Abstract
Ribosomally synthesized peptides are generally limited to l-amino acid building blocks. Given the advantageous properties of peptides containing d-amino acids such as stabilization of certain turns and against proteolytic degradation, methods to introduce d-stereocenters are valuable. Here we report the first in vitro reconstitution and characterization of a dehydrogenase that carries out the asymmetric reduction of dehydroalanine. NpnJA reduces dehydroalanine to d-Ala using NAPDH as cosubstrate. The enzyme displays high substrate tolerance allowing introduction of d-Ala into a range of non-native substrates. In addition to the in vitro reactions, we describe five examples of using Escherichia coli as biosynthetic host for d-alanine introduction into ribosomal peptides. A deuterium-label-based coupled-enzyme assay was used to rapidly determine the stereochemistry of the newly installed alanine.
The d-amino acids impart a range of favorable properties onto peptides, such as reduced susceptibility to proteolysis,1−4 induction of turns,5 and self-assembly into higher order structures.6 Unfortunately, the ribosomal machinery is generally specific for l-amino acids.7,8 Hence, preparation of d-amino acid-containing peptides has been limited mostly to chemical synthesis. In the past decade or so, a number of enzymes have been reported that post-translationally introduce d-stereocenters into peptides, including epimerases that convert l-amino acids into d-amino acids via a deprotonation–protonation mechanism.9,10 Such enzymes have been isolated from frog,11 spider,9,12 platypus,13 and sponges.10 The substrate specificity of the enzymes from frog and platypus has been investigated, showing that they act specifically on the second amino acid from the N-terminus,13,14 whereas the spider enzyme acts on the third residue from the C-terminus.9 These isomerases have additional disadvantages for synthetic purposes in that they thus far have been isolated from the native organisms and hence have limited supply, and they tend to give a thermodynamic mixture of products.
A very different means of introducing d-stereocenters was recently reported that involves radical chemistry.15 During biosynthesis of proteusin natural products in cyanobacteria, radical-S-adenosyl methionine proteins catalyze the irreversible epimerization of multiple residues in their substrate peptides.10 Thus, these enzymes offer considerable potential for introducing d-amino acids into ribosomally synthesized peptides, although their sequence specificity has not yet been investigated in detail.
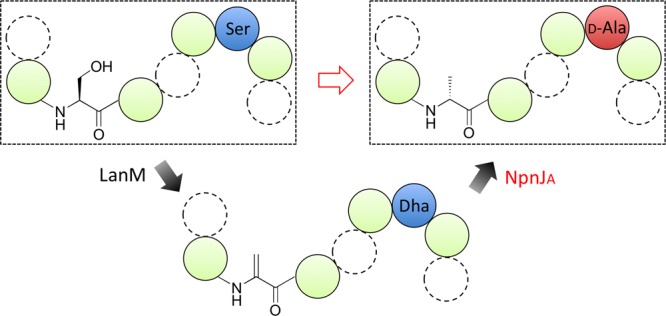
A final method to introduce d-amino acids into ribosomally synthesized peptides involves enzymes found in lanthipeptide biosynthesis. Like the proteusins, lanthipeptides are ribosomally synthesized and post-translationally modified peptides (RiPPs).16 They contain thioether residues termed lanthionine (Lan) and methyllanthionine (MeLan) that are formed by addition of a Cys residue to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively.17 These dehydro amino acids are formed by dehydration of Ser and Thr residues, respectively (Figure 1). A small subclass of lanthipeptides contains d-amino acids that are formed by hydrogenation of Dha and/or Dhb. The resulting d-Ala and d-amino butyric acid residues have been found in lactosin S,18 lacticin 3147,19,20 and carnolysin.21 The enzymes that carry out the reduction reactions are encoded in their biosynthetic gene clusters and fall into two dehydrogenase classes that collectively have been termed LanJ enzymes.16 The protein LtnJ involved in lacticin 3147 biosynthesis is a member of the zinc-dependent dehydrogenases and was the first functionally identified dehydro amino acid reductase.20 Our bioinformatics analysis shows that the enzyme in carnolysin biosynthesis (CrnJ) belongs to the flavin oxidoreductase superfamily. CrnJ-like homologues are also found in other lanthipeptide biosynthetic clusters that have not been fully characterized, such as lactosin S (Figure S1). We propose here to designate these two enzyme classes LanJA and LanJB, respectively. The proteins involved in the biosynthesis of lacticin 3147 and carnolysin have been successfully used in heterologous expression systems,21−23 but thus far, their activity has not been reconstituted in vitro and their substrate specificity has not been investigated in great detail. We recently reported the presence of a LanJA homologue in the cyanobacterium Nostoc punctiforme PCC 73102,24 and in this work, we reconstituted the activity of this NpnJA enzyme in vitro and show that its substrate specificity is promiscuous such that it can be used to introduce d-Ala into a variety of peptides.
Figure 1.
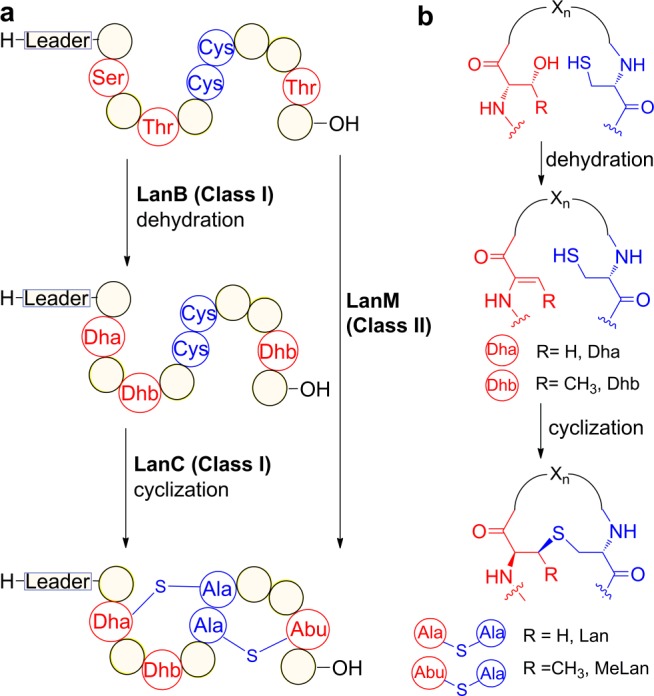
(a) Post-translational maturation of class I and II lanthipeptides. (b) Generation of (methyl)lanthionine motifs.
Lanthipeptides are generated from a precursor peptide containing an N-terminal leader sequence and a C-terminal core peptide. The leader peptide guides the dehydration and cyclization reactions (Figure 1). For class I lanthipeptides, the dehydration and cyclization processes are carried out by two distinct enzymes LanB and LanC, whereas for class II lanthipeptides, a single bifunctional enzyme LanM catalyzes both reactions (Figure 1a).
The gene cluster in N. punctiforme PCC 73102 is unusual in that it contains several Ser/Thr-rich substrate peptides that do not contain Cys residues. These Cys-free peptides appear to have co-evolved with a LanM enzyme (NpnM) that has an intact dehydration site but lacks several residues that are critical for cyclization activity.24 We predicted that this biosynthetic machinery might lead to the production of linear d-amino acid-containing peptides. However, previous attempts in our laboratory and that of others to express NpnJA in E. coli or Lactococcus lactis were unsuccessful.23,24
In this study, we obtained soluble and active NpnJA by expression in E. coli with maltose-binding protein (MBP) attached to its N-terminus. The dehydrated substrate NpnA3 was obtained by co-expression of the N-terminally hexahistidine tagged peptide (His6-NpnA3) in E. coli with NpnM and purification by Ni2+-affinity chromatography. This procedure resulted in formation of one Dha and three Dhb residues in His6-NpnA3 as determined by matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) (Figure 2a). Incubation of MBP-NpnJAin vitro with dehydrated His6-NpnA3 in the presence of NADPH resulted in formation of a product that had increased in mass by 2 Da as shown by MALDI-TOF MS (Figure 2a). NADH was not accepted as cosubstrate (Figure S2). Tandem MS analysis demonstrated that the Dha residue was selectively reduced (Figure 2b). In order to determine the stereochemistry of the newly formed alanine, endoproteinase LysC was used to remove the leader peptide, and the resulting purified peptide was hydrolyzed in 6 M HCl and derivatized using Marfey’s reagent.25 Liquid chromatography (LC)-MS analysis revealed the presence of l- and d-Ala stereoisomers in a 2:1 ratio. Since the peptide after LysC digestion contains two gene-encoded l-Ala residues, the new Ala that originated from Ser has the d-configuration (Figure 2c). Interestingly, MBP-NpnJA was also able to reduce the dehydrated core peptide in the absence of the leader peptide (Figure S3). Such leader-independent activity has also been reported for some other tailoring enzymes during RiPP biosynthesis16 and indicated that NpnJA might have relatively relaxed substrate specificity.
Figure 2.
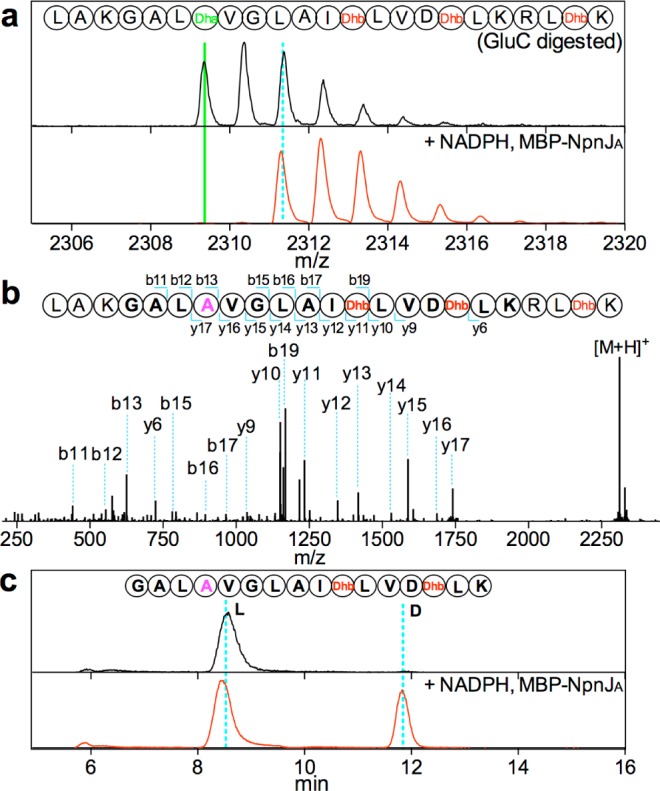
(a) MALDI-TOF mass spectra showing the 2 Da increase (solid green to dashed blue line) upon treatment of dehydrated NpnA3 with MBP-NpnJA and NADPH and subsequent treatment with GluC. (b) ESI-Tandem MS confirming that the Dha was selectively reduced to Ala (magenta). (c) Extracted ion chromatogram of the derivatized Ala showing the presence of a 2:1 ratio of l-Ala to d-Ala in the LysC-digested core peptide fragment (sequence shown in top panel), indicating the newly generated Ala had the d-configuration.
Next, we investigated whether MBP-NpnJA would be able to generate d-Ala at non-native positions. The three Thr residues in the NpnA3 core peptide were individually mutated to Ser, and the mutant peptides (T19S, T23S, and T28S) were co-expressed with NpnM to generate dehydroalanines at these positions. In vitro incubation of the dehydrated peptides with MBP-NpnJA resulted in reduction at Dha19 and Dha28, but not Dha23 as determined by tandem MS (Figure S4). The observation that Dha23 was not reduced could potentially be a consequence of the N-terminal flanking Asp22 residue. For instance, a deactivating effect of flanking charged residues was reported in the case of the homologous enzyme LtnJ in L. lactis.26 Indeed, by mutating Asp22 to Ala, Dha23 was reduced by MBP-NpnJA (Figure S4). To further test the influence from the flanking residues, a series of dehydrated NpnA3 mutant peptides was tested in vitro (Table S1). In addition to the hydrophobic residues that flank Dha in wt NpnA3, these mutants demonstrated that NpnJA also tolerates a large number of other flanking residues. Thus, the enzyme is very flexible with respect to the positional specificity and flanking residues. It is selective for reduction of Dha residues and, unlike the LanJB enzyme CrnJ,21 does not reduce Dhb.
Given the recently described ability of the sequence of lanthipeptide substrates to determine the stereochemical outcome of enzymatic reactions,27,28 it was important to verify that Dha reduction at non-native positions still resulted in d-Ala. At this point, a challenge arose when we tried to characterize the stereochemistry of the newly formed Ala residues using the traditional l/d ratio-based strategy: As a consequence of the presence of small amounts of partially dehydrated intermediates that could not be separated from the fully dehydrated and reduced core peptide by HPLC under the conditions used (Figure S4), the relative amount of l-Ala is increased in the final product, which makes it difficult to determine the l/d ratio in the product. To differentiate the newly generated Ala from the existing Ala residues, we developed an in vitro coupled-enzyme assay in D2O using deuterium-labeled cofactor (NADP2H) and an engineered phosphite dehydrogenase (PTDH) as cofactor regenerating catalyst (Figure 3a).29,30 Incubation of the Dha-containing substrate together with MBP-NpnJA, PTDH, NADP+, and deuterium-labeled phosphite in D2O, resulted in deuterium labeling at the α and β carbons of the newly formed alanine, which, after hydrolysis and derivatization, could be readily analyzed independently from the unlabeled l-Ala residues by LC/MS (Figure 3b). With this method, we confirmed the d-configuration of the newly formed alanine at the non-native positions (Figures 3c and S4j).
Figure 3.
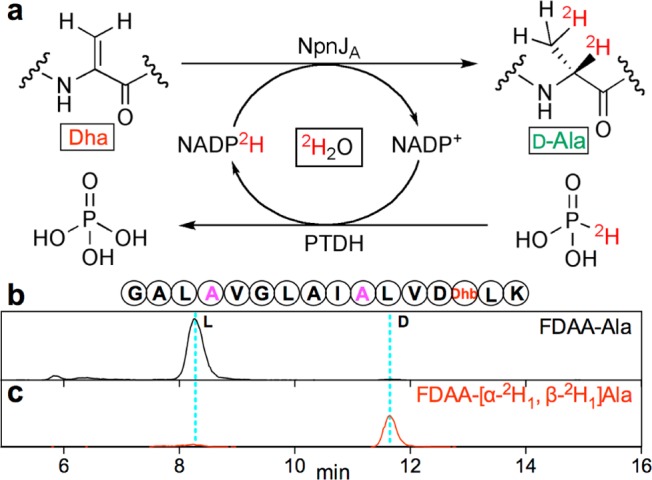
(a) Strategy for selective labeling of Ala produced by MBP-NpnJA to determine stereochemistry. After the coupled-enzyme assay on the NpnA3-T19S substrate, the purified peptide was hydrolyzed, and the amino acids derivatized by Marfey’s reagent and analyzed by LC-MS. Extracted ion chromatogram of the gene-encoded Ala (b) or labeled Ala (c) confirmed that the Ala formed by MBP-NpnJA (magenta) had the d-configuration. See Supporting Information (SI) for mass spectra.
Encouraged by the apparent substrate tolerance of MBP-NpnJA, we next investigated if the enzyme would be able to introduce d-Ala into non-native substrate sequences. Previously, Kuipers et al. demonstrated the use of LtnJ with non-native substrates in L. lactis, but many of these peptides resulted in low conversions.22 Since introduction of d-amino acids would be most conveniently done in E. coli, we evaluated its use as biosynthetic host with NpnJA.
To further explore the utility of NpnJA, we focused on lacticin 3147 (Figure 4).31,32 It belongs to a subclass of lanthipeptides that consist of two post-translationally modified peptides, which act synergistically to exhibit antimicrobial activity, and as mentioned it contains d-Ala. Because we were unable to express LtnJ solubly in E. coli, we evaluated the potential use of NpnJA as an LtnJ replacement to prepare lacticin 3147 in E. coli.
Figure 4.
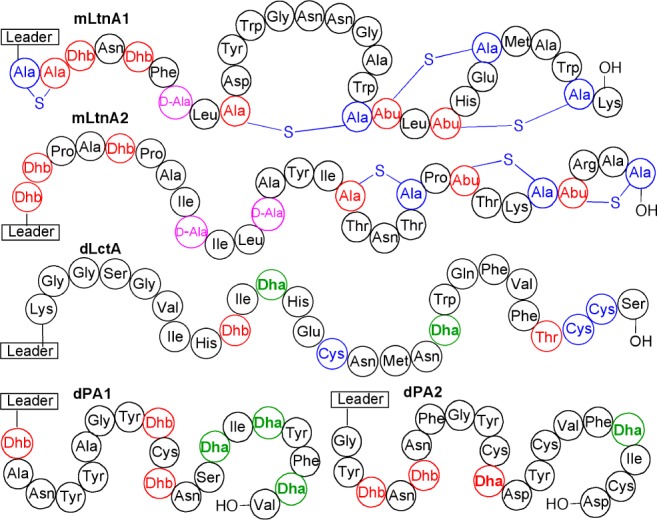
Authentic d-Ala residues (magenta) in LtnA peptides were introduced by MBP-NpnJA. Dha residues were also reduced by MBP-NpnJA in a variety of other peptides as indicated in green font. The full-length LanA peptides with the same modifications as produced by their native producer strains are termed mLanA for modified LanA, e.g., mLtnA1 and mLtnA2. The full-length dehydrated peptides with only dehydrations (no cyclizations) in their core peptide are termed dLanA for dehydrated LanA (i.e., dLctA, dPA1, and dPA2). For the sequence of leader sequences, see the SI supporting figures. Red Dhb/Dha residues were not reduced.
Co-expression of MBP-NpnJA with the precursor peptides (LtnA1 and LtnA2) and the corresponding synthetases (LtnM1 and LtnM2) in E. coli resulted in seven or eight-fold dehydrated, mostly cyclized and reduced products in which one and two d-Ala were introduced into the desired positions of LtnA1 and LtnA2, respectively (Figures 4 and S5). These observations again show the positional tolerance of NpnJA and the selectivity for reducing Dha over Dhb. These findings also open up the possibility for production of lacticin 3147 in E. coli, which paves the way for isotopic labeling and nonproteinogenic amino acid incorporation33 to study the mode of action of this two-component lantibiotic.
We investigated the substrate tolerance with three more examples where no lanthionine rings are formed. First, the precursor peptide of lacticin 481 was dehydrated with the dehydratase domain of its synthetase LctM (residues 1–543),34 and MBP-NpnJA was able to reduce both Dha residues formed in the core peptide (Figures 4 and S6), illustrating that they were not transformed into a lanthionine residue. With respect to using the methodology for nonlanthipeptide substrates, two dehydratases that have been shown to display very high substrate tolerance toward such peptides would be preferred over NpnM, which has limited substrate tolerance. Both NisB35,36 and ProcM from the prochlorosin biosynthetic pathway exhibit great substrate tolerance.37,38 We chose to focus on the latter with two randomized peptide substrates. Each substrate contained two or three Ser residues at various positions and was fused to the C-terminus of the prochlorosin leader sequence (peptides PA1 and PA2, Figure 4). By co-expression of each peptide with MBP-NpnJA and a mutant ProcM with decreased cyclization activity,39 we were able to dehydrate these peptides and also reduce the resulting dehydroalanines (Figures 4 and S6). The d-Ala containing final products were then obtained by proteolytic removal of the leader sequence.
In summary, we present the first successful in vitro reconstitution and characterization of a dehydroalanine reductase. NpnJA exhibits remarkable substrate tolerance and high conversions. Collectively, our results with all investigated peptides demonstrate that the enzyme tolerated Leu, Val, Ile, Lys, Arg, Phe, Ala, Trp, Asn, and His as flanking residues, Dha residues were reduced independent of the presence of a leader peptide, and reductions were observed at 13 different positions along the backbone. Hence, the enzyme can be coupled with substrate tolerant lanthipeptide dehydratases for engineering d-alanine residues into various peptides.
Acknowledgments
This work was supported by the US National Institutes of Health (GM 58822 to W.A.v.d.D.). We thank Prof. Martinez-Cuesta for providing the pBAC105 plasmid containing lacticin 3147 biosynthetic genes, Dr. Weixin Tang for cloning the plasmids for co-expression of Ltn3147A1/A2 and LtnM1/M2, and Dr. John E. Hung for helpful discussion about PTDH.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b05207.
Experimental details and supporting figures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Jungheim L. N.; Shepherd T. A.; Baxter A. J.; Burgess J.; Hatch S. D.; Lubbehusen P.; Wiskerchen M.; Muesing M. A. J. Med. Chem. 1996, 39, 96. 10.1021/jm950576c. [DOI] [PubMed] [Google Scholar]
- Tugyi R.; Uray K.; Ivan D.; Fellinger E.; Perkins A.; Hudecz F. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 413. 10.1073/pnas.0407677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink R.; Arkema-Meter A.; Baudoin I.; Post E.; Kuipers A.; Nelemans S. A.; Akanbi M. H.; Moll G. N. J. Pharmacol. Toxicol. Methods 2010, 61, 210. 10.1016/j.vascn.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Chatterjee S.; Gu Z. Q.; Dunn D.; Tao M.; Josef K.; Tripathy R.; Bihovsky R.; Senadhi S. E.; O’Kane T. M.; McKenna B. A.; Mallya S.; Ator M. A.; Bozyczko-Coyne D.; Siman R.; Mallamo J. P. J. Med. Chem. 1998, 41, 2663. 10.1021/jm980035y. [DOI] [PubMed] [Google Scholar]
- Imperiali B.; Fisher S. L.; Moats R. A.; Prins T. J. J. Am. Chem. Soc. 1992, 114, 3182. 10.1021/ja00035a002. [DOI] [Google Scholar]
- Fernandez-Lopez S.; Kim H. S.; Choi E. C.; Delgado M.; Granja J. R.; Khasanov A.; Kraehenbuehl K.; Long G.; Weinberger D. A.; Wilcoxen K. M.; Ghadiri M. R. Nature 2001, 412, 452. 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- Banik S. D.; Nandi N. Top. Curr. Chem. 2012, 333, 255. 10.1007/128_2012_369. [DOI] [PubMed] [Google Scholar]
- Fujino T.; Goto Y.; Suga H.; Murakami H. J. Am. Chem. Soc. 2013, 135, 1830. 10.1021/ja309570x. [DOI] [PubMed] [Google Scholar]
- Heck S. D.; Faraci W. S.; Kelbaugh P. R.; Saccomano N. A.; Thadeio P. F.; Volkmann R. A. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 4036. 10.1073/pnas.93.9.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka B. I.; Vagstad A. L.; Helf M. J.; Gugger M.; Kegler C.; Freeman M. F.; Bode H. B.; Piel J. Angew. Chem., Int. Ed. 2014, 53, 8503. 10.1002/anie.201400478. [DOI] [PubMed] [Google Scholar]
- Jilek A.; Mollay C.; Tippelt C.; Grassi J.; Mignogna G.; Mullegger J.; Sander V.; Fehrer C.; Barra D.; Kreil G. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 4235. 10.1073/pnas.0500789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata Y.; Watanabe T.; Teramoto T.; Inoue A.; Kawakami Y.; Nishizawa Y.; Katayama K.; Kuwada M. J. Biol. Chem. 1995, 270, 16719. 10.1074/jbc.270.28.16719. [DOI] [PubMed] [Google Scholar]
- Bansal P. S.; Torres A. M.; Crossett B.; Wong K. K.; Koh J. M.; Geraghty D. P.; Vandenberg J. I.; Kuchel P. W. J. Biol. Chem. 2008, 283, 8969. 10.1074/jbc.M709762200. [DOI] [PubMed] [Google Scholar]
- Jilek A.; Mollay C.; Lohner K.; Kreil G. Amino Acids 2012, 42, 1757. 10.1007/s00726-011-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. F.; Gurgui C.; Helf M. J.; Morinaka B. I.; Uria A. R.; Oldham N. J.; Sahl H. G.; Matsunaga S.; Piel J. Science 2012, 338, 387. 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K.-D.; Fischbach M. A.; Garavelli J. S.; Göransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Müller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J. T.; Rebuffat S.; Ross R. P.; Sahl H.-G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Süssmuth R. E.; Tagg J. R.; Tang G. L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. Nat. Prod. Rep. 2013, 30, 108. 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knerr P. J.; van der Donk W. A. Annu. Rev. Biochem. 2012, 81, 479. 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- Skaugen M.; Nissenmeyer J.; Jung G.; Stevanovic S.; Sletten K.; Abildgaard C. I. M.; Nes I. F. J. Biol. Chem. 1994, 269, 27183. [PubMed] [Google Scholar]
- Martin N. I.; Sprules T.; Carpenter M. R.; Cotter P. D.; Hill C.; Ross R. P.; Vederas J. C. Biochemistry 2004, 43, 3049. 10.1021/bi0362065. [DOI] [PubMed] [Google Scholar]
- Cotter P. D.; O’Connor P. M.; Draper L. A.; Lawton E. M.; Deegan L. H.; Hill C.; Ross R. P. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 18584. 10.1073/pnas.0509371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohans C. T.; Li J. L.; Vederas J. C. J. Am. Chem. Soc. 2014, 136, 13150. 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- van Heel A. J.; Mu D.; Montalban-Lopez M.; Hendriks D.; Kuipers O. P. ACS Synth. Biol. 2013, 2, 397. 10.1021/sb3001084. [DOI] [PubMed] [Google Scholar]
- Suda S.; Lawton E. M.; Wistuba D.; Cotter P. D.; Hill C.; Ross R. P. J. Bacteriol. 2012, 194, 708. 10.1128/JB.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Yang X.; Wang H.; van der Donk W. A. ACS Chem. Biol. 2014, 9, 2686. 10.1021/cb500622c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan R.; Bruckner H. Amino Acids 2004, 27, 231. 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- Mu D.; Montalban-Lopez M.; Deng J.; Kuipers O. P. Appl. Environ. Microbiol. 2015, 81, 3679. 10.1128/AEM.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.; van der Donk W. A. Nat. Chem. Biol. 2013, 9, 157. 10.1038/nchembio.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.; Jimenez-Oses G.; Houk K. N.; van der Donk W. A. Nat. Chem. 2015, 7, 57. 10.1038/nchem.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrtis J. M.; White A.; Metcalf W. W.; van der Donk W. A. Angew. Chem., Int. Ed. 2002, 41, 3257.. [DOI] [PubMed] [Google Scholar]
- Johannes T. W.; Woodyer R. D.; Zhao H. Biotechnol. Bioeng. 2007, 96, 18. 10.1002/bit.21168. [DOI] [PubMed] [Google Scholar]
- Ryan M. P.; Jack R. W.; Josten M.; Sahl H. G.; Jung G.; Ross R. P.; Hill C. J. Biol. Chem. 1999, 274, 37544. 10.1074/jbc.274.53.37544. [DOI] [PubMed] [Google Scholar]
- Martinez-Cuesta M. C.; Buist G.; Kok J.; Hauge H. H.; Nissen-Meyer J.; Pelaez C.; Requena T. J. Appl. Microbiol. 2000, 89, 249. 10.1046/j.1365-2672.2000.01103.x. [DOI] [PubMed] [Google Scholar]
- Shi Y.; Yang X.; Garg N.; van der Donk W. A. J. Am. Chem. Soc. 2011, 133, 2338. 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.-H.; Tang W.; Lukk T.; Yu Y.; Nair S. K.; van der Donk W. A. eLife 2015, 4, e07607. 10.7554/eLife.07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink R.; Wierenga J.; Kuipers A.; Kluskens L. D.; Driessen A. J. M.; Kuipers O. P.; Moll G. N. Appl. Environ. Microbiol. 2007, 73, 1792. 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll G. N.; Kuipers A.; Rink R. Antonie van Leeuwenhoek 2010, 97, 319. 10.1007/s10482-010-9418-4. [DOI] [PubMed] [Google Scholar]
- Li B.; Sher D.; Kelly L.; Shi Y.; Huang K.; Knerr P. J.; Joewono I.; Rusch D.; Chisholm S. W.; van der Donk W. A. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 10430. 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Yu Y.; Velásquez J. E.; van der Donk W. A. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 18361. 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Mukherjee S.; van der Donk W. A. J. Am. Chem. Soc. 2015, 137, 5140. 10.1021/jacs.5b01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


