Smart hydrogels1a susceptible to volume changes induced by external stimuli such as pH,1b antigen–antibody recognition,1c,d and temperature1e,f have been extensively evaluated as materials for applications including bioactive surfaces, bioreactors, microfluidics, diagnostics, and drug delivery systems.
Recently, conformational changes accompanying protein–ligand recognition were used to trigger the volume change of smart hybrid hydrogels.2 Calmodulin (CaM) mutants were used as cross-linkers, and hydrogel volume changes were mediated by a conformational shift of CaM upon binding of ligands (phenothiazine2a or trifluoperazine2b,c).
Here we report a new design of enzyme-based hybrid hydrogels that employs substrate–enzyme (adenylate kinase3–ATP) interaction to induce a conformational change with concomitant decrease of the hydrogel volume. The uniqueness in our design is the combination of biorecognition with a catalyzed chemical reaction—the transfer of a phosphate group. There are numerous enzymes that undergo conformational changes following the binding of a substrate in the active site.4 Thus, this is a new paradigm for hybrid hydrogel design, where a variety of chemical reactions can be combined with biorecognition and transform nanoscale conformational changes into macroscopic motion.
Escherichia coli adenylate kinase (AKe; EC 2.7.4.3) is a 214 residue, three domain bacterial enzyme (transferase), which catalyzes the phosphoryl transfer reaction Mg2+ · ATP + AMP ↔ Mg2+ · ADP + ADP. Upon the binding of a substrate (or inhibitor), AKe undergoes a large conformational change. The bulky lid domain closes over the active site to shield it from water to avoid substrate hydrolysis and to facilitate the transfer of a phosphate group3 (Figure 1).
Figure 1.
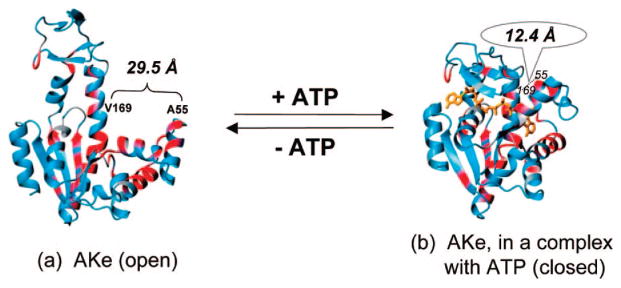
Ribbon diagram of the structures of AKe in two conformational states: open state (a) and closed state (b). Adapted from ref 3h.
AKe was incorporated into defined hydrogel structures through thiol-maleimide reaction. Synthetic N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers with pendant maleimide groups5 were chosen as the backbone. To create two attachment points, a triple mutant of adenylate kinase, AKtm (C77S, A55C, V169C), was designed, engineered, and purified. The wild-type enzyme has one cysteine at position 77. The distance between Cα-atoms of the residues 55 and 169 decreases from 29.5 Å in the apoenzyme to 12.4 Å when forming the enzyme–substrate complex.3a,b The SH groups at positions 55 and 169 are easily accessible as shown by reactivity studies involving 26 AK double mutants.3g The enzymatic activity of AKtm was found to be similar to AKe (Supporting Information).
Hybrid HPMA-based hydrogels were made in a mold by cross-linking of HPMA copolymer P with AKtm (Gels 1 and 5) or AKtm and an additional cross-linking agent, dithiothreitol (DTT) (Gels 2 and 3). Gel 4 (control) was prepared by cross-linking P with DTT only (Figure 2; Supporting Information).
Figure 2.
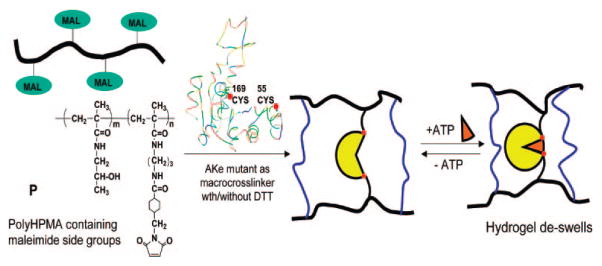
HPMA-based hydrogels cross-linked with AKtm/DTT and the macroscopic motion of the hydrogels triggered by substrate recognition.
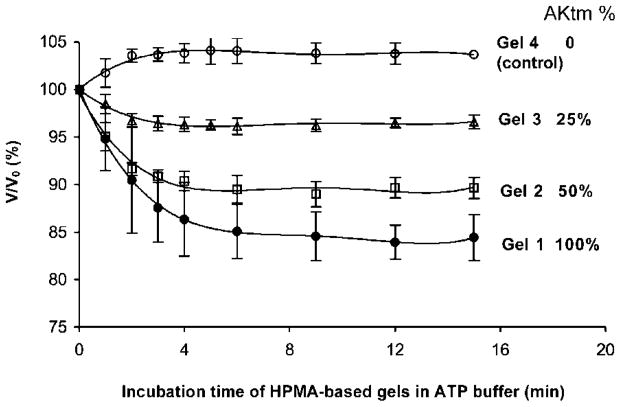
Importantly, when the hydrogels were exposed to ATP (substrate), a volume change occurred. Gels 1–4 were incubated in 50 mM MgCl2, 50 mM Tris-HCl, pH 7.5 (ATP-free buffer) until equilibrium was reached. Exposure to buffer containing 4 mM ATP induced the conformational transition from an “open” to “closed” conformation, resulting in hydrogel volume change. The shrinkage of ~5–17% was proportional to the AKtm content in the hydrogels. Gel 4, cross-linked with DTT only, did not deswell (Figure 3).
Figure 3.
Deswelling ratios of Gels 1, 2, 3, 4 prepared with 100%, 50%, 25% of AKtm, and 100% DTT in molar percentage of cross-linking agents, respectively. Assuming isotropic swelling, the V/V0 (%) was calculated as (L/L0),3 where L is the one-dimensional size of the hydrogel in buffer containing substrate, and L0 is the size of the original hydrogel equilibrated in ATP-free buffer.
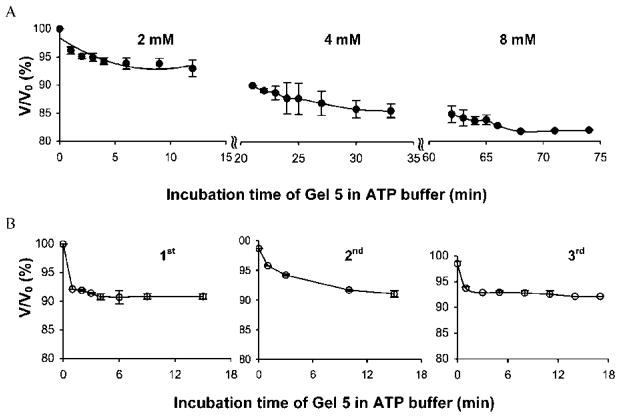
The degree of deswelling of Gel 5 (100% AKtm) increased with increasing concentration of the substrate (Figure 4A). When the concentration of substrate was ≥8 mM ATP, the volume changes were not significant (data not shown) reflecting the access to the Vmax area.6
Figure 4.
(A) Deswelling of Gel 5 incubated in 2 mM, 4 mM, and 8 mM ATP sequentially. (B) Three cycles of deswelling of Gel 5 in 4 mM ATP buffer.
Repeated exposure of Gel 5 to 4 mM ATP buffer followed by a washing step in ATP-free buffer demonstrated the reproducibility of hydrogel swelling alterations and of the AKtm conformational changes (Figure 4B). It is worth noting that, unlike in protein–ligand binding systems, the reversible reswelling occurred during a quite short washing period (~30 min; see Supporting Information). This demonstrates another unique potential of smart materials that are based on enzyme–substrate recognition: to provide a fast response after being exposed to substrate and ultimately endow hydrogels with self-oscillating characteristics. This bodes well for the application of these gels in drug delivery, microfluidic valves, and mechanical actuators.
To verify the covalent attachment of AKtm to the 3-D network structure, Gel 6 (100% AKtm) was prepared using AKtm labeled with Alexa Fluor 488 (Invitrogen, Carlsbad, CA). The images of Gels 5 and 6 obtained under both UV light and visible light clearly demonstrate homogeneous incorporation of AKtm into the hydrogel structure (Figure 5). No fluoro-labeled protein in the medium was detectable by spectrofluorometry. Measurement of SH content in the incubation buffer (50 mM Tris-HCl pH 7.5) using Ellman’s assay also suggested full incorporation of AKtm into hydrogels.
Figure 5.
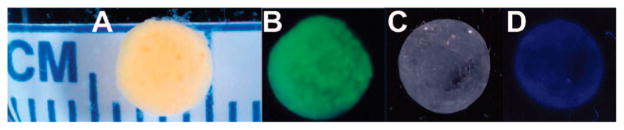
Images of hybrid hydrogels cross-linked with fluorescently labeled AKtm (Gel 6) and with AKtm (Gel 5). (A) Gel 6 under visible light; (B) Gel 6 under UV light; (C) Gel 5 under visible light; (D) Gel 5 under UV light.
Finally, experiments were performed to prove that translating substrate recognition into mechanical motion is a generally applicable approach. To this end, four-arm poly(ethylene glycol) (PEG; 10 kDa) based hybrid hydrogels were designed. Maleimide terminated 4-arm PEG was cross-linked with 50% AKtm + 50% DTT (Supporting Information). The volume change of Gel 7 observed after addition of 4 mM of ATP was ~12% (Figure 6).
Figure 6.
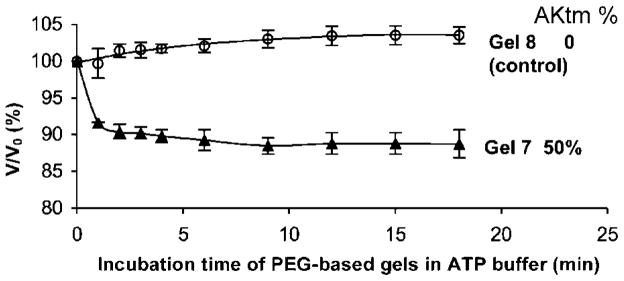
Deswelling ratios of PEG-based hydrogels. Gel 7 was prepared with 50% AKtm and 50% DTT as cross-linkers, Gel 8 with 100% DTT.
We are aware that the structure and corresponding volume change are far from the best possible. However, with the exception of hydrogels prepared by copolymerization,2c the observed volume changes (<20%) were similar to those for hydrogels containing calmodulin mutants,2a,b or those where volume change was mediated by antigen–antibody recognition.1c Optimization of the cross-linking process will undoubtedly result in hydrogels with larger volume changes.
In conclusion, hydrogels based on HPMA copolymer and 4-arm PEG have been synthesized via a thiol-maleimide coupling reaction, by conjugation of reactive polymers with an AKe mutant. The triple mutant, AKtm, contained two thiol groups at positions with a large spatial displacement following substrate-recognition-mediated conformational change. Upon substrate binding, the nanoscale conformational change translated into macroscale mechanical motion.
Supplementary Material
Acknowledgments
The research was supported in part by NIH Grant R01 EB005288 and by University of Utah Research Foundation Project No. 51003289. We thank Dr. E. Haas, Bar-Ilan University, for the kind gift of the double mutant plasmid (C77S, A55C)-AKe.
Footnotes
Supporting Information Available: Experimental details of the synthesis, purification and enzymatic activity of AKtm, the synthesis of hydrogels, the labeling of AKtm, and swelling. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Kopeček J, Yang J. Polym Int. 2007;56:1078. [Google Scholar]; (b) Kopeček J, Vacík J, Lím D. J Polym Sci A-1. 1971;9:2801. [Google Scholar]; (c) Miyata T, Asami N, Uragami T. Nature. 1999;399:766. doi: 10.1038/21619. [DOI] [PubMed] [Google Scholar]; (d) Lu Z, Kopečková P, Kopeček J. Macromol Biosci. 2003;3:296. [Google Scholar]; (e) Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]; (f) Wang C, Stewart RJ, Kopeček J. Nature. 1999;397:419. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 2.(a) Ehrick JD, Deo SK, Browning TW, Bachas LG, Madou MJ, Daunert S. Nat Mater. 2005;4:298. doi: 10.1038/nmat1352. [DOI] [PubMed] [Google Scholar]; (b) Murphy WL, Dillmore WS, Modica J, Mrksich M. Angew Chem, Int Ed. 2007;46:3066. doi: 10.1002/anie.200604808. [DOI] [PubMed] [Google Scholar]; (c) Sui Z, King WJ, Murphy WL. Adv Mater. 2007;19:3377. [Google Scholar]
- 3.(a) Müller CW, Schlauderer GJ, Reinstein J, Schulz GE. Structure. 1996;4:147. doi: 10.1016/s0969-2126(96)00018-4. [DOI] [PubMed] [Google Scholar]; (b) Müller CW, Schulz GE. J Mol Biol. 1992;224:159. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]; (c) Sinev MA, Sineva EV, Ittah V, Haas E. Biochemistry. 1996;35:6425. doi: 10.1021/bi952687j. [DOI] [PubMed] [Google Scholar]; (d) Schlauderer GJ, Proba K, Schulz GE. J Mol Biol. 1996;256:223. doi: 10.1006/jmbi.1996.0080. [DOI] [PubMed] [Google Scholar]; (e) Müller-Dieckmann HJ, Schulz GE. J Mol Biol. 1995;246:552. doi: 10.1006/jmbi.1994.0104. [DOI] [PubMed] [Google Scholar]; (f) Haas E. IEEE J Sel Top Quant Electr. 1996;2:1088. [Google Scholar]; (g) Jacob MH, Amir D, Ratner V, Gussakowsky E, Haas E. Biochemistry. 2005;44:13664. doi: 10.1021/bi051205t. [DOI] [PubMed] [Google Scholar]; (h) Ådén J, Wolf-Watz M. J Am Chem Soc. 2007;129:14003. doi: 10.1021/ja075055g. [DOI] [PubMed] [Google Scholar]
- 4.Gerstein M, Krebs W. Nucleic Acids Res. 1998;26:4280. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Xu C, Kopečková P, Kopeček J. Macromol Biosci. 2006;6:201. doi: 10.1002/mabi.200500208. [DOI] [PubMed] [Google Scholar]
- 6.Whitesides GM, Lamotte A, Adalsteinsson O, Colton CK. Methods Enzymol. 1976;44:887. doi: 10.1016/s0076-6879(76)44064-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.