al., 2012; Willert et al., 2002] and this commands further investigation.
To our knowledge, this is the first study to establish that the glucocorticoid receptor (GR; NR3C1 in array results) is negatively regulated by androgen action in muscle satellite cells. While both AR and GR are members of the nuclear hormone receptor superfamily, the actions of their agonists (androgen and glucocorticoid, respectively) are opposing. Glucocorticoids, signaling through the GR, have profound catabolic effects on muscle [Chen et al., 1997; Crawford et al., 2003] via induction of several atrogenes, including atrogin-1 [Sandri et al., 2004] and FoxO transcription factors [Imae et al., 2003] and MuRF-1 [Schakman et al., 2013]. Androgens have been shown to offset the catabolic effects of liganded-GR, however limited evidence exists regarding mechanism [Crawford et al., 2003; Creutzberg and Schols, 1999; Creutzberg et al., 2003]. Studies showing AR-mediated down regulation of GR in the CNS (hippocampus [Kerr et al., 1996] and motor neurons [Blanco et al., 2002]) provide possible mechanism behind AR and GR interaction. Our results provide support of this mechanism with NARA-mediated down regulation of GR in satellite cells and rat skeletal muscle. Furthermore, results from this study showing NARA-mediated acute and sustained down regulation of GR augment recent evidence describing GR down regulation in muscle after a 42 day dosing regimen with an AR-agonist [Ye et al., 2014]. Combined, these data provide evidence of the anti-catabolic actions of androgen and justify further investigation.
The mechanisms involved in muscle hypertrophy often overlap with those important for muscle regeneration. Limited molecular evidence exists that directly examines the association between androgen and muscle regeneration. Furthermore, muscle regeneration requires activation of muscle satellite cells, a process which we demonstrate herein is regulated by androgen. Characterization of a clinically relevant model of muscle injury and repair using the gene response signature provided an improved understanding of muscle regeneration. The mode of injury (3-point injection of cardiotoxin) has been shown to be very efficient in recapitulating the specific pathologies associated with muscle trauma within the entire muscle (e.g., inflammation, tissue necrosis, macrophage infiltration, and satellite cell activation) [Garry et al., 2000; Yaden et al., 2014a; Yaden et al., 2014b]. A thorough description of muscle regeneration was achieved in the androgen normal animals with transient increases observed in HGF and CXCR4 expression. Similar to the rat muscle growth (re-supplementation) model, we believe HGF increases seen in the injury model provided the activation signal for satellite cells. Furthermore, CXCR4 likely initiated chemotactic signals for guiding the movement of activated satellite cells to the site of injury. Concurrently, IGF-1 expression was observed to be significantly lower immediately following injury (vs. Control), though by day 7 it trended higher (vs control), suggesting an initial delay in the MSC differentiation signal. Likewise, FST expression was initially decreased (vs. control), though it gradually recovered expression levels comparable to controls. Recent evidence indicates FST may control inflammation during the regeneration process [Yaden et al., 2014a], and therefore this initial down regulation may be critical for the pro-inflammatory signaling needed for proper muscle repair. The significant TNF-α increase observed on day 1 clearly represented the pro-inflammatory response, which then subsided as the muscle switched to an anti-inflammatory, pro-regeneration program.
With our understanding of muscle regeneration under androgen normal status, we aimed to better understand its direct role. Given that aging and other disease states are characterized by inadequate circulating androgen levels, an understanding of muscle recovery in the absence of androgen is also critical. Crucial knowledge is obtained in studying the effect of androgen via supplementation, but there is also great value to be derived from investigating models of androgen depletion. Using a clinically relevant model, this study showed removal of androgens (via gonadectomy) resulted in delayed muscle recovery and regeneration. The gene expression signature derived from LASCs was modified in androgen-null mice upon muscle insult and the regeneration process. While FST and IGF-1 signaling between WT and GDX appears consistent regardless of androgen-status, the initial lack of CXCR4 response in GDX mice suggests impaired motility of satellite cells to the site of injury. Interestingly HGF expression was greater in the GDX vs. WT mice, suggesting overcompensation for the lack of satellite cell response (i.e. migration) to the site of injury. The sustained increase in TNF-α expression past day 3 and into day 5, indicated that the delayed degeneration may be partly attributable to increased inflammation derived from sustained fatty infiltration. Likewise, histology of the androgen-depleted muscle highlighted increased fat infiltration. This warrants further investigation regarding androgen action on inflammatory pathways and associated cells (e.g. neutrophils, macrophages, and FAPs).
Previous work from the Bhasin lab [Serra et al., 2013] has shown that testosterone (T) supplementation in orchidectomized mice resulted in increased number, and larger, embryonic-MyHC+ fibers 4 days post injury. Concurrently, an increase in BrdU+/NCAM+ myoblasts was observed at 4 days post injury. However, by 9 days, there were no differences in regeneration markers observed between vehicle- and T-treated, suggesting the effects of androgen signaling occur during the early stages of regeneration, thereby supporting enhanced recovery. In order to ultimately confirm the connection between androgen signaling and MSCs in the early phases of muscle regeneration, a NARA-treated injury model was utilized in our studies. The results (Figure 5) suggest that androgen signaling indeed activates satellite cell proliferation early in the injury response. The NARA-driven augmentation in Pax7 and MyoD mRNA levels in GDX-CTX vs. intact-CTX animals confirms the early role of androgen signaling during regeneration. Based on the in vitro and in vivo analysis provided herein, the described androgen-driven increase in MSC proliferation would yield to amplified differentiation, ultimately supporting enhanced regeneration.
Our approach to developing a gene signature of biomarkers for androgen-mediated activation of satellite cells has notable advantages and limitations. We utilized a novel in vitro system in conjunction with in vivo models to understand examples of disease (muscle atrophy and injury). Nonetheless, inherent limitations with cultured satellite cell lines not displaying a true quiescent state is an unavoidable caveat when working with these cells ex vivo. In addition, conclusions from in vivo evidence were obtained using separate muscles, the perineal levator ani (LA) and gastrocnemius. In vivo, these muscles have differing functional demands. Like other muscle groups (e.g., hindlimb vs. forelimb), they display differing response to stimuli (e.g., denervation, injury, hormonal stimulation). In vivo, the response to androgen is greater in the LA versus the gastrocnemius [Ho et al., 2004], though the reasons for this remain poorly understood. However, Braga et al. [Braga et al., 2012] demonstrated that MSCs isolated from these respective muscles had a similar response to androgen stimulation in vitro. The discrepancy between in vivo and in vitro results in that study, along with those seen in the current analysis (LASC and LA vs. gastrocnemius results), likely has multiple explanatory factors including neural and vascular inputs, and/or cell-cell interactions.
In summary, this paper provides novel insight by establishing a signature of androgen-mediated gene regulation in 1) muscle satellite cells, 2) an in vivo model of androgen-mediated muscle recovery and growth, and 3) an in vivo model of muscle regeneration. Based on our findings, we propose a mechanism of androgen action in muscle regeneration which begins with increased satellite cell activation, proliferation and mobilization driven by increases in HGF, IGF-1, FST, and CXCR4. The androgen-mediated differentiation of satellite cells occurs in a time-dependent manner and is driven by decreases in HGF and increases in IGF-1, FST and CXCR4. Satellite cell engagement into myogenesis and incorporation into muscle follows, along with pro-anabolic, anti-catabolic and anti-inflammatory actions driven, in part, by androgen-mediated decreases in HGF, CXCR4, GR, and TNF-α, along with increases in IGF-1 and FST. The gene signature described can be utilized to further understand androgen action in translational applications of muscle regeneration.
Supplementary Material
Supplementary zip file supplied by authors.
Acknowledgements:
The authors thank Shawn T. Estrem and Hui-Rong Qian (Eli Lilly & Company) for their technical assistance with organization and statistical analysis of the microarray dataset.
Abbreviations:
- AR
androgen receptor
- ARE
androgen response element
- CHX
cyclohexamide CTX, cardiotoxin
- CXCR4
chemokine (c-x-c motif) receptor 4
- DAPI
4’,6-diamidino-2-phenylindole
- DM
differentiation media
- DMSO
dimethyl sulfoxide
- ECIS
electrical cell impedance sensing
- FDR
false positive rate
- FITC
fluorescein isothiocyanate
- FST
follistatin
- GDX
gonadectomy
- GR
glucocorticoid receptor
- HBSS
Hank’s balanced salt solution
- H&E
hematoxylin and eosin
- HGF
hepatocyte growth factor
- IGF-1
insulin-like growth factor-1
- IHC
immunohistochemistry
- IPA
Ingenuity Pathway Analysis
- LA
levator ani
- LASC
levator ani satellite cell
- MRI
magnetic resonance imaging
- MSC
muscle satellite cells
- NARA
non-steroidal androgen receptor agonist
- PCA
principal components analysis PCR, polymerase chain reaction
- RNA
ribonucleic acid
- RT-PCR
real-time polymerase chain reaction
- RT-qPCR
real-time quantitative polymerase chain reaction
- T
testosterone
- TNF-α
tumor necrosis factor α
- Veh
vehicle
Footnotes
JGM, BCY, HB, KC, PS, HUB and VK are all owners of Lilly Stock (LLY) as part of their status as employees at Eli Lilly & Company.
References
- Acosta S., Dizeyi N., Feinstein R., Pierzynowski S., Abrahamsson P. A. (2004). Long-term testosterone stimulation induces hyperplasia in the guinea-pig prostate. Prostate Cancer Prostatic Dis 7, 227-231. 10.1038/sj.pcan.4500744 [DOI] [PubMed] [Google Scholar]
- Allbrook D. B., Han M. F., Hellmuth A. E. (1971). Population of muscle satellite cells in relation to age and mitotic activity. Pathology 3, 223-243. [DOI] [PubMed] [Google Scholar]
- Allen R. E., Sheehan S. M., Taylor R. G., Kendall T. L., Rice G. M. (1995). Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J Cell Physiol 165, 307-312. 10.1002/jcp.1041650211 [DOI] [PubMed] [Google Scholar]
- Basaria S., Coviello A. D., Travison T. G., Storer T. W., Farwell W. R., Jette A. M., Eder R., Tennstedt S., Ulloor J., Zhang A., et al. (2010). Adverse events associated with testosterone administration. N Engl J Med 363, 109-122. 10.1056/NEJMoa1000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. H., Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57, 289-300. [Google Scholar]
- Blanco C. E., Peltz A., Staley R., Kim F. (2002). Effects of pharmacologic androgen treatment duration on glucocorticoid receptor alpha immunoreactivity of lumbosacral motor neurons in the male rat. Neuroscience 115, 941-949. 10.1016/S0306-4522(02)00338-X [DOI] [PubMed] [Google Scholar]
- Braga M., Bhasin S., Jasuja R., Pervin S., Singh R. (2012). Testosterone inhibits transforming growth factor-beta signaling during myogenic differentiation and proliferation of mouse satellite cells: potential role of follistatin in mediating testosterone action. Mol Cell Endocrinol 350, 39-52. 10.1016/j.mce.2011.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion D. R. (1984). The muscle satellite cell: a review. Int Rev Cytol 87, 225-251. 10.1016/S0074-7696(08)62444-4 [DOI] [PubMed] [Google Scholar]
- Celotti F., Negri Cesi P. (1992). Anabolic steroids: a review of their effects on the muscles, of their possible mechanisms of action and of their use in athletics. J Steroid Biochem Mol Biol 43, 469-477. 10.1016/0960-0760(92)90085-W [DOI] [PubMed] [Google Scholar]
- Chen S., Wang J., Yu G., Liu W., Pearce D. (1997). Androgen and glucocorticoid receptor heterodimer formation. A possible mechanism for mutual inhibition of transcriptional activity. J Biol Chem 272, 14087-14092. 10.1074/jbc.272.22.14087 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zajac J. D., MacLean H. E. (2005). Androgen regulation of satellite cell function. J Endocrinol 186, 21-31. 10.1677/joe.1.05976 [DOI] [PubMed] [Google Scholar]
- Clark M. R., Jury E. I., Krishnan V. V., Stark L. (1975). Computer simulation of biological models using the Inners approach. Comput Biol Med 5, 263-282. 10.1016/0010-4825(75)90028-1 [DOI] [PubMed] [Google Scholar]
- Corona G., Rastrelli G., Forti G., Maggi M. (2011). Update in testosterone therapy for men. J Sex Med 8, 639-654; quiz 655. 10.1111/j.1743-6109.2010.02200.x [DOI] [PubMed] [Google Scholar]
- Crawford B. A., Liu P. Y., Kean M. T., Bleasel J. F., Handelsman D. J. (2003). Randomized placebo-controlled trial of androgen effects on muscle and bone in men requiring long-term systemic glucocorticoid treatment. J Clin Endocrinol Metab 88, 3167-3176. 10.1210/jc.2002-021827 [DOI] [PubMed] [Google Scholar]
- Creutzberg E. C., Schols A. M. (1999). Anabolic steroids. Curr Opin Clin Nutr Metab Care 2, 243-253. 10.1097/00075197-199905000-00008 [DOI] [PubMed] [Google Scholar]
- Creutzberg E. C., Wouters E. F., Mostert R., Pluymers R. J., Schols A. M. (2003). A role for anabolic steroids in the rehabilitation of patients with COPD? A double-blind, placebo-controlled, randomized trial. Chest 124, 1733-1742. 10.1378/chest.124.5.1733 [DOI] [PubMed] [Google Scholar]
- Czerwinska A. M., Streminska W., Ciemerych M. A., Grabowska I. (2012). Mouse gastrocnemius muscle regeneration after mechanical or cardiotoxin injury. Folia Histochem Cytobiol 50, 144-153. 10.5603/FHC.2012.0021 [DOI] [PubMed] [Google Scholar]
- Dayton W. R., White M. E. (2008). Cellular and molecular regulation of muscle growth and development in meat animals. J Anim Sci 86, E217-225. 10.2527/jas.2007-0456 [DOI] [PubMed] [Google Scholar]
- Doumit M. E., Cook D. R., Merkel R. A. (1996). Testosterone up-regulates androgen receptors and decreases differentiation of porcine myogenic satellite cells in vitro. Endocrinology 137, 1385-1394. [DOI] [PubMed] [Google Scholar]
- Finkle W. D., Greenland S., Ridgeway G. K., Adams J. L., Frasco M. A., Cook M. B., Fraumeni J. F., Jr, Hoover R. N. (2014). Increased risk of non-fatal myocardial infarction following testosterone therapy prescription in men. PLoS One 9, e85805. 10.1371/journal.pone.0085805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Levi R., Leshem Y., Aoki S., Nakamura T., Halevy O. (1998). Hepatocyte growth factor plays a dual role in regulating skeletal muscle satellite cell proliferation and differentiation. Biochim Biophys Acta 1402, 39-51. 10.1016/S0167-4889(97)00124-9 [DOI] [PubMed] [Google Scholar]
- Garry D. J., Meeson A., Elterman J., Zhao Y., Yang P., Bassel-Duby R., Williams R. S. (2000). Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc Natl Acad Sci U S A 97, 5416-5421. 10.1073/pnas.100501197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds M. D., White J. D., Rosenthal N., Bogoyevitch M. A. (2002). The role of stem cells in skeletal and cardiac muscle repair. J Histochem Cytochem 50, 589-610. 10.1177/002215540205000501 [DOI] [PubMed] [Google Scholar]
- Hamann L. G., Higuchi R. I., Zhi L., Edwards J. P., Wang X. N., Marschke K. B., Kong J. W., Farmer L. J., Jones T. K. (1998). Synthesis and biological activity of a novel series of nonsteroidal, peripherally selective androgen receptor antagonists derived from 1,2-dihydropyridono[5,6-g]quinolines. J Med Chem 41, 623-639. 10.1021/jm970699s [DOI] [PubMed] [Google Scholar]
- Hazra R., Jimenez M., Desai R., Handelsman D. J., Allan C. M. (2013). Sertoli cell androgen receptor expression regulates temporal fetal and adult Leydig cell differentiation, function, and population size. Endocrinology 154, 3410-3422. 10.1210/en.2012-2273 [DOI] [PubMed] [Google Scholar]
- Herbst K. L., Bhasin S. (2004). Testosterone action on skeletal muscle. Curr Opin Clin Nutr Metab Care 7, 271-277. 10.1097/00075197-200405000-00006 [DOI] [PubMed] [Google Scholar]
- Ho M. H., Bhatia N. N., Bhasin S. (2004). Anabolic effects of androgens on muscles of female pelvic floor and lower urinary tract. Curr Opin Obstet Gynecol 16, 405-409. 10.1097/00001703-200410000-00009 [DOI] [PubMed] [Google Scholar]
- Hughes I. A., Lim H. N., Martin H., Mongan N. P., Dovey L., Ahmed S. F., Hawkins J. R. (2001). Developmental aspects of androgen action. Mol Cell Endocrinol 185, 33-41. 10.1016/S0303-7207(01)00622-0 [DOI] [PubMed] [Google Scholar]
- Imae M., Fu Z., Yoshida A., Noguchi T., Kato H. (2003). Nutritional and hormonal factors control the gene expression of FoxOs, the mammalian homologues of DAF-16. J Mol Endocrinol 30, 253-262. 10.1677/jme.0.0300253 [DOI] [PubMed] [Google Scholar]
- Ivanga M., Labrie Y., Calvo E., Belleau P., Martel C., Pelletier G., Morissette J., Labrie F., Durocher F. (2009). Fine temporal analysis of DHT transcriptional modulation of the ATM/Gadd45g signaling pathways in the mouse uterus. Mol Reprod Dev 76, 278-288. 10.1002/mrd.20949 [DOI] [PubMed] [Google Scholar]
- Joubert Y., Tobin C. (1995). Testosterone treatment results in quiescent satellite cells being activated and recruited into cell cycle in rat levator ani muscle. Dev Biol 169, 286-294. 10.1006/dbio.1995.1144 [DOI] [PubMed] [Google Scholar]
- Kadi F., Bonnerud P., Eriksson A., Thornell L. E. (2000). The expression of androgen receptors in human neck and limb muscles: effects of training and self-administration of androgenic-anabolic steroids. Histochem Cell Biol 113, 25-29. 10.1007/s004180050003 [DOI] [PubMed] [Google Scholar]
- Kamanga-Sollo E., Pampusch M. S., Xi G., White M. E., Hathaway M. R., Dayton W. R. (2004). IGF-I mRNA levels in bovine satellite cell cultures: effects of fusion and anabolic steroid treatment. J Cell Physiol 201, 181-189. 10.1002/jcp.20000 [DOI] [PubMed] [Google Scholar]
- Kamanga-Sollo E., White M. E., Hathaway M. R., Weber W. J., Dayton W. R. (2011). Effect of trenbolone acetate on protein synthesis and degradation rates in fused bovine satellite cell cultures. Domest Anim Endocrinol 40, 60-66. 10.1016/j.domaniend.2010.08.007 [DOI] [PubMed] [Google Scholar]
- Kawano H., Sato T., Yamada T., Matsumoto T., Sekine K., Watanabe T., Nakamura T., Fukuda T., Yoshimura K., Yoshizawa T., et al. (2003). Suppressive function of androgen receptor in bone resorption. Proc Natl Acad Sci U S A 100, 9416-9421. 10.1073/pnas.1533500100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. E., Beck S. G., Handa R. J. (1996). Androgens selectively modulate C-fos messenger RNA induction in the rat hippocampus following novelty. Neuroscience 74, 757-766. 10.1016/0306-4522(96)00219-9 [DOI] [PubMed] [Google Scholar]
- Kogan I., Goldfinger N., Milyavsky M., Cohen M., Shats I., Dobler G., Klocker H., Wasylyk B., Voller M., Aalders T., et al. (2006). hTERT-immortalized prostate epithelial and stromal-derived cells: an authentic in vitro model for differentiation and carcinogenesis. Cancer Res 66, 3531-3540. 10.1158/0008-5472.CAN-05-2183 [DOI] [PubMed] [Google Scholar]
- Lepper C., Partridge T. A., Fan C. M. (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639-3646. 10.1242/dev.067595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Sutton J. C., Nirschl A., Zou Y., Wang H., Sun C., Pi Z., Johnson R., Krystek S. R., Jr, Seethala R., et al. (2007). Discovery of potent and muscle selective androgen receptor modulators through scaffold modifications. J Med Chem 50, 3015-3025. 10.1021/jm070312d [DOI] [PubMed] [Google Scholar]
- Machida S., Booth F. W. (2004). Insulin-like growth factor 1 and muscle growth: implication for satellite cell proliferation. Proc Nutr Soc 63, 337-340. 10.1079/PNS2004354 [DOI] [PubMed] [Google Scholar]
- Manfredi M. C., Bi Y., Nirschl A. A., Sutton J. C., Seethala R., Golla R., Beehler B. C., Sleph P. G., Grover G. J., Ostrowski J., Hamann L. G. (2007). Synthesis and SAR of tetrahydropyrrolo[1,2-b][1,2,5]thiadiazol-2(3H)-one 1,1-dioxide analogues as highly potent selective androgen receptor modulators. Bioorg Med Chem Lett 17, 4487-4490. 10.1016/j.bmcl.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Mathelier A., Zhao X., Zhang A. W., Parcy F., Worsley-Hunt R., Arenillas D. J., Buchman S., Chen C. Y., Chou A., Ienasescu H., et al. (2014). JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42, D142-147. 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. (1961). Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9, 493-495. 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. J., Banisadr G., Bhattacharyya B. J. (2008). CXCR4 signaling in the regulation of stem cell migration and development. J Neuroimmunol 198, 31-38. 10.1016/j.jneuroim.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. (2005). Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064-2067. 10.1126/science.1114758 [DOI] [PubMed] [Google Scholar]
- Mooradian A. D., Morley J. E., Korenman S. G. (1987). Biological actions of androgens. Endocr Rev 8, 1-28. 10.1210/edrv-8-1-1 [DOI] [PubMed] [Google Scholar]
- Motohashi N., Asakura A. (2014). Muscle satellite cell heterogeneity and self-renewal. Front Cell Dev Biol 2:1., eCollection. 10.3389/fcell.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N., Asakura Y., Asakura A. (2014). Isolation, culture, and transplantation of muscle satellite cells. J Vis Exp 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. M., Lawson J. A., Mathew S. J., Hutcheson D. A., Kardon G. (2011). Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625-3637. 10.1242/dev.064162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nnodim J. O. (2001). Testosterone mediates satellite cell activation in denervated rat levator ani muscle. Anat Rec 263, 19-24. 10.1002/ar.1072 [DOI] [PubMed] [Google Scholar]
- O'Connell M. D., Wu F. C. (2014). Androgen effects on skeletal muscle: implications for the development and management of frailty. Asian J Androl 16, 203-212. 10.4103/1008-682X.122581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled A., Wald O., Burger J. (2012). Development of novel CXCR4-based therapeutics. Expert Opin Investig Drugs 21, 341-353. 10.1517/13543784.2012.656197 [DOI] [PubMed] [Google Scholar]
- Rakhilin S., Turner G., Katz M., Warden R., Irelan J., Abassi Y. A., Glass D. J. (2011). Electrical impedance as a novel biomarker of myotube atrophy and hypertrophy. J Biomol Screen 16, 565-574. 10.1177/1087057111401392 [DOI] [PubMed] [Google Scholar]
- Saini A., Al-Shanti N., Stewart C. E. (2006). Waste management - cytokines, growth factors and cachexia. Cytokine Growth Factor Rev 17, 475-486. 10.1016/j.cytogfr.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647-3656. 10.1242/dev.067587 [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399-412. 10.1016/S0092-8674(04)00400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schakman O., Kalista S., Barbe C., Loumaye A., Thissen J. P. (2013). Glucocorticoid-induced skeletal muscle atrophy. Int J Biochem Cell Biol 45, 2163-2172. 10.1016/j.biocel.2013.05.036 [DOI] [PubMed] [Google Scholar]
- Schultz E., Gibson M. C., Champion T. (1978). Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J Exp Zool 206, 451-456. 10.1002/jez.1402060314 [DOI] [PubMed] [Google Scholar]
- Serra C., Tangherlini F., Rudy S., Lee D., Toraldo G., Sandor N. L., Zhang A., Jasuja R., Bhasin S. (2013). Testosterone improves the regeneration of old and young mouse skeletal muscle. J Gerontol A Biol Sci Med Sci 68, 17-26. 10.1093/gerona/gls083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Bhasin S., Braga M., Artaza J. N., Pervin S., Taylor W. E., Krishnan V., Sinha S. K., Rajavashisth T. B., Jasuja R. (2009). Regulation of myogenic differentiation by androgens: cross talk between androgen receptor/ beta-catenin and follistatin/transforming growth factor-beta signaling pathways. Endocrinology 150, 1259-1268. 10.1210/en.2008-0858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha-Hikim I., Cornford M., Gaytan H., Lee M. L., Bhasin S. (2006). Effects of testosterone supplementation on skeletal muscle fiber hypertrophy and satellite cells in community-dwelling older men. J Clin Endocrinol Metab 91, 3024-3033. 10.1210/jc.2006-0357 [DOI] [PubMed] [Google Scholar]
- Sinha-Hikim I., Roth S. M., Lee M. I., Bhasin S. (2003). Testosterone-induced muscle hypertrophy is associated with an increase in satellite cell number in healthy, young men. Am J Physiol Endocrinol Metab 285, E197-205. 10.1152/ajpendo.00370.2002 [DOI] [PubMed] [Google Scholar]
- Sinha-Hikim I., Taylor W. E., Gonzalez-Cadavid N. F., Zheng W., Bhasin S. (2004). Androgen receptor in human skeletal muscle and cultured muscle satellite cells: up-regulation by androgen treatment. J Clin Endocrinol Metab 89, 5245-5255. 10.1210/jc.2004-0084 [DOI] [PubMed] [Google Scholar]
- Stadler G., Chen J. C., Wagner K., Robin J. D., Shay J. W., Emerson C. P., Jr, Wright W. E. (2011). Establishment of clonal myogenic cell lines from severely affected dystrophic muscles - CDK4 maintains the myogenic population. Skelet Muscle 1, 12. 10.1186/2044-5040-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift-Gallant A., Monks D. A. (2013). Androgen receptor expression in satellite cells of the neonatal levator ani of the rat. Dev Neurobiol 73, 448-454. 10.1002/dneu.22066 [DOI] [PubMed] [Google Scholar]
- Tidball J. G. (2005). Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol 288, R345-353. 10.1152/ajpregu.00454.2004 [DOI] [PubMed] [Google Scholar]
- Tidball J. G. (2011). Mechanisms of muscle injury, repair, and regeneration. Compr Physiol 1, 2029-2062. [DOI] [PubMed] [Google Scholar]
- Tingus S. J., Carlsen R. C. (1993). Effect of continuous infusion of an anabolic steroid on murine skeletal muscle. Med Sci Sports Exerc 25, 485-494. [PubMed] [Google Scholar]
- Turner N. J., Badylak S. F. (2012). Regeneration of skeletal muscle. Cell Tissue Res 347, 759-774. 10.1007/s00441-011-1185-7 [DOI] [PubMed] [Google Scholar]
- Turner R. T., Wakley G. K., Hannon K. S. (1990). Differential effects of androgens on cortical bone histomorphometry in gonadectomized male and female rats. J Orthop Res 8, 612-617. 10.1002/jor.1100080418 [DOI] [PubMed] [Google Scholar]
- Vanderschueren D., Van Herck E., Suiker A. M., Visser W. J., Schot L. P., Chung K., Lucas R. S., Einhorn T. A., Bouillon R. (1993). Bone and mineral metabolism in the androgen-resistant (testicular feminized) male rat. J Bone Miner Res 8, 801-809. 10.1002/jbmr.5650080705 [DOI] [PubMed] [Google Scholar]
- Venken K., De Gendt K., Boonen S., Ophoff J., Bouillon R., Swinnen J. V., Verhoeven G., Vanderschueren D. (2006). Relative impact of androgen and estrogen receptor activation in the effects of androgens on trabecular and cortical bone in growing male mice: a study in the androgen receptor knockout mouse model. J Bone Miner Res 21, 576-585. 10.1359/jbmr.060103 [DOI] [PubMed] [Google Scholar]
- Vigen R., O'Donnell C. I., Baron A. E., Grunwald G. K., Maddox T. M., Bradley S. M., Barqawi A., Woning G., Wierman M. E., Plomondon M. E., et al. (2013). Association of testosterone therapy with mortality, myocardial infarction, and stroke in men with low testosterone levels. JAMA 310, 1829-1836. 10.1001/jama.2013.280386 [DOI] [PubMed] [Google Scholar]
- Wang Q., Carroll J. S., Brown M. (2005). Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19, 631-642. 10.1016/j.molcel.2005.07.018 [DOI] [PubMed] [Google Scholar]
- Willert J., Epping M., Pollack J. R., Brown P. O., Nusse R. (2002). A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol 2, 8. 10.1186/1471-213X-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaden B. C., Croy J. E., Wang Y., Wilson J. M., Datta-Mannan A., Shetler P., Milner A., Bryant H. U., Andrews J., Dai G., Krishnan V. (2014a). Follistatin: a novel therapeutic for the improvement of muscle regeneration. J Pharmacol Exp Ther 349, 355-371. 10.1124/jpet.113.211169 [DOI] [PubMed] [Google Scholar]
- Yaden B. C., Wang Y. X., Wilson J. M., Culver A. E., Milner A., Datta-Mannan A., Shetler P., Croy J. E., Dai G., Krishnan V. (2014b). Inhibition of activin A ameliorates skeletal muscle injury and rescues contractile properties by inducing efficient remodeling in female mice. Am J Pathol 184, 1152-1166. 10.1016/j.ajpath.2013.12.029 [DOI] [PubMed] [Google Scholar]
- Ye F., McCoy S. C., Ross H. H., Bernardo J. A., Beharry A. W., Senf S. M., Judge A. R., Beck D. T., Conover C. F., Cannady D. F., et al. (2014). Transcriptional regulation of myotrophic actions by testosterone and trenbolone on androgen-responsive muscle. Steroids 87C, 59-66. 10.1016/j.steroids.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Gao W., Kearbey J. D., Xu H., Chung K., He Y., Marhefka C. A., Veverka K. A., Miller D. D., Dalton J. T. (2003). Pharmacodynamics of selective androgen receptor modulators. J Pharmacol Exp Ther 304, 1334-1340. 10.1124/jpet.102.040840 [DOI] [PubMed] [Google Scholar]
- Zhang X., Sui Z. (2013). Deciphering the selective androgen receptor modulators paradigm. Expert Opin Drug Discov 8, 191-218. 10.1517/17460441.2013.741582 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary zip file supplied by authors.