Abstract
Fragile X syndrome (FXS) is the most common form of inherited cognitive impairment. It is caused by developmental inactivation of the FMR1 gene and the absence of its encoded protein FMRP, which plays pivotal roles in brain development and function. In FXS embryos with full FMR1 mutation, FMRP is expressed during early embryogenesis and is gradually downregulated at the third trimester of pregnancy. FX-human embryonic stem cells (FX-hESCs), derived from FX human blastocysts, demonstrate the same pattern of developmentally regulated FMR1 inactivation when subjected to in vitro neural differentiation (IVND). In this study, we used this in vitro human platform to explore the molecular mechanisms downstream to FMRP in the context of early human embryonic neurogenesis. Our results show a novel role for the SOX superfamily of transcription factors, specifically for SOX2 and SOX9, which could explain the reduced and delayed neurogenesis observed in FX cells. In addition, we assess in this study the “GSK3β theory of FXS” for the first time in a human-based model. We found no evidence for a pathological increase in GSK3β protein levels upon cellular loss of FMRP, in contrast to what was found in the brain of Fmr1 knockout mice. Our study adds novel data on potential downstream targets of FMRP and highlights the importance of the FX-hESC IVND system.
Introduction
Fragile X syndrome (FXS) is the most common form of inherited intellectual disability [1]. It is a neurodevelopmental disorder characterized by abnormal neural plasticity, cognitive impairment, autism, and epilepsy. FXS is caused by silencing of the FMR1 gene and the consequent absence of its protein, fragile x mental retardation protein (FMRP). FMR1 is inactivated because of a dynamic mutation composed of a CGG-triplet repeat expansion in the 5′-untranslated region of the gene [2]. In human fetuses affected by the full mutation, FMR1 is gradually downregulated during embryonic development [3] and its consequent adverse effects on brain function suggest a role for FMRP in early neurogenesis, including maintenance and differentiation of neural progenitor cells [4].
Several in vivo and in vitro models have been established to investigate FXS pathology. Fmr1 knockout (KO) animals do not express Fmr1 at any stage of development [5,6] and even in conditional KO mice [7], the natural disease progression, which includes gradual FMRP downregulation, is not fully recapitulated. Human in vitro models include postmortem adult neurons [8], adult neural progenitors [9], or fetal neural progenitor cells [10–12]. These FX cells show only mild differences in their morphology and gene expression from normal human controls[10], but show significant differences from their Fmr1−/− mice counterparts [11,13]. Collectively, these studies suggest that the role of FMRP in early neurogenesis could be significantly different between human and mouse.
Human embryonic stem cells (hESCs) are a powerful tool in disease modeling because of their ability to proliferate indefinitely in culture, while maintaining their potential to differentiate into all cell types in the body [14,15]. We have previously derived male FX-hESC lines carrying the full mutation at the FMR1 gene [16,17]. We have shown that undifferentiated FX-hESCs express FMR1 and FMRP, and that this expression is gradually inactivated only later during differentiation, mimicking the natural progression of the disease. Surprisingly, although full FMR1 inactivation was detectable only in mature FX-neurons, in vitro neural differentiation (IVND) of FX-hESCs resulted in aberrant expression of several key neural genes already at early stages of neurogenesis, indicating that partial downregulation of FMR1 is enough to induce neurodevelopmental abnormalities [17].
Similarly, others found abnormal expression of neural genes in human neural precursor cells (hNPCs) harvested from FXS fetuses [10] and in hNPCs differentiated from FX-human-induced pluripotent stem cells (hiPSCs) generated from fibroblasts of FXS patients [18,19]. However, the functional consequences of these findings and the exact molecular mechanism regulating abnormal human neurogenesis in FXS remain unclear.
In our previous study, we showed a deficit in SOX1 expression in FX-hESCs undergoing IVND, concomitant with reduced and delayed development of neural rosettes (NRs) [17]. The SOX superfamily of transcription factors is regarded as “master switches” in human embryonic development, including the formation of the nervous system [20,21]. Members of the SOXB1 subgroup (SOX1, SOX2, and SOX3) were found to be important in directing the early development of neural tissue, whereas SOX9 and SOX21 were involved in late neuronal development. Interestingly, Sox1−/− mice are characterized by epilepsy [22], which is also known to affect 20%–25% of FXS patients. In addition, patients with SOX3 deficiency show symptoms similar to those observed in FXS patients, characterized by intellectual disability [23,24]. SOX9 is known to play key roles in neural crest development, chondrogenesis, and testis development [25], which are also affected in FXS individuals. Collectively, these studies, together with our previous findings, hint at a potential role for SOX genes in FXS pathology during human embryonic development.
Other possible mechanisms explaining the deficits observed in FXS pathology have been proposed. Studies conducted on Fmr1−/− mice have consistently shown that lack of FMRP results in an abnormal increase in glycogen synthase kinase 3β (GSK3β) mRNA and protein levels [26]. Although GSK3β plays key roles in several molecular pathways, it has been proposed that its involvement in FXS neuropathology is mediated through the canonical Wnt/β-catenin signaling pathway [27,28], a critical signaling pathway for embryonic neural development as well as for adult neurogenesis [29]. These studies showed that ablation of FMRP in vivo reduced the capacity of murine adult neural stem cells (aNSCs) to differentiate into hippocampal neurons because of an increase in GSK3β and a consequent reduction in β-catenin and its downstream neuronal transcription factors, reducing neuronal yield and increasing the relative number of glia cells in the hippocampus. However, the role of Wnt signaling and the functional connection between FMRP and GSK3β have not been tested yet during embryonic neurogenesis. This is important because although adult neurogenesis is confined to the replenishment of specific neuronal populations in the brain (mainly the hippocampus and the olfactory bulb), embryonic neurogenesis is responsible for the formation of the entire nervous system. Furthermore, this question has never been addressed in an FXS human-based model. In this study, for the first time, the relationship between FMRP, GSK3β, and the canonical Wnt signaling pathway is explored in the context of human embryonic neurogenesis.
Materials and Methods
hESC lines
The use of spare IVF-derived embryos following pre-implantation genetic diagnosis for the derivation of hESCs was approved by the Israeli National Ethics Committee (7/04-043). Three male FX-hESC lines were studied: HEFX1, SZ-FX6, and Lis_FX6, entitled here as FX1, FX2, and FX3, respectively. FX1 and FX3 were established by us, whereas FX2 was kindly provided by Dr. Eiges (Shaare Tzedek Medical Center) and their full characterization was already published [16,17]. Four nonaffected hESC lines were used: HUES-6, HUES-16, HUES-13, and HUES-64, kindly provided by Dr. Melton (Harvard University). A summary of all hESC lines used in this study is provided in Supplementary Table S1 (Supplementary materials are available online at www.liebertpub.com/scd). Cells were cultured on Matrigel (BD), in the hESC medium supplemented with basic fibroblast growth factor (bFGF) (8 ng/mL; R&D), as previously reported [17,30,31].
IVND of hESCs
IVND of hESCs was conducted as previously described [17]. IVND included three steps: (a) formation of neuroectoderm aggregates in suspension, grown in the presence of noggin (250 ng/mL; PeproTech) for 4 days and in bFGF (20 ng/mL) for 4 additional days; (b) development of attached NRs in the presence of sonic hedgehog (Shh, 200 ng/mL; PeproTech) for 10 days; and (c) mechanical excision of NRs and creation of neurospheres (NS) in suspension for 12 days supplemented with bFGF (20 ng/mL). The neural induction medium (NIM) consisted of Dulbecco's modified Eagle's medium:F12 (LifeTech.), 0.5% B27 (LifeTech.), 1% N2 (LifeTech.), 1% GlutaMAX (LifeTech.), 1% nonessential amino acids (BioInd.), and 0.1 mg/mL Primocin (InvivoGen).
Derivation of hNPCs
Stable lines of hNPCs were derived from each of the hESC lines used (Supplementary Table S1), following previously described protocols [32–34], with slight modifications. In brief, at day 18 of IVND (see IVND of hESCs), the attached cells surrounding excised NRs were dissociated using TrypLE (LifeTech.) and passaged on Matrigel (BD)-coated polystyrene wells in NIM, supplemented with bFGF (20 ng/mL).
Neuronal differentiation
To induce neuronal differentiation of floating NS (after 30 days of IVND), we collected aggregates by gentle centrifugation and replated on Poly-d-Lysine/Laminin (Sigma)-coated glass coverslips and the medium was switched from NIM to neuronal differentiation medium (NDM), supplemented with brain-derived neurotrophic factor (BDNF), glia-derived neutrophic factor (GDNF) and neurotrophin-3 (NT-3; 10 ng/mL; PreproTech). The NDM consisted of Neurobasal (LifeTech.), 1% B27 (LifeTech.), 1% N2 (Life Tech.), 1% GlutaMAX (LifeTech.), 1% nonessential amino acids (BioInd.), and 0.1 mg/mL Primocin (InvivoGen). To induce neuronal differentiation of hNPCs, cells were first dissociated using TrypLE (LifeTech.), collected by centrifugation, and allowed to spontaneously form aggregates in conical tubes for ∼30 min at 37°C, followed by the same process described for NS.
Gene transcription analysis
Relative transcription levels were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR), as previously described [17]. RNA was extracted (RNeasy; Qiagen), reversed transcribed using the SuperScript-III kit (Invitrogen), and analyzed using SYBR green (ABgene) in Rotor-Gene 6000 Series (Corbett). The housekeeping gene GAPDH was used as a control for ΔΔCt analysis. All qRT-PCR assays included nontemplate control, nonhuman cells (mouse embryonic fibroblasts), and human-FXS white blood cells. All primers (Sigma) are listed in Supplementary Table S2.
Immunostaining assay
Immunostaining was performed as previously described [17]. Cells were fixated using Cytofix (BD). Incubation with primary antibodies was performed overnight at 4°C, detected using Cy2/Cy3-conjugated secondary antibodies, applied for 1 h at RT, in the dark. For the full list of antibodies, see Supplementary Table S3. Nuclei were stained using DAPI (Sigma). Coverslips were mounted using Fluoromount G (Southern Biotech). Cells were imaged using an inverted fluorescent microscope (Olympus). All conditions were similar for all lines in all experiments. For characterization of hNPCs, each line was stained for each marker twice. For establishing the MAP2/GFAP ratio, each experiment was performed in triplicate and images were taken from five different fields/coverslip and >50 cells per field were analyzed by manual counting of positively (Cy2/Cy3) stained cells relative to DAPI.
Western blot analysis
Protein was extracted using a lysis buffer (Promega), and 25–30 μg of protein was loaded on a 7.5% separating gel using the Mini Trans-Blot Cell (Bio-Rad). Nitrocellulose membranes were stained with primary antibodies and detected with HRP-conjugated secondary antibodies. The full list of antibodies for western blot (WB) is shown in Supplementary Table S4. Protein bands were detected using EZ-ECL (BioInd.) and imaged using a chemiluminescent camera.
Manipulation of gene expression
Silencing of FMR1 and SOX2 expression in hNPCs was carried out using custom-made siRNA or its scrambled negative control (Stealth RNAi; LifeTech.) at 20 nM, using the Lipofectamine RNAi Max reagent (LifeTech.). siRNA sequences are provided in Supplementary Table S2. Overexpression of FMR1 in hNPCs was performed using a CMV-FMR1 cDNA plasmid. Transfection was carried out using Lipofectamine 2000 (LifeTech.) and selection with G418 (InvivoGen), at 100 μg/mL. Transient overexpression of SOX9 in hNPCs was performed using an SOX9 cDNA plasmid under the Tet-On system of induction. Doxycycline (1 μg/mL) was added 24 h after transfection for a total of 48 h.
Wnt modulators
The GSK3β inhibitor CHIR99021 (Tocris) and the tankyrase inhibitor XAV939 (Selleckchem) were dissolved in DMSO of 20 mM stock solutions and stored at −80°C. Working concentration for both inhibitors was 3 μM.
Cell patch clamp
Electrophysiological recordings were conducted as previously described [17]. In brief, neurons on glass coverslips were transferred to a recording chamber in a standard recording medium, containing (in mM)10 HEPES, 4 KCl, 2 CaCl2, 1 MgCl2, 139 NaCl, and 10 d glucose (340 mOsm, pH 7.4). Cells were patch clamped with pipettes containing (in mM) 136 K-gluconate,10 KCl, 5 NaCl, 10 HEPES, 0.1 EGTA, 0.3 Na-GTP, 1 Mg-ATP, and 5 phosphocreatine, pH 7.2 (pipette tip resistance was 5–8 MΩ). Action potentials were evoked (in current-clamp mode) by injecting depolarizing current pulses. Membrane potential was held at −60 mV. Signals were amplified with a Multiclamp 700B amplifier and recorded with Clampex 9.2 software (Axon Instruments). Data were analyzed using Clampfit-9 and SigmaPlot.
Migration assay
Outgrowths of NS were cut using an ultrasharp splitting microblade (Bioniche). NS were imaged using an inverted light microscope (Olympus) before the cut and at 0, 2, 4, 8, 16, and 24 h following the cut. Time course of regrowth was reconstructed from the images using Cell^A (Olympus) and ImageJ (NIH) software, and the number of cells populating the cut area was manually quantified and normalized to the area analyzed (in μm2). All cultures were grown in NDM previous to the assay, and growth factors were added immediately before the cut (20 ng/mL for BDNF, GDNF, and NT-3, and 200 ng/mL for Shh and Noggin).
Statistical analysis
Statistical analysis (Student's t-test and ANOVA) was performed using SPSS, SigmaPlot, and online GraphPad QuickCalcs (www.graphpad.com/quickcalcs).
Results
Delayed neurodevelopment of FX-NS
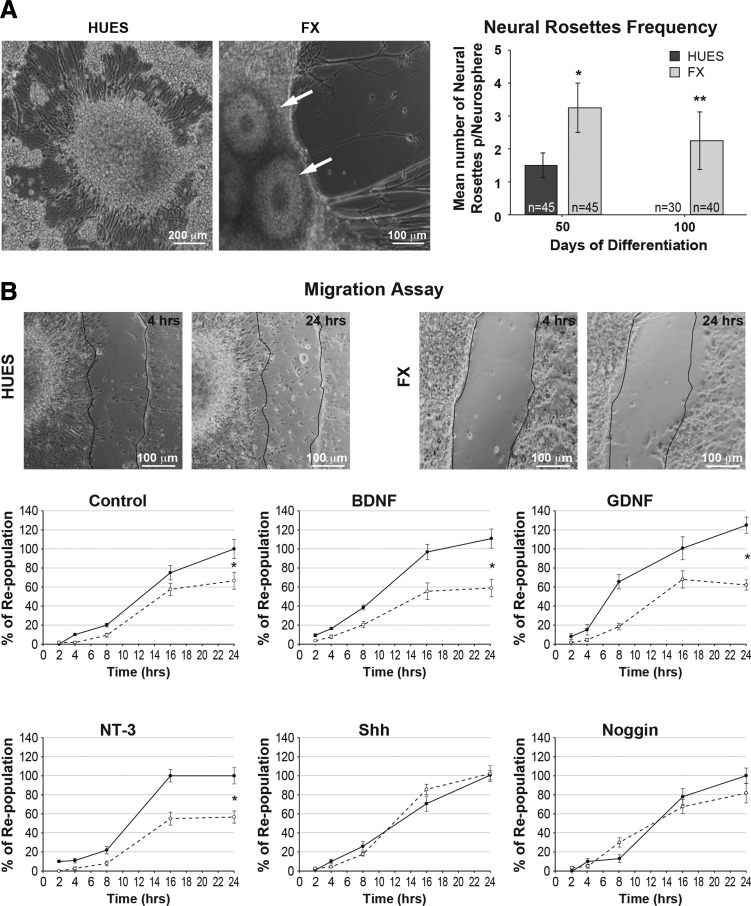
This study was performed on four control hESC lines (HUES; Melton Lab) and three FX-hESC lines (Supplementary Table S1 for full details and references) and is a direct continuation of our previous report, in which, for the first time, we induced IVND in FX-hESCs [17]. In that first report, we described a reduced and delayed development of NRs during IVND of FX-hESC lines. In this study, after the initial 30 days of IVND and production of floating NS (see the Materials and Methods section), we replated HUES and FX NS and followed their neuronal differentiation for an additional 70 days, to further characterize the phenotypic differences in their neurodevelopmental status. We observed that attached NS produced neuroblasts and neurons sprouting radially from the NS, in both FX and HUES lines (Fig. 1A). However, we also noticed that some of the attached NS exhibited transient NR formations. Importantly, the frequency of NRs on FX-NS was significantly higher and remained for longer periods of time (Fig. 1A). By day 100, NRs were still present only in FX cultures and completely disappeared from control cultures. Since NRs are considered as primitive neuroectodermal structures reminiscent of neural plate or neural tube formations [35,36], we suggest that this finding provides another demonstration of delayed embryonic neurogenesis in FXS. We have previously shown that delayed development of FX-NRs could be rescued by Shh, a key morphogen in early neural development. Similarly, two recent studies conducted on FX-hiPSCs suggest that abnormal neurite development and axonal guidance cues could be linked to FXS pathology [37,38]. To further show in a functional context that these FX cultures are enriched with immature precursor cells and to explore possible effects of the FMR1 mutation on neuronal development and functionality, we devised a migration assay, in which the periphery of plated NS was subjected to microcuts, and the repopulation rate of the injury area, probably by newly formed neuroblasts (as previously observed, see Telias et al. [17]), was measured during the next 24 h (Fig. 1B). Our results show that FX cells were significantly less efficient in repopulating the injury area, in control conditions (ie, no growth factors added to the medium). However, in the presence of the early neural morphogens, Noggin and Shh, the repopulation rate of FX cells had significantly increased to a level similar to that measured for HUES cells. The addition of BDNF, GDNF, and NT-3, growth factors known to affect the differentiation and migration of neurons in the postnatal and adult brain [39,40], did not have any differential effect on the repopulation rate of FX, compared to that of HUES. These results suggest that indeed the developmental delay observed during early IVND of FX-hESCs is maintained during late stages of neuronal differentiation. Furthermore, these results also suggest that molecular pathways activated by early morphogens such as Noggin and Shh could be involved in FXS neurodevelopmental pathology.
FIG. 1.
Analysis of long-term neural differentiation of FX-human embryonic stem cells (hESCs). (A) Representative images of neurospheres (NS) developed from HUES-hESCs (left) and FX-hESCs (middle). Neural rosettes (NRs) in FX-NS are indicated by white arrows. The number of NRs per NS was quantified at days 50 and 100 of in vitro neural differentiation (IVND) (20 and 70 days after NS replating, respectively). The experiment was repeated twice in HUES-13-hESC line and in all three FX-hESC lines. n = total number of NS analyzed as indicated at the bottom of each bar. All values are mean ± SEM (*P < 0.05; **P < 0.01; t-test). (B) Representative images of HUES-NS (left) and FX-NS (right) lines, 4 and 24 h following microblade cut. Experiments were performed in HUES-13-hESC line and in all three FX-hESC lines, repeated four times for control (baseline difference between HUES and FX), and twice for each treatment. Quantified values are the percentage of repopulation relative to control, at 2, 4, 8, 16, and 24 h following the cut with or without BDNF, GDNF, NT-3 (all 20 ng/mL), Shh, and Noggin (both 200 ng/mL). HUES-NS are shown in continuous lines; FX-NS are shown in dotted lines. All values are mean ± SEM (n = 2–3/line; *P < 0.05; t-test). Statistical analysis result is shown for the last time point (24 h) only.
Derivation and characterization of hNPCs
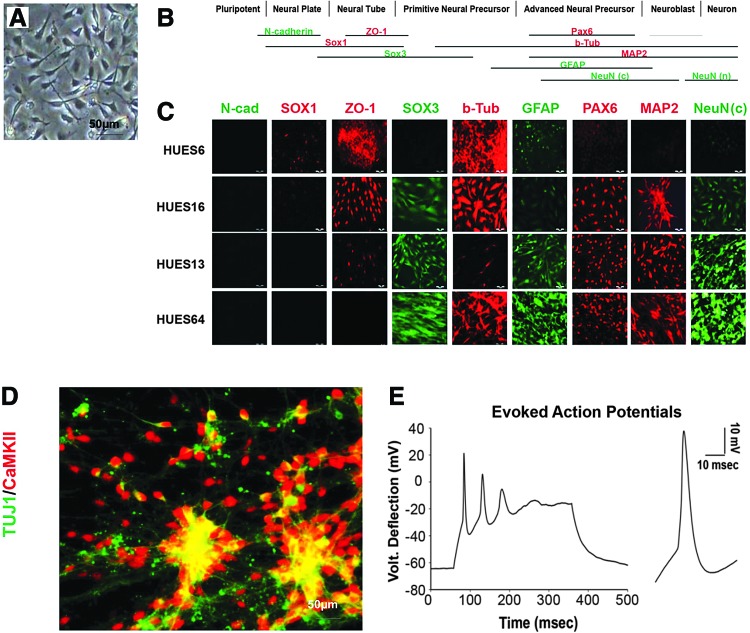
On the basis of our previous published results and the extended phenotypic observations described in Figure 1, we hypothesized that FX hNPCs will exhibit a more persistent primitive phenotype than the HUES counterparts. To investigate this hypothesis, we derived stable self-renewing hNPC lines from the FX and HUES hESC lines described above, following published protocols (see the Materials and Methods section for details). The derived cells, in all lines (FX and HUES alike), had small triangular somata and one or two short projections (Fig. 2A). The morphology of these hNPCs, their rate of growth, and the potential for terminal neuronal differentiation were not altered for at least 10 passages in all lines, with passaging typically occurring once a week in a 1:5 dilution. To characterize the neural status of these hNPCs, we analyzed the protein expression of nine different neural genes, each corresponding to a specific stage during neural development (Fig. 2B). We found that HUES-hNPC lines expressed neural genes characteristic of primitive and advanced hNPCs, depending on the specific line (Fig. 2C). While HUES-6- and HUES-16-derived hNPCs express high levels of early neural genes (especially ZO-1), HUES-13- and HUES-64-derived hNPCs express also more advanced neural genes (especially cytoplasmic NeuN). To demonstrate that these are bona fide hNPCs, we induced their neuronal differentiation, and ∼20–30 days later, mature neurons were obtained that stained positive for the neuronal cytoskeleton protein Tuj1 and for the cortical neuronal marker CaMK-II (Fig. 2D). Furthermore, these neurons were electrically active, firing bursts of action potentials, as demonstrated by electrophysiological recordings performed in the current-clamp mode (Fig. 2E). Action potentials exhibited threshold, peak, repolarization, and after-hyperpolarization, as expected from a firing neuron.
FIG. 2.
Molecular characterization of HUES-hNPC lines. (A) Representative images of hNPCs (bright field, derived from HUES-13 hESC line). (B) Schematic presentation of the neurodevelopmental genes expressed in the different hNPC lines, used for immunostaining analysis. The neural genes are scaled according to their corresponding expression during different neurodevelopmental stages. (C) Representative immunofluorescence images of the expression of the neural genes are presented in (B), for the four HUES cell lines used in this study. (D) Neurons derived from HUES-13 hNPCs (repeated for all lines, HUES and FX alike) show positive staining for Tuj1 (green) and CamK-II (red). (E) Electrophysiological recordings in neurons derived from HUES-13 hNPCs (repeated for all lines, HUES and FX alike, n = 3–4 cells/line). Current-clamp recordings show the voltage deflection (mV) generated by a burst of three consecutive evoked action potentials and details of a single action potential. Cells were held at ∼−60 mV and a current of ∼100 pA was injected. Color images available online at www.liebertpub.com/scd
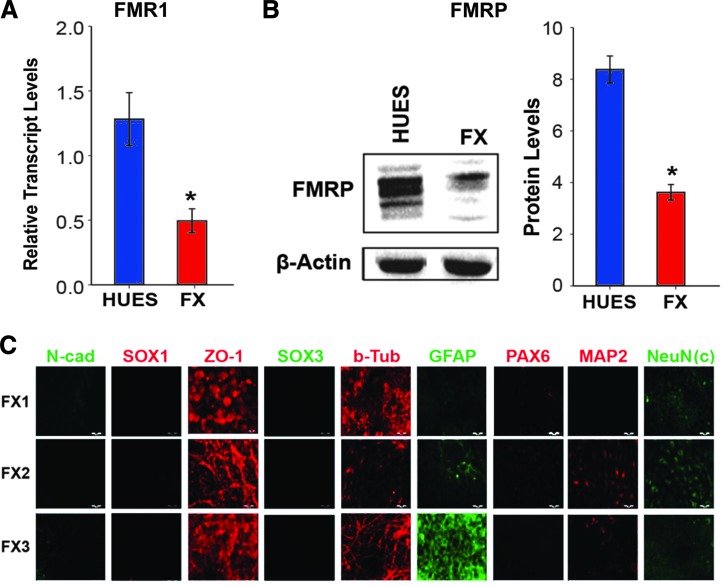
Next, we characterized the hNPCs obtained from FX-hESC lines (Fig. 3). We found that FMR1 expression and FMRP levels were >50% lower in FX-hNPCs than in HUES lines (Fig. 3A, B; Supplementary Fig. S1). The fact that the levels of FMR1 expression and FMRP translation in FX-hNPCs are significantly downregulated, but not completely inactivated, is in accordance with our previous studies, in which we showed that downregulation of FMR1 in FX-hESCs is a developmentally regulated process, and that complete inactivation in vitro occurs only in terminally differentiated cells [16,17]. In accordance with our previous observations regarding the relative immaturity of FX-NS, we found that FX-hNPCs express high levels of the early neural tube gene ZO-1, but only low levels of PAX6, MAP2, and cytoplasmic-NeuN that correspond to more advanced stages of neurogenesis (Fig. 3C). Taken together, these results suggest that partial inactivation of FMR1 is sufficient to retain FX-hNPCs in a more primitive neural status, demonstrating that FMRP plays an important role in early human embryonic neurogenesis.
FIG. 3.
Molecular characterization of FX-hNPC lines. (A) Relative transcript levels of FMR1 in HUES-derived hNPC lines (blue) and in FX-derived hNPC lines (red). The expression of FMR1 in each HUES and FX line was tested in two different samples, one obtained from a low-passage hNPC culture (2–4) and the other from a high-passage hNPC culture (7–10). Values were normalized to GAPDH. All values are mean ± SEM (n = 2/line, *P < 0.05; t-test). (B) Protein levels of FMRP in HUES and FX-hNPCs, as described for A. Quantification from western blots was carried out using optical densitometry. Values were normalized to β-actin levels. All values are mean ± SEM (n = 2/line, *P < 0.05; t-test). (C) Representative immunofluorescence images of the expression of the neural genes presented in Fig. 2B, for the three FX cell lines used in this study. Color images available online at www.liebertpub.com/scd
Expression of SOX genes in FX-hNPCs
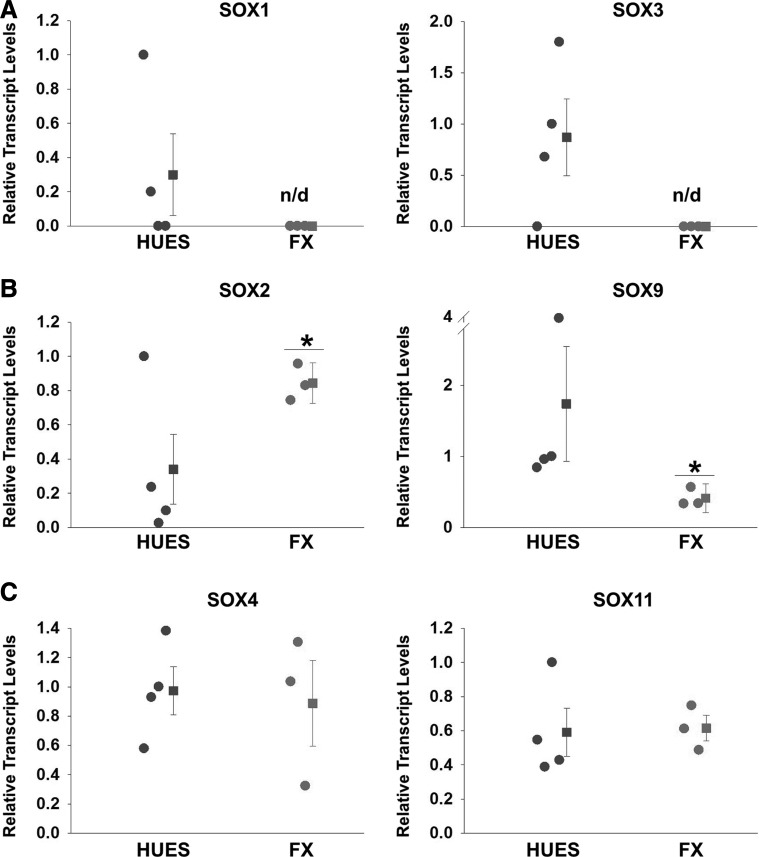
One of the molecular pathways activated by morphogens such as Shh and Noggin is the expression of the SOX superfamily of transcription factors, which in turn drive the progression of neural development [20,21,41–43]. We have previously shown that FX-hESCs fail to upregulate SOX1 during IVND, concomitant with FMR1 downregulation [17]. It has been established that several SOX genes act in tandem to regulate human embryonic development in general and the formation of the nervous system in particular [20,21]. Therefore, we hypothesized that similar to SOX1, the expression of other closely related members of the SOX superfamily of transcriptions factors could be affected by downregulation of FMR1 during IVND of FX-hESCs. We therefore analyzed the transcriptional activation of seven SOX genes in our hNPC lines (Fig. 4). The results show significant downregulation of SOX1, SOX3, and SOX9 in FX-hNPCs, and a significant increase in the expression of SOX2 (Fig. 4A, B). In contrast, the expression of SOX4 and SOX11 (Fig. 4C), known to activate neuronal genes in the developing and adult brain, did not differ between FX and HUES-hNPCs. The late neuronal gene SOX21 was not detected in any of the lines (data not shown). Taken together, these data suggest that SOX1, SOX2, SOX3, and SOX9 may be involved in the aberrant neurogenesis observed in FXS. In the following experiments, we decided to focus on the role of SOX2 and SOX9 in FXS, since their expression was significantly altered in FX-hNPCs, compared to that in HUES counterparts, and was expressed in all lines examined in contrast to SOX1 and SOX3, for which expression was not detected in some of the HUES-hNPC lines (see also Fig. 2C).
FIG. 4.
Expression of SOX superfamily of transcription factors in hNPCs. Relative transcript levels of SOX genes in hNPCs: (A) SOX1 and SOX3, (B) SOX2 and SOX9, (C) SOX4 and SOX11. The mean values for individual lines are shown as circles and the mean for each group is shown as squares. The expression of each HUES and FX line was tested in two samples, one obtained from a low-passage hNPC culture (2–4) and the other from a high-passage hNPC culture (7–10). Values were normalized to GAPDH. All values are mean ± SEM (n = 2/line, *P < 0.05; n/d, not detected).
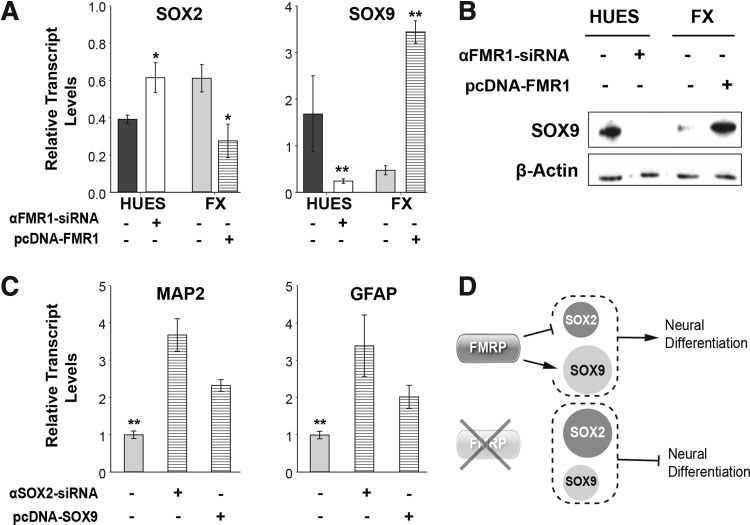
To test the hypothesis that FMRP levels affect the transcriptional activation of SOX2 and SOX9, we performed either knockdown of FMR1 expression in HUES-hNPCs using siRNA, or overexpression of FMR1 in FX-hNPCs. Knockdown of FMR1 expression in HUES-hNPCs significantly increased SOX2 and reduced SOX9 expression, similar to the levels observed in FX-hNPCs. Concomitantly, overexpression of FMR1 in FX-hNPCs reduced SOX2 and increased SOX9 to levels comparable to those in HUES-hNPCs (Fig. 5A, B). It is important to mention here that overexpression of FMRP in FX-hNPCs did not reactivate SOX1 or SOX3 expression (data not shown). These results suggest that the expression of SOX2 and SOX9 is downstream to FMRP during early neural development, probably affecting the outcome of neural and neuronal differentiation. To test this hypothesis, we knocked down SOX2 expression and overexpressed SOX9 in FX-hNPCs (Fig. 5C) and measured their neural status by analyzing the relative expression of GFAP and MAP2, as previously described [17,28,44]. Our results show that SOX2 inhibition and SOX9 overexpression in FX-hNPCs resulted in a significant increase in both MAP2 and GFAP levels, reflecting a progression of these hNPCs from a primitive to a more advanced neural status. Taken together, these results suggest that FMRP may affect neural development by inhibiting SOX2 and activating SOX9 (Fig. 5D). In FMR1-affected cells, FMRP is downregulated during early neurogenesis, causing an abnormal increase in SOX2 and an abnormal decrease in SOX9, leading to delayed and reduced neural development, although this interaction may be indirect.
FIG. 5.
Proposed molecular mechanism behind abnormal neurogenesis in FXS. (A) Relative transcript levels of SOX2 and SOX9 following manipulation of FMR1 expression. FMR1 knockdown (“αFMR1-siRNA,” white) was carried out in HUES-13 and HUES-64 hNPCs, in three different experiments (20 nM, 48–72 h). Overexpression of FMR1 (“pcDNA-FMR1,” horizontal strips) was carried out in a FX3-hNPC line subclone, in which stable transfection was achieved by antibiotic selection (two different samples obtained from different passages were included in the analysis). Values were normalized to GAPDH. All values are mean ± SEM (n = 2–3/line, *P < 0.05; **P < 0.01; t-test). (B) Representative images of western blot analysis of SOX9 levels in hNPCs, corresponding to manipulation of FMR1 expression as described in (B). (C) Relative transcript levels of MAP2 and GFAP expression in FX-hNPC lines following siRNA-mediated knockdown of SOX2 (“αSOX2-siRNA”) or transiently induced overexpression of SOX9 (“pcDNA-SOX9”). Values were normalized to GAPDH. All values are mean ± SEM (n = 2–3/line, **P < 0.01; ANOVA, t-test). (D) Proposed molecular mechanism regulating abnormal human in vitro neurogenesis in FXS. FMRP inhibits SOX2 and enhances SOX9 to promote neural development. FMR1 downregulation in FXS leads to a decrease in FMRP, an increase in SOX2, and a decrease in SOX9, which result in reduced and delayed neural differentiation.
Expression of GSK3β and β-catenin in FX-hNPCs
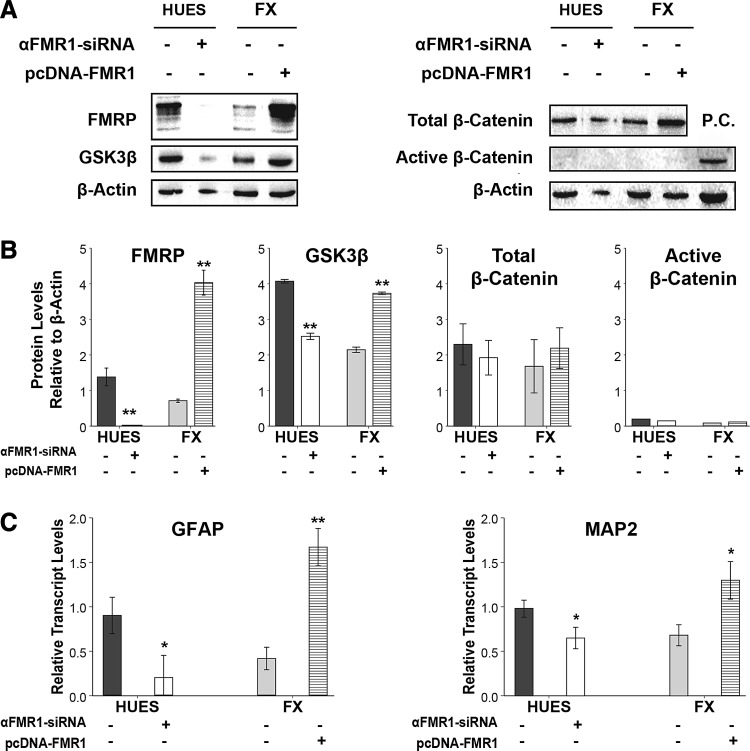
These findings, on the possible involvement of SOX2 and SOX9 during abnormal embryonic neurodevelopment of FX-cells, do not exclude the possibility that other molecular mechanisms could be involved as well. A major finding in brain samples of Fmr1−/− mice is the abnormally high levels of GSK3β, a key kinase shown to play critical roles in several molecular pathways, including MAPK, Cyclin, Akt, and the canonical Wnt/β-catenin signaling pathway [26,45–47]. The role of GSK3β, as part of the canonical Wnt/β-catenin signaling pathway, was shown to be important during adult neurogenesis in the murine hippocampus [29] and in FXS pathology in Fmr1−/− mice [27,28]. Ablation of FMRP in aNSCs leads to an increase in GSK3β protein levels, causing a phosphorylation-dependent decrease in β-catenin activation and an increase in β-catenin degradation, impairing neuronal differentiation. We therefore tested the outcome of FMR1 expression in hNPCs on the protein levels of GSK3β and β-catenin (Fig. 6A, B). Strikingly, our results show that in FX-hNPCs, in which FMR1 expression is decreased, GSK3β protein levels were reduced rather than increased, compared to control. Furthermore, knockdown of FMR1 in HUES-hNPCs reduced GSK3β protein levels, and overexpression of FMR1 in FX-hNPCs increased GSK3β protein levels. However, total levels of β-catenin were unaffected by changes in FMRP levels in hNPCs, and no significant levels of dephosphorylated (ie, active) β-catenin were detected in any of the samples—HUES and FX-hNPCs alike—suggesting that basal levels of Wnt activation are very low in hNPCs and are not affected by FMR1 expression. Taken together, these results suggest that FMRP positively regulates the expression of GSK3β, but it does not affect the expression of β-catenin or its functional activation in these cells. Importantly, we also show that FMR1 expression alone, without the involvement of the Wnt signaling pathway, is enough to significantly increase GFAP and MAP2 expression in hNPCs (Fig. 6C), which is reduced by FMR1 knockdown and increased by FMR1 overexpression, further demonstrating the notion that FMRP regulates the progression of neural differentiation in a Wnt-independent manner.
FIG. 6.
Analysis of the canonical Wnt/β-catenin signaling pathway during neural differentiation of FX-hESCs. (A) Western blot analysis of FMRP, GSK3β, total β-catenin, active (dephosphorylated) β-catenin, and β-actin protein levels in HUES and FX-hNPCs. Representative images show baseline levels for HUES and FX-hNPCs, as well as following 48 h of siRNA-mediated knockdown of FMR1 (“αFMR1-siRNA”) in HUES-hNPCs and constitutive overexpression of FMR1 (“pcDNA-FMR1”) in FX-hNPCs. Positive control (PC): protein extracted from the hESC line Lis25_FAP previously known to express active β-catenin. (B) Optical densitometry quantification of protein levels. FMR1 knockdown (“αFMR1-siRNA,” white) was carried out in three different experiments in two control hNPC lines (HUES-13 and HUES-64; 20 nM siRNA for 48–72 h). Overexpression of FMR1 (“pcDNA-FMR1,” horizontal strips) was carried out in an FX3-hNPC line subclone, in which stable transfection was achieved for at least five passages by antibiotic selection (four different samples obtained from two different passages were included in the analysis). Values were normalized to β-actin levels. All values are mean ± SEM (n = 2–3/line, **P < 0.01; t-test). (C) Relative transcript levels of GFAP (left) and MAP2 (right) in hNPCs following manipulation of FMR1 expression as described in A. Values were normalized to GAPDH. All values are mean ± SEM (n = 2–3/line; *P < 0.05; **P < 0.01; t-test).
Modulation of GSK3β and β-catenin in FX neurons
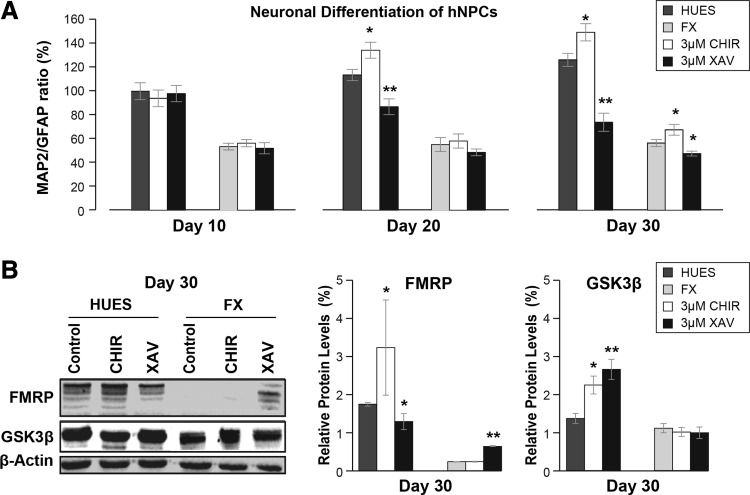
Since the role of Wnt signaling in FXS was shown in adult neurogenesis, we decided to test its possible involvement in later stages of differentiation of FX-hNPCs into FX neurons. We thus subjected hNPCs to 30 days of neuronal differentiation in the presence of Wnt signaling modulators: a GSK3β inhibitor (CHIR99021; “CHIR”), which reduces GSK3β-mediated phosphorylation of β-catenin, thus increasing Wnt/β-catenin activity [48], and a specific tankyrase inhibitor (XAV939, “XAV”), which stabilizes Axin levels in the cells, thus decreasing Wnt/β-catenin activity in a GSK3β-independent manner [49]. The MAP2/GFAP bioassay was implemented to measure the efficiency of neuronal neurogenesis, as has been done previously by us and others [17,28,44]. In control cultures (ie, no CHIR or XAV added), the efficiency of neuronal differentiation was significantly lower for FX lines than for HUES lines, as expected (Fig. 7A). In addition, we here show that CHIR-mediated inhibition of GSK3β had a neuralizing effect [50], significantly increasing the MAP2/GFAP ratio compared to control, whereas inhibition of β-catenin by XAV decreased the MAP2/GFAP ratio, in both HUES and FX lines alike. Congruently, in CHIR-treated cultures and controls, neuronal differentiation of FX-hNPCs resulted in full inactivation of FMR1 and complete absence of FMRP, reflecting successful neuronal differentiation (Fig. 7B). In contrast, neuronal differentiation in the presence of XAV resulted in partial FMRP expression even after 30 days, demonstrating differentiation inhibition as suggested by the decreased MAP2/GFAP ratio. Most importantly, after 30 days of neuronal differentiation, a reduction in FMRP (XAV-treated FX-cultures) or its complete absence (control and CHIR-treated FX-cultures) was not associated with an abnormal increase in GSK3β. In conclusion, our results show that in hNPCs, loss of FMRP does not correlate with an abnormal increase in GSK3β, and that drug-mediated inhibition of GSK3β in FX-hNPCs undergoing neuronal differentiation does not lead to an improvement in this process, compared to controls. These results demonstrate that the relationship between FMRP and GSK3β is both stage specific and species specific.
FIG. 7.
Effect of Wnt modulators on neuronal differentiation of FX hNPCs. (A) Neuron-to-glia ratio (MAP2/GFAP) during 30 days of neuronal differentiation of hNPC lines (HUES in dark gray; FX in light gray) at 10-day intervals in the presence of CHIR99021 (“CHIR,” 3 μM, white) or XAV939 (“XAV,” 3 μM, black). Relative number of MAP2- and/or GFAP-positive cells was quantified relative to DAPI staining, in all hNPC lines, in three randomly selected fields per experiment, containing at least 50 cells per field. Two independent experiments were performed in all four HUES-hNPC lines and in all three FX-hNPC lines. Values were normalized to control (% of control). All values are mean ± SEM (n = 2/line; *P < 0.05; **P < 0.01; ANOVA and t-test). (B) Western blot analysis of FMRP and GSK3β at day 30 of hNPC neuronal differentiation, as described in A. Representative images of western blot analysis are shown for HUES-13 and FX1 lines (left) and optical densitometry quantification of protein levels (right). Quantified data were obtained from all four HUES-hNPC lines and all three FX-hNPC lines. Values were normalized to β-actin levels. All values are mean ± SEM (n = 2/line; *P < 0.05; **P < 0.01; two-way ANOVA).
Discussion
The developmentally regulated inactivation of FMR1 in FXS human embryos is a dynamic process that starts with full expression of FMR1 in the pluripotent state even with an existing full mutation. Thus, the expression of FMR1 in affected humans progressively diminishes until it is completely lost in mature neurons. Using FX-hESCs, we can recapitulate this developmentally regulated inactivation, focusing on in vitro derived FX-hNPCs and mature neurons, at which stages FMR1 is partially and then fully repressed. FMRP is a multitarget RNA-binding protein [51]. In healthy individuals, FMRP is upregulated during development of the neural lineage [52,53], suggesting that it is involved in progression of neural development, and that at different stages of neurodevelopment, FMRP regulates different targets. Indeed, several developmental functions have been found for FMRP linked to maintenance and differentiation of embryonic and adult stem cells [4]. In this study, we have shown that a partial reduction in FMRP expression was enough to induce a delayed and less effective neural differentiation. Therefore, the molecular mechanisms downstream to FMRP are of high interest for the role they could play in neurogenesis and in FXS pathology.
In this study, we show evidence supporting the hypothesis that abnormal neurogenesis in FX-hNPCs could be the result of an aberrant expression of SOX2 and SOX9. Transcription factors of the SOX superfamily are well known for their important role in embryogenesis in general and neurogenesis in particular, but their possible role in FXS neurodevelopmental pathology has not been studied yet. SOX2 plays a critical role in the regulation of early neural development [41]. It has been shown that SOX2 is important in the maintenance of pluripotency, but it also promotes neural progenitor ground state by inhibiting further neuronal differentiation [54]. More recently, it has been shown that SOX2 expression in astrocytes effectively reprograms these cells to the neural progenitor/precursor stage [55]. Our results show that even partial reduction in FMRP levels is sufficient to induce an increase in SOX2, promoting a delay in neurodevelopment, explaining the more primitive neural phenotype observed in FX-hNPCs than in controls. In addition, we found that SOX9 expression is positively affected by FMRP levels. SOX9 is involved in regulation of cell fate in several lineages: neural tissue [56], including neural crest [25], and glia [57], testis [58], and chondrocytes [59], which are all known to be affected in FXS. Indeed, FXS patients exhibit mental retardation and autism (an effect on the nervous system), macroorchidism (in males, testis), hyperextensible joints (chondrocytes), and stereotypic faces (neural crest). Furthermore, in a recent independent study, SOX9 has been found as a candidate gene for autism in human samples [60]. Our proposed mechanism suggests that FMRP contributes to neural development by inhibiting SOX2 and activating SOX9. Therefore, in FXS, when FMRP is absent, SOX2 is abnormally increased and SOX9 is abnormally decreased, leading to delayed and impaired neurodevelopment. We conclude that exploring the role of the SOX gene superfamily in FXS pathology could prove to be valuable in future diagnostic and therapeutic strategies for FXS patients.
A major finding in FXS research based on murine models is the involvement of GSK3β, a kinase that plays key roles in several molecular pathways, including the canonical Wnt/β-catenin signaling [26,28]. Specifically, it has been found that the absence of FMRP leads to an abnormal increase in GSK3β protein levels. However, the “GSK3β theory of FXS” has never been tested in any human-based model. In this study, we have found that (i) loss of FMRP in hNPCs correlates with a decrease in GSK3β, (ii) loss of FMRP in neurons derived from hNPCs does not correlate with a significant change in GSK3β levels, and (iii) drug-mediated inhibition of GSK3β activity did not improve neuronal differentiation of hNPCs. These differences between mice and human-based research on FXS pathology could be attributed to several factors, such as species specificity; the different developmental stages of the studied specimen (aNSCs in Fmr1−/− mice vs. human ESCs and NPCs), in which different FMRP targets may play different roles; the full CGG expansion in the FMR1 gene that is present in FX-hESCs and FX-hNPCs, but not in animals; or the inherent variability between in vivo and in vitro systems. The results shown in this study are not enough to discharge the “GSK3β theory of FXS,” and more research is needed. However, they hint that the possible involvement of GSK3β in FXS pathology needs to be revisited and validated in human-based models.
Suitable human models for the study of the developmental aspects of FXS pathology are very limited [10,11]. The use of hiPSCs to model neurodevelopmental disorders is a relatively new strategy [15]. In the context of FXS research, the use of FX-hESCs as opposed to FX-hiPSCs can greatly affect the results obtained. The current literature clearly indicates that reprogramming of fibroblasts from FXS patients does not erase the methylation pattern of an expanded FMR1 allele, while FX-hESCs have mostly unmethylated FMR1 loci [16,17,61–66]. In accordance, the results obtained from these two models can vary significantly. Using FX-hESCs, we previously found no difference between FX and control cells in the initial outgrowth of neurites from neural precursors and neuroblasts, and rather significant abnormalities at the electrophysiological level in neurons, at the end of differentiation [17]. In contrast, induction of neural differentiation in FX-hiPSCs, in which FMR1 is silenced at all developmental stages, produced a significant defect in initial neurite outgrowth [37]. We previously reported on the aberrant patterns of expression for SOX1, NOTCH1, and PAX6 during IVND of FX-hESCs and reduced neuronal yields compared to controls [17]; in this study, we provide data suggesting a role for SOX2 and SOX9, and excluding a role for GSK3β. Using FX-hiPSCs, others have found a significant reduction in SOX1 expression and a similar phenotype of reduced neuronal yields [18], suggesting that these phenotypes are strongly correlated with a reduction in FMR1 expression in both models. Finally, a recent study using FX-hiPSCs suggested a role for the transcription repressor REST in FXS neuropathology [19], but these results have not yet been tested in any other model.
In summary, in this study, we have uncovered a new mechanism suggested to play a role in the neurodevelopmental abnormalities characterizing FXS in humans, involving specific members of the SOX superfamily, especially SOX2 and SOX9. However, more research is required to unveil the complete sequence of molecular events that mediate FMRP function.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Douglas Melton (Harvard University) and Dr. Rachel Eiges (Shaare Tzedek Medical Center) for HUES lines and SZFX6, respectively. We thank Dr. Rina Arbesfeld (Tel Aviv University) for fruitful discussions on the Wnt signaling pathway, Dr. Gary Bassell (Emory University) for the CMV-FMR1 plasmid, and Javier Ganz and Dr. Daniel Offen (Rabin Medical Center) for the inducible SOX9 plasmid. We thank Dr. Yael Kalma and Dr. Rachel Eiges for revisions and comments on the manuscript. This study was funded by a doctoral scholarship from the National Network of Excellence in Neurosciences (TEVA Pharmaceutics Ltd.) and by a grant from the Chief Scientist Office of the Israel Ministry of Health (300000-6237). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Penagarikano O, Mulle JG. and Warren ST. (2007). The pathophysiology of fragile X syndrome. Annu Rev Genomics Hum Genet 8:109–129 [DOI] [PubMed] [Google Scholar]
- 2.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 [DOI] [PubMed] [Google Scholar]
- 3.Willemsen R, Bontekoe CJ, Severijnen LA. and Oostra BA. (2002). Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet 110:601–605 [DOI] [PubMed] [Google Scholar]
- 4.Li Y. and Zhao X. (2014). Fragile X proteins in stem cell maintenance and differentiation. Stem Cells 32:1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan L, Dockendorff TC, Jongens TA. and Dreyfuss G. (2000). Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol 20:8536–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Broeder MJ, van der Linde H, Brouwer JR, Oostra BA, Willemsen R. and Ketting RF. (2009). Generation and characterization of FMR1 knockout zebrafish. PLoS One 4:e7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mientjes EJ, Nieuwenhuizen I, Kirkpatrick L, Zu T, Hoogeveen-Westerveld M, Severijnen L, Rife M, Willemsen R, Nelson DL. and Oostra BA. (2006). The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol Dis 21:549–555 [DOI] [PubMed] [Google Scholar]
- 8.Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, et al. (2001). Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet 98:161–167 [DOI] [PubMed] [Google Scholar]
- 9.Schwartz PH, Tassone F, Greco CM, Nethercott HE, Ziaeian B, Hagerman RJ. and Hagerman PJ. (2005). Neural progenitor cells from an adult patient with fragile X syndrome. BMC Med Genet 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharyya A, McMillan E, Wallace K, Tubon TC, Jr., Capowski EE. and Svendsen CN. (2008). Normal neurogenesis but abnormal gene expression in human fragile X cortical progenitor cells. Stem Cells Dev 17:107–117 [DOI] [PubMed] [Google Scholar]
- 11.Castren M, Tervonen T, Karkkainen V, Heinonen S, Castren E, Larsson K, Bakker CE, Oostra BA. and Akerman K. (2005). Altered differentiation of neural stem cells in fragile X syndrome. Proc Natl Acad Sci U S A 102:17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castren M. (2006). Differentiation of neuronal cells in fragile X syndrome. Cell Cycle 5:1528–1530 [DOI] [PubMed] [Google Scholar]
- 13.Braun K. and Segal M. (2000). FMRP involvement in formation of synapses among cultured hippocampal neurons. Cereb Cortex 10:1045–1052 [DOI] [PubMed] [Google Scholar]
- 14.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. and Jones JM. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147 [DOI] [PubMed] [Google Scholar]
- 15.Telias M. and Ben-Yosef D. (2014). Modeling neurodevelopmental disorders using human pluripotent stem cells. Stem Cell Rev 10:494–511 [DOI] [PubMed] [Google Scholar]
- 16.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N. and Ben-Yosef D. (2007). Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1:568–577 [DOI] [PubMed] [Google Scholar]
- 17.Telias M, Segal M. and Ben-Yosef D. (2013). Neural differentiation of fragile X human embryonic stem cells reveals abnormal patterns of development despite successful neurogenesis. Dev Biol 374:32–45 [DOI] [PubMed] [Google Scholar]
- 18.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF. and Haggarty SJ. (2011). Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile x syndrome. PLoS One 6:e26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halevy T, Czech C. and Benvenisty N. (2014). Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Reports 4:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiefer JC. (2007). Back to basics: Sox genes. Dev Dyn 236:2356–2366 [DOI] [PubMed] [Google Scholar]
- 21.Wegner M. and Stolt CC. (2005). From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci 28:583–588 [DOI] [PubMed] [Google Scholar]
- 22.Malas S, Postlethwaite M, Ekonomou A, Whalley B, Nishiguchi S, Wood H, Meldrum B, Constanti A. and Episkopou V. (2003). Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience 119:421–432 [DOI] [PubMed] [Google Scholar]
- 23.Laumonnier F, Ronce N, Hamel BC, Thomas P, Lespinasse J, Raynaud M, Paringaux C, Van Bokhoven H, Kalscheuer V, et al. (2002). Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet 71:1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamel BC, Smits AP, Otten BJ, van den Helm B, Ropers HH. and Mariman EC. (1996). Familial X-linked mental retardation and isolated growth hormone deficiency: clinical and molecular findings. Am J Med Genet 64:35–41 [DOI] [PubMed] [Google Scholar]
- 25.Lee YH. and Saint-Jeannet JP. (2011). Sox9 function in craniofacial development and disease. Genesis 49:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Portis S, Giunta B, Obregon D. and Tan J. (2012). The role of glycogen synthase kinase-3 signaling in neurodevelopment and fragile X syndrome. Int J Physiol Pathophysiol Pharmacol 4:140–148 [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Murthy AC, Zhang L, Johnson EB, Schaller EG, Allan AM. and Zhao X. (2011). Inhibition of GSK3 beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet 1:681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, et al. (2010). Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet 6:e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M. and Gage FH. (2009). Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci 12:1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frumkin T, Malcov M, Telias M, Gold V, Schwartz T, Azem F, Amit A, Yaron Y. and Ben-Yosef D. (2010). Human embryonic stem cells carrying mutations for severe genetic disorders. In Vitro Cell Dev Biol Anim 46:327–336 [DOI] [PubMed] [Google Scholar]
- 31.Telias M, Segal M. and Ben-Yosef D. (2014). Electrical maturation of neurons derived from human embryonic stem cells. F1000Res 3:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang SC, Wernig M, Duncan ID, Brustle O. and Thomson JA. (2001). In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19:1129–1133 [DOI] [PubMed] [Google Scholar]
- 33.Hicks AU, Lappalainen RS, Narkilahti S, Suuronen R, Corbett D, Sivenius J, Hovatta O. and Jolkkonen J. (2009). Transplantation of human embryonic stem cell-derived neural precursor cells and enriched environment after cortical stroke in rats: cell survival and functional recovery. Eur J Neurosci 29:562–574 [DOI] [PubMed] [Google Scholar]
- 34.Banda E. and Grabel L. (2015). Directed differentiation of human embryonic stem cells into neural progenitors. Methods Mol Biol 1307:289–298 [DOI] [PubMed] [Google Scholar]
- 35.Elkabetz Y. and Studer L. (2008). Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb Symp Quant Biol 73:377–387 [DOI] [PubMed] [Google Scholar]
- 36.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V. and Studer L. (2008). Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev 22:152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L. and Bhattacharyya A. (2014). iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev 23:1777–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halevy T, Czech C. and Benvenisty N. (2015). Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Reports 4:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castren ML. and Castren E. (2014). BDNF in fragile X syndrome. Neuropharmacology 76 Pt C:729–736 [DOI] [PubMed] [Google Scholar]
- 40.Erickson JT, Brosenitsch TA. and Katz DM. (2001). Brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor are required simultaneously for survival of dopaminergic primary sensory neurons in vivo. J Neurosci 21:581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyagi S, Kato H. and Okuda A. (2009). Role of SoxB1 transcription factors in development. Cell Mol Life Sci 66:3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuda Y, Ogura E, Kondoh H. and Kamachi Y. (2010). B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet 6:e1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trowe MO, Zhao L, Weiss AC, Christoffels V, Epstein DJ. and Kispert A. (2013). Inhibition of Sox2-dependent activation of Shh in the ventral diencephalon by Tbx3 is required for formation of the neurohypophysis. Development 140:2299–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briggs JA, Sun J, Shepherd J, Ovchinnikov DA, Chung TL, Nayler SP, Kao LP, Morrow CA, Thakar NY, et al. (2013). Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology. Stem Cells 31:467–478 [DOI] [PubMed] [Google Scholar]
- 45.Mines MA, Yuskaitis CJ, King MK, Beurel E. and Jope RS. (2010). GSK3 influences social preference and anxiety-related behaviors during social interaction in a mouse model of fragile X syndrome and autism. PLoS One 5:e9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franklin AV, King MK, Palomo V, Martinez A, McMahon LL. and Jope RS. (2014). Glycogen synthase kinase-3 inhibitors reverse deficits in long-term potentiation and cognition in fragile X mice. Biol Psychiatry 75:198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen T, Lu JS, Song Q, Liu MG, Koga K, Descalzi G, Li YQ. and Zhuo M. (2014). Pharmacological rescue of cortical synaptic and network potentiation in a mouse model for fragile X syndrome. Neuropsychopharmacology 39:1955–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, et al. (2003). Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52:588–595 [DOI] [PubMed] [Google Scholar]
- 49.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620 [DOI] [PubMed] [Google Scholar]
- 50.Esfandiari F, Fathi A, Gourabi H, Kiani S, Nemati S. and Baharvand H. (2012). Glycogen synthase kinase-3 inhibition promotes proliferation and neuronal differentiation of human-induced pluripotent stem cell-derived neural progenitors. Stem Cells Dev 21:3233–3243 [DOI] [PubMed] [Google Scholar]
- 51.Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, et al. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492:382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhakar AL, Dolen G. and Bear MF. (2012). The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci 35:417–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abitbol M, Menini C, Delezoide AL, Rhyner T, Vekemans M. and Mallet J. (1993). Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet 4:147–153 [DOI] [PubMed] [Google Scholar]
- 54.Bylund M, Andersson E, Novitch BG. and Muhr J. (2003). Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci 6:1162–1168 [DOI] [PubMed] [Google Scholar]
- 55.Niu W, Zang T, Smith DK, Vue TY, Zou Y, Bachoo R, Johnson JE. and Zhang CL. (2015). SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Reports 4:780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stolt CC. and Wegner M. (2010). SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol 42:437–440 [DOI] [PubMed] [Google Scholar]
- 57.Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM. and Deneen B. (2012). Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74:79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang T, Hou CC, She ZY. and Yang WX. (2013). The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol Biol Rep 40:2187–2194 [DOI] [PubMed] [Google Scholar]
- 59.Wuelling M. and Vortkamp A. (2011). Chondrocyte proliferation and differentiation. Endocr Dev 21:1–11 [DOI] [PubMed] [Google Scholar]
- 60.Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G. and Scherer SW. (2011). Gene and miRNA expression profiles in autism spectrum disorders. Brain Res 1380:85–97 [DOI] [PubMed] [Google Scholar]
- 61.Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD. and Jaffrey SR. (2014). Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343:1002–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Avitzour M, Mor-Shaked H, Yanovsky-Dagan S, Aharoni S, Altarescu G, Renbaum P, Eldar-Geva T, Schonberger O, Levy-Lahad E, Epsztejn-Litman S. and Eiges R. (2014). FMR1 epigenetic silencing commonly occurs in undifferentiated fragile X-affected embryonic stem cells. Stem Cell Reports 3:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alisch RS, Wang T, Chopra P, Visootsak J, Conneely KN. and Warren ST. (2013). Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med Genet 14:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Esch CE, Ghazvini M, Loos F, Schelling-Kazaryan N, Widagdo W, Munshi ST, van der Wal E, Douben H, Gunhanlar N, et al. (2014). Epigenetic characterization of the FMR1 promoter in induced pluripotent stem cells from human fibroblasts carrying an unmethylated full mutation. Stem Cell Reports 3:548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bar-Nur O, Caspi I. and Benvenisty N. (2012). Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. J Mol Cell Biol 4:180–183 [DOI] [PubMed] [Google Scholar]
- 66.Urbach A, Bar-Nur O, Daley GQ. and Benvenisty N. (2010). Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 6:407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.