Abstract
The cell surfaces of bacteria are replete with diverse glycoconjugates that play pivotal roles in determining how bacteria interact with the environment and the hosts that they colonize. Studies to advance our understanding of these interactions rely on the availability of chemically defined glycoconjugates that can be selectively modified under orthogonal reaction conditions to serve as discrete ligands to probe biological interactions, in displayed arrays and as imaging agents. Herein, enzymes in the N-linked protein glycosylation (Pgl) pathway of Campylobacter jejuni are evaluated for their tolerance for azide-modified UDP-sugar substrates, including derivatives of 2,4-diacetamidobacillosamine and N-acetylgalactosamine. In vitro analyses reveal that chemoenzymatic approaches are useful for the preparation of undecaprenol diphosphate-linked glycans and glycopeptides with site-specific introduction of azide functionality for orthogonal labeling at three specific sites in the heptasaccharide glycan. The uniquely modified glycoconjugates represent valuable tools for investigating the roles of C. jejuni cell surface glycoconjugates in host pathogen interactions.
Cell-surface glycoconjugates are common participants in the interactions among mammalian cells and between pathogenic microorganisms and the host cells that they infect.1,2 For bacterial glycolipids and glycoproteins, the surge of information from whole genome sequencing and mass spectrometry-based analysis has accelerated the pace at which new glycans are discovered and highlights the importance of these entities in bacterial pathogenesis3 and symbiosis.4 In this context, methods for the preparation and modification of complex glycans and glycoconjugates, with defined modification sites, are critical for parsing the molecular basis for crucial biological responses. Bacterial glycans present particular challenges for study due to the prevalence of highly modified “non-standard” carbohydrates, such as 2,4-diacetamidobacillosamine (diNAcBac) and pseudaminic acid, embedded within diverse glycan architectures,5,6 which exacerbates the task of chemical synthesis requiring that tailored methods for the generation of each unique glycan must be developed to study the roles of carbohydrates in each organism. Chemoenzymatic methods provide an important complement to chemical synthesis enabling access to defined materials for biological studies including the generation of glycan arrays,7 molecular imaging probes,8 and vaccines.9
Many of the challenges are exemplified in the N-linked protein glycosylation system of Campylobacter jejuni,10 a widespread human enteropathogen. C. jejuni requires N-glycosylation to adhere to, invade, and colonize target host cells.11−13 Glycosylation is accomplished by enzymes of the protein glycosylation (Pgl) pathway through stepwise assembly of a heptasaccharide onto an undecaprenol diphosphate (Und-PP) carrier, followed by transfer of the glycan to an acceptor protein by the oligosaccharyl transferase, PglB. Enzymes in the Pgl pathway use complex polyprenol-linked substrates to produce glycopeptide and glycoprotein products. To study these enzymes and characterize the glycan functions, reliable preparative approaches, including opportunities for introduction of bioorthogonal chemical handles for appending reporter molecules are needed. The chemical synthesis of the C. jejuni N-linked glycan is extremely labor-intensive14 and would require significant repurposing for the assembly of variants that include uniquely modified carbohydrates; therefore, we set out to establish the practicality and limitations of a general chemoenzymatic approach using native and modified nucleotide sugar donors.
Herein we present a systematic approach for the production of defined glycans with bioorthogonal conjugation handles representing intermediates and products in the C. jejuni pathway. Since the bacterial gene clusters encoding enzymes in the biosynthesis and utilization of polyprenol diphosphate-linked glycans in N- and O-linked bacterial protein glycosylation can now be identified using bioinformatics approaches,15,16 we anticipate that this study will provide a guide for the application of parallel approaches for the preparation of glycoconjugate targets from other pathogenic and symbiotic bacteria.
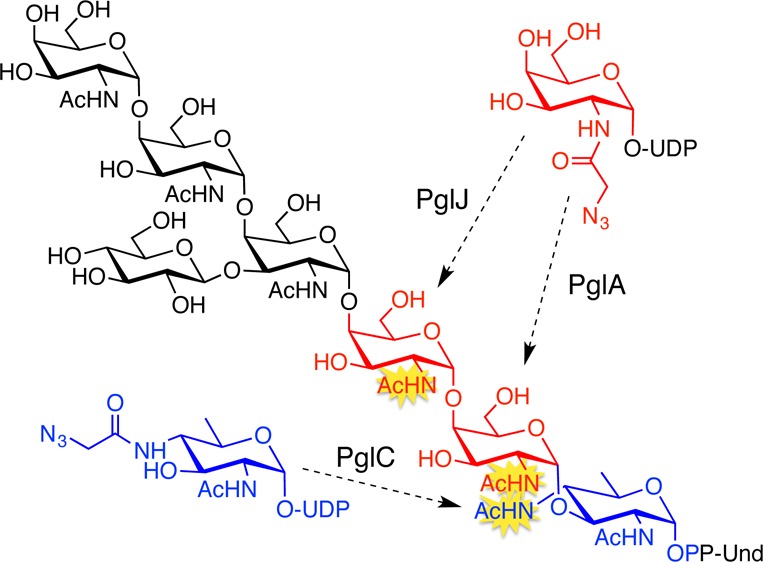
Previously, we demonstrated that the C. jejuni N-glycan can be prepared on an analytical scale, using enzymes in the Pgl pathway.17 In this pathway, PglC, a phosphoglycosyltransferase (PGT), catalyzes the first membrane-committed step transferring phospho-diNAcBac from UDP-diNAcBac to Und-P to afford Und-PP-diNAcBac (Figure 1). The glycosyltransferases (GTs) PglA and PglJ elaborate the glycan by adding a single GalNAc each, and PglH18 adds three more α-1,4-GalNAcs. The native branched heptasaccharide is completed by the glucosyl transferase, PglI. After translocation to the periplasm, the glycan is transferred to proteins by the oligosaccharyl transferase, PglB. There is precedent for tolerance of azide-modified sugars as substrates for GTs when the azide is incorporated into N-acetamido sugars.19 Of the seven carbohydrates that comprise the C. jejuni glycan, six feature N-acetyl groups that can potentially be modified with azide chemical handles. The first three enzymes, PglC, PglA, and PglJ, would deliver an azide-modified carbohydrate into a single, site-specific position. In contrast, the polymerase activity of PglH18 could potentially result in insertion of azide-modified carbohydrates in place of the three terminal GalNAc residues of the glycan. Therefore, incorporating an azide-modified carbohydrate into the glycan with any of the first three enzymes in the pathway would be advantageous due to the potential for unique positional control.
Figure 1.
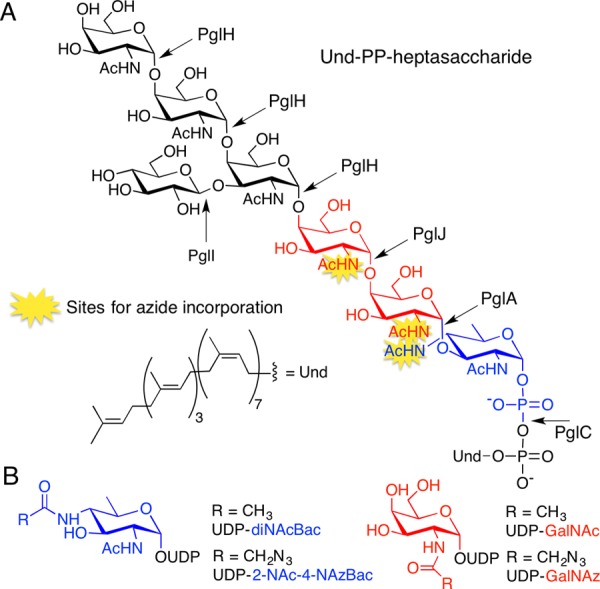
(A) Structure of the C. jejuni Und-PP-heptasaccharide highlighting azide incorporation sites. (B) UDP-sugars in this study.
To study the tolerance of Pgl enzymes for unnatural azido sugars, Pgl pathway enzymes (PglC, PglA, PglJ, PglH, PglI, and PglB) were prepared as previously described,17,20 and substrates were assembled using a combination of chemical and enzymatic approaches. First, polyprenols (C50–60) derived from the leaves of Rhus typhina were subject to phosphorylation with phosphoramidite (FmO)2PNiPr2, followed by oxidation and deprotection to afford the corresponding polyprenol phosphates.21,22 UDP-sugars (Figure 1B) were prepared by chemoenzymatic approaches. Specifically, UDP-diNAcBac was generated enzymatically,23 while UDP-2-NAc-4-NAzBac, a new azide-modified UDP-sugar, was prepared from UDP-4-amino-4,6-dideoxy-GlcNAc (an intermediate in UDP-diNAcBac biosynthesis) through selective chloroacetylation followed by azide substitution. The C-2 position of UDP-diNAcBac was not considered for azide modification because a C-2 N-acetyl group is critical for catalysis in the PglB reaction.24 UDP-GalNAz was prepared as previously described.25,26 With these enzymes and substrates in hand, the tolerance of each of the GTs was evaluated by comparing the relative turnover of the native UDP-sugar and the corresponding azide-derivative. In each case product quantitation was tailored to provide the most accurate measure of activity. The tolerance of PglC for UDP-2-NAc-4-NAzBac relative to the native substrate was examined by comparing activity with the UDP-sugars in the presence of PglC and Und-P for 60 min (Figure 2A), followed by product isolation and hydrolysis of Und-PP-monosaccharide. For quantification, the free monosaccharide was labeled with 8-aminopyrene-1,3,6-trisulfonate (APTS) and analyzed by capillary electrophoresis.27 The azide derivative was well tolerated by PglC with product formation corresponding to ∼60–65% of the natural substrate (Figure 2A). However, the conversion was low for both, UDP-diNAcBac and UDP-2-NAc-4-NAzBac at ∼14% and 8%, respectively. In contrast, a coupled reaction using PglC and PglA, afforded much higher conversions (>60%, data not shown) suggesting that addition of PglA can overcome the unfavorable equilibrium of PglC to increase flux through the two enzymes.
Figure 2.
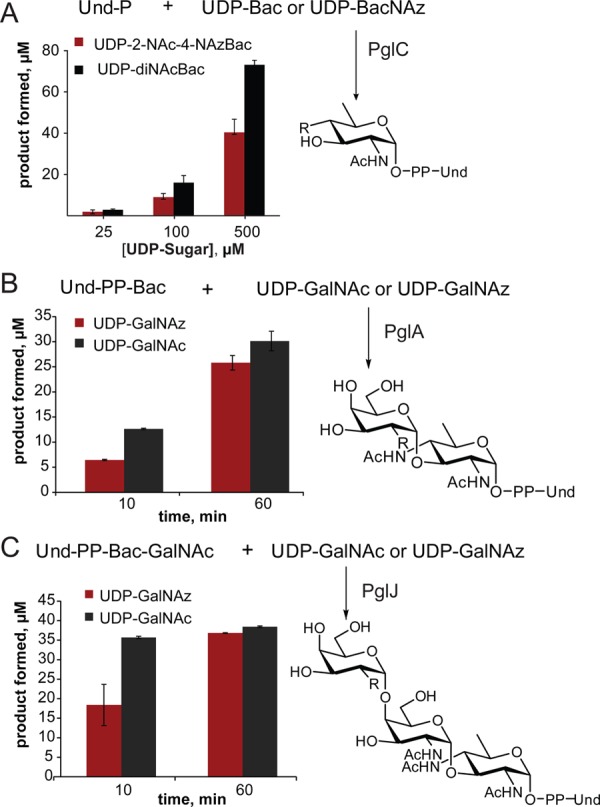
Kinetic analysis of PglC, PglA, and PglJ comparing natural and azide-modified substrates (R = NHAc or NHAz). (A) PglC (10 nM) was evaluated at increasing concentrations of UDP-Bac and UDP-2-NAc-4-NAzBac (UDP-BacNAz). (B) Time course for PglA (10 nM) turnover with 200 μM UDP-GalNAc or UDP-GalNAz. (C) Time course for PglJ (90 nM) turnover with 100 μM UDP-GalNAc and UDP-GalNAz.
The specificity of PglA for UDP-GalNAc and UDP-GalNAz was compared using a coupled assay with PglC to make the Und-PP-diNAcBac starting material in situ. Reactions were performed with an excess of the UDP-sugars and were quenched at 10 and 60 min, after which the products were isolated by liquid/liquid extraction. The products were then hydrolyzed, and the resulting disaccharide was labeled with 2-aminobenzamide for analysis by normal-phase HPLC.28 The azide-modified UDP-sugar substrate is well tolerated by PglA with product formation comparable to the native substrate after 60 min (Figure 2B).
To assess the activity of PglJ, first, Und-PP-diNAcBac-[3H]-GalNAc was prepared using PglC and PglA with Und-P, UDP-diNAcBac, and UDP-[6-3H]-GalNAc. The resulting Und-PP-disaccharide was incubated with PglJ, in the presence of either UDP-GalNAc or UDP-GalNAz (Figure 2C). For each substrate the reaction was quenched at 10 and 60 min and purified by HPLC. Product formation was quantified by scintillation counting. While 50% less product was formed in the presence of UDP-GalNAz relative to UDP-GalNAc after 10 min, both substrates showed >90% conversion after 60 min, indicating that PglJ efficiently incorporates an azido sugar into the Und-PP-trisaccharide. The site selectivity of the process was supported by MS analysis and by activity of the product with PglH (vide infra).
In contrast to promiscuity of PglC, PglA, and PglJ, PglH, failed to react in the presence of UDP-GalNAz. In this case, the tolerance of PglH was tested by incubating enzyme with UDP-GalNAc and UDP-GalNAz and [3H]-labeled Und-PP-diNAcBac-GalNAc-GalNAc trisaccharide. Reactions were analyzed by normal-phase HPLC and no azide-containing products were observed, even in the presence of a 10-fold excess of UDP-GalNAz after 4h (data not shown). Therefore, under these conditions, PglH does not accept UDP-GalNAz as a substrate, underscoring the need for systematic analysis of each enzyme and substrate combination to assess the substrate tolerance.
These studies establish the compatibility of PglC, PglA, and PglJ with UDP-azido-sugars. To build on these results, PglC, PglA, and PglJ were then used in combination to prepare three uniquely modified Und-PP-trisaccharides (Scheme 1). The syntheses utilized strategic quenching and isolation steps to ensure that the azide-modified carbohydrate was added only at a single position. The products were confirmed using HPLC and MS analysis (Figure S1). Successful synthesis of Und-PP-trisaccharides with azide-modified carbohydrates in the first two sites of the glycan (Figure 2A,B) demonstrates that PglA and PglJ elaborate azide-modified glycan precursors. Finally, to establish that Und-PP-diNAcBac-GalNAc-GalNAz (Figure 2C) can be elaborated into the native heptasaccharide, the synthesis was completed using PglH and PglI (Figure 3). It has previously been reported that the terminal GalNAc of the C. jejuni glycan is a key determinant in binding to the human macrophage galactose-type lectin receptor, thereby mediating interactions between C. jejuni and host cells.29 The ability to preserve this binding partnership in the full glycan, while introducing bioorthogonal chemical handles into carbohydrates with less dominant roles in glycan-host cell receptor interactions, highlights potential future applications of these reagents.
Scheme 1. Chemoenzymatic Synthesis of Azide-Modified Und-PP-trisaccharides.
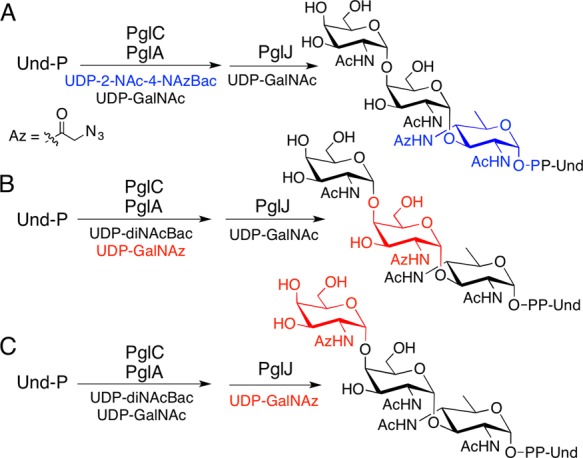
(A) Synthesis of Und-PP-2NAc-4-NAzBac-(GalNAc)2. (B) Synthesis of Und-PP-diNAcBac-GalNAz-GalNAc. (C) Synthesis of Und-PP-diNAcBac-GalNAc-GalNAz.
Figure 3.
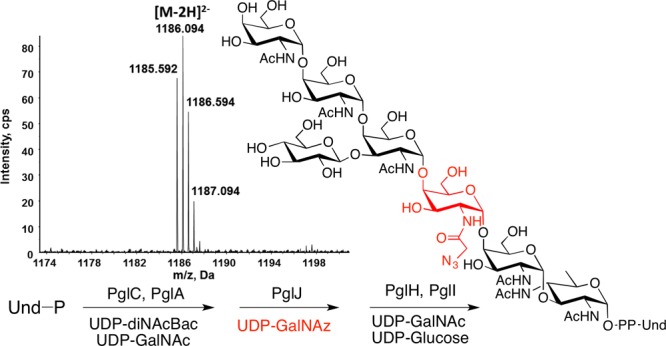
Enzymatic synthesis of Und-PP-linked heptasaccharide with incorporation of an azide-modified carbohydrate (red) in the third site of the glycan. Inset: Mass spectrum showing the doubly deprotonated [M – 2H]2– ion of the product.
In order for this system to be applicable to the biosynthesis of glycopeptides and potentially glycoproteins, the tolerance of PglB, for the azide-modified Und-PP-glycans was then investigated. The ability of PglB to transfer each of these azide-modified trisaccharides to a peptide substrate30 was quantified using [3H]-labeled substrates. The efficiency of peptide glycosylation by PglB is between 75 and 90% of native levels when the modified carbohydrate is distal to the aglycone; however, glycosylation is somewhat more impacted, at about 60%, when the azide-modified carbohydrate is proximal to the diphosphate leaving group (Figure 4, Figure S2). It has previously been shown that synthetic substrates, including highly truncated (C20) polyprenol diphosphate derivatives of 6-azido-GlcNAc and GlcNAz (Az = COCH2N3), failed to serve as substrates in peptide glycosylation by PglB.31 However, in that case, the lack of activity was attributed to the cumulative effect of unnatural moieties in both components of the polyprenol diphosphate-sugar substrate. Here, we demonstrate that when native polyprenols are used in conjunction with azide-modified glycans, PglB catalyzes glycosylation of peptides with conversions comparable to those of the unmodified substrate. These results combined with previous studies utilizing PglB to synthesize glycoproteins32 set the stage for synthesis of diverse glycoproteins containing unnatural carbohydrates using this system.
Figure 4.
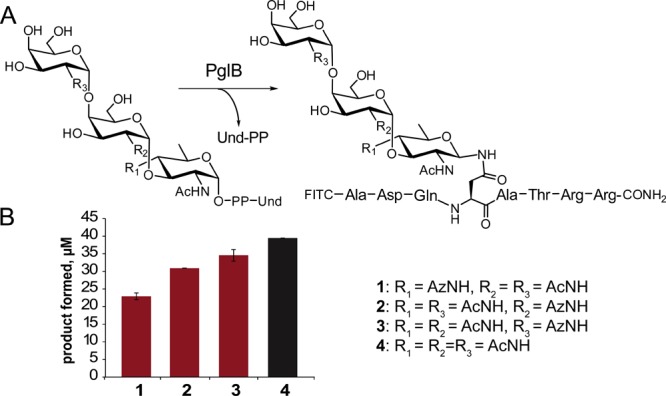
(A) PglB-catalyzed peptide glycosylation with azido-glycan substrates (50 μM Und-PP-trisaccharide, 250 μM peptide). (B) Production of azide-modified glycopeptide products.
Finally, to validate the utility of these compounds for orthogonal conjugation, copper-catalyzed click reactions were performed. The Und-PP-diNAcBac-GalNAc-GalNAz was conjugated to an acetylene-545 fluorophore using copper-catalyzed azide–alkyne cycloaddition and characterized by MS after purification (see Figure S3A). In addition, the glycopeptide, modified with a terminal GalNAz, was reacted with acetylene-PEG4-biotin also using copper-catalyzed azide–alkyne cycloaddition, and characterized directly without purification (Figure S3B).
Together, these studies reveal that azido sugar substrates are well tolerated by several of the C. jejuni Pgl enzymes. Unfortunately, attempts to apply in vivo metabolic labeling in C. jejuni, using exogenous acetylated monosaccharide derivatives, proved unsuccessful. This was likely due to inefficient processing of the precursors to the corresponding UDP-sugars, which would involve deacetylation, C-1 phosphorylation, and uridinylation. Indeed it is reported that GalNAz-1-P is not a substrate for the homologous E. coli GlmU.26 Additionally, in vitro kinetic studies show that diNAcBac-1-P is not a substrate for the C. jejuni GlmU (SI Methods and Table S1). Therefore, given the lack of promiscuity in the bacterial GlmU enzymes,26 further studies with the corresponding azide were not pursued. Together this suggests that further attempts to incorporate azide-modified sugars into the C. jejuniN-glycan would require engineering of the C. jejuni UDP-sugar biosynthesis pathways. The studies presented herein now define that metabolic labeling in C. jejuni should be feasible from the perspective of the Pgl pathway enzymes, and therefore, future focus can turn to specifically engineering C. jejuni to enable conversion of cell permeable sugar derivatives into UDP-GlcNAz and UDP-2-NAc-4-NAzBac. This general concept has precedent in mammalian cell culture systems.33,34
This communication defines the tolerance of the Pgl enzymes toward azide-modified carbohydrates and shows that chemoenzymatic approaches are valuable for the preparation of Und-PP-glycans and glycopeptides with site-specific introduction of azide functionality for orthogonal labeling. The uniquely labeled glycoconjugates promise to be extremely valuable for understanding the roles of C. jejuni cell surface glycoconjugates. As a priority, sufficient quantities can be readily prepared for study of mammalian cell adhesion and invasion, in the preparation of glycan arrays, and in biophysical studies to investigate substrate binding to the Pgl pathway enzymes using fluorescence and luminescence resonance energy transfer.
Acknowledgments
This research was supported by NIH GM-039334 (to B.I.), GM-069338 and EY023666 (to Z.G.), and an NSERC Fellowship to G.W. We also acknowledge the MIT BIF, Dr. Rabideau and Prof. Pentelute for MS analysis, Prof. George Peng Wang for Agx1 and NahK plasmids, and Dr. Musial-Siwek for PglB.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b07146.
Figures and experimental methods (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Varki A. Glycobiology 1993, 3, 97–130. 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I.; Schmidt M. A. Mol. Microbiol. 2002, 45, 267–276. 10.1046/j.1365-2958.2002.03030.x. [DOI] [PubMed] [Google Scholar]
- Tytgat H. L.; Lebeer S. Microbiol. Mol. Biol. Rev. 2014, 78, 372–417. 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock L. E. Cell Host Microbe 2009, 5, 522–526. 10.1016/j.chom.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Nothaft H.; Szymanski C. M. Nat. Rev. Microbiol. 2010, 8, 765–778. 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Tra V. N.; Dube D. H. Chem. Commun. (Cambridge, U. K.) 2014, 50, 4659–4673. 10.1039/c4cc00660g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell S. R.; Arthur C. M.; McBride R.; Berger O.; Razi N.; Heimburg-Molinaro J.; Rodrigues L. C.; Gourdine J. P.; Noll A. J.; von Gunten S.; Smith D. F.; Knirel Y. A.; Paulson J. C.; Cummings R. D. Nat. Chem. Biol. 2014, 10, 470–476. 10.1038/nchembio.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Li Z.; Duan X.; Yi W. J. Am. Chem. Soc. 2014, 136, 12536–12539. 10.1021/ja5054225. [DOI] [PubMed] [Google Scholar]
- Astronomo R. D.; Burton D. R. Nat. Rev. Drug Discovery 2010, 9, 308–324. 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski C. M.; Logan S. M.; Linton D.; Wren B. W. Trends Microbiol. 2003, 11, 233–238. 10.1016/S0966-842X(03)00079-9. [DOI] [PubMed] [Google Scholar]
- Hendrixson R. R.; DiRita V. J. Mol. Microbiol. 2004, 52, 471–484. 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- Karlyshev A. V.; Everest P.; Linton D.; Cawthraw S.; Newell D. G.; Wren B. W. Microbiology 2004, 150, 1957–1964. 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- Kelly J.; Jarrell H.; Millar L.; Tessier L.; Fiori L. M.; Lau P. C.; Allan B.; Szymanski C. M. J. Bacteriol. 2006, 188, 2427–2434. 10.1128/JB.188.7.2427-2434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M. N.; Ishiwata A.; Ito Y. Tetrahedron 2007, 63, 8181–8198. 10.1016/j.tet.2007.05.126. [DOI] [Google Scholar]
- Kumar M.; Balaji P. V. Mol. BioSyst. 2011, 7, 1629–1645. 10.1039/c0mb00259c. [DOI] [PubMed] [Google Scholar]
- Chauhan J. S.; Bhat A. H.; Raghava G. P.; Rao A. PLoS One 2012, 7, e40155. 10.1371/journal.pone.0040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover K. J.; Weerapana E.; Imperiali B. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 14255–14259. 10.1073/pnas.0507311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutman J. M.; Imperiali B. Biochemistry 2009, 48, 2807–2816. 10.1021/bi802284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D. H.; Bertozzi C. R. Curr. Opin Chem. Biol. 2003, 7, 616–625. [DOI] [PubMed] [Google Scholar]
- Jaffee M. B.; Imperiali B. Biochemistry 2011, 50, 7557–7567. 10.1021/bi201018d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewska E.; Sasak W.; Mankowski T.; Jankowski W.; Vogtman T.; Krajewska I.; Hertel J.; Skoczylas E.; Chojnacki T. Acta Biochim Pol 1994, 41, 221–260. [PubMed] [Google Scholar]
- Watanabe Y.; Nakamura T.; Mitsumoto H. Tetrahedron Lett. 1997, 38, 7407–7410. 10.1016/S0040-4039(97)85781-4. [DOI] [Google Scholar]
- Olivier N. B.; Chen M. M.; Behr J. R.; Imperiali B. Biochemistry 2006, 45, 13659–13669. 10.1021/bi061456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M.; Feldman M. F.; Callewaert N.; Kowarik M.; Clarke B. R.; Pohl N. L.; Hernandez M.; Vines E. D.; Valvano M. A.; Whitfield C.; Aebi M. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 7088–7093. 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L.; Guan W.; Kitaoka M.; Shen J.; Xia C.; Chen W.; Wang P. G. Chem. Commun. (Cambridge, U. K.) 2009, 2944–2946. 10.1039/b904853g. [DOI] [PubMed] [Google Scholar]
- Guan W.; Cai L.; Wang P. G. Chem. - Eur. J. 2010, 16, 13343–13345. 10.1002/chem.201002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro L. A.; Salas-Solano O. Anal. Chem. 2008, 80, 3838–3845. 10.1021/ac800152h. [DOI] [PubMed] [Google Scholar]
- Maury D.; Couderc F.; Czaplicki J.; Garrigues J. C.; Poinsot V. Biomed. Chromatogr. 2010, 24, 343–346. 10.1002/bmc.1299. [DOI] [PubMed] [Google Scholar]
- van Sorge N. M.; Bleumink N. M.; van Vliet S. J.; Saeland E.; van der Pol W. L.; van Kooyk Y.; van Putten J. P. Cell. Microbiol. 2009, 11, 1768–1781. 10.1111/j.1462-5822.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- Chen M. M.; Glover K. J.; Imperiali B. Biochemistry 2007, 46, 5579–5585. 10.1021/bi602633n. [DOI] [PubMed] [Google Scholar]
- Liu F.; Vijayakrishnan B.; Faridmoayer A.; Taylor T. A.; Parsons T. B.; Bernardes G. J.; Kowarik M.; Davis B. G. J. Am. Chem. Soc. 2014, 136, 566–569. 10.1021/ja409409h. [DOI] [PubMed] [Google Scholar]
- Hug I.; Zheng B.; Reiz B.; Whittal R. M.; Fentabil M. A.; Klassen J. S.; Feldman M. F. J. Biol. Chem. 2011, 286, 37887–37894. 10.1074/jbc.M111.287755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M.; Carrico I. S.; Ganguli A. S.; Yu S. H.; Hangauer M. J.; Hubbard S. C.; Kohler J. J.; Bertozzi C. R. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3141–3146. 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. H.; Boyce M.; Wands A. M.; Bond M. R.; Bertozzi C. R.; Kohler J. J. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 4834–4839. 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


