Abstract
Genes underlying signal reception should evolve to maximize signal detection in a particular environment. In animals, opsins, the protein component of visual pigments, are predicted to evolve according to this expectation. Fireflies are known for their bioluminescent mating signals. The eyes of nocturnal species are expected to maximize detection of conspecific signal colors emitted in the typical low-light environment. This is not expected for species that have transitioned to diurnal activity in bright daytime environments. Here we test the hypothesis that opsin gene sequence plays a role in modifying firefly eye spectral sensitivity. We use genome and transcriptome sequencing in four firefly species, transcriptome sequencing in six additional species, and targeted gene sequencing in 28 other species to identify all opsin genes present in North American fireflies and to elucidate amino acid sites under positive selection. We also determine whether amino acid substitutions in opsins are linked to evolutionary changes in signal mode, signal color, and light environment. We find only two opsins, one long wavelength and one ultraviolet, in all firefly species and identify 25 candidate sites that may be involved in determining spectral sensitivity. In addition, we find elevated rates of evolution at transitions to diurnal activity, and changes in selective constraint on LW opsin associated with changes in light environment. Our results suggest that changes in eye spectral sensitivity are at least partially due to opsin sequence. Fireflies continue to be a promising system in which to investigate the evolution of signals, receptors, and signaling environments.
Keywords: insects, molecular evolution, adaptation, comparative biology, Lampyridae, binding pocket
Introduction
The diversity of visual signals in nature is a long-standing enigma in evolutionary biology. Natural selection favors signals and receptors that maximize the detection of signals against environmental “noise”, leading to the expectation that signals, receptors, and the environments in which signals are displayed will be evolutionarily linked, a process known as “sensory drive” (Endler 1992). One prediction of this framework, as applied to visual signals, is that visual receptors will be “tuned” to best detect signals in a specific light environment. This “spectral tuning” hypothesis can be extended to the genes underlying reception, generating the prediction that genes involved in tuning vision should evolve in response to selection pressure from signals, light environments, or both.
Vision in most animals is mediated by opsin proteins in the eye. Opsins are signaling proteins that contain a conserved lysine residue which binds a vitamin A-derived chromophore that is required for light absorption (Palczewski et al. 2000). An opsin with a bound chromophore, collectively termed a visual pigment, maximally absorbs light at a particular wavelength, λmax (Yokoyama 2008). Differences in opsin amino acid sequence and/or chromophore type can change λmax (Yokoyama 2000). Amino acid sites in the opsin at which changes in sequence alter λmax are generally located in regions that interact with the chromophore, termed the chromophore binding pocket (Wilkie et al. 2000).
The range of wavelengths that an organism can detect is affected by opsin copy number. Many animal lineages have several visual opsin paralogs that are classified based on the wavelengths of light they detect. For example, the common ancestor of all insects is thought to have contained three visual opsins, one sensitive to long wavelengths (LW; λmax from 600 to 480 nm), one sensitive to blue wavelengths (B; λmax from 480 nm to 400 nm), and one sensitive to ultraviolet wavelengths (UV; λmax from 400 to 300 nm)(Briscoe & Chittka 2001). Selection on visual spectral sensitivity may affect both the number of opsin paralogs and their peak sensitivities. For example, nocturnal, cave-dwelling, and low-light-living species tend to show reduced selective constraint on or the loss-of-function of one or more opsin copies (e.g. beetles: Jackowska et al. 2007; flying squirrels: Carvalho et al. 2006).
Fireflies are a diverse, globe-spanning family of beetles that allow for testing of the associations between signals, light environments, and visual receptor evolution (e.g. Biggley et al. 1967; Lall, Seliger et al. 1980). This family includes many species renowned for their nocturnal lighted mating displays. Generally, flying males flash to stationary females, who respond with flashes of their own (Lloyd 1966). While fireflies are perhaps best known for their species-specific variation in flash pattern (e.g. Lloyd 1966), their flashes also differ in color, with peak emission wavelengths ranging from green (554 nm) to yellow (580 nm) across North American species (Seliger et al. 1964; Biggley et al. 1967; Lall, Seliger, et al. 1980). Nocturnal taxa are also active at different times and in different habitats (Lloyd 1966). These conditions alter the light environment in which signals are displayed (considered in Endler 1993; Théry et al. 2008). In addition, there are several diurnal lineages that have independently lost adult light signals and instead use long-distance pheromones to identify and locate mates (Stanger-Hall et al. 2007; Stanger-Hall & Lloyd 2015). Thus, fireflies provide the opportunity to examine the evolution of signals and signal reception in response to changes in signal color, light environment, and signal mode (nocturnal, light signals vs. diurnal, no light signals).
Fireflies also vary in visual reception. Previous work provided physiological evidence that firefly vision is tuned to detect conspecific light signals with the recorded peak spectral sensitivity closely matching the peak emission wavelength of the conspecific light signal (Lall, Chapman, et al. 1980; Lall 1981; Lall et al. 1982; Eguchi et al. 1984; Cronin et al. 2000). These data suggested that there are three expressed opsins in fireflies: one LW, one B, and one UV, similar to the hypothesized situation in the ancestral insect. However, this prediction has not been supported to date—only two expressed opsins, one LW and one UV, were found in the dusk-active Little Asian Firefly, Luciola cruciata (Oba & Kainuma 2009).
Here we examine whether there is evidence that opsin sequence and copy number contribute to the reported spectral sensitivity of the firefly eye. To accomplish this goal we first elucidate the molecular evolution of visual opsins across 38 species of North American fireflies. We then test for relationships between selective constraint, amino acid sequence, and signaling ecology in a phylogenetic context. We predict that at least three opsins, one LW, one UV, and one B, determine the spectral sensitivity of firefly eyes. Further, we gather evidence for the potential role of opsins in spectral tuning in several ways. (1) We develop a set of candidate functional sites by testing for selection and constraint across the opsin molecule. We expect to find evidence of positive selection at a subset of amino acids that may influence λmax. (2) We examine the location of candidate sites, which we predict will be in positions that change λmax, specifically the chromophore binding pocket. (3) We examine both selective constraint and amino acid sequence variation in relationship to potential sources of selection. Positive selection and amino acid substitutions at candidate sites should correlate with changes in signaling ecology, including ambient light, conspecific light emissions, or both. These patterns should be most apparent between nocturnal and diurnal taxa since these species differ the most in both signal (light signal vs. no light signal) and light environment (night vs. day). To generate our dataset and test our predictions, we used high-throughput RNA and genomic sequencing to identify putative opsins, and then employed tests of selection and ancestral state reconstruction to investigate their molecular evolutionary history.
Materials and Methods
Specimen and Data collection
Firefly specimens were caught by hand and the date, time, temperature, locality, habitat type, and, when possible, flash pattern were recorded. Light emission spectra from 1–5 individuals per population per species were measured using a portable spectrophotometer (Text S1). Specimens were initially identified to species by a combination of ecological, morphological, and behavioral characteristics (Green 1956, 1957; Lloyd 1966, 1969; Fender 1966; Luk et al. 2011). To confirm species identification, genomic DNA was extracted from legs or thorax using a DNeasy Blood and Tissue kit (QIAGEN), and 647 bp of cytochrome c oxidase I (COI) were amplified and sequenced (Stanger-Hall et al. 2007; Stanger-Hall & Lloyd 2015).
RNA Sequencing
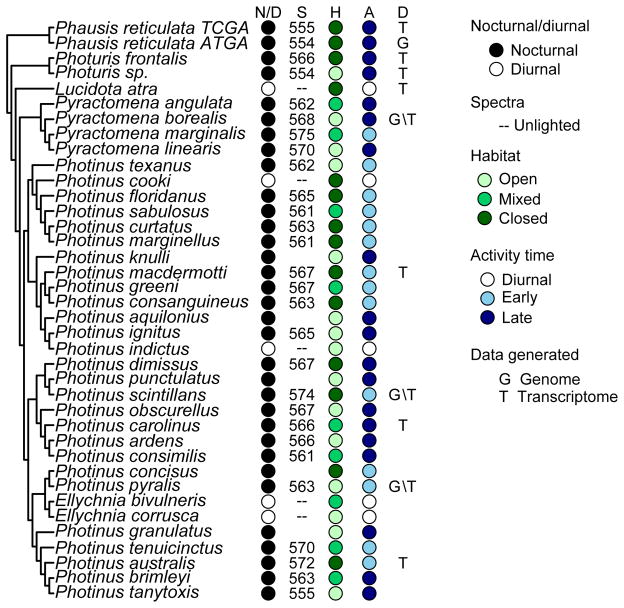
We selected 10 firefly species for RNA sequencing based on taxonomic divergence, divergence in signaling mode and emission wavelength, and the availability of specimens (Figure 1; Table S1). Males were used since females of many species are difficult to locate in the field. For each of the 10 species, RNA was isolated from 1–6 heads harvested during the active period and immediately frozen in liquid nitrogen or stored in RNAlater (LifeTechnologies). Bodies of specimens selected for RNA and genomic sequencing, and all other specimens used in PCR amplification, were preserved in 95% ethanol at −80°C until extraction. RNA was also isolated from the adult light organs of Photinus pyralis and Photinus macdermotti, and from the larval light organ of Pn. pyralis based on specimen availability.
Figure 1.
Total RNA from all samples was extracted using Trizol (LifeTechnologies) and treated with DNase prior to library construction. TruSeq RNA libraries (Illumina) were constructed at the Georgia Genomics Facility (Athens, GA) with an average insert size of 150 bp. Samples were individually barcoded and pooled before submission to BGI (Hong Kong) for 100 bp, paired-end sequencing in one lane of Illumina HiSeq2000 v3.
Genomic Sequencing
Sequencing of genomic DNA was performed on a taxonomically diverse subset of species (Pn. pyralis, Pn. scintillans, Pyractomena borealis, and Phausis reticulata) to determine opsin copy number. Genomic DNA was phenol-chloroform extracted (Sambrook et al. 1989) from thorax (Pn. pyralis, Py. borealis) or whole body (Pn. scintillans, Pa. reticulata) and treated with RNAseA before library preparation. Truseq DNA libraries (Illumina) with an average insert size of 300 bp were constructed at the Georgia Genomics Facility before submission to BGI for sequencing. All four species were individually barcoded, pooled, and sequenced in one lane of Illumina HiSeq2000 v3 100 bp paired-end reads for 10x (Pn. scintillans, Py. borealis, Pa. reticulata) and 25x (Pn. pyralis) coverage.
Opsin Identification
Following sequencing, Illumina reads were assessed for quality using FastQC v0.10.1 (Babraham Bioinformatics 2012) and then trimmed, adapters removed, and filtered for quality using the fastqmcf program in the eautils package v1.1.2 (Aronesty 2011; parameters: -m 13, -C 1000000, -x 0.01, -q 20, -w 4). Transcriptomes were assembled de novo using the Trinity pipeline v20121005 with default parameters (Grabherr et al. 2011). Candidate opsin transcripts were identified by querying the previously published amino acid sequences of L. cruciata opsins (GenBank: LW, AB300328; UV, AB300329; Oba & Kainuma 2009) against the assembled transcriptomes using tBLASTn (Altschul et al. 1990; default parameters) and then querying the results against the NCBI nucleotide database using BLASTn (evalue: 1e-06). Transcriptome components with the greatest identity to insect opsins were then aligned in Geneious R6 (Biomatters Ltd 2013) using Muscle (Edgar 2004). Alignments were manually reviewed for sequence similarity and the presence of an open reading frame (ORF) longer than 300 bp (100 amino acids). Expression levels were obtained by aligning trimmed mRNA sequencing reads to the assembled transcriptome components using bowtie (Langmead et al. 2009) and quantifying expression with RSEM (Li & Dewey 2011). Putative opsin wavelength sensitivity was inferred from homology to experimentally validated insect LW and UV opsin sequences and confirmed using a neighbor-joining phylogeny (Figure S4).
Finally, to verify the presence of putative opsins and examine opsin copy number variation across the firefly genomes, L. cruciata mRNA sequence for UV and LW opsins were queried against the four libraries of genomic DNA sequences using the dc-megablast program for blastn. These sequencing reads were matched to their paired-ends using faSomeRecords (UCSC Genome Browser) and assembled into contigs in Sequencher (Gene Codes Corp. 2011) using the clean data assembly algorithm (minimum match percentage = 85, minimum overlap = 10). Following assembly, each contig was BLASTed against the NCBI nt database using the megablast program for blastn (default parameters) and contigs with top hits to opsin were retained. Contigs were then “walked out” to encompass ~1 kb on either side of the coding sequence (CDS) using the process described above substituting the ends of assembled contigs as queries.
To corroborate opsin sequences identified bioinformatically, degenerate primers were designed from transcriptomic and genomic sequences and used to amplify LW and UV opsin from genomic DNA. After putative LW and UV opsins were verified in the genomes of the 10 individuals used to obtain transcriptome data, the primers were then used to amplify opsins from the genomic DNA of 28 additional species for which habitat, activity period, and spectra data were available (Figure 1; Table S1). Information on primer sequences, PCR cycling conditions, and Sanger sequencing is given in Table S2. Sequences were assembled de novo, annotated for intron-exon boundaries using homology to transcriptome sequences, and introns removed to obtain the CDS in Geneious. All CDS were then aligned using Muscle in Geneious and manually reviewed. The final alignment for LW opsin was 1,032 bp in length and represented amino acids 18-378, ending 18 amino acids upstream of the stop codon. The final alignment for UV opsin was 1,137 bp in length, included the start site, and ended 6 amino acids upstream of the stop codon. Amino acids at either end of the opsins are not known to be involved with the chromophore and their exclusion from the final alignments should not affect tests of our hypotheses. Homology models were created in SwissModel (Arnold et al. 2006) using squid rhodopsin (Todarodes pacificus, PDB: 2z73A; Murakami & Kouyama 2008) as a template (Text S2). Chromophore binding pocket sites were identified by visual inspection of Van der Waals forces in each model. Amino acid sites are numbered in reference to the full-length Pn. pyralis amino acid sequences.
Species phylogeny
Analysis of opsin sequences was performed on species tree topologies achieved by extending the three-locus dataset described in Stanger-Hall and Lloyd (2015) to include taxa outside Photinus. Briefly, wingless (WG, 420 bp), rudimentary (CAD, 594 bp), and COI (1272 bp) sequences were obtained from nine additional taxa (Table S3) and tree construction procedures from Stanger-Hall and Lloyd (2015) applied to the extended dataset. This analysis resulted in two slightly different, highly supported tree topologies, depending on taxon sampling. Subsequent analyses of positive selection were performed on both topologies and resulted in similar findings. Figures presented in the main text display the topology consistent with that presented in Stanger-Hall and Lloyd (2015). Details of the phylogenetic methods, comparison of the species phylogeny to a phylogeny generated from the opsin sequences, and the robustness of the phylogeny to the model of sequence evolution, number of taxa, and the method of construction are discussed in the Supporting Information (Text S3).
Identifying positively selected sites
The two tree topologies with branch lengths were used to examine rates of molecular evolution across branches and sites using PAML4 (Yang 2007). Phausis reticulata, a North American species shown to be basal to all other North American taxa (Stanger-Hall et al. 2007), was used as the outgroup in the opsin analyses since a complete dataset containing all of the loci used in our phylogeny and the opsins was not available from any other beetle taxon. We used the metric ω, the ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site, dN/dS, as a measure of positive selection. An ω value that is less than, equal to, or greater than 1 is indicative of purifying selection, no selection (neutrality), or positive selection, respectively. The branch models tested included M0 (one ω over all branches and sites), two-rate (one ω for branches with transitions to diurnal and one for branches without transitions), and a free-ratio model (each branch has its own ω). The site models tested were: M1a (neutral) with a class of sites evolving under purifying selection and a class of sites evolving neutrally; M2a (selection) with 3 classes of sites, one under purifying selection, one evolving neutrally, and one evolving under positive selection; M3 (discrete) with 3 classes of sites, similar to M2a, except that the sites classes are constrained to have successively greater ω values rather than a specific ω; M7 (beta) with 10 classes of sites evolving with 0 < ω < 1, sampled according to a beta distribution; and M8 (beta), similar to the M7 model, with the addition of a class of sites with ω greater than 1. Nested models were compared using likelihood ratio tests (LRTs). In models that included a positively selected class of sites, Bayes Empirical Bayes analysis (Yang et al. 2005) was performed to identify sites under selection.
Fitmodel v20140407 (Guindon et al. 2004) was used to evaluate sites under positive selection along branches without defining a branch of interest a priori. Fitmodel accomplishes this by allowing sites to switch between ω classes. The M2a and M3 selection models can then be tested while incorporating different switching models. Equal switching models estimate a single switching rate among the ω classes, while biased switching models estimate a separate switching rate between each pair of classes. Fitmodel has been shown to outperform standard methods for detecting selection at sites along branches, especially when the foreground branches defined in a standard analyses do not represent the branches along which there has been selection (Lu & Guindon 2014).
Testing for correlations with signal and ecological traits
We tested for correlations between measures of opsin evolution for each lineage and four explanatory signal and ecological traits: 1) signaling mode (nocturnal/light or diurnal/no light), 2) spectra (mean male peak emission wavelength), 3) habitat (open, mixed, or closed) based on the amount of canopy cover in the signaling environment, and 4) activity start time (early or late) based on when the first individual was observed signaling relative to sunset (Lall, Seliger, et al. 1980). All 38 North American taxa from which we were able to obtain LW and UV opsin sequences were used to test signal mode. Only nocturnal taxa were used to test emission spectra, habitat, and activity time because the spectral tuning hypothesis for fireflies is predicted for light-signaling taxa only and adults of diurnal species do not produce light. Data for signal and ecological traits was gathered from both the literature (Seliger et al. 1964; Biggley et al. 1967; Otte & Smiley 1977; Cicero 1983; Stanger-Hall & Lloyd 2015) and field measurements. Where possible, the spectra, habitat, and activity time values for the population where the specimen(s) used for opsin sequencing was caught were used in the final dataset (Text S1). Investigation of spectra was limited to the 28 species for which we were able to obtain emission data. Male average peak emission wavelengths were used because female data were not available in either the literature or in our collected dataset across a sufficient number of species.
Signaling and ecological traits were tested for phylogenetic signal using Blomberg’s K (Blomberg et al. 2003) in the picante package in R (Kembel et al. 2010; Table S4). All traits except for signaling mode showed significant or nearly significant phylogenetic signal; therefore, analyses were performed on values calculated using phylogenetic independent contrasts (Felsenstein 1985) using the ape package (Paradis et al. 2004) in R. In contrast to the other traits, nocturnal/diurnal activity was treated as a discrete variable, reconstructed using maximum parsimony, and measures of opsin evolution compared between branches with different signal modes.
The measures of opsin evolution examined included: number of amino acid substitutions, number of parallel (same amino acid to same amino acid) and convergent (different amino acid to same amino acid) substitutions, dN, whole protein ω values, and site-specific ω values. The number and types (parallel or convergent) of amino acid substitutions between nodes was determined using ancestral sequence reconstruction. Amino acid alignments were first tested for an appropriate model of evolution using ProtTest v3.3 (best models, LW: LG; UV: JTT; Darriba et al. 2011), then ancestral states at each site were reconstructed and visualized using the phangorn package in R (Schliep 2011; Kenaley et al. 2014). Absolute numbers and types of substitutions were parsed from phangorn output using a custom R script. Distributions for numbers of parallel and convergent amino acid substitutions occurring in nocturnal lineages were generated by bootstrapping the parsed dataset in R for nocturnal branches only (1000 reps). Estimates of selective constraint along branches and at sites along branches were taken from PAML free-ratio and fitmodel M2aS1 (LW) or M3S2 (UV) model output, respectively. These models were shown to be the best fit for the data using LRTs.
We explored the relationships between several response and explanatory variables using partial correlations to control for divergence (branch length) in SPSS v22 (IBM Corp. 2013). The response variables examined were the number of amino acid substitutions along lineages, difference in dN, cumulative nonsynonymous substitution rate, and difference in selective constraint (ω) between taxa for each opsin. The explanatory variables were change in peak emission wavelength, habitat, and activity time. Before analysis, variables were assessed for normality in JMP Pro 10 (SAS Institute Inc.). Change in peak emission wavelength was normally distributed, while all other variables did not conform to normality even after transformation. Accordingly, we proceeded with the analysis under the assumption that partial correlation analysis is robust to departures from normality (Voortman & Druzdzel 2008).
Results
Transcriptome and genome sequencing reveal two opsin genes
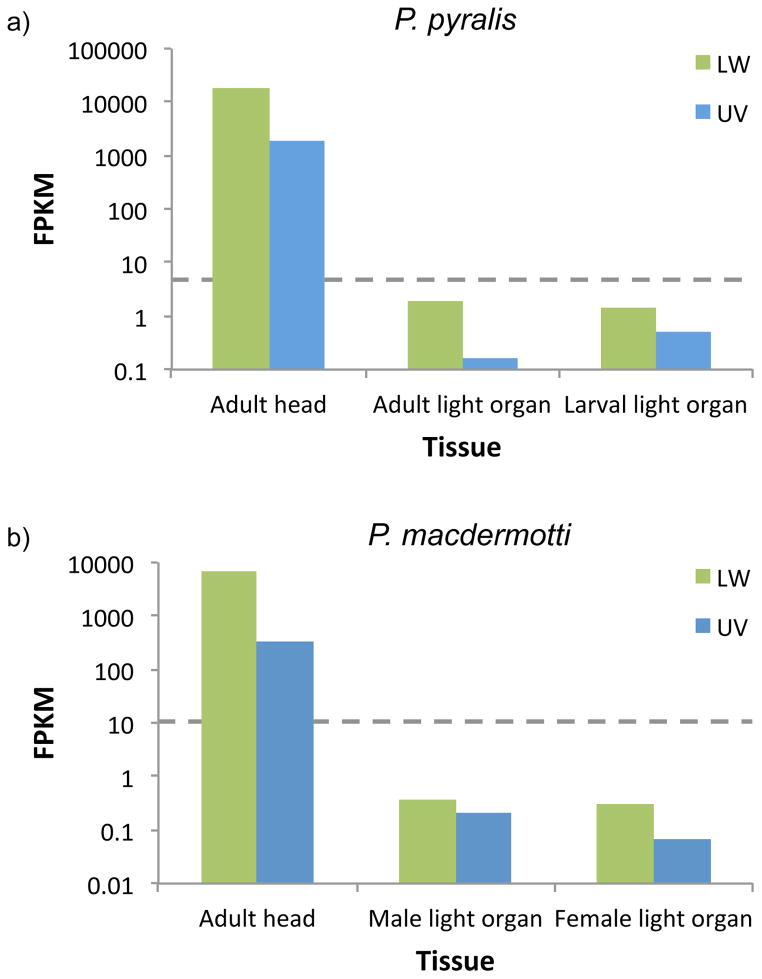
RNAseq of heads from 10 species yielded 172 million reads totaling 34.5 Gb (Table 1). Several transcripts showed homology to known insect opsins using BLAST. However, only two transcripts, one with homology to LW opsin and one with homology to UV opsin, had additional evidence to suggest that they encode bona fide opsin proteins: both had complete open reading frames (ORFs) and could be recapitulated from whole-genome shotgun Illumina reads. Comparing opsins assembled from genomic reads and the putative opsin transcripts revealed that firefly LW opsin genes are 1247–1935 bp in length (including introns), encode 344–381 amino acids, and contain 5 exons (Figure S1a). The firefly UV opsins are 1404–1525 bp in length, encode 385–386 amino acids, and have 6 exons (Figure S1b). The lengths of both opsins are within the range of other sequenced insect opsins. Both candidate opsins also have the structural characteristics of other described opsins, including 7 transmembrane domains and the conserved lysine at K324 (LW) and K323 (UV) where the chromophore is bound (Figure S2). In addition, the full-length LW and UV opsin candidates are highly expressed in heads; in all 10 species they are in the top 5% of expressed genes (e.g. Figure S3). In contrast, opsins are expressed at 10,000-fold lower levels in adult and larval light organs compared to heads in the two species for which we have data, Pn. pyralis and Pn. macdermotti (Figure 2). No other putative visual opsin transcripts identified in the RNAseq data could be reconstructed from genomic reads. However, we did detect a distantly related c-opsin in several species (data not shown). Arthropod c-opsins diverged from arthropod visual opsins in an ancient split and are thought to function in circadian rhythm regulation (Arendt 2003; Porter et al. 2012). In species where we did not detect full-length ORFs of c-opsin, we were able to detect members of the extended G protein-coupled receptor gene family, indicating that our analysis would have detected additional visual opsins if they were present. A neighbor-joining phylogeny of insect opsin sequences showed that the putative LW and UV opsins fall in clades with LW and UV, but not B, opsins from other insect species (Figure S4).
Table 1.
Transcriptome assembly metrics
| Species | Number of Reads | % lost to QC | Total # of components (# comps 1 – 4 kb) | Trinity Assemblies | # similar to opsin | # BLAST to opsin | # putative opsins highly expressed | |
|---|---|---|---|---|---|---|---|---|
| Mean length | Median length | |||||||
| Photinus pyralis | 39,069,471 | 1.7 | 77,811 (27,766) | 1715 | 881 | 107 | 10 | 2 |
| Photuris sp. | 12,359,044 | 1.4 | 54,415 (17,746) | 1395 | 653 | 34 | 6 | 2 |
| Photinus scintillans | 15,456,064 | 1.8 | 63,440 (20,865) | 1440 | 666 | 39 | 5 | 2 |
| Lucidota atra | 11,194,970 | 1.8 | 56,584 (19,259) | 1440 | 718 | 73 | 10 | 2 |
| Photinus carolinus | 7,472,450 | 1.6 | 43,254 (13,950) | 1338 | 615 | 25 | 2 | 2 |
| Photinus macdermotti | 41,783,634 | 4.9 | 76,743 (30,730) | 1701 | 1022 | 80 | 12 | 2 |
| Photinus australis | 13,145,094 | 1.7 | 43,963 (14,649) | 1140 | 606 | 32 | 4 | 2 |
| Photuris frontalis | 11,373,006 | 1.3 | 56,427 (17,855) | 1278 | 589 | 34 | 4 | 2 |
| Pyractomena borealis | 11,431,732 | 1.6 | 52,416 (18,463) | 1544 | 792 | 60 | 5 | 2 |
| Phausis reticulata | 9,245,174 | 0.02 | 67,843 (21,132) | 1297 | 581 | 47 | 12 | 2 |
Figure 2.
LW and UV opsin survey in fireflies
Using transcriptome and genome sequences, we designed degenerate primers and used them to amplify and sequence genomic LW and UV opsins from a total of 38 species representing a diversity of ecological and signaling traits. Across all taxa in our dataset, within both LW and UV opsins, there was 86–94% identity at the amino acid level with the previously published L. cruciata opsin sequences, and ~70 % identity with other beetle opsins. There was no evidence for recent gene duplications of LW or UV opsin in any of the lineages examined: coverage of opsins in genomic sequences was within that expected for single-copy genes, there was no evidence of multiple flanking regions in either the transcriptome or genomic datasets, and double peaks in Sanger-sequence chromatograms were consistent with heterozygosity rather than duplication.
LW and UV opsins show evidence of positive selection at specific sites
Using PAML (Yang 2007), we found evidence of strong purifying selection (ω <1) over the entire gene for both LW and UV opsins (M0, one ratio model; LW: ω = 0.07, UV: ω = 0.05; Table 2). In both cases, models that included a small proportion of sites under positive selection over the entire phylogeny (M2a, M8) were a better fit than the M0 model (Table 2). Bayes Empirical Bayes analysis under the M8 model identified seven sites with ω > 1 in LW opsin (Table 3); of these, the ω values were statistically greater than neutrality (ω = 1) for three sites, 181, 235, and 188, indicating positive selection. In UV opsin, Bayes Empirical Bayes analysis under the M8 model identified seven sites with ω > 1, but none were statistically greater than 1.
Table 2.
Results of PAML analysis for LW and UV opsin genes
| Gene | Parametersb | |||||||
|---|---|---|---|---|---|---|---|---|
| Modela | lnL | ω0/p (p0) | ω1/q (p1) | ω2/ωP (p2) | Null | LRT | df | |
| LW opsin | M0 (one rate) | −6550.53 | 0.07 | |||||
| Branch | H1 (N vs D)c | −6548.44 | 0.07 | 0.10 | M0 | 4.17* | 1 | |
| Free | −6432.33 | see Figure S11 | M0 | 236.40* | 37 | |||
| Sites | M1a (neutral) | −6348.90 | 0.04 (0.75) | 1 (0.25) | M0 | 403.26* | 1 | |
| M2a (selection) | −6302.40 | 0.04 (0.91) | 1 (0.09) | 999d (0.0003) | M1a | 92.99* | 2 | |
| M3 (discrete) | −6285.76 | 0.02 (0.84) | 0.49 (0.16) | 999d (0.0003) | M0 | 529.54* | 4 | |
| M7 (beta) | −6329.70 | 0.14 | 0.48 | |||||
| M8 (beta, selection) | −6277.28 | 0.15 | 1.22 | 243.12 (0.001) | M7 | 104.85* | 2 | |
|
| ||||||||
| UV opsin | M0 (one rate) | −7241.40 | 0.05 | |||||
| Branch | H1 (N vs D)c | −7236.91 | 0.05 | 0.09 | M0 | 8.98* | 1 | |
| Free | −7151.25 | see Figure S11 | M0 | 180.31* | 37 | |||
| Sites | M1a (neutral) | −7111.53 | 0.04 (0.80) | 1 (0.20) | M0 | 259.74* | 1 | |
| M2a (selection) | −7060.18 | 0.04 (0.94) | 1 (0.06) | 999d (0.0004) | M1a | 102.71* | 2 | |
| M3 (discrete) | −7031.77 | 0.02 (0.83) | 0.34 (0.17) | 999d (0.0003) | M0 | 419.26* | 4 | |
| M7 (beta) | −7093.36 | 0.14 | 0.62 | |||||
| M8 (beta, selection) | −7028.47 | 0.18 | 2.08 | 443.45 (0.0006) | M7 | 129.78* | 2 | |
Best models within a nested set of models (branch, site: selection, discrete, beta) are shown in bold
For branch models, ω0 (background branches) and ω1 (branches of interest); for site models M1a-M3, ωx and px (proportion of sites) in each class (0, 1, 2); for site models M7 and M8, p and q describe the shape of the beta distribution, ωP is the value of ω for the positively selected site class, and p2 gives the proportion of sites in this class.
Nocturnal versus diurnal branches
A value of 999 indicates an ω2 greater than 1, but unable to be precisely estimated
p < 0.05
Table 3.
Sixteen candidate sites with evidence for positive selection and/or function
| Gene | Sitea | Identified in Model(s) | Locationb | Change at homologous site | |
|---|---|---|---|---|---|
| Evidence for Phenotypic Effect | Site, Organism, Opsin | ||||
| LW | T108 | M2a, M8 | NC | Shift from 530 to 550 nm | 63, Heliconius, LWc |
| Cline with latitude | 112, Liminitis LWd | ||||
| A181* | M2a, M8, M2aS1 | H4 | Shift between 530 and 510 nm | 136, Vanessa, LWc | |
| Shift from 530 to 550 nm | 136, Heliconius, LWc | ||||
| Shift between 530 and 575 nm | 136, Papilio, LWc | ||||
| Positively selected in parasitic wasps | 198, Fig wasp, LWe | ||||
| L188* | M2a, M8, M2aS1 | NC | Shift between 530 and 575 nm | 143, Papilio, LWc | |
| Shift from 530 to 550 nm | 143, Heliconius, LWc | ||||
| Faster evolution after duplication | 143, Bombus, LWf | ||||
| V235* | M2a, M8, M2aS1 | H5 | |||
| A248 | M8 | H5 | |||
| F307 | M2aS1 | H6 | |||
| T309 | M8 | EC | |||
| A356 | M8 | T | Positively selected in dim-light foragers | 229, Halictidae, LWg | |
|
| |||||
| UV | L59 | M2a, M8 | NC | Positively selected after duplication | 60, Heliconius, UVh |
| S133 | M3S2 | BP | S to A shifts absorption (450–437 nm) | 116, Pieris rapae, B and Vi | |
| V218 | M8 | NC | |||
| A267 | M8 | IC | |||
| A268 | M8 | IC | |||
| S299 | M8 | BP | |||
| T321 | M2a, M8 | NC | |||
| T373 | M8 | T | |||
Site number and amino acid in reference to Pn. pyralis
NC: near chromophore, but not in contact; BP: binding pocket; H: helix, far away from binding pocket; EC: extracellular loop; IC: intracellular loop; T: tail
Ancestral state reconstruction (Briscoe 2001)
Within-species cline with latitude. Hypothesized involvement in thermal stability. (Frentiu et al. 2015)
Tests of positive selection (Wang et al. 2013)
Tests of positive selection (Spaethe and Briscoe 2004)
Tests of positive selection (Tierney et al. 2011)
Tests of positive selection (Briscoe et al. 2010)
Site-directed mutagenesis (Wakakuwa et al. 2010)
significant at p=0.05, BEB analysis in PAML M8 model.
To examine selection on sites across branches of the phylogeny, we employed a fitmodel analysis (Guindon et al. 2004). The best models for LW opsin based on LRTs were M2aS1 (selection model with equal switching; ω0 = 0.00, ω1 = 1, ω2 = 2.75), and M3S2 (discrete model with biased switching; ω0 = 0.000, ω1 = 0.003, ω2 = 9.79; Table 4), with one exception based on the specific tree topology that was used (Text S3). The four sites detected by the M2aS1 models included all three of the positively selected sites identified in the PAML site-model analysis, though the posterior probabilities did not exceed 0.95. The M3S2 model identified most of the 104 variable sites in LW as under positive selection on at least one branch of the phylogeny. However, this model was too complex for our data because the posterior probabilities for sites being in the positively selected class were below the recommended cut-off of 0.9 (maximum: 0.84), the estimates of ω0 and ω1 were nearly equal, and the switching parameter was zero. Thus, we further investigated only the eight sites with evidence for ω > 1 across the PAML M2a, PAML M8, and fitmodel M2aS1 analyses (Table 3).
Table 4.
Results of Fitmodel analysis for LW and UV opsin genes
| Modela | Parametersc | |||||||
|---|---|---|---|---|---|---|---|---|
| Switchingb | lnL | ω0 (p0) | ω1 (p1) | ω2 (p2) | Null | LRT | df | |
| LW opsin | ||||||||
| M0 (one rate) | none | −6190.41 | 0.08 (1) | n/a | n/a | n/a | ||
| M1a (neutral) | none | −6309.16 | 0 (0.71) | 1 (0.29) | n/a | M0 | −37.49 | 1 |
| M2a (selection) | none | −6150.19 | 0.04 (0.91) | 1 (0.09) | 20 (0.001) | M1a | 317.95* | 2 |
| M2aS1 (selection) | equal | −6086.95 | 0.00 (0.92) | 1 (0.07) | 2.75 (0.01) | M2a | 129.48* | 1 |
| M2aS2 (selection) | biased | −6086.15 | 0.00 (0.91) | 1 (0.08) | 2.04 (0.01) | M2aS1 | 1.59 | 2 |
| M3 (discrete) | none | −6120.35 | 0.00 (0.65) | 0.14 (0.28) | 0.75 (0.07) | M1a | 377.63* | 3 |
| M3S1 (discrete) | equal | −6084.41 | 0.00 (0.87) | 0.41 (0.09) | 1.65 (0.04) | M3 | 71.87* | 1 |
| M3S2 (discrete) | biased | −6068.79 | 0.00d (0.10) | 0.00d (0.88) | 9.79 (0.01) | M3S1 | 31.25* | 2 |
|
| ||||||||
| UV opsin | ||||||||
| M0 (one rate) | none | −6936.73 | 0.06 (1) | n/a | n/a | n/a | ||
| M1a (neutral) | none | −7058.89 | 0 (0.73) | 1 (0.27) | n/a | M0 | −244.31 | 1 |
| M2a (selection) | none | −6859.71 | 0.04 (0.94) | 1 (0.06) | 20 (0.004) | M1a | 398.35* | 2 |
| M2aS1 (selection) | equal | −6816.10 | 0.01 (0.88) | 0.30 (0.06) | 1e (0.06) | M2a | 87.23* | 1 |
| M2aS2 (selection) | biased | −6824.05 | 0.01 (0.92) | 1 (0.07) | 18.04 (0.01) | M2aS1 | −15.91 | 2 |
| M3 (discrete) | none | −6824.91 | 0.00 (0.61) | 0.10 (0.33) | 0.54 (0.06) | M1a | 467.96* | 3 |
| M3S1 (discrete) | equal | −6817.10 | 0.01 (0.89) | 0.59 (0.11) | 17.37 (0.0005) | M3 | 15.62* | 1 |
| M3S2 (discrete) | biased | −6813.45 | 0.01 (0.89) | 0.49 (0.11) | 20 (0.0009)f | M3S1 | 7.29* | 2 |
Best models within each nested model set (selection, discrete) as given by LRTs shown in bold. Selection models are constrained so that ω1 = 1, while discrete models are constrained to have ω0 < ω1 < ω2.
Switching scheme for each model. None = sites do not switch between ω classes, equal = sites have equal rates of switching between ω classes, biased = unequal rates of switching between ω classes.
Estimated parameters for each ω class under the model. ωx is the estimated omega for the 3 classes of sites (0, 1, 2). px is the estimated proportion of sites in each class.
In this case, while the biased switching model was favored significantly over the equal switching model, ω0 was estimated to be = ω1, and the switching parameters was low (0, data not shown) due to the fact that these classes were virtually indistinguishable in evolutionary rate. The interpretation of this result is that most of the molecule is under constraint, while a small proportion of sites (0.01) is in the positively selected class (ω2).
ω2 seems to violate the constraints of the selection model. Fitmodel estimates the ω values for each class with the constraint that ω1 = 1. However, if the estimated ω2 is smaller than 1 at the end of analysis, then the omega values are re-ordered so that ω2 is always has the largest ω value, in this case 1.
The estimated proportion of sites is small compared to the alignment length (385 codons). Accordingly, we further examined mutations at the single site identified as in ω2 by this model (see main text).
significant at p < 0.05
For UV opsin, fitmodel supported models M2aS1 (ω0 = 0.01, ω1 = 0.3, ω2 = 1) and M3S2 (discrete with biased switching; ω0 = 0.01, ω1 = 0.5, ω2 = 20; Table 4), again with one exception based on the specific topology used (Text S3). Even though M2aS1 is a selection model, it did not estimate any sites with ω > 1. In contrast, the M3S2 model identified one site, 133, as being in the ω2 class (ω = 20) on three of the 73 branches in the phylogeny. Though the posterior probabilities of site 133 being in the ω2 class on these branches were low (range: 0.52–0.58), investigation of nonsynonymous and synonymous nucleotide changes showed that this site had multiple nonsynonymous hits in these lineages. Consequently, we considered this site, along with the seven sites identified in the PAML analysis, in further analysis (Table 3).
Most positively selected sites are outside the chromophore binding pocket
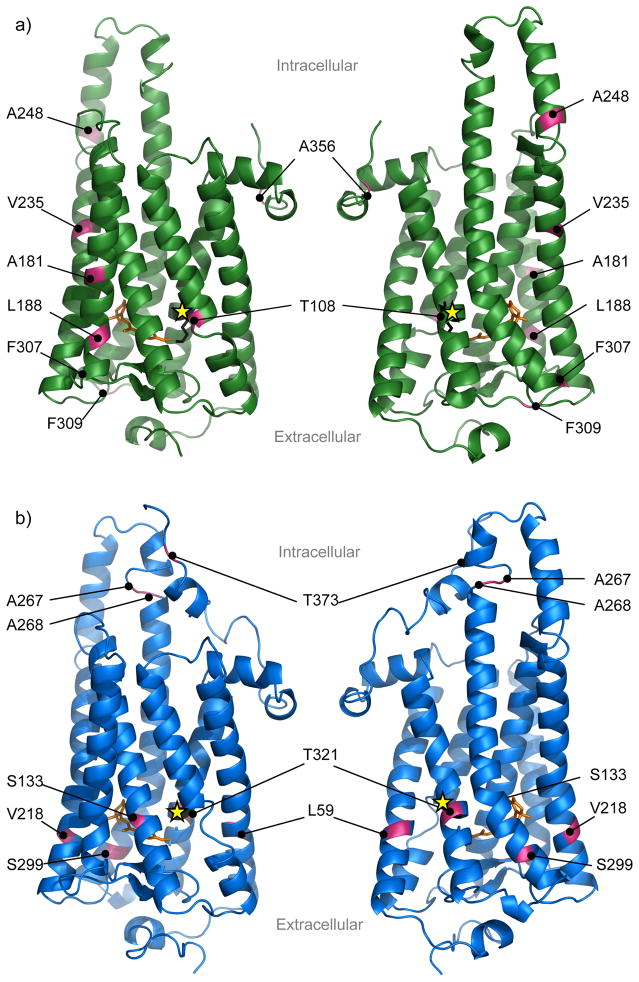
We used homology modeling to estimate the tertiary structure of firefly opsins and identify sites in the chromophore binding pocket. To do this, we compared the sequence of firefly LW and UV opsins separately to Japanese Common Squid (T. pacificus) opsin, the closest species with a known protein tertiary structure (Murakami & Kouyama 2008). In total, we identified 24 sites in each opsin protein that are likely to interact with or have potential long-range effects on the chromophore. Twenty-two of the 24 binding pocket sites identified in LW opsin were invariant across the 38 species (Table S5). None of the eight sites with ω > 1 in LW opsins were identified as a binding pocket site (Figure 3). In UV opsin, 19 of 24 binding pocket sites were invariant and two of the eight positively selected sites, 133 and 299, were identified as binding pocket sites (Figure 3, Table S5).
Figure 3.
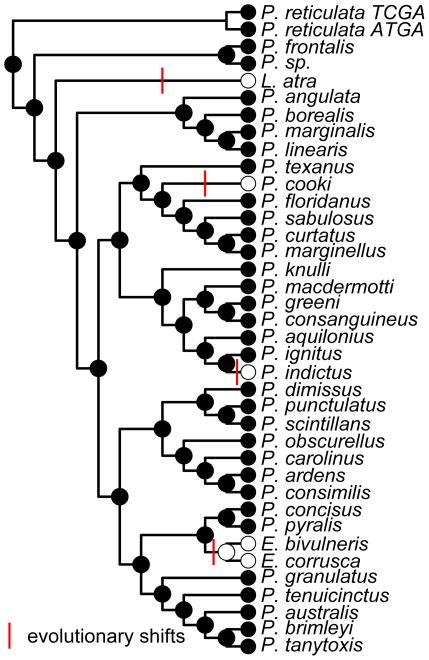
Amino acid changes are linked to signaling mode
We used ancestral state reconstruction to identify branches where evolutionary transitions between signaling modes (nocturnal, light/diurnal, no light) occurred. Maximum parsimony reconstruction supported a model with a nocturnal ancestor and four independent transitions to diurnal activity (Figure 4). PAML branch analysis supported models with elevated whole-molecule ω along branches shifting to diurnal activity relative to branches remaining nocturnal in both LW and UV opsins (Table 2; LW: 0.10 vs 0.07; UV: 0.09 vs 0.05) indicating positive or relaxed purifying selection. In LW opsin, all four sites identified in fitmodel were under positive selection along at least one of the branches with a transition from nocturnal to diurnal activity. However, these sites were also positively selected along some nocturnal branches. In UV opsin, the single site identified by fitmodel was positively selected along the branch leading to the most recent common ancestor (MRCA) of Ellychnia, a diurnal clade of fireflies, as well as on the two branches within the group.
Figure 4.
To better understand the relationship between signal mode and amino acid substitutions at sites of interest (positively selected, binding pocket, and other variable sites), we reconstructed ancestral amino acid sequences across the phylogeny using maximum likelihood. Significantly more changes occurred on diurnal branches than expected based on their phylogenetic representation (Fisher’s exact text (one-tailed); LW: diurnal: 53 changes on 6 branches, nocturnal: 177 changes on 68 branches, p=0.0024; UV: diurnal: 67 changes, nocturnal: 137 changes, p = 0.0001). In addition, there were significantly greater numbers of parallel and convergent amino acid substitutions between pairs of diurnal lineages than pairs of nocturnal lineages for LW opsin (parallel: diurnal: 1.6 changes, nocturnal: 0.35 changes, p=0.004; convergent: diurnal: 0.13 changes, nocturnal: 0.006 changes, p=0.015), but not for UV opsin (parallel: diurnal: 0.47 changes, nocturnal: 0.04 changes, p=0.34; convergent: diurnal: 0 changes, nocturnal: 0.001 changes).
LW opsin
One parallel (A248V) and one convergent (S/T309A) change were noted at two of the eight positively selected sites in LW opsin in diurnal lineages. The number of changes was significantly higher in diurnal than nocturnal lineages for convergent changes (diurnal: 0.067 changes, nocturnal: 0.004 changes, p=0.046), but not for parallel changes (diurnal: 0.067 changes, nocturnal: 0.012 changes, p=0.14). However, the specific amino acid changes that occurred were not unique to diurnal lineages. There were no parallel or convergent amino acid changes in the LW opsin binding pocket; however, two other sites had amino acid changes exclusive to diurnal lineages (L59I in Ld. atra and El. corrusca; T/I301V in Ld. atra and MRCA Ellychnia).
UV opsin
Parallel changes in the diurnal lineages occurred at two of the eight positively selected sites in UV opsin (V59L and V218A), significantly higher than the number of parallel changes on nocturnal branches (parallel: diurnal: 0.13, nocturnal: 0.02, p=0.029), but the specific amino acid changes that occurred were not unique to diurnal lineages. There was one parallel change in the UV binding pocket that was unique to diurnal lineages (F128Y), but this was not significantly different from the number of parallel changes that occurred in nocturnal lineages. There were no convergent changes at positively selected or binding pocket sites in UV opsin on diurnal lineages; however two binding pocket sites showed single amino acid changes exclusive to diurnal lineages (G129A El. bivulneris, S203T Ld. atra). Two additional sites showed parallel changes unique to diurnal lineages (I68V and D214N in Ld. atra and El. corrusca).
In summary, the transition from nocturnal to diurnal activity is associated with an increase whole-molecule ω in both LW and UV opsins. A total of 25 positively selected, binding pocket, parallel, or convergent amino acid substitutions are excellent candidates for functional studies. Descriptions of specific candidate sites and figures showing ancestral sequence reconstructions are given in the Supporting Information (Text S4; Figures: S5–S7 (LW) and S8–S10 (UV)).
Changes in selective constraint linked to transitions in ecological and signaling traits
For each of the nocturnal species we used data on male peak emission wavelength, habitat in which signals are displayed, and activity start time to examine whether molecular evolution of opsins is correlated with changes in signal color or light environment. We found a significant positive correlation between the change in selective constraint (ω) in LW opsin and change in activity time along branches after accounting for divergence between taxa (pr = 0.54, p=0.01). This relationship was not statistically significant for UV opsin (Table 5). No significant correlations were found between response variables (number of amino acid substitutions, dN, whole molecule ω values, site-specific ω values) and signal emission color or habitat.
Table 5.
Partial correlations between change in selective constraint (ω) and change ecological and signaling traits
| Δ ωa | Δ traitb | prc | p | df |
|---|---|---|---|---|
| LW | Spectra | 0.11 | 0.64 | 19 |
| Habitat | −0.23 | 0.31 | ||
| Activity | 0.54 | 0.01* | ||
| UV | Spectra | 0.23 | 0.29 | 21 |
| Habitat | −0.26 | 0.23 | ||
| Activity | 0.24 | 0.27 |
Change in ω values between branches obtained from PAML free-ratio branch models. Branches with ω > 900 (signifying a branch on which there were no synonymous substitutions) were removed prior to analysis.
Change in traits determined using PIC values.
Partial correlation coefficient after controlling for branch length
Discussion
The visual opsins of North American fireflies
Vision in North American fireflies is due to two opsins, one LW and one UV opsin. It is surprising that we did not detect a B opsin among the 10 transcriptomes and 4 genomes sequenced, since this opsin was present in the common ancestor of insects and previous studies on firefly eye sensitivity suggest the presence of blue-sensitive photoreceptors (Lall, Chapman, et al. 1980; Lall 1981; Lall et al. 1982; Eguchi et al. 1984; Lall et al. 1988; Booth et al. 2004). Similarly, soldier beetles in the family Cantharidae, a sister family to Lampyridae, also exhibit sensitivity to blue wavelengths (Horridge 1979).
We are confident that we would have detected a B opsin if present given our ability to detect c-opsins as well as other divergent gene family transcripts. It is possible that we did not detect B opsins because we sequenced only species that lack them, because they are exclusively expressed at times that we did not sample, or because they are expressed at very low levels. However, we are confident that none of these possibilities apply for the following reasons: First, we sequenced the transcriptome of Photuris frontalis, the species with the strongest evidence for blue-sensitivity (Lall et al. 1988) and found no B opsin. Second, we found no evidence for any other visual opsins, including a B opsin, in the four genomic data sets. Third, the sequencing depth of the head tissue transcriptomes was sufficient to identify very rare transcripts. For example, we identified luciferase transcripts, the light-producing enzyme putatively expressed only in the light organ, at a level that was three orders of magnitude lower than the LW and UV opsins. Fourth, a recently-published study did not find a B opsin in the transcriptomes of nine firefly species (Martin et al. 2015). Based on our data, we conclude that fireflies have lost the hypothesized ancestral insect B opsin paralog.
In other beetle species there is evidence for both the presence (Scarabidae: Théry et al. 2008; Curculionidae: Groberman & Borden 1982; Coccinelidae: Lin 1993) and absence (Elateridae: Lall et al. 2000; Lall et al. 2010; Tenebrionidae: Yinon 1970) of blue sensitivity. In some cases, loss of blue sensitivity indicates a loss of B opsin (e.g. Tribolium castaneum: Jackowska et al. 2007); however, it is unknown to what extent loss and retention of blue sensitivity is coupled to loss or retention of the B opsin across beetle taxa. Without a B opsin, sensitivity to blue wavelengths in Photuris and other species may be explained by additional photosensitizing pigments that extend the range of wavelengths either LW or UV opsin are able to detect (Lall et al. 1982), similar to “antenna” pigments in deep-sea dragonfish (Douglas et al. 2000). Direct measures of λmax are needed to confirm the phylogenetically inferred spectral absorbances of each of the firefly opsins.
The functions of LW and UV opsins
The presence of only two opsins in fireflies and their inferred absorbances (LW and UV) has implications for possible sources of selection on their amino acid sequences. Vision in adult fireflies is likely used for the detection of conspecific and heterospecific light signals, avoidance of obstacles during locomotion, and timing the onset of activity. In contrast, vision probably plays a minor role for larvae, which are generally active at night and are often below leaves or in the soil. All firefly larvae are bioluminescent, but they appear to use their light an aposematic signal to predators rather than for communication with conspecifics (McLean et al. 1972). The evolution of visual pigments is thus likely to be primarily driven by selection in adults.
Unlike in some other insect lineages (e.g. Briscoe & Chittka 2001), there has been no lineage-specific diversification of either LW or UV opsins in fireflies. Since there is no known UV component of firefly light emissions (Eguchi et al. 1984), these results are concordant with monochromatic detection of firefly light signals using the LW opsin (Lall et al. 2000; Lall & Worthy 2000). UV opsin may aid in navigation during flight, especially in species active at twilight in open habitats because UV and blue wavelengths are enriched in these conditions (Cronin et al. 2000), or it may be important for detecting polarized light (Dacke et al. 2004). UV opsin may also be involved in detecting the threshold of ambient light that cues initiation of evening flashing activity (Lall 1993, 1994).
Spectral tuning and the molecular evolution of LW and UV opsins
We examined the evolution of LW and UV opsins by testing for positive selection within each opsin across 38 North American species. To investigate the spectral tuning hypothesis, we further determined whether positively selected sites in each opsin occurred in locations that are functionally relevant to tuning (the chromophore binding pocket). Visual opsins generally exhibit purifying selection across the entire molecule suggesting functional constraints (e.g. Terai et al. 2006; Briscoe et al. 2010; Shen et al. 2010; Sivasundar & Palumbi 2010; Audzijonyte et al. 2012; Weadick & Chang 2012; Meredith et al. 2013; Kenaley et al. 2014). For both LW and UV opsins the entire molecule showed evidence of selective constraint. However, 16 sites across both opsins exhibited ω > 1 in at least one of our tests of selection. Three of these sites (LW: 181, 188, and 235) were significantly elevated, and seven others were either in or near the chromophore binding pocket and/or have been identified in other insect studies (Table 3). Contrary to our expectations, only two of these 16 sites were predicted to alter λmax based on their location in the binding pocket in the homology model. The low number of λmax–altering sites was not caused by fewer identified binding pocket sites; we predicted 24 sites, similar to other insect studies (Wakakuwa et al. 2010).
The lack of overlap between positively selected sites and binding pocket sites suggests that either (a) the homology model is inaccurate or, if the homology model is correct, that (b) sites may be under selection for spectral tuning through long-range effects or (c) selection targets other opsin functions. Z-scores indicated that our homology model was under-performing relative to models of other genes generated in SwissModel, likely due to low identity between firefly opsins and the most similar available template, squid opsin (Text S2). However, six of the positively selected sites that we identified are common to studies of selection and spectral tuning in other insect opsins (Briscoe 2001; Briscoe et al. 2010; Wakakuwa et al. 2010; Tierney et al. 2011). The presence of elevated rates of evolution at these sites across butterflies, bees, and fireflies suggests that, even if our homology model is inaccurate, there may be similar selective pressures at work across these diverse lineages. Besides spectral tuning of λmax, it is possible that selection for other functions includes breadth of sensitivity (Lall, Seliger, et al. 1980), thermal stability (Endler 1992, Frentiu et al. 2015), protein folding, chromophore uptake, or interactions with downstream molecules, some of which may affect λmax (Mackin et al. 2014; Schott et al. 2014). For example, site 356 in LW opsin, one of the positively selected sites, is likely not involved in spectral tuning since it is located in helix 8, parallel to the membrane, and instead probably interacts with the downstream G protein.
These functional hypotheses require a better crystal structure for homology modeling, preferably from a firefly species, to corroborate the position of amino acid sites relative to the chromophore and assess the potential effects of identified mutations on opsin structure and function. These will then need to be functionally verified by empirically measuring the effects of amino acid substitutions at the sites identified in this study.
The influence of light signaling on opsin evolution
If the predictions of sensory drive, specifically tuning of genes underlying visual reception, apply to fireflies, we expect to see changes at amino acid sites in opsins associated with adult light signaling, especially at sites predicted to alter λmax. If these changes are driven purely by selection for conspecific signal detection, we expect to find these patterns specifically in LW opsin.
We find several lines of evidence supporting an association between changes in opsin sequence and changes in signal mode. (1) ω values were higher, indicating faster evolution, in lineages of diurnal species relative to nocturnal species. Since ω for both LW and UV opsin in diurnal lineages is less than 1, this could be due to either positive or relaxed purifying selection (Bielawski & Yang 2003). Surprisingly, UV opsin and LW opsin showed similar patterns, suggesting that both opsins have important functions to fireflies transitioning to a diurnal lifestyle. (2) There were more parallel and convergent amino acid changes among diurnal lineages than nocturnal lineages in LW opsins, though not in UV opsins. This was primarily driven by comparisons between Ld. atra and Ellychnia lineages. The lack of amino acid substitutions in the other two diurnal taxa can be explained by recent divergence (branch lengths) from their nocturnal sister taxa (Stanger-Hall & Lloyd 2015). (3) A binding pocket site in UV opsin, site 133, was under positive selection in diurnal Ellychnia lineages. The S133A substitution is of particular interest because it is a polar to nonpolar change and such changes at binding pocket sites are known to affect λmax (Briscoe 2008). In addition, site-directed mutagenesis found that a S133A substitution blue shifted absorbance in a butterfly B opsin (Wakakuwa et al. 2010).
While this evidence supports that some variable sites in LW and at least one binding pocket site in UV opsin are involved in spectral tuning associated with a change in signal mode (i.e. the transition to diurnal, unlighted, pheromone signals), the other sites identified in LW and UV opsin may or may not be involved in spectral tuning. The fact that we did not find a correlation for UV opsin suggests that sources of selection may be different for LW vs UV opsin, as expected based on their presumed different functions. Activity time rather than habitat may best reflect the quality and amount of light available in the environment and future work should seek to quantify light environments in both spectrum and intensity to examine the relationship with LW opsin more closely.
Contrary to our expectations, we did not find evidence of spectral tuning of LW opsin to male emission wavelength. There was a positive correlation between number of amino acid changes in LW opsin and change in emission spectra; however, both variables were correlated with branch length and the relationship became nonsignificant when branch length was taken into account. Interestingly, specific substitutions at two of the candidate sites are associated with shifts in opsin absorbance in other taxa that are in the same direction as the emission spectrum shifts identified in the present study (Table S6). All three nocturnal lineages that had T108M substitutions were red-shifted in emission relative to their reconstructed ancestor. This site, analogous to V63M in Heliconius, is associated with a red shift in opsin absorbance in butterflies (Briscoe 2001). In addition, V188I, associated with a red shift in emission in fireflies, is analogous to V143I in Heliconius, also associated with a red shift in opsin absorbance. Neither of these sites are identified as in the binding pocket based on homology modeling, yet are associated with shifts in opsin absorbance.
There are several possible explanations for our overall negative finding while individual substitutions have some evidence for functional effects on absorbance. First, there may not be enough large phylogenetically-independent transitions in spectra to give us sufficient power to detect a relationship. Sampling more taxa would help to alleviate this problem. Second, the presence of selective constraint across the entire molecule may make single amino acid substitutions difficult to detect as driven by selection. Third, the homology model may not identify all the amino acid sites that are candidates for changing λmax, or it may be combinations of changes at different sites that tune opsins (Yokoyama et al. 2008). Finally, a lack of concordance between opsin amino acid sequence, spectral sensitivity, and light environment has been observed in other species (e.g. Mysis: Audzijonyte et al. 2012), suggesting that the paradigm of opsin spectral tuning does not hold universally and that other mechanisms should be examined.
Other mechanisms of spectral tuning
Our data suggest that changes in opsin sequence, some with potential to affect λmax, occur at large transitions in signaling characteristics, especially the transitions from nocturnal lighted signaling to diurnal pheromone signaling. However, they do not explain the observed tight correlation between firefly visual sensitivities and male emission wavelengths. Fine-tuning of spectral sensitivity may instead be due to other molecules that interact with the opsins or with photons of light.
One candidate molecule that may be involved is the chromophore. While most insect lineages use a single chromophore in their visual pigment, Asian firefly species possess both A1 and A3 chromophore types (Gleadall et al. 1989). These two types are similar in chemical structure; however, alternate chromophores are known to change the spectral absorbance of their paired opsin (Briscoe & Chittka 2001). The difference in λmax with different chromophores is typically large (35–40 nm), much greater than the difference in LW spectral sensitivity observed across all firefly species (26 nm), suggesting that the use of alternate chromophores may not explain fine-tuning. It is also possible that the observed blue sensitivity without a B opsin may be caused by the use of different chromophores.
Alternatively, fine-tuning may involve screening pigments that absorb specific wavelengths, thus modifying the spectrum of light that reaches the opsins (Seliger et al. 1982a, b; Cronin et al. 2000). Our data give ambivalent support for predictions of LW opsin spectral absorbance based on this hypothesis (Text S5). Nevertheless, screening pigments have been found in 12 North American firefly species, with the exception of Ld. atra, the only diurnal species examined to date (Seliger et al. 1982b; Lall et al. 1988; Cronin et al. 2000). Screening pigments may be less constrained than opsins, allowing rapid evolution of eye sensitivity. It is also possible that screening pigments may be responsible for blue sensitivity. If screening pigments in the rhabdom mask light in the UV spectrum they could effectively convert UV receptors into blue receptors, much like the “sunscreen” pigments in mantis shrimp (Bok, et al. 2014). Future work will develop the link between opsin sequence and absorbance, chromophore usage, and screening pigments in relation to both blue sensitivity and the spectral tuning hypothesis.
Conclusion
We have demonstrated that across firefly species, there is a single LW opsin, a single UV opsin, and no B opsin. Within both LW and UV, there is evidence for parallel and convergent amino acid changes at transitions from the use of nocturnal light signals to diurnal pheromone signals, and within LW opsin, evidence for greater changes in selective constraint with greater changes in activity time. In addition, there is evidence for positive selection at six sites that have been identified in other insect orders as under positive selection and/or of functional importance (LW sites 108, 181, 188, 356; UV sites 59, 133; Table 3, S6). This study represents a first step in testing the molecular basis for spectral tuning in fireflies with a comparative dataset. These data provide candidate sites and mutations for future functional testing. Recent advances in insect opsin expression systems (e.g. Frentiu et al. 2015) will aid in this effort. Given the tight correspondence between signal phenotypes and spectral sensitivity (e.g. Lall 1981), the diversity of species in both signaling and ecological traits, the existing and expandable phylogeny, and emerging genomic resources, fireflies continue to be a rich study system for investigating the evolution of signaling and signal reception in a comparative context.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (GRF to S.E.S and DDIG DEB-1311315 to D.W.H. and S.E.S.) and the NIGMS of the National Institute of Health (award number T32GM007103 to S.E.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank: Allegheny National Forest (permit to Lynn Faust), Great Smoky Mountains National Park (permit to Kathrin Stanger-Hall), Raphael de Cock, Lynn Faust, Sara Lewis, Michael Marsh (University of Georgia), Jerry McCollum (Charles H. Wharton Conservation Center), David McNaughton (Fort Indiantown Gap), Jenna Pallansch, the Sander Family, and Dorset Trapnell for collection assistance; Megan Behringer, Zachary Wood, and Kelly Dyer for bioinformatics support, homology-modeling advice, and manuscript comments respectively; and Kathrin Stanger-Hall for collection assistance, manuscript comments, and inspiring the study.
Footnotes
Author Contributions
S.E.S. collected specimens, performed transcriptome, genomic, and Sanger sequencing, data analysis, and wrote the initial draft of the manuscript. D.W.H. developed methods to measure light emissions and contributed to data analysis. Both authors contributed to revisions of the initial manuscript.
Data accessibility
Raw reads: Reads from both transcriptomic and genomic Illumina sequencing are available on the NCBI Sequence Read Archive (Bioproject SRP061172; Table S7).
Opsin sequences: GenBank accession numbers for all opsin sequences used in the analysis are available in Table S1. Alignments for LW and UV opsin are shown in Supporting Information and included in fasta format on Dryad (DOI: http://dx.doi.org/10.5061/dryad.q878c).
Phylogeny: GenBank accession numbers for all opsin sequences used in the analysis are available in Table S3.
Scripts: R scripts for parsing phangorn datasets are available upon request.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. International Journal of Developmental Biology. 2003;47:563–571. [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Aronesty E. ea-utils: Command-line tools for processing biological sequencing data. 2011 http://code.google.com/p/ea-utils.
- Audzijonyte A, Pahlberg J, Viljanen M, Donner K, Väinölä R. Opsin gene sequence variation across phylogenetic and population histories in Mysis (Crustacea: Mysida) does not match current light environments or visual-pigment absorbance spectra. Molecular Ecology. 2012;21:2176–2196. doi: 10.1111/j.1365-294X.2012.05516.x. [DOI] [PubMed] [Google Scholar]
- Biggley WH, Lloyd JE, Seliger HH. The spectral distribution of firefly light II. Journal of General Physiology. 1967;50:1681–1692. doi: 10.1085/jgp.50.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. Maximum likelihood methods for detecting adaptive evolution after gene duplication. Journal of Structural and Functional Genomics. 2003;3:201–212. [PubMed] [Google Scholar]
- Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Bok MJ, Porter ML, Place AR, Cronin TW. Biological sunscreens tune polychromatic ultraviolet vision in mantis shrimp. Current Biology. 2014;24:1636–1642. doi: 10.1016/j.cub.2014.05.071. [DOI] [PubMed] [Google Scholar]
- Booth D, Stewart AJA, Osorio D. Colour vision in the glow-worm Lampyris noctiluca (L.) (Coleoptera: Lampyridae): evidence for a green-blue chromatic mechanism. Journal of Experimental Biology. 2004;207:2373–2378. doi: 10.1242/jeb.01044. [DOI] [PubMed] [Google Scholar]
- Briscoe AD. Functional diversification of Lepidopteran opsins following gene duplication. Molecular Biology and Evolution. 2001;18:2270–2279. doi: 10.1093/oxfordjournals.molbev.a003773. [DOI] [PubMed] [Google Scholar]
- Briscoe AD. Reconstructing the ancestral butterfly eye: focus on the opsins. Journal of Experimental Biology. 2008;211:1805–1813. doi: 10.1242/jeb.013045. [DOI] [PubMed] [Google Scholar]
- Briscoe AD, Bybee SM, Bernard GD, Yuan F, Sison-Mangus MP, Reed RD, Warren AD, Llorente-Bousquets J, Chiao C-C. Positive selection of a duplicated UV-sensitive visual pigment coincides with wing pigment evolution in Heliconius butterflies. Proceedings of the National Academy of Sciences. 2010;107:3628–3633. doi: 10.1073/pnas.0910085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe AD, Chittka L. The evolution of color vision in insects. Annual Reviews of Entomology. 2001;46:471–510. doi: 10.1146/annurev.ento.46.1.471. [DOI] [PubMed] [Google Scholar]
- Carvalho L, Cowing JA, Wilkie SE, Bowmaker JK, Hunt DM. Shortwave visual sensitivity in tree and flying squirrels reflects changes in lifestyle. Current Biology. 2006;16:R81–R83. doi: 10.1016/j.cub.2006.01.045. [DOI] [PubMed] [Google Scholar]
- Cicero JM. Lek assembly and flash synchrony in the Arizona firefly Photinus knulli Green (Coleoptera: Lampyridae) Coleopterists Bulletin. 1983;37:318–342. [Google Scholar]
- Cronin TW, Jarvilehto M, Weckstrom M, Lall AB. Tuning of photoreceptor spectral sensitivity in fireflies (Coleoptera: Lampyridae) Journal of Comparative Physiology A. 2000;186:1–12. doi: 10.1007/s003590050001. [DOI] [PubMed] [Google Scholar]
- Dacke M, Byrne MJ, Scholtz CH, Warrant EJ. Lunar orientation in a beetle. Proceedings of the Royal Society B: Biological Sciences. 2004;271:361–365. doi: 10.1098/rspb.2003.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RH, Mullineaux CW, Partridge JC. Long-wave sensitivity in deep-sea stomiid dragonfish with far-red bioluminescence: evidence for a dietary origin of the chlorophyll-derived retinal photosensitizer of Malacosteus niger. Philosophical Transactions of the Royal Society B: Biological Sciences. 2000;355:1269–1272. doi: 10.1098/rstb.2000.0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi E, Nemoto A, Meyer-Rochow VB, Ohba N. A comparative study of spectral sensitivity curves in three diurnal and eight nocturnal species of Japanese fireflies. Journal of Insect Physiology. 1984;30:607–612. [Google Scholar]
- Endler JA. Signals, signal conditions, and the direction of evolution. American Naturalist. 1992;139:S125–S153. [Google Scholar]
- Endler JA. The color of light in forests and Its implications. Ecological Monographs. 1993;63:2–27. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Fender KM. The genus Phausis in America north of Mexico (Coleoptera-Lampyridae) Northwest Science. 1966;40:83–95. [Google Scholar]
- Frentiu FD, Yuan F, Savage WK, Bernard GD, Mullen SP, Briscoe AD. Opsin clines in butterflies suggest novel roles for insect photopigments. Molecular Biology and Evolution. 2015;32:368–379. doi: 10.1093/molbev/msu304. [DOI] [PubMed] [Google Scholar]
- Gleadall IG, Hariyama T, Tsukahara Y. The visual pigment chromophores in the retina of insect compound eyes, with special reference to the Coleoptera. Journal of Insect Physiology. 1989;35:787–795. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. Revision of the nearctic species of Pyractomena (Coleoptera: Lampyridae) Wasmann Journal of Biology. 1957;15:237–284. [Google Scholar]
- Green J. Revision of the Nearctic species of Photinus (Lampyridae: Coleoptera) Proceedings of the California Academy of Sciences. 1956;28:561–613. [Google Scholar]
- Groberman LJ, Borden JH. Electrophysiological response of Dendroctonus pseudotsugae and Ips paraconfusus (Coleoptera: Scolytidae) to selected wavelength regions of the visible spectrum. Canadian Journal of Zoology. 1982;60:2180–2189. [Google Scholar]
- Guindon Sp, Rodrigo AG, Dyer KA, Huelsenbeck JP. Modeling the site-specific variation of selection patterns along lineages. Proceedings of the National Academy of Sciences. 2004;101:12957–12962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horridge GA. The eye of the soldier beetle Chauliognathus pulchellus (Cantharidae) Proceedings of the Royal Society B: Biological Sciences. 1979;203:361–378. doi: 10.1098/rspb.1979.0003. [DOI] [PubMed] [Google Scholar]
- Jackowska M, Bao R, Liu Z, McDonald EC, Cook TA, Friedrich M. Genomic and gene regulatory signatures of cryptozoic adaptation: Loss of blue sensitive photoreceptors through exapansion of long wavelength-opsin expression in the red flour beetle Tribolium castaneum. Frontiers in Zoology. 2007;4:24. doi: 10.1186/1742-9994-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Kenaley CP, Devaney SC, Fjeran TT. The complex evolutionary history of seeing red: molecular phylogeny and the evolution of an adaptive visual system in deep-sea dragonfishes (Stomiiformes: Stomiidae) Evolution. 2014;68:996–1013. doi: 10.1111/evo.12322. [DOI] [PubMed] [Google Scholar]
- Lall A, Chapman R, Trouth CO, Holloway J. Spectral mechanisms of the compound eye in the firefly Photinus pyralis (Coleoptera: Lampyridae) Journal of Comparative Physiology. 1980;135:21–27. [Google Scholar]
- Lall A, Lord E, Trouth CO. Vision in the firefly Photuris lucicrescens (Coleoptera: Lampyridae): Spectral sensitivity and selective adaptation in the compound eye. Journal of Comparative Physiology. 1982;147:195–200. [Google Scholar]
- Lall AB. Electroretinogram and the spectral sensitivity of the compound eyes in the firefly Photuris versicolor (Coleoptera-Lampyridae): A correspondence between green sensitivity and species bioluminescence emission. Journal of Insect Physiology. 1981;27:461–468. [Google Scholar]
- Lall AB. Action spectra for the initiation of bioluminescent flashing activity in males of twilight-active firefly Photinus scintillans (Coleoptera: Lampyridae) Journal of Insect Physiology. 1993;39:123–127. [Google Scholar]
- Lall AB. Spectral cues for the regulation of bioluminescent flashing activity in the males of twilight-active firefly Photinus scintillans (Coleoptera: Lampyridae) in nature. Journal of Insect Physiology. 1994;40:359–363. [Google Scholar]
- Lall AB, Cronin TW, Carvalho AA, de Souza JM, Barros MP, Stevani CV, Bechara EJ, Ventura DF, Viviani VR, Hill AA. Vision in click beetles (Coleoptera: Elateridae): pigments and spectral correspondence between visual sensitivity and species bioluminescence emission. Journal of Comparative Physiology A. 2010;196:629–638. doi: 10.1007/s00359-010-0549-x. [DOI] [PubMed] [Google Scholar]
- Lall AB, Seliger HH, Biggley WH, Lloyd JE. Ecology of colors of firefly bioluminescence. Science. 1980;210:560–562. doi: 10.1126/science.210.4469.560. [DOI] [PubMed] [Google Scholar]
- Lall AB, Strother GK, Cronin TW, Seliger HH. Modification of spectral sensitivities by screening pigments in the compound eyes of twilight-active fireflies (Coleoptera: Lampyridae) Journal of Comparative Physiology A. 1988;162:23–33. doi: 10.1007/BF01342700. [DOI] [PubMed] [Google Scholar]
- Lall AB, Ventura DSF, Bechara EJH, de Souza JM, Colepicolo-Neto P, Viviani VR. Spectral correspondence between visual spectral sensitivity and bioluminescence emission spectra in the click beetle Pyrophorus punctatissimus (Coleoptera: Elateridae) Journal of Insect Physiology. 2000;46:1137–1141. doi: 10.1016/s0022-1910(99)00224-3. [DOI] [PubMed] [Google Scholar]
- Lall AB, Worthy KM. Action spectra of the female’s response in the firefly Photinus pyralis (Coleoptera: Lampyridae): evidence for an achromatic detection of the bioluminescent optical signal. Journal of Insect Physiology. 2000;46:965–968. doi: 10.1016/s0022-1910(99)00206-1. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzburg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Identification of photoreceptor locations in the compound eye of Coccinella septempunctata Linnaeus (Coleoptera, Coccinellidae) Journal of Insect Physiology. 1993;39:555–562. [Google Scholar]
- Lloyd JE. Miscellaneous Publications. Ann Arbor, Michigan: Museum of Zoology, University of Michigan; 1966. Studies on the flash communication system in Photinus fireflies. [Google Scholar]
- Lloyd JE. Flashes, behavior and additional species of Nearctic Photinus fireflies (Coleoptera: Lampyridae) Coleopterists Bulletin. 1969;23:29–40. [Google Scholar]
- Lu A, Guindon S. Performance of standard and stochastic branch-site models for detecting positive selection among coding sequences. Molecular Biology and Evolution. 2014;31:484–495. doi: 10.1093/molbev/mst198. [DOI] [PubMed] [Google Scholar]
- Luk S, Marshall SA, Branham MA. The fireflies of Ontario (Coleoptera: Lampyridae) Canadian Journal of Arthropod Identification. 2011;16:1–105. [Google Scholar]
- Mackin KA, Roy RA, Theobald DL. An empirical test of convergent evolution in rhodopsins. Molecular Biology and Evolution. 2014;31:85–95. doi: 10.1093/molbev/mst171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Lord N, Branham M, Bybee S. Review of the firefly visual system (Coleoptera: Lampyridae) and evolution of the opsin genes underlying color vision. Organisms Diversity & Evolution. 2015:1–14. preprint. [Google Scholar]
- McLean M, Buck J, Hanson FE. Culture and larval behavior of Photurid fireflies. American Midland Naturalist. 1972;87:133–145. [Google Scholar]
- Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. Rod monochromacy and the coevolution of Cetacean retinal opsins. PLoS Genetics. 2013;9:e1003432. doi: 10.1371/journal.pgen.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- Oba Y, Kainuma T. Diel changes in the expression of long-wavelength sensitive and ultraviolet-sensitive opsin genes in the Japanese firefly, Luciola cruciata. Gene. 2009;436:66–70. doi: 10.1016/j.gene.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Otte D, Smiley J. Synchrony in Texas fireflies with a consideration of male interaction models. Biology of Behavior. 1977;2:143–158. [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW, Robinson PR. Shedding new light on opsin evolution. Proceedings of the Royal Society B: Biological Sciences. 2012;279:3–14. doi: 10.1098/rspb.2011.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott RK, Refvik S, Hauser FE, López-Fernández H, Chang BSW. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Molecular Biology and Evolution. 2014;31:1149–1165. doi: 10.1093/molbev/msu064. [DOI] [PubMed] [Google Scholar]
- Seliger HH, Buck JB, Fastie WG, McElroy WD. The spectral distribution of firefly light. Journal of General Physiology. 1964;48:95–104. doi: 10.1085/jgp.48.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger HH, Lall AB, Lloyd JE, Biggley WH. The colors of firefly bioluminescence—I. Optimization model. Photochemistry and Photobiology. 1982a;36:673–680. [Google Scholar]
- Seliger HH, Lall AB, Lloyd JE, Biggley WH. The colors of firefly bioluminescence—II. Experimental evidence for the optimization model. Photochemistry and Photobiology. 1982b;36:681–688. [Google Scholar]
- Shen YY, Liu J, Irwin DM, Zhang YP. Parallel and convergent evolution of the dim-light vision gene RH1 in bats (Order: Chiroptera) PLoS ONE. 2010;5:e8838. doi: 10.1371/journal.pone.0008838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A, Palumbi SR. Parallel amino acid replacements in the rhodopsins of the rockfishes (Sebastes spp.) associated with shifts in habitat depth. Journal of Evolutionary Biology. 2010;23:1159–1169. doi: 10.1111/j.1420-9101.2010.01977.x. [DOI] [PubMed] [Google Scholar]
- Stanger-Hall KF, Lloyd JE, Hillis DM. Phylogeny of North American fireflies (Coleoptera: Lampyridae): Implications for the evolution of light signals. Molecular Phylogenetics and Evolution. 2007;45:33–49. doi: 10.1016/j.ympev.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Stanger-Hall KF, Lloyd JE. Flash signal evolution in Photinus fireflies: Character displacement and signal exploitation in a visual communication system. Evolution. 2015;69:666–682. doi: 10.1111/evo.12606. [DOI] [PubMed] [Google Scholar]
- Terai Y, Seehausen O, Sasaki T, Takahashi K, Mizoiri S, Sugawara T, Sato T, Watanabe M, Konijnendijk N, Mrosso HDJ, et al. Divergent selection on opsins drives incipient speciation in lake victoria cichlids. PLoS Biology. 2006;4:e433. doi: 10.1371/journal.pbio.0040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M, Pincebourde S, Feer F. Dusk light environment optimizes visual perception of conspecifics in a crepuscular horned beetle. Behavioral Ecology. 2008;19:627–634. [Google Scholar]
- Tierney SM, Sanjur O, Grajales GG, Santos LM, Bermingham E, Wcislo WT. Photic niche invasions: phylogenetic history of the dim-light foraging augochlorine bees (Halictidae) Proceedings of the Royal Society B: Biological Sciences. 2011;279:794–803. doi: 10.1098/rspb.2011.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voortman M, Druzdzel MJ. Insensitivity of constraint-based causal discovery algorithms to violations of the assumption of multivariate normality. In: Wilson David, Chad Lane H., editors. FLAIRS Conference. AAAI Press; 2008. pp. 690–695. [Google Scholar]
- Wakakuwa M, Terakita A, Koyanagi M, Stavenga DG, Shichida Y, Arikawa K. Evolution and mechanism of spectral tuning of blue-absorbing visual pigments in butterflies. PLoS ONE. 2010;5:e15015. doi: 10.1371/journal.pone.0015015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Xiao J-H, Bian S-N, Niu L-M, Murphy RW, Huang D-W. Evolution and expression plasticity of opsin genes in a fig pollinator, Ceratosolen solmsi. PLoS ONE. 2013;8:e53907. doi: 10.1371/journal.pone.0053907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS. Complex patterns of divergence among green-sensitive (RH2a) African cichlid opsins revealed by clade model analyses. BMC Evolutionary Biology. 2012;12:206. doi: 10.1186/1471-2148-12-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie SE, Robinson PR, Cronin TW, Poopalasundaram S, Bowmaker JK, Hunt DM. Spectral tuning of avian violet- and ultraviolet-sensitive visual pigments. Biochemistry. 2000;39:7895–7901. doi: 10.1021/bi992776m. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes Empirical Bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yinon U. The visual mechanisms of Tenebrio molitor: some aspects of the spectral response. Journal of Experimental Biology. 1970;53:221–229. doi: 10.1242/jeb.53.1.221. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Progress in Retinal and Eye Research. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Evolution of dim-light and color vision pigments. Annual Review of Genomics and Human Genetics. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Yang H, Starmer WT. Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics. 2008;179:2037–2043. doi: 10.1534/genetics.108.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.