Abstract
Lifeguard (LFG) is a transmembrane protein which is highly expressed in tissues of the hippocampus and the cerebellum, especially during postnatal development. This protein is responsible for the protection of neurons against Fas-induced apoptosis, and the same effect can be seen in tumor cells derived from mastocarcinoma. However, the molecular function of LFG and its regulation in the carcinogenesis of human breast cells remains to be elucidated. In the present study, we investigated the connection of the interaction of LFG within an array analysis of over 9,000 different proteins. Results showed an interaction between the proteins tripartite motif-containing 21 (TRIM21) and LFG and a negative regulatory effect of TRIM21 towards LFG on the protein level. Furthermore, Fas-induced apoptosis decreased upon the addition of TRIM21 to the cultured cells. These results revealed TRIM21 to be a negative modulator of LFG in cells of mastocarcinoma in vitro. For all analyses, MDA-MB-231 cells were used. The interaction of TRIM21 and LFG was analyzed by co-immunoprecipitation. To examine changes in regulatory processes, western blot analyses, real-time PCR, activity of apoptotic process and flow cytometric analyses were carried out.
Keywords: Lifeguard, TRIM21, apoptosis, mastocarcinoma
Introduction
One of the many possible causes in the development of cancer is the dysfunction in the regulation of apoptosis. This mechanism usually serves as a means of eliminating damaged or degenerated cells within the human body. If the tight regulation of this balance between regeneration and elimination is malfunctioning, the consequences are a loss of tissue or neoplasms. A certain protein highly expressed in breast cancer cells leads to a dysfunction in the regulation of apoptosis.
The transmembrane protein Lifeguard (LFG) was first described by Somia et al in 1999. Its localization is predicted to be in the endoplasmatic reticulum, or the plasma membrane, with ubiquitous expression except for in the placenta and pancreas. Lifeguard has been identified as an inhibitor of Fas-induced apoptosis. Immunoprecipitation analysis showed its interaction with Fas-receptor and the Fas-antibody, which mimics the Fas-ligand, but not with the protein FADD (1). Still, the mechanism by which LFG blocks the signal pathway that starts with the ligand-induced clustering of Fas-receptors, and ends with the final activation of caspase-8 and -3, is unknown (2,3). For structures of the central nervous system in adults, this is an advantage as the predicted function of the neural membrane protein 35, a structural homolog of LFG found in rats, provides protection of tissues in these regions (4). In other organs, however, the shift of the apoptotic homeostasis by the expression of LFG can have fatal consequences. In previous publications, it has been demonstrated that there is a correlation between the expression level of LFG and the increasing degree of malignity of breast cancer (5). Furthermore, it was shown that the effect of the alkyl phospholipid perifosine was reduced in cells expressing LFG (6).
Tripartite motif-containing 21 (TRIM21) is usually associated with autoimmune diseases such as systemic lupus erythematodes (SLE), although its role in the pathomechanism is not yet revealed (7). The physiological functions of TRIM21 are mainly attributed to the area of the innate immune system concerning the defense against viral pathogens but it is also associated with signaling pathways concerning for example cell division by inhibiting the activation of the transcription factor NF-κB (8,9). This transcription factor family is significantly involved in cell proliferation and has therefore been associated with different kinds of cancer where it favors fast tumor growth and development of metastasis (10).
Materials and methods
Cell lines and culture condition
The human breast carcinoma cell line MDA-MB-231 was obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and grown in Dulbecco's modified Eagle's medium (DMEM; PAA, Cölbe, Germany) supplemented with 10% FCS (Biochrom, Berlin, Germany) and 50 mg/ml penicillin-streptomycin. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. The cells were subcultured every 2 to 3 days by treatment with a 0.25% Trypsin/0.53 mM ethylenediamine-tetraacetic acid (EDTA) solution.
Protein-array
The ProtoArray Human Protein Microarray v4.0 containing 8,268 human proteins, reagents and other equipment were from Invitrogen (Carlsbad, CA, USA). Assays including appropriate controls were performed according to the instructions of the manufacturer. In short, protein microarray slides were blocked with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.1% Tween-20 before incubation with 10 μM of recombinant LFG-Protein (1:500; Abnova, Taipei, Taiwan) and Alexa Fluor 647-conjugated antihuman IgG (1.0 μg/ml buffer). The arrays were dried and scanned using an Axon GenePix 400B fluorescent microarray scanner. GenePix 6.0 software was used to align the scanned image to the template as well as to determine the pixel intensities for each spot on the array. The reported pixel intensity was calculated as the average of duplicate signals after background subtraction. Prospector software (Invitrogen), which is based on M-statistics were used for statistical analysis of the microarray data.
TRIM21 vectors
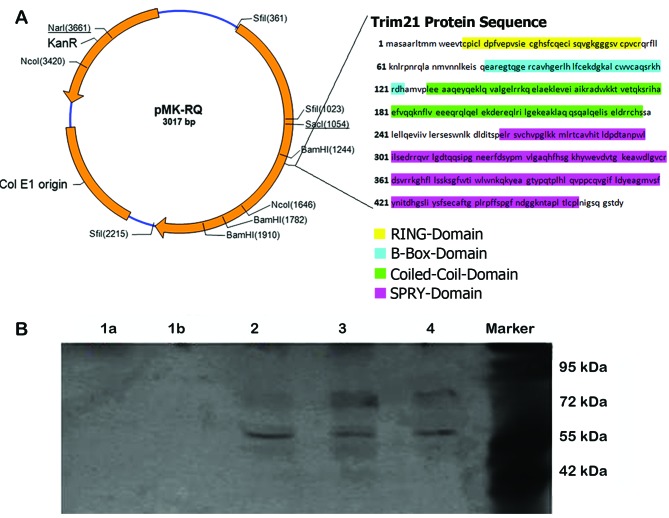
To identify the essential domain for the interaction between TRIM21 and LFG, different vectors, using pCMV6-AC-GFP vector (NM_003141; OriGene, Cambridge, UK) were cloned containing the sequences of TRIM21 with the deletion of an entire domain. The vector map as well as the sequences of the individual TRIM21 domains are shown in Fig. 3.
Figure 3.
TRIM21 domain. (A) (Left) Vector map of the used pMK-RQ vector (Life Technologies), (right) amino acid sequence of TRIM21 and the sequences of an individual domains. (B) Western blot analysis after native 12.5% polyacrylamide gel. Detection of TRIM21 with Li-Cor 800 anti-goat, detection of LFG with Li-Cor-680 anti-rabbit. Sample 1a (PRY domain), sample 1b (SPRY domain), sample 2 (coiled-coil domain), sample 3 (B-box domain), sample 4 (RING domain), marker (M). The samples 2–4 show clearly visible bands of ~80 and 60 kDa. The bands at 80 kDa are larger but slightly less visible as the bands at the height of 60 kDa. In samples 1a and 1b no bands are visible.
Co-immunoprecipitation
MDA-MB-231 were seeded in 100 mm culture plates and transfected with vectors coding for proteins LFG (pTriEx-1.1-FAIM2, NM-0112306; GeneArt, Regensburg, DE, USA) and TRIM21 (pCMV6-AC-GFP-Trim21, NM_003141; OriGene) by using FuGENE 6 (Promega, Madison, WI, USA) according to manufacturer's instructions. The implementation of the co-immunoprecipitation was performed according to the protocol of the Co-Immunoprecipitation kit of Dynabeads, Inc. A total of 42 μg LFG antibody were added to 6 mg beads and shaken overnight at 37°C. After several washes with the extraction and LWF buffer of Dynabeads, Inc., 100 μl of the LFG antibody coupled beads suspension were transferred to 50 μl of one cell lysate, containing one TRIM21 protein with a deletion of one domain. These approaches had to be incubated overnight at 4°C. The associated proteins were added in 800 μl SB buffer and subsequently used in western blot analysis.
Co-localization
For every glass slide, 5×104 MDA-MB-231 were cultivated and transfected with vectors coding for the proteins LFG and TRIM21 for 24 h after which time the cells were fixed in 4% paraformaldehyde for 20 min and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, Steinheim, Germany) for 4 min. Slides were then incubated with rabbit polyclonal anti-hLFG antibody (1:100 dilution) and goat polyclonal anti-hTrim21 (Santa Cruz Biotechnology, Inc.) antibodies at 37°C for 1 h, washed three times in ice-cold PBS, and incubated with Alexa Fluor 488 and Alexa Fluor 680 conjugated goat anti-rabbit and chicken anti-goat secondary antibody (each in 1:600 dilution; Invitrogen) at 37°C for 30 min. After three washing steps with PBS, the sample was dried and covered with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA), an antifade reagent containing the DNA-staining dye 4′,6′-diamidino-2-phenylindole (DAPI). Images were attained using Zeiss Axiovert 200M fluorescence microscope, equipped with the appropriate barrier filters.
Small interfering RNA
MDA MB231 cells were transfected with siRNA LFG-650 5′-cctcctacccttccaatatgt-3 (Sirion, Munich, Germany) and the appropriate control vector. The algorithm used by Sirion for the siRNA design was optimized for maximum gene specificity, and KD efficiency. Subsequent virus rescue and production were carried out in HEK 293 cells. Virus purification was performed using the ViraBind™ Adenovirus Miniprep kit (Cell Biolabs, Inc., San Diego, CA, USA). The cells were seeded at 2×104 cells/cm2 and incubated at 37°C in a humidified atmosphere with 5% CO2 for 48 h before being analyzed.
Western blot analysis
Each sample (20 μl) was dissolved in 7.5 μl loading buffer. The samples were applied with a protein marker on a native polyacrylamide gel, which had been equilibrated in a forerun with 0,5× TBE as running buffer for 30 min at 25 mA. The subsequent run with the samples was at 25 mA for 3 h. The blotting onto a PVDF membrane (Millipore Corp., Bedford, MA, USA), which had been previously activated in 99% ethanol, was carried out overnight at 40 V and 80 mA. Immunoblotting was performed with polyclonal antibodies: anti-FAIM2 (1:200 dilution), anti-Trim21 (1:200 dilution) (both from Santa Cruz Biotechnology, Inc.) and anti-actin (1:200 dilution; Abcam, Cambridge, UK). As a secondary antibody, Odyssey 600 anti-rabbit and Odyssey 800 anti-goat (Invitrogen) were used for the quantification of protein expression levels, and signals were obtained using the Odyssey Infrared Imaging System and software (Li-Cor Biosciences, Lincoln, NE, USA).
RT-PCR analysis
Total RNA was extracted using the NucleoSpin RNA II kit (MN Macherey-Nagel, Düren, Germany), and then 1 μg of RNA was reverse transcribed into cDNA and amplified using the iScript™ cDNA kit (Bio-Rad Laboratories, Hercules, CA, USA). The following reverse (R) and forward (F) primers were used: trim21-F, 5′-gaccatggctccctcatcta-3′ and trim21-R, 5′-agggttagaggggcgtgtt-3′; β2-microglobulin-F, 5′-atgagtatgcctgccgtgtga-3′ and β2-microglobulin-R, 5′-ggcatcttcaaacctccatg-3′.
Real-time polymerase chain reaction (PCR) was carried out in 20 μl samples with 5 ng cDNA, 1 mM of each forward and reverse primer and a 2× SYBR-Green Sensi-Mix DNA kit (Quantace, London, UK). Relative gene expression was determined by normalization of the fluorescence intensity to β2-microglobulin gene expression. Amplification cycles were as follows: 35 cycles at 95°C for 10 sec, 60°C or 30 sec, 70°C+0.2°C for 15 min.
Caspase assay
Activation of caspase-3/7 was determined using the Apo-One homogeneous caspase-3/7 assay (Promega) following the protocol provided by the manufacturer. Briefly, 1×104 MDA-MB-231 breast cancer cells were seeded per well of a 96-well plate, and incubated with 1 μg TRIM21 human recombinant protein (ProSpec; Ness Ziona, Tel Aviv, Israel) every 24 h. After 48 h, 100 ng of agonistic anti-Fas (clone CH11; Abcam) were added and after 24 h, incubated cells were mixed with 100 μl of Apo-One homogeneous caspase-3/7 reagent. Following incubation at room temperature for 2 h, caspase-3/7 activation was estimated from sample fluorescence at the excitation wavelength of 485 nm and an emission wavelength of 530 nm using a fluorescence plate reader Tecan GENios (Tecan Schweiz AB, Zurich, Switzerland).
Cell cycle analysis by flow cytometry
For distinct cell cycle phase distribution, ~106 MDA-MB-231 breast cancer cells were analysed. The cells were cultivated by the addition of different concentrations of TRIM21 as human recombinant protein within culture medium. Thus, the cells were harvested and fixed in 70% (v/v) ice-cold ethanol and kept at 4°C for 24 h. After washing, cells were resuspended in 1 ml PBS containing 10 μg/ml RNase and 1 mg/ml propidium iodide (PI; Sigma-Aldrich); they were then incubated for 1 h at 37°C in the dark. Thereafter, cells were analysed on a FACSCalibur flow cytometer (Becton-Dickinson, San Jose, CA, USA).
NF-κB-array
cDNA plate array analysis is a plate-based hybridization profiling technique that is used for monitoring the expression of dozens of genes through reverse transcription of mRNA into cDNA. MDA-MB-231 were cultivated for 48 h with the addition of 2.0 μg human recombinant Trim21 every 24 h. The control was cultivated without supplements. For this analysis, total RNA was isolated from the cell samples, using a NucleoSpin RNA II kit (MN Macherey-Nagel). RNA samples (8 μg) were analyzed by microarray analysis using an NF-κB pathway-regulated cDNA plate array (Signosis, Sunnyvale, CA, USA) according to the manufacturer's instructions. Each well on the plate contained a cDNA probe for one of the 24 NF-κB pathway-regulated genes. After reverse transcription, in situ hybridization, blocking, and extensive washing, the wells were incubated with streptavidin-HRP, and the resulting chemiluminescence was measured within 5 min using a luminometer (Tecan Schweiz AB). As a gene of reference, β-actin was chosen and the calculations were carried out using qbase+ software (Biogazelle).
Immunohistochemistry
Breast tissue slides (US Biomax, Inc., Rockville, MD, USA) were deparaffinized in xylene, and transferred through two changes of 100% ethanol. For antigen retrieval, the slides were pressure cooked in 6.5 mM sodium citrate (pH 6.0). To reduce non-specific background staining, slides were incubated for 30 min in 0.3% BSA/1× Tris-buffered saline. Slides were incubated at 4°C overnight with hTrim21 goat primary antibodies (1:100 dilution) (Santa Cruz Biotechnology, Inc.) and mouse primary cdc6 antibodies (1:100) (MoBiTec GmbH, Göttingen, Germany). The slides were washed twice for 5 min with PBS, and incubated for 30 min with goat anti-rabbit Li-Cor-680 and anti-mouse Li-Cor-800CW conjugated secondary antibody. Signals were detected by using the Li-Cor Infra-Red imaging system.
Results
Analyses of interaction
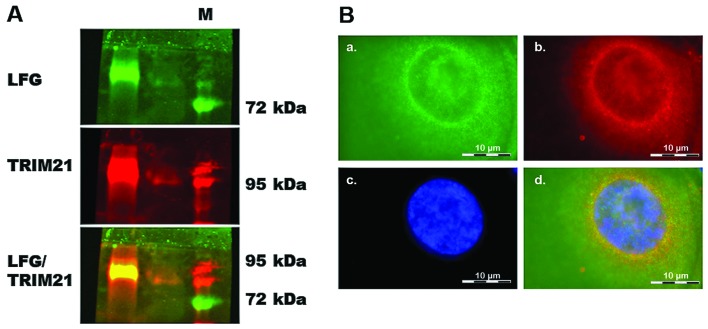
To find a protein that could be used to target LFG, >9,000 proteins were tested for their interaction in an array-analysis (Fig. 1). The protein-array was incubated with the human recombinant protein LFG which was detected by fluorescence labeled antibodies. The signal intensity was therefore proportional to the concentration of bound LFG-protein. The signal of the interaction of LFG with TRIM21 clearly stands out as a white signal, as seen in the detailed display in Fig. 1. The Z-score confirms that the interaction of TRIM21 is around 2.5 times stronger than the second strongest signal (Table I). To analyze if the interaction occured within the environment of a living cell as well, a co-immunoprecipitation was carried out using MDA-MB-231, which had been transfected with vectors coding for the proteins LFG and TRIM21 (Fig. 3) to achieve a clearer result. After cell disruption, LFG was isolated from the lysate using magnetic beads coated with the specific LFG antibody. The protein eluted from the beads was analyzed in a native PAGE followed by western blot analysis. Detection by fluorescence labeled antibodies revealed a green signal for LFG as well as a red signal for TRIM21 (Fig. 2A). The yellow signal visible in the overlay of the single signals shows that both proteins can be found in the same position on the blot-membrane, although not being of the same molecular weight. Furthermore, the LFG and TRIM21 antibodies were used to detect the proteins within intact, but PFA-fixated MDA-MB-231, that were previously transfected with vectors coding for the two proteins (Fig. 2B). In this fluorescence microscope image, the green signals of LFG are visible predominantly in close proximity to the cell core (Fig. 2B-a–c), very similar to the ring of red labeled TRIM21 (Fig. 2B-b and c). The core was stained with DAPI and therefore is visible as a blue structure (Fig. 2B-c). In the overlay, the yellow signal shows the positions in which both proteins are localized in the same place. These combined signals also display a ring around the core with fewer signals within the cytoplasm.
Figure 1.
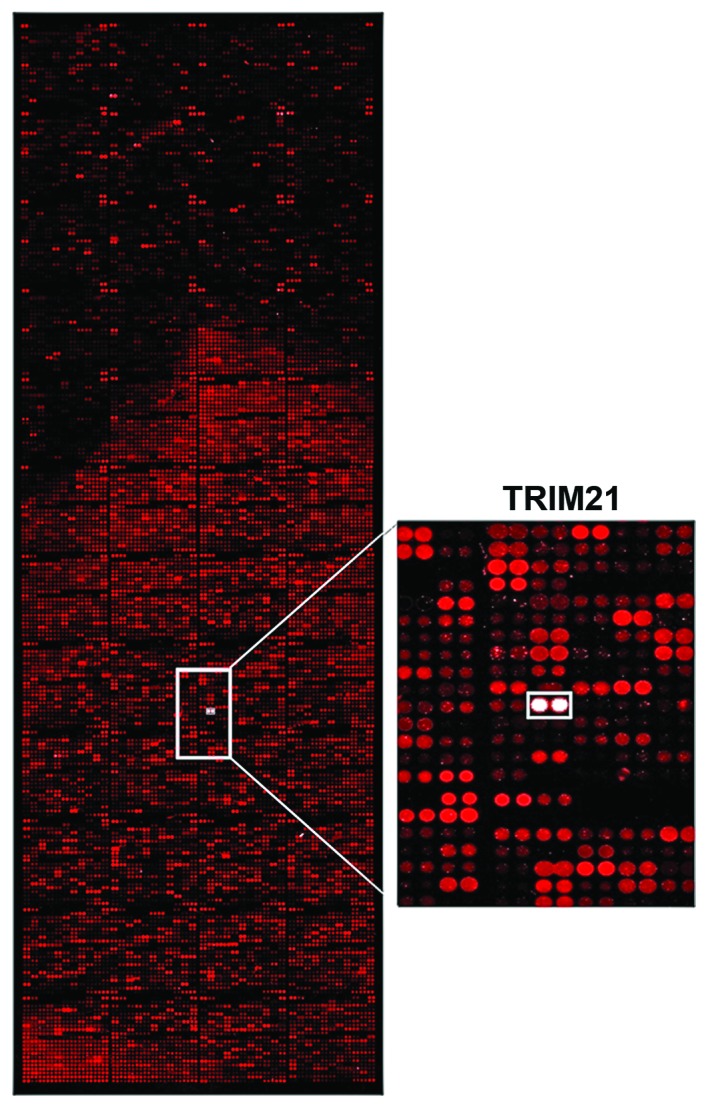
Analysis of the interaction. Array analysis of the interaction of over 9,000 different proteins with LFG. Recombinant LFG protein (10 μM) was used and detected by specific first antibody (FAIM2; Santa Cruz Biotechnology, Inc.) and fluorescence labeled second antibody (Alexa Fluor 546). The display window shows the signal stimulated by the interaction of LFG with TRIM21 (white signal).
Table I.
Z-score of protein-protein interaction array-analysis.
| Z-factor | Z-score | CI P-value | CV | Significance call | GenePix Flags | Description |
|---|---|---|---|---|---|---|
| 0.87066 | 3.585.224 | 0.0000008 | 0.04300 | Hit | 0 | Tripartite motif-containing 21 (TRIM21) |
| 0.87066 | 3.369.140 | 0.0000009 | 0.04300 | Hit | 0 | Tripartite motif-containing 21 (TRIM21) |
| 0.98147 | 1.299.203 | 0.0000059 | 0.00512 | Hit | 0 | General transcription factor II-I |
| 0.98147 | 1.289.273 | 0.0000059 | 0.00512 | Hit | 0 | General transcription factor II-I |
| 0.79642 | 1.071.535 | 0.0000084 | 0.06428 | Hit | 0 | Potassium voltage-gated channel, shaker-related subfamily, β member 1 (KCNAB1), transcript variant 1 |
| 0.87427 | 1.045.647 | 0.0000088 | 0.03992 | Hit | 0 | Similar to FRG1 protein (FSHD region gene 1 protein) (MGC72104) |
| 0.53896 | 1.044.110 | 0.0000088 | 0.15190 | Hit | 0 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ polypeptide (YWHAZ), transcript variant 1 |
| 0.78119 | 1.018.105 | 0.0000093 | 0.07059 | Hit | 0 | 14-3-3 protein ζ/δ |
| 0.04732 | 986.543 | 0.0000098 | 0.31200 | Hit | 0 | Nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 (NUDT16L1) |
| 0.87427 | 984.061 | 0.0000099 | 0.03992 | Hit | 0 | Similar to FRG1 protein (FSHD region gene 1 protein) (MGC72104) |
| 0.79642 | 971.767 | 0.0000101 | 0.06428 | Hit | 0 | Potassium voltage-gated channel, shaker-related subfamily, β member 1 (KCNAB1), transcript variant 1 |
| 0.78119 | 914.082 | 0.0000113 | 0.07059 | Hit | 0 | 14-3-3 protein ζ/δ |
| 0.53896 | 826.844 | 0.0000136 | 0.15190 | Hit | 0 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ polypeptide (YWHAZ), transcript variant 1 |
| 0.74035 | 809.350 | 0.0000142 | 0.08360 | Hit | 0 | Potassium voltage-gated channel, shaker-related subfamily, β member 2 (KCNAB2), transcript variant 1 |
| 0.63579 | 789.136 | 0.0000148 | 0.11980 | Hit | 0 | Small nuclear ribonucleoprotein polypeptide C (SNRPC) |
| 0.47616 | 783.226 | 0.0000150 | 0.17161 | Hit | 0 | WW domain containing transcription regulator 1 (WWTR1) |
| 0.74035 | 710.528 | 0.0000180 | 0.08360 | Hit | 0 | Potassium voltage-gated channel, shaker-related subfamily, β member 2 (KCNAB2), transcript variant 1 |
| 0.63579 | 654.025 | 0.0000208 | 0.11980 | Hit | 0 | Small nuclear ribonucleoprotein polypeptide C (SNRPC) |
| 0.43427 | 651.306 | 0.0000210 | 0.18583 | Hit | 0 | UBX domain containing 3 (UBXD3) |
Figure 2.
Co-immunoprecipitation. (A) Native western blot analysis after co-immunoprecipitation of LFG and TRIM21. LFG was specifically isolated from the cell lysate of MDA-MB-231 after transfection with vectors coding for LFG and TRIM21 (24 h) by use of antibody coated magnetic beads (Dynabeads; Invitrogen). Detection of LFG (FAIM2) and TRIM21 (Ssa1/2) (both from Santa Cruz Biotechnology, Inc.) by specific first antibodies and fluorescence labeled second antibodies. LFG is visible as a green signal (Alexa Fluor 546), TRIM21 is visible as a red signal (Alexa Fluor 488), and combined signals are visible as a yellow signal. (B) Analysis of LFG and TRIM21 protein in the MDA-MB-231 breast cancer cells 24 h after transfection with vectors coding for LFG and TRIM21, by specific first and fluorescence labeled second antibodies. a, Green signal for LFG; b, red signal for TRIM21; c, blue signal for DAPI-stained core; d, overlay of the single signals with yellow signals in places where green and red signals appear in the same spot.
Co-immunoprecipitation
The co-immunoprecipitation was performed to investigate a potential complex formation. The samples represent cell lysates from cells expressing the modified TRIM21 proteins as a result of transfection. To illustrate the bond between LFG and the different TRIM21 proteins, they had to be extracted by the co-immunoprecipitation in a pure form and without foreign protein interaction. By running a western blot analysis with fluorescent antibodies, the complex could be detected afterwards. A lack of a complex formation can therefore be assumed as a lack of the essential domain, whereby the interaction-depended domain can be identified. The samples 2–4 show clearly visible bands of ~80 and 60 kDa. The bands at 80 kDa are larger but slightly less visible than the bands at the height of 60 kDa. In the samples 1a and 1b no bands are visible (Fig. 3B).
Analysis of expression
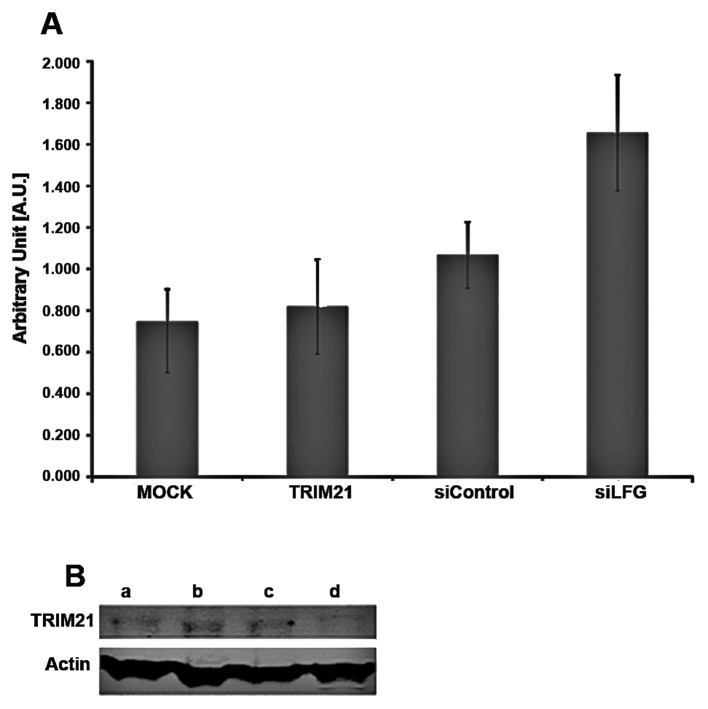
To investigate whether these two proteins influence the expression of each other, MDA-MB-231 cells were cultivated under different conditions. The expression of TRIM21 RNA-level was detected by real-time PCR using 1.0 μg total RNA for transcription to cDNA (Fig. 4A) and the amounts of LFG and TRIM21 protein were detected using PAGE followed by western blot analysis, using 25 μg total protein (Fig. 4B). The samples were treated identically for both analyses. For siLFG (Fig. 4A), MDA-MB-231 were transfected with an adenoviral vector coding for siRNA against LFG, and an empty vector was used for transfection in sample siControl. Untreated cells were used for MOCK, while cells of the sample TRIM21 were cultivated with culture medium containing TRIM21-recombinant protein. Samples MOCK, TRIM21 and siControl only differed slightly in RNA expression levels for TRIM21 (Fig. 4A). A distinct difference can be seen between the samples, MOCK showing the weakest, and siLFG producing the strongest signal. The order of increasing signal intensity of TRIM21 is MOCK, TRIM21, siControl and siLFG.
Figure 4.
Trim21 and LFG expression. (A) Results of real-time PCR-analysis of Trim21-expression after 24 h under different culture conditions. Mock, Untreated MDA-MB-231 as negative control, Trim21, 2.0 μl recombinant human Trim21 protein added to culture medium; siControl, adenoviral transfection of MDA-MB-231 with empty vector; siLFG, adenoviral transfection of MDA-MB-231 with vector coding for siRNA against LFG. (B) Expression analysis of LFG protein under different culture conditions by western blot analysis using 25 μg of total protein. a, Adenoviral transfection of MDA-MB-231 with vector coding for siRNA against LFG (24 h). b, Adenoviral transfection of MDA-MB-231 with empty vector. c, Untreated MDA-MB-231 as negative control. d, MDA-MB231 cultivated with 2.0 μg recombinant Trim21-protein in culture medium.
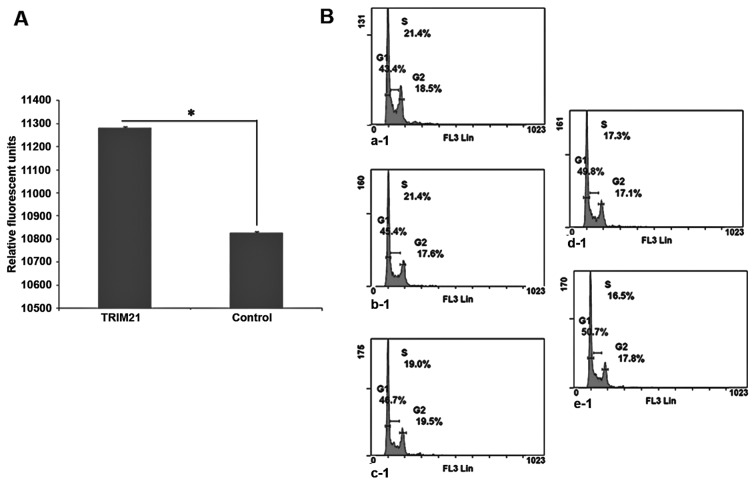
The detection of the proteins LFG, TRIM21 and actin after western blot analysis was carried out using fluorescence labeled antibodies. The protein actin was used as a housekeeping gene and showed comparable band intensities throughout the different samples (Fig. 4B). In the western blot analysis, the band intensity of TRIM21 in the gel differed only slightly between the samples a, siLFG; b, siControl; and c, MOCK whereas the band of sample d, TRIM21, is weaker. The bands of the protein LFG are strongest in samples b, siControl and c, MOCK and less in a, siLFG. The weakest band, however, results from sample d, TRIM21. To analyze if the apoptotic protection provided by LFG is influenced by TRIM21, MDA-MB-231 cell were cultivated with medium containing TRIM21 recombinant protein, and with medium not containing the protein for 48 h. After the addition of the agonistic anti-Fas-antibody, and further incubation for 24 h, the caspase-activity was tested using Apo-One (Promega). As the caspases cleave the substrate the degradation product is detectable by emitted fluorescence. Therefore, the signal intensity is proportional to the activity of the caspase-3 in the sample. The results show a much higher caspase-3 activity in the samples cultivated in the presence of TRIM21 recombinant protein (Fig. 5A). The signal intensity of the Control is reduced by approximately half compared to the sample with TRIM21 recombinant protein.
Figure 5.
Caspase-3-activity and cell cycle-analysis. (A) Analysis of caspase-3-activity in MDA-MB-231 cells. Cultivation for 48 h under different conditions and addition of 100 ng agonistic anti-Fas (clone CH11; Abcam). Measurement of caspase-3-activity after 24 h. TRIM21, addition of 1.0 μg/ml recombinant human TRIM21-protein every 24 h. Control, untreated MDA-MB-231 cultivated for 48 h. Detection of LFG, TRIM21 and actin by specific first antibodies and fluorescence labeled second antibodies. (B) Plots of cell cycle-analysis of MDA-MB-231 after 24 h cultivation in presence of different concentration of recombinant human TRIM21-protein after staining with propidium iodide. a-1, Cultivation of MDA-MB-231 without addition of TRIM21-recombinant protein. b-1, Addition of 0.25 μg/ml TRIM21-recombinant protein to the culture medium. c-1, Addition of 0.5 μg/ml TRIM21-recombinant protein to the culture medium. d-1, Addition of 0.75 μg/ml TRIM21-recombinant protein to the culture medium. e-1, Addition of 1.0 μg/ml TRIM21-recombinant protein to the culture medium.
It was necessary that yet another influence on MDA-MB-231 cells by TRIM21 should be analyzed in a cell cycle analysis. Therefore, a defined amount of cells were cultivated in the presence of different concentrations of TRIM21-recombinant protein within the culture medium for 24 h. Subsequently, the cells were separated from the culture flasks and fixed with 70% ethanol and stained with PI. The analysis was carried out using flow cytometry. On the basis of the plots in Fig. 5B a steady increase in the amount of cells in the G0/G1-phase can be seen, whereas, the percentage of cells in S-phase is steadily decreasing.
TRIM21-expression and its influence on cancer cells
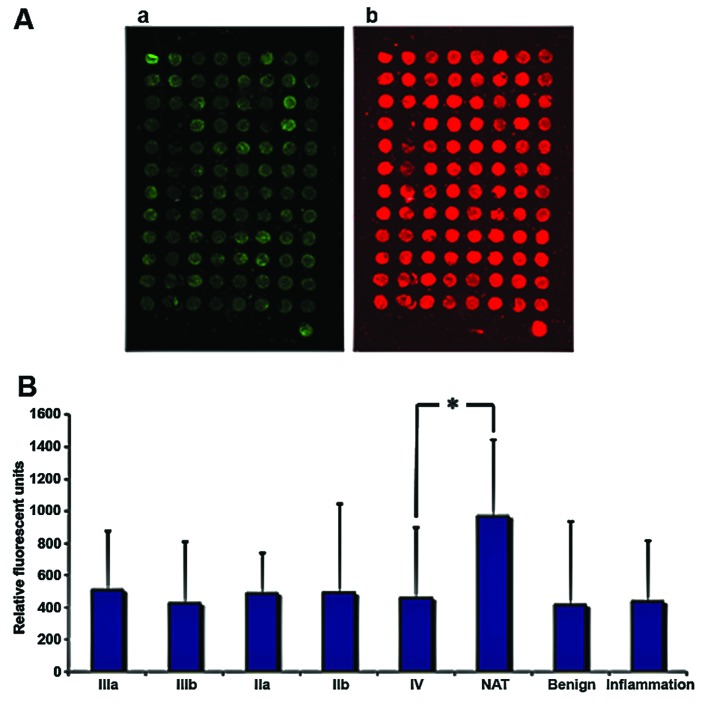
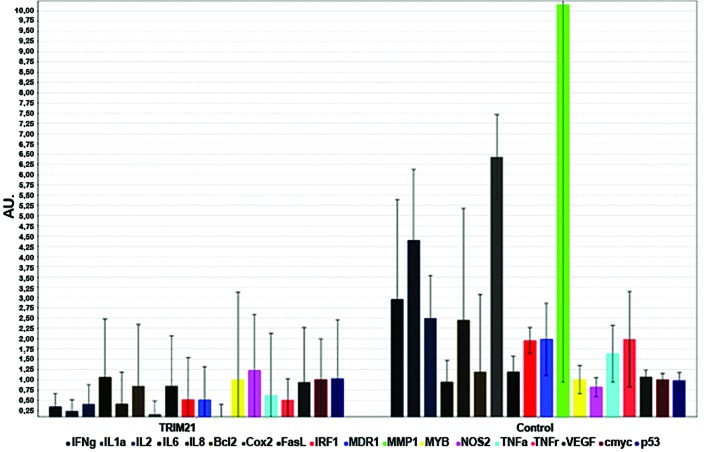
To examine how TRIM21 is expressed in breast cancer tissue, samples from tumors with different degrees of malignity were obtained from US Biomax, Inc. (Table II) and the protein TRIM21 was detected via fluorescence labeled antibodies. The samples were also stained with the nucleic acid stain Syto-60 as a reference. The expression of TRIM21 does not differ significantly throughout the varying degrees of malignity, but all samples derived from tumors or inflamed tissue show approximately half the signal intensity compared to the native tissue sample (Fig. 6). To analyze if TRIM21 protein, has the same inhibiting effect of activation when added to the medium as NF-κB in breast cancer cells, as shown by Niida et al (9), a real-time PCR was carried out using MDA-MB-231 cells cultivated with TRIM21 recombinant protein and no further supplements for the negative control. The expression of the most frequently NF-κB-regulated genes were detected, and an overall decrease can be seen in Fig. 7.
Table II.
Specification Sheet of breast cancer tissue array-analysis.
| Pos | No. | Gender | Age (years) | Organ | Pathology diagnosis | Grade | Stage | TNM | ER | PR | HER2 | Typea |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 1 | F | 42 | Breast | Normal breast tissue (hyalinosis) | - | - | - | N/A | - | N/A | NAT |
| A2 | 2 | F | 42 | Breast | Normal breast tissue | - | - | - | - | - | 0 | NAT |
| A3 | 3 | F | 28 | Breast | Plasma cell mastitis | - | - | - | - | - | 0 | Inflammation |
| A4 | 4 | F | 28 | Breast | Plasma cell mastitis | - | - | - | - | - | 0 | Inflammation |
| A5 | 5 | F | 32 | Breast | Plasma cell mastitis | - | - | - | +, 4% | +, 5% | 0 | Inflammation |
| A6 | 6 | F | 32 | Breast | Plasma cell mastitis | - | - | - | - | - | 0 | Inflammation |
| A7 | 7 | F | 35 | Breast | Plasma cell mastitis | - | - | - | - | - | 0 | Inflammation |
| A8 | 8 | F | 35 | Breast | Plasma cell mastitis | - | - | - | - | - | 0 | Inflammation |
| A9 | 9 | F | 28 | Breast | Adenosis | - | - | - | +, 40% | +, 40% | 0 | Benign |
| A10 | 10 | F | 28 | Breast | Adenosis | - | - | - | - | - | 0 | Benign |
| A11 | 11 | F | 49 | Breast | Adenosis | - | - | - | ++, 30% | +, 35% | 0 | Benign |
| A12 | 12 | F | 49 | Breast | Adenosis | - | - | - | ++, 88% | 0,0085 | 0 | Benign |
| B1 | 13 | F | 44 | Breast | Adenosis | - | - | - | ++, 8% | ++, 70% | 1+ | Benign |
| B2 | 14 | F | 44 | Breast | Adenosis | - | - | - | +, 5% | ++, 3% | 0 | Benign |
| B3 | 15 | F | 58 | Breast | Fibroadenoma | - | - | - | - | - | 0 | Benign |
| B4 | 16 | F | 58 | Breast | Fibroadenoma | - | - | - | - | - | 0 | Benign |
| B5 | 17 | F | 22 | Breast | Fibroadenoma | - | - | - | ++, 10% | ++, 10% | 0 | Benign |
| B6 | 18 | F | 22 | Breast | Fibroadenoma | - | - | - | 0,0096 | +++, 70% | 0 | Benign |
| B7 | 19 | F | 54 | Breast | Fibroadenoma | - | - | - | - | - | 0 | Benign |
| B8 | 20 | F | 54 | Breast | Fibroadenoma | - | - | - | - | - | 0 | Benign |
| B9 | 21 | F | 34 | Breast | Invasive ductal carcinoma | 1 | IIb | T2N1M0 | - | - | 1+ | Malignant |
| B10 | 22 | F | 34 | Breast | Invasive ductal carcinoma | 1 | IIb | T2N1M0 | - | - | 1+ | Malignant |
| B11 | 23 | F | 43 | Breast | Invasive ductal carcinoma | 1 | IIb | T3N0M0 | - | - | 2+ | Malignant |
| B12 | 24 | F | 43 | Breast | Invasive ductal carcinoma | 1 | IIb | T3N0M0 | - | - | 2+ | Malignant |
| C1 | 25 | F | 40 | Breast | Invasive ductal carcinoma | 1 | IIb | T2N1M0 | N/A | N/A | N/A | Malignant |
| C2 | 26 | F | 40 | Breast | Invasive ductal carcinoma | 1 | IIb | T2N1M0 | +, 60% | - | 0 | Malignant |
| C3 | 27 | F | 50 | Breast | Invasive ductal carcinoma | 1 | IIb | T3N0M0 | - | - | 1+ | Malignant |
| C4 | 28 | F | 50 | Breast | Invasive ductal carcinoma | 1 | IIb | T3N0M0 | - | - | 1+ | Malignant |
| C5 | 29 | F | 37 | Breast | Invasive ductal carcinoma | 1 | IIa | T2N0M0 | - | - | 0 | Malignant |
| C6 | 30 | F | 37 | Breast | Invasive ductal carcinoma | 1 | IIa | T2N0M0 | - | - | 0 | Malignant |
| C7 | 31 | F | 46 | Breast | Invasive ductal carcinoma | 1 | IIIa | T3N2M0 | - | - | 2+ | Malignant |
| C8 | 32 | F | 46 | Breast | Invasive ductal carcinoma (fibrous tissue and blood vessel) | - | IIIa | T3N2M0 | - | - | 1+ | Malignant |
| C9 | 33 | F | 69 | Breast | Invasive ductal carcinoma (fibrofatty tissue and blood vessel) | - | IIa | T2N0M0 | - | - | 0 | Malignant |
| C10 | 34 | F | 69 | Breast | Invasive ductal carcinoma | 1 | IIa | T2N0M0 | - | - | 0 | Malignant |
| C11 | 35 | F | 52 | Breast | Invasive ductal carcinoma | 1 | IIIb | T4N1M0 | - | - | 1+ | Malignant |
| C12 | 36 | F | 52 | Breast | Invasive ductal carcinoma | 1 | IIIb | T4N1M0 | - | - | 1+ | Malignant |
| D1 | 37 | F | 65 | Breast | Invasive ductal carcinoma | 2 | IV | T3N1M1 | - | - | 1+ | Malignant |
| D2 | 38 | F | 65 | Breast | Invasive ductal carcinoma | 2 | IV | T3N1M1 | - | - | 1+ | Malignant |
| D3 | 39 | F | 45 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | +, 50% | ++, 90% | 0 | Malignant |
| D4 | 40 | F | 45 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | +, 65% | ++, 95% | 0 | Malignant |
| D5 | 41 | F | 37 | Breast | Invasive ductal carcinoma | 1 | IIIa | T3N1M0 | - | - | 1+ | Malignant |
| D6 | 42 | F | 37 | Breast | Invasive ductal carcinoma | 1 | IIIa | T3N1M0 | - | - | 1+ | Malignant |
| D7 | 43 | F | 55 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 0 | Malignant |
| D8 | 44 | F | 55 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 0 | Malignant |
| D9 | 45 | F | 55 | Breast | Invasive ductal carcinoma | 2 | IIIb | T4N1M0 | - | - | 1+ | Malignant |
| D10 | 46 | F | 55 | Breast | Invasive ductal carcinoma | 2 | IIIb | T4N1M0 | - | - | 1+ | Malignant |
| D11 | 47 | F | 75 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 0 | Malignant |
| D12 | 48 | F | 75 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 0 | Malignant |
| E1 | 49 | F | 41 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | +, 8% | 0 | Malignant |
| E2 | 50 | F | 41 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | +, 5% | 0 | Malignant |
| E3 | 51 | F | 44 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 2+ | Malignant |
| E4 | 52 | F | 44 | Breast | Invasive ductal carcinoma (sparse) | 1 | IIb | T2N1M0 | - | - | 1+ | Malignant |
| E5 | 53 | F | 37 | Breast | Invasive ductal carcinoma | 2 | IIb | T3N0M0 | - | - | 1+ | Malignant |
| E6 | 54 | F | 37 | Breast | Invasive ductal carcinoma | 2 | IIb | T3N0M0 | - | - | 1+ | Malignant |
| E7 | 55 | F | 61 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | ++, 90% | - | 0 | Malignant |
| E8 | 56 | F | 61 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | ++, 95% | 0,20% | 0 | Malignant |
| E9 | 57 | F | 38 | Breast | Invasive ductal carcinoma (sparse) | 2 | IIb | T3N0M0 | - | - | 0 | Malignant |
| E10 | 58 | F | 38 | Breast | Invasive ductal carcinoma | 2 | IIb | T3N0M0 | - | - | 0 | Malignant |
| E11 | 59 | F | 65 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | +++, 100% | +++, 97% | 0 | Malignant |
| E12 | 60 | F | 65 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | +++, 99% | +++, 99% | 0 | Malignant |
| F1 | 61 | F | 53 | Breast | Invasive ductal carcinoma (sparse) | 2 | IIa | T2N0M0 | - | - | 1+ | Malignant |
| F2 | 62 | F | 53 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | - | 1+ | Malignant |
| F3 | 63 | F | 32 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 1+ | Malignant |
| F4 | 64 | F | 32 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 0 | Malignant |
| F5 | 65 | F | 56 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 1+ | Malignant |
| F6 | 66 | F | 56 | Breast | Invasive ductal carcinoma | 2 | IIb | T2N1M0 | - | - | 1+ | Malignant |
| F7 | 67 | F | 52 | Breast | Invasive ductal carcinoma (sparse) | 2 | IIIa | T2N2M0 | +++, 98% | - | 0 | Malignant |
| F8 | 68 | F | 52 | Breast | Invasive ductal carcinoma | 2 | IIIa | T2N2M0 | ++, 75% | - | 0 | Malignant |
| F9 | 69 | F | 37 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| F10 | 70 | F | 37 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| F11 | 71 | F | 42 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | - | 1+ | Malignant |
| F12 | 72 | F | 42 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | - | 1+ | Malignant |
| G1 | 73 | F | 54 | Breast | Invasive ductal carcinoma | 2 | IIb | T3N0M0 | - | - | 2+ | Malignant |
| G2 | 74 | F | 54 | Breast | Invasive ductal carcinoma | 2 | IIb | T3N0M0 | - | - | 2+ | Malignant |
| G3 | 75 | F | 40 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| G4 | 76 | F | 40 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| G5 | 77 | F | 59 | Breast | Invasive ductal carcinoma | 2 | IIIa | T2N2M0 | - | - | 1+ | Malignant |
| G6 | 78 | F | 59 | Breast | Invasive ductal carcinoma | 2 | IIIa | T2N2M0 | - | - | 1+ | Malignant |
| G7 | 79 | F | 42 | Breast | Invasive ductal carcinoma | 3 | IIIb | T4N0M0 | - | - | 0 | Malignant |
| G8 | 80 | F | 42 | Breast | Invasive ductal carcinoma | 3 | IIIb | T4N0M0 | - | - | 0 | Malignant |
| G9 | 81 | F | 54 | Breast | Invasive ductal carcinoma | 3 | IIa | T2N0M0 | +++, 100% | ++, 5% | 0 | Malignant |
| G10 | 82 | F | 54 | Breast | Invasive ductal carcinoma | 3 | IIa | T2N0M0 | +++, 100% | +++, 3% | 0 | Malignant |
| G11 | 83 | F | 60 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| G12 | 84 | F | 60 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| H1 | 85 | F | 49 | Breast | Invasive ductal carcinoma | 2 | IIIa | T2N2M0 | +, 80% | +, 50% | 3+ | Malignant |
| H2 | 86 | F | 49 | Breast | Invasive ductal carcinoma | 2 | IIIa | T2N2M0 | +, 50% | +, 40% | 1+ | Malignant |
| H3 | 87 | F | 27 | Breast | Invasive ductal carcinoma | 2 | IIIb | T4N2M0 | - | - | 2+ | Malignant |
| H4 | 88 | F | 27 | Breast | Invasive ductal carcinoma | 2 | IIIb | T4N2M0 | - | - | 2+ | Malignant |
| H5 | 89 | F | 49 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | - | 2+ | Malignant |
| H6 | 90 | F | 49 | Breast | Invasive ductal carcinoma | 2 | IIa | T2N0M0 | - | - | 2+ | Malignant |
| H7 | 91 | F | 71 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| H8 | 92 | F | 71 | Breast | Invasive ductal carcinoma | 3 | IIb | T2N1M0 | - | - | 0 | Malignant |
| H9 | 93 | F | 50 | Breast | Invasive lobular carcinoma | - | IIIa | T2N2M0 | +++, 90% | ++, 10% | 0 | Malignant |
| H10 | 94 | F | 50 | Breast | Invasive lobular carcinoma | - | IIIa | T2N2M0 | +++, 85% | ++, 1% | 0 | Malignant |
| H11 | 95 | F | 42 | Breast | Invasive lobular carcinoma | - | IIb | T2N1M0 | ++, 60% | +++, 60% | 0 | Malignant |
| H12 | 96 | F | 42 | Breast | Invasive lobular carcinoma | - | IIb | T2N1M0 | ++, 65% | +++, 40% | 0 | Malignant |
| - | - | M | 42 | Adrenal gland | Pheochromocytoma (tissue marker) | - | Malignant |
For precise diagnosis, refer to pathology description. The grade 1–3 (or I–III) in Pathology Diagnosis is equivalent to well-differentiated, moderately-differentiated or poorly differentiated, respectively, under microscope. Grade 1 or well-differentiated, cells appear normal and are not growing rapidly. Grade 2 or moderately-differentiated: cells appear slightly different than normal. Grade 3 or poorly differentiated, cells appear abnormal and tend to grow and spread more aggressively. Grade 4 or undifferentiated, *(for certain tumors), features are not significantly distinguishing to make it look any different from undifferentiated cancers which occur in other organs. Staining scoring: −, no staining; +−, borderline staining; +, weak staining; ++, moderate staining; +++, strong staining; %, percentage of positive cells. TNM grading: T, primary tumor; Tx, primary tumor cannot be assessed; T0, no evidence of primary tumor; Tis, carcinoma in situ; intraepithelial or invasion of lamina propria; T1, tumor invades submucosa; T2, tumor invades muscularis propria; T3, tumor invades through muscularis propria into subserosa or into non-peritonealized pericolic or perirectal tissues; T4, tumor directly invades other organs or structures and/or perforate visceral peritoneum; N, regional lymph nodes; Nx, regional lymph nodes cannot be assessed; N0, no regional lymph node metastasis; N1, metastasis in 1 to 3 regional lymph nodes; N2, metastasis in 4 or more regional lymph nodes; M, distant metastasis; Mx, distant metastasis cannot be assessed; M0, no distant metastasis; M1, distant metastasis.
Figure 6.
Arrays. (A) Tissue samples of breast tumors having different degrees of malignity (US Biomax, Inc.) (a and b). Specific antibody against Trim21 and fluorescence labeled second antibody (Li-Cor-800), and staining of nucleic acid with Syto-60. Detection was carried out using Odyssey (Li-Cor Biosciences). (B) Bar chart of the fluorescence intensity of the tissue samples, degrees of malignity (IIa, IIb, IIIa, IIIb and IV); *P<0.05 vs. control (NAT). NAT, native tissue sample. Benign, benign breast tumor. Inflammation, inflamed tissue sample.
Figure 7.
NF-κB array. Bar chart of gene expression of common NF-κB regulated genes after real-time PCR-analysis. Trim21, MDA-MB-231 cultivated with 2.0 μg/ml recombinant human Trim21-protein for 24 h. Control, MDA-MB-231 cells cultivated for 24 h under standard conditions.
Discussion
We demonstrated here that LFG interacts with TRIM21, an E3-ubiquitin ligase that can ubiquitinate itself, IRF transcription factors, and p27 cell cycle inhibitor (12–14). The strong interaction which was detectable in the array-analysis, could be confirmed by co-immunoprecipitation whereby isolating LFG from the cell lysate of MDA-MB-231 cells. A complex of LFG and TRIM21 could be detected, as both proteins are detectable in the same position on the blot membrane, although being of different molecular weight. By detecting the proteins in intact and fixed MDA-MB-231 cells, the localization of the complexes could be detected predominantly in close proximity to the cell core.
With the co-immunoprecipitation and subsequent western blot analysis, the result shown in Fig. 3A were obtained. The bands at the size of ~80 kDa of the samples 2–4 are evidence of the complex formation between TRIM21 (52 kDa) and LFG (35 kDa). Thus, an interaction between the two proteins is still possible despite a deletion of the domains 2–4 of TRIM21. In the samples 1a and 1b, the band is missing which indicates a missed complex formation. Thereby, the domains 1a and 1b can be explained as essential domains for the connection between TRIM21 and LFG. Further, one additional band occurs in the samples 2–4 at the size of ~60 kDa. It is possible that a part separated from the protein complex. In a study of Dastagir et al (15) in 2014 another form of LFG was examined, the β-isoform. This is a shorter form of Lifeguard and is also expressed in breast cancer cell lines. The study even showed that the smaller isoform of the Lifeguard enacts in an equally important role in the development of breast cancer as LFG itself (15). Hence, the band at the size of 60 kDa possibly indicates a bond between TRIM21 and β isoform of Lifeguard. One could verify this hypothesis by using a native gel instead of an SDS gel so that the proteins are examined individually. The samples 1a and 1b shown in this experiment are the samples containing the TRIM21 protein without the C-terminal SPRY domain and the PRY subdomain. Approximately 100 human proteins have this ~140–200 amino acid domain, wherein it is contained in 40 of the 66 human TRIM proteins. A possible role for this domain has been discovered by Nisole et al (8) in 2005. However, a closer examination of the TRIM5α protein of the primate turned out that it is able to intervene in the replication cycle of various DNA and RNA viruses. As a consequence, the reverse transcription of the viral genome is suppressed. This antiviral activity is, however, completely lost by exchanging a proline in the amino acid sequence of the SPRY domain, so this domain constitutes the basis of the inteference between TRIM5α and viruses. This function dependent amino acid is not only located in the SPRY domain, but also in his PRY subdomain. Such a mutation is the possible reason for the weak bond between the human TRIM5α protein and the capsid of human immunodeficiency virus-1 (HIV-1), whereby a possible human immunity against the virus cannot be ensured. Thus, both the SPRY, as well as the PRY domain seem to be crucial for an interaction with different types of viruses (8). Furthermore, the E3-ubiquitin ligase activity is not only by the RING domain, but also seems to be dependent of the SPRY domain. The TRIM27 protein binds to the NOD2 protein that recruits a K48-dependent ubiquitination, followed by a subsequent degradation in the proteasome. These antiviral activities, as well as the already illustrated interaction between TRIM21 and the Fc region of IgG using the SPRY and PRY domain indicate an essential role of SPRY and PRY domain in the immune response. The binding of TRIM21 with LFG using the SPRY and PRY domain points out a potential new function of these domains. Until now, it was assumed that the function of these domains is limited on the immune response, whereby the evolutionary benefit by the mutation of the domain in human TRIM proteins and thus the loss a possible immunity to different viruses was a mystery. But the latest results of the working group Bucan allude to a possible new function of TRIM21 protein in humans: The interaction with Lifeguard caused sensitivity to the induced apoptosis at the SPRY and PRY domain playing a regulatory role (8).
To find out if this interaction affects the expression of LFG and TRIM21, real-time PCR and western blot analysis were carried out. Concerning TRIM21, an increase of expression on the RNA level in cells with down regulated LFG-expression was detected. However, the differences were rather small and could not be detected on the protein level in western blot analysis, which results from the low expression of TRIM21 in MDA-MB-231 cells. For LFG however, a more distinct difference in expression could be seen as the expression of LFG within the sample cultivated in the presence of TRIM21 protein is even lower than in the sample transfected with the siRNA against LFG. In the following caspase-3-activity analysis, the anti-apoptotic effect of LFG in the presence of TRIM21 was examined. Results showed a much higher caspase-3-activity in cells cultivated with TRIM21, which could result from an increased LFG-expression due to TRIM21. As Jauharoh et al (11) predicted, TRIM21 acts as a mediator for the proapoptotic signals. The increase in caspase-activity in our analysis cannot be definitely attributed to a decrease of LFG-expression. The results obtained by Jauharoh et al (11) can, however, not be directly compared to our analysis, as they used cell flow cytometric detection of Annexin IV as evidence for the apoptotic processes. Furthermore, they used HeLa cells for the analysis, which express TRIM21 in significantly higher levels than MDA-MB-231, and a Fas-antibody was added in combination with INFγ, not merely as a supplement.
A positive effect of TRIM21 concerning the field of oncology could be shown by cell cycle analysis. Within this test a correlation between TRIM21 concentration and a higher amount of resting cells in G0/G1-phase and a decrease of cells in S-phase could be shown. A contrary effect of TRIM21 was predicted by Sabile et al (12), who described TRIM21 as an essential mediator of ubiquitinylation for phosphorylated p27, a tumor-suppressor which prevents cells from entering S-phase. To examine how TRIM21 is actually expressed in breast tumors throughout different degrees of malignity, corresponding tissue samples were obtained from US Biomax, Inc. and TRIM21 was detected using fluorescence labeled antibodies against TRIM21. The expression levels of TRIM21 do not differ distinctly throughout the different degrees of malignity of the inflamed tissue sample but all of these samples show only around half of the expression levels, compared to native tissue. The results we obtained are, however, more consistent with the fact that TRIM21 inhibits activation of NF-κB, a family of transcription factors which are mainly responsible for the activation and promotion of cell division. That the protein form of TRIM21 can be transported into the cell could be shown by the NF-κB array. The inhibiting effect of TRIM21 towards NF-κB becomes clear due to the decrease in expression of the analyzed NF-κB-regulated genes. A possible mode of regulation of LFG expression by TRIM21 could be achieved by inhibition of NF-κB-activation. As the expression of LFG is regulated partly by NF-κB, its inhibition by TRIM21, as shown by Niida et al (9), could follow the schema displayed in. The reduced activity of NF-κB would therefore result in a reduced expression of LFG. This hypothesis however, does not involve an interaction between the two proteins.
The obtained results reveal TRIM21 as a potential instrument to affect the anti-apoptotic effect of LFG, which keeps tumor cells alive and even protects them from certain drugs (Bucan et al (6). To reveal the detailed characteristics of their interaction, and the regulating effect that TRIM21 seems to have on the expression of LFG, further analyses are required.
Acknowledgements
The study was funded by the Niedersächsische Krebs-gesellschaft. The authors are grateful to Andrea Lazaridis for excellent technical assistance.
References
- 1.Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM. LFG: An anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci USA. 1999;96:12667–12672. doi: 10.1073/pnas.96.22.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walczak H, Krammer PH. The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp Cell Res. 2000;256:58–66. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- 3.Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schweitzer B, Taylor V, Welcher AA, McClelland M, Suter U. Neural membrane protein 35 (NMP35): A novel member of a gene family which is highly expressed in the adult nervous system. Mol Cell Neurosci. 1998;11:260–273. doi: 10.1006/mcne.1998.0697. [DOI] [PubMed] [Google Scholar]
- 5.Bucan V, Reimers K, Choi CY, Eddy MT, Vogt PM. The anti-apoptotic protein lifeguard is expressed in breast cancer cells and tissues. Cell Mol Biol Lett. 2010;15:296–310. doi: 10.2478/s11658-010-0009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucan V, Choi CY, Lazaridis A, Vogt PM, Reimers K. Silencing of anti-apoptotic transmembrane protein lifeguard sensitizes solid tumor cell lines MCF-7 and SW872 to perifosine-induced cell death activation. Oncol Lett. 2011;2:419–422. doi: 10.3892/ol.2011.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa A, Zhou W, Ek M, Hedlund M, Brauner S, Popovic K, Horvath L, Wallerskog T, Oukka M, Nyberg F, et al. The Sjogren's syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- 8.Nisole S, Stoye JP, Saïb A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 9.Niida M, Tanaka M, Kamitani T. Downregulation of active IKK beta by Ro52-mediated autophagy. Mol Immunol. 2010;47:2378–2387. doi: 10.1016/j.molimm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Pettaway CA, Uehara H, Bucana CD, Fidler IJ. Blockade of NF-kappaB activity in human prostate cancer cells is associated with suppression of angiogenesis, invasion, and metastasis. Oncogene. 2001;20:4188–4197. doi: 10.1038/sj.onc.1204535. [DOI] [PubMed] [Google Scholar]
- 11.Jauharoh SNA, Saegusa J, Sugimoto T, Ardianto B, Kasagi S, Sugiyama D, Kurimoto C, Tokuno O, Nakamachi Y, Kumagai S, et al. SS-A/Ro52 promotes apoptosis by regulating Bcl-2 production. Biochem Biophys Res Commun. 2012;417:582–587. doi: 10.1016/j.bbrc.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Sabile A, Meyer AM, Wirbelauer C, Hess D, Kogel U, Scheffner M, Krek W. Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol Cell Biol. 2006;26:5994–6004. doi: 10.1128/MCB.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozato K, Shin DM, Chang TH, Morse HC., III TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, III, et al. Cutting edge: Autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 15.Dastagir N, Lazaridis A, Dastagir K, Reimers K, Vogt PM, Bucan V. Role of lifeguard β-isoform in the development of breast cancer. Oncol Rep. 2014;32:1335–1340. doi: 10.3892/or.2014.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]