Abstract
Background
The striated muscle Z-line, a multiprotein complex at the boundary between sarcomeres, plays an integral role in maintaining striated muscle structure and function. Multiple Z-line-associated proteins have been identified and shown to play an increasingly important role in the pathogenesis of human cardiomyopathy. Cypher and its close homologue, Enigma homolog protein (ENH), are 2 Z-line proteins previously shown to be individually essential for maintenance of postnatal cardiac function and stability of the Z-line during muscle contraction, but dispensable for cardiac myofibrillogenesis and development.
Methods and Results
The current studies were designed to test whether Cypher and ENH play redundant roles during embryonic development. Here, we demonstrated that mice lacking both ENH and Cypher exhibited embryonic lethality and growth retardation. Lethality in double knockout embryos was associated with cardiac dilation and abnormal Z-line structure. In addition, when ENH was ablated in conjunction with selective ablation of either Cypher short isoforms (CypherS), or Cypher long isoforms (CypherL), only the latter resulted in embryonic lethality.
Conclusions
Cypher and ENH redundantly play an essential role in sustaining Z-line structure from the earliest stages of cardiac function, and are redundantly required to maintain normal embryonic heart function and embryonic viability.
Keywords: Cypher, embryonic lethality, Enigma homolog protein, Z-line
In striated muscle, the Z-line forms the boundary between sarcomeres and contains multiprotein complexes essential for muscle development and maintenance of mature fibers.1–8 Recently, a large number of Z-line-associated proteins have been identified, and mutations in many of these have been linked to cardiac and skeletal myopathies in humans and mice.1,2,9,10 Further characterization of the roles of Z-line proteins in disease is essential for providing insights into treatment strategies for Z-line-associated human muscle myopathies. One such protein is Cypher, a PDZ and LIM domain containing protein that was first cloned in 1999.11,12 Both conventional and cardiac conditional Cypher knockout mice display premature lethality associated with severe dilated cardiomyopathy and heart failure. Global and cardiac-restricted knockout mice showed severely disorganized and fragmented Z-lines in cardiac muscle.13,14 Surprisingly, no abnormalities were detected in the Z-lines of the noncontracting diaphragm muscle before birth, indicating that Cypher is necessary for maintenance of Z-line integrity during the stress of muscle contraction, but is not essential for Z-line formation.14 These observations were later supported by the observation of a similar phenotype in the morpholino knockdown of Cypher in zebrafish.15 Furthermore, mutations in the human Cypher gene ortholog, ZASP, have been identified in patients with different forms of cardiomyopathy and myofibrillar myopathies.9,16,17
Enigma homolog protein (ENH), a close homologue of Cypher, is also a member of the PDZ-LIM domain protein family, highly expressed in the heart, and localized at the Z-line in striated muscle.10,18–20 Although there are no cardiomyopathies yet associated with ENH mutations in humans, we previously reported that global loss of ENH in mice (ENH−/−) is associated with dilated cardiomyopathy and disorganization of cardiac Z-lines.21 Furthermore, ENH forms a protein complex with Cypher short isoform (CypherS) and Calsarcin-1 at the Z-line and, as a result, CypherS and Calsarcin-1 proteins are specifically downregulated in ENH−/− mouse hearts.21
Although both Cypher and ENH play important roles in maintaining Z-line structure and heart function, no obvious heart developmental defects were observed in either Cypher−/− or ENH−/− global knockout mice, suggesting a possible functional overlap between these 2 Enigma family members during embryonic heart development. To investigate possible redundancy between Cypher and ENH in the Z-line during embryonic development, we crossed Cypher knockout mice with ENH knockout mice. Our results demonstrated that no Cypher and ENH double knockout (dKO) mice survived past embryonic day 11.5 (E11.5). In addition, dKO embryos exhibited a severely abnormal cardiac phenotype by E9.5. Immunostaining clearly showed abnormal Z-line organization in hearts of dKO embryos. Further analyses also showed functional redundancy between ENH and Cypher long isoform (CypherL), but not CypherS.
Methods
Animal Models
The generation of global ENH, global Cypher, CypherS, and CypherL knockout mouse models has been described previously.14,21,22 Genotyping of isolated embryos, neonates, and mice was performed using previously described primer sets.14,21,22 All experiments described in this study were completed using mice of a mixed 129/SvJ and Black Swiss background. All procedures were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at the University of California, San Diego.
In Situ Hybridization
Whole-mount in situ hybridization was performed as previously described.23 Briefly, embryos isolated at different stages were fixed in pre-chilled 4% paraformaldehyde in phosphate-buffered saline at 4°C overnight, and subsequently subjected to RNA in situ analyses. The ENH RNA in situ probe was generated using the following primer set: ENH-Forward: CACATACCCACTCACAGTGATG and ENH-Reverse: GAATAGATGTAAATACATTGAAGG. For histological analyses, following whole-mount in situ hybridization, embryos were embedded in paraffin and 8-μm transverse sections were prepared using a microtome.
Immunohistochemistry
Fixed embryos were embedded in OCT Tissue-Tek (Thermo Fisher Scientific, Waltham, MA), and sectioned (8 μm) using a Leica CM 3050S cryostat (Leica Microsystems, Bannockburn, IL). Sections were blocked in a solution containing 2% horse serum, 0.2% Triton X-100, and 5% bovine serum albumin in phosphate-buffered saline, followed by incubation with primary antibody overnight at 4°C. The following primary antibodies were used: Cypher24 (1:300), CD31 (1:300; BD Biosciences), sarcomeric alpha(α)-actinin (1:300; Sigma-Aldrich), myomesin (1:300; Santa Cruz Biotechnology), cleaved caspase 3 (1:300; Cell Signaling Technology), and phospho-histone H3 S10 (1:300; Millipore). Subsequently, sections were incubated with fluorescently labeled secondary antibodies. Confocal microscopy was performed using a Zeiss Axioplan 2 upright confocal microscope (Carl Zeiss Inc, Thornwood, NY).
Morphological and Histological Analysis
Female mice with copulation plugs were considered to be at embryonic day 0.5 (E0.5). Timed pregnant mice were sacrificed at various stages of gestation. Embryos were harvested, examined, imaged, and fixed for 1 to 24 hours in 4% paraformaldehyde in phosphate-buffered saline. Genotyping of isolated embryos was performed using individual embryo yolk sac extracts. Following fixation, embryos were embedded in paraffin, transversely sectioned, and analyzed by staining with hematoxylin and eosin as previously described.13,25,26
Transmission Electron Microscopy
Embryos isolated at E9.5 were immediately immersed in fixative (2% paraformaldehyde, 2% glutaraldehyde in phosphate buffered saline) and incubated overnight. Embryos were then washed in 0.15 mol/L sodium cacodylate buffer and postfixed in 2% osmium tetroxide and 0.8% potassium ferrocyanide in 0.15 mol/L sodium cacodylate buffer for 1 hour. Subsequently, embryos were stained overnight in 2% uranyl acetate, dehydrated, and embedded in Durcupan resin (EMD, Gibbstown, NJ) according to standard procedures. Ultrathin sections (60 to 70 nm) were stained with a solution of 2% uranyl acetate and lead citrate. Electron micrographs were recorded using a JEOL-1200EX transmission electron microscope operated at an accelerating voltage of 80 kV. Transmission electron microscopy was performed at the National Center for Microscopy and Imaging Research, University of California, San Diego.
Results
Cypher and ENH Are Highly Expressed in the Myocardium During Early Heart Development
In previous studies, we have demonstrated that Cypher is highly expressed in the early developing heart.11,14 To investigate specific cell types expressing Cypher in embryonic heart, we performed immunostaining analysis of E9.5 embryos using antibodies to Cypher, and CD31, a marker of endothelium (Figure 1A through 1D). Cypher expression was only detected in myocardium, not in endocardium. To determine the expression pattern of ENH during murine development, embryos staged between E8.5 and E10.5 were analyzed. Owing to lack of antibodies specific to ENH for immunofluorescence studies, we performed whole-mount RNA in situ hybridization utilizing a probe specific for ENH transcripts. At E8.5 to E9.5, ENH was highly expressed and detected exclusively in the heart (Figure 1E and 1F, and S1A and S1B), extending to the head region by E10.5 (Figure S1C and S1D). Histological analysis of E9.5 transverse sections following whole-mount RNA in situ hybridization for ENH further revealed that expression of ENH was restricted to myocardium, and was not detected in endocardium (Figure 1G and 1H).
Figure 1.
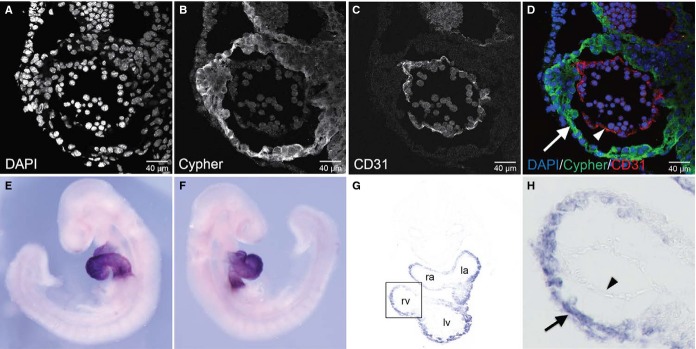
Cypher and ENH are specifically expressed in the myocardium at E9.5. A through D, Immunofluorescence analysis of a transverse section of the heart of a wild-type embryo at E9.5, staining for (A) DAPI, (B) Cypher, and (C) CD31. D, Color-merged image of DAPI (blue), Cypher (green), and CD31 (red). Arrow, myocardium; arrowhead, endocardium. E and F, Whole-mount RNA in situ hybridization analysis of ENH expression in a wild-type embryo (right and left lateral views) at E9.5 using an ENH-specific probe. G, Histological analysis of a transverse section of the heart from the embryo depicted in (E and F). H, High-magnification view of boxed area in (G). Arrow indicates myocardium; arrowhead, endocardium; DAPI, 4’,6-diamidino-2-phenylindole; ENH, Enigma homolog protein; la, left atrium; lv, left ventricle; ra, right atrium; rv, right ventricle.
Ablation of Cypher and ENH Results in Embryonic Lethality
Both Cypher and ENH knockout mice develop postnatal dilated cardiomyopathy,13,14,21 with differences in severity, suggesting a unique role for Cypher and ENH in the adult heart. Since there is no obvious heart developmental defect in either Cypher−/− or ENH−/− global knockout mice, yet Cypher and ENH are specifically expressed in the early developing heart11,14 (Figure 1), we speculated that there is a possible functional overlap between these 2 Enigma family members during embryonic heart development. To address this question, we generated ENH−/−/Cypher−/− double knockout (dKO) mice by crossing ENH−/− with Cypher+/− mice. No viable ENH−/−/Cypher−/− mutants were obtained in litters from ENH−/−/Cypher+/− intercrosses at weaning, indicating that dKO of Cypher and ENH results in embryonic lethality.
To determine the stage of embryonic lethality, timed pregnancies were performed, and embryos were isolated at various stages of gestation. Genotyping revealed the presence of ENH−/−/Cypher−/− embryos up until E10.5, but not beyond. The genotype distribution fit the expected Mendelian ratios at E8.5 to E9.5; however, at E10.5 fewer live dKO embryos than predicted were recovered (Table1). Collectively these data reveal that dKO of Cypher and ENH leads to lethality of embryos between E9.5 and E11.5.
Table 1.
Genotypes of Offspring From ENH−/−/Cypher+/− Intercrosses at Developmental Stages Embryonic Day (E)8.5 to 21 dpn, Show Embryonic Lethality of ENH−/−/Cypher−/− Double Knockout Mice Between E9.5 and E11.5
| Genotype | Developmental Stage | Expected | |||||
|---|---|---|---|---|---|---|---|
| E8.5 | E9.0 | E9.5 | E10.5 | E11.5 | 21 dpn | ||
| ENH−/−/Cypher+/+ | 28.6% (n=12) | 27.4% (n=14) | 26.7% (n=16) | 34% (n=14) | 36.8% (n=7) | 32.4% (n=25) | 25% |
| ENH−/−/Cypher+/− | 47.6% (n=20) | 47% (n=24) | 46.6% (n=28) | 54% (n=22) | 63.2% (n=12) | 67.5% (n=52) | 50% |
| ENH−/−/Cypher−/− | 23.8% (n=10) | 25.5% (n=13) | 26.7% (n=16) | 12% (n=5) | 0% (n=0) | 0% (n=0) | 25% |
dpn indicates days postnatal; ENH, Enigma homolog protein.
ENH−/−/Cypher−/− Mutant Embryos Display Aberrant Embryonic Development and Cardiac Morphogenesis
We observed no difference in the gross morphology of ENH−/−/Cypher+/− and ENH−/−/Cypher+/+ embryos during embryonic development or in the postnatal phenotype (data not shown). Thus, ENH−/−/Cypher+/− or ENH−/−/Cypher+/+ embryos were used interchangeably as controls for all experiments. In the absence of both ENH and Cypher, embryonic development appeared normal at E9.0, as dKO embryos were comparable to control embryos in size and morphology (Figure 2A through 2F). However, by E9.5, although variability in the severity of the phenotype was observed, almost all dKO embryos displayed dilation of the heart with an abnormally thin myocardium transparent in appearance (Figure 2G through 2L). By E10.5, ENH and Cypher dKO embryos were severely growth retarded and heart defects were evident (Figure 2M through 2R). Many of the ENH−/−/Cypher−/− hearts failed to beat and all the dKO embryos at this stage displayed severe pericardial effusion indicative of heart failure. Previously, we have shown that Cypher−/− mice develop dilated cardiomyopathy as early as postnatal day 1 (P1).14 Thus, to exclude the possibility that the morphological abnormalities observed in dKO embryos at E9.5 and E10.5 were due to the ablation of Cypher alone, we investigated the morphology of Cypher−/− embryos at the same stages of embryonic development (Figure S2). Consistent with our hypothesis that a functional overlap exists between Cypher and ENH during embryonic development, we did not observe any phenotypic abnormalities in Cypher−/− knockout embryos when compared to their wild-type littermates at E9.5 to E10.5.
Figure 2.
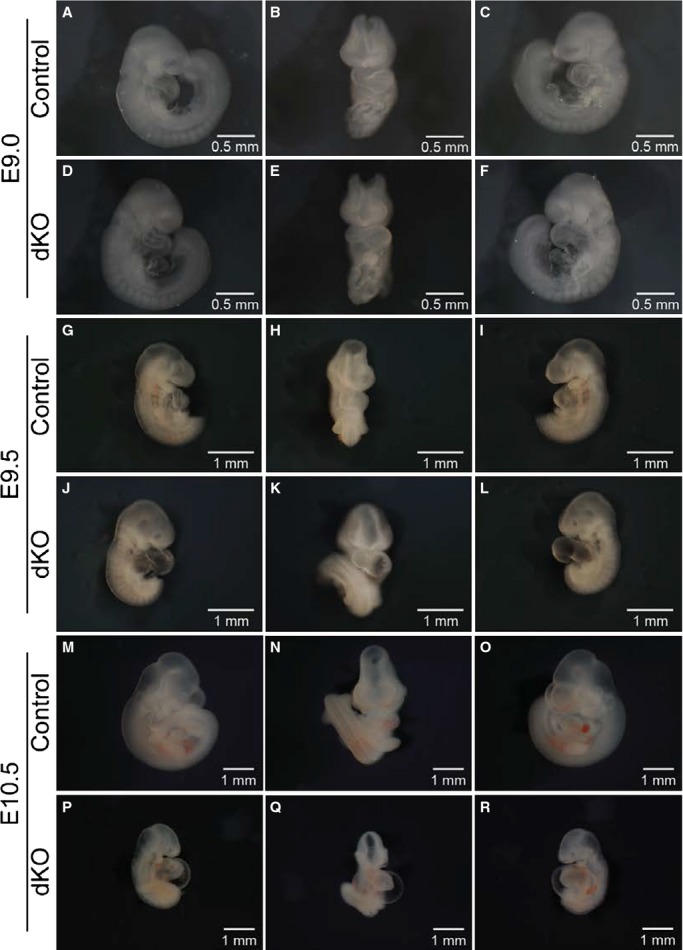
ENH−/−/Cypher−/− embryos display aberrant embryonic development and cardiac morphogenesis at E9.5 to E10.5. Whole-mount microscopic assessment of control and ENH−/−/Cypher−/− dKO embryos (right lateral, anterior, and left lateral views) at different developmental stages. A through C, Control embryo and (D through F), somite-matched ENH−/−/Cypher−/− dKO embryo at E9.0. G through I, Control embryo and (J through L), somite-matched dKO embryo at E9.5. M through O, Control embryo and (P through R), dKO embryo at E10.5. dKO indicates double knockout; ENH, Enigma homolog protein.
To further investigate the phenotype of ENH and Cypher dKO embryos, heart sections of embryos at E9.5 were analyzed by hematoxylin and eosin staining (Figure 3). Consistent with the observed whole-mount morphological changes, histological analysis demonstrated severe dilation of the heart chambers. The compact zone in the ENH−/−/Cypher−/− embryos was considerably thinner and appeared to lack well-defined trabeculations in comparison to control embryos at the same stage of development. There were also marked changes in the development of the outflow tract and the atrioventricular canal in the dKO embryos.
Figure 3.
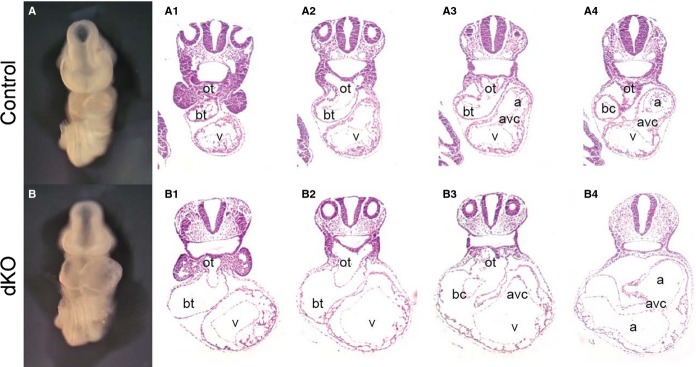
ENH−/−/Cypher−/− embryos display severe cardiac morphological abnormalities at E9.5. Whole-mount (anterior view) and histological H&E staining analysis of consecutive (1 to 4) transverse sections of the heart of a (A) control embryo, and (B) a somite-matched ENH−/−/Cypher−/− dKO embryo at E9.5. a indicates atrium; avc, atrioventricular canal; bc, bulbus-cordis; bt, bulbus-truncus; dKO, double knockout; ENH, Enigma homolog protein; H&E, hematoxylin and eosin; ot, outflow tract; v, ventricle.
To determine whether the aberrant thinning of the myocardium observed in dKO embryos was a result of increased cardiomyocyte apoptosis or decreased cardiomyocyte proliferation, confocal immunofluorescence microscopy using antibodies to cleaved caspase 3 (cCASP3), an apoptotic marker, and a marker of mitosis, phospho-histone H3 (pHH3) was performed (Figure S3). No abnormalities or differences in cardiomyocyte apoptosis or proliferation were detected between mutant and control embryos.
ENH−/−/Cypher−/− Cardiac Muscle Displays Disorganized Z-Lines
Cypher13,14 and ENH21,27 have both been shown to localize to the Z-line. Moreover, Cypher−/− and ENH−/− mutant mouse hearts display disorganized Z-lines, although this phenotype is not directly linked to their lethality.13,14,21 Since deletion of several sarcomeric proteins has been shown to cause aberrant sarcomere structure leading to embryonic lethality,28–30 we speculated that double ablation of Cypher and ENH would result in sarcomere abnormalities. To investigate, we performed immunostaining analyses of E9.0 to E9.5 cardiac muscle for sarcomeric α-actinin, and myomesin, an M-line protein that is associated with thick filaments (Figure 4A through 4D). ENH−/−/Cypher−/− mice showed severely disorganized and fragmented Z-lines in cardiac muscle, illustrated by the absence of α-actinin striations (Figure 4A and 4B). However, no detectable defects in M-line structure were observed in dKO hearts when compared with control littermate hearts (Figure 4C and 4D). To further examine the structure of the Z-line in the hearts of dKO mice at E9.5, transmission electron microscopy was performed (Figure 4E and 4F). Consistently, Z-lines in ENH−/−/Cypher−/− cardiac muscle were poorly organized with loosely arranged myofibrils between the 2 Z-lines when compared with controls. These data showed that deletion of ENH and Cypher together impaired the compact and ordered features of the Z-line structure.
Figure 4.
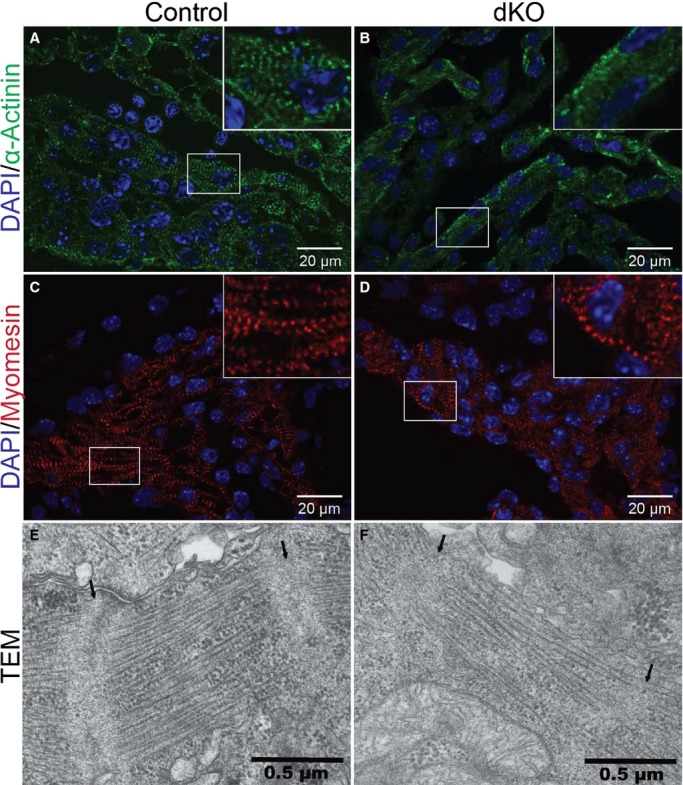
Disorganized Z-lines in ENH−/−/Cypher−/− mouse cardiac muscle. Immunofluorescence analysis of a transverse section of the heart of (A and C) control and (B and D) somite-matched ENH−/−/Cypher−/− dKO embryos at E9.0 to E9.5. Z- and M-lines were stained using antibodies against (A and B) α-actinin, and (C and D) myomesin, respectively. DNA is stained with DAPI (blue). High-magnification views of boxed areas are shown in the inset. E and F, Representative TEM images of a transverse section of the heart of (E) control and (F) somite-matched dKO embryos at E9.5. Z-lines are indicated by arrows. dKO indicates double knockout; DAPI, 4’,6-diamidino-2-phenylindole; ENH, Enigma homolog protein; TEM, transmission electron microscopy.
ENH−/−/CypherL−/− Mutants Die Before Birth While ENH−/−/CypherS−/− Mutants Survive Postnatally
We have previously shown that Cypher’s long (CypherL) and short (CypherS) isoforms have distinct roles in the regulation of cardiac structure and function.22 Selective ablation of CypherL resulted in partial neonatal lethality. Surviving CypherL knockout (CypherL−/−) mice displayed defects in Z-line ultrastructure, cardiac fibrosis, calcification, and developed late-onset dilated cardiomyopathy leading to premature adult mortality. In contrast, mice deficient in CypherS (CypherS−/−) were viable and had no phenotypic abnormalities.22 To investigate potential unique and redundant roles of Cypher isoforms and ENH in cardiac development, we crossed CypherL−/− or CypherS−/− mice with ENH−/− mice. ENH−/−/CypherS−/− mutants displayed no embryonic lethality and survived to adulthood (Table2), while ENH−/−/CypherL−/− mutants were embryonic lethal, with no ENH−/−/CypherL−/− embryos surviving past E12.5 (Table3). The observed phenotypes suggest that while Cypher’s short isoforms are dispensable, in the absence of ENH, the long isoforms of Cypher are indispensable for normal cardiac function. Thus, ENH and CypherL have redundant roles in heart development.
Table 2.
Genotypes of Offspring From ENH−/−/CypherS+/− Intercrosses at Developmental Stages Embryonic Day (E)8.5 to 21 dpn, Show Expected Mendelian Ratios
| Genotype | Developmental Stage | Expected | |||||
|---|---|---|---|---|---|---|---|
| E8.5 | E9.5 | E10.5 | E11.5 | E12.5 | 21 dpn | ||
| ENH−/−/CypherS+/+ | 26.9% (n=7) | 22.2% (n=6) | 23.8% (n=5) | 30% (n=6) | 16.7% (n=3) | 26% (n=31) | 25% |
| ENH−/−/CypherS+/− | 53.8% (n=14) | 55.5% (n=15) | 52.3% (n=11) | 50% (n=10) | 55.6% (n=10) | 51.3% (n=61) | 50% |
| ENH−/−/CypherS−/− | 19.2% (n=5) | 22.2% (n=6) | 23.8% (n=5) | 20% (n=4) | 27.8% (n=5) | 22.7% (n=27) | 25% |
dpn indicates days postnatal; ENH, Enigma homolog protein.
Table 3.
Genotypes of Offspring From ENH−/−/CypherL+/− Intercrosses at Developmental Stages Embryonic Day (E)8.5 to 21 dpn, Show Embryonic Lethality of ENH−/−/CypherL−/− Double Knockout Mice at E12.5
| Genotype | Developmental Stage | Expected | ||||||
|---|---|---|---|---|---|---|---|---|
| E8.5 | E9.5 | E10.5 | E11.5 | E12.5 | E14.5 | 21 dpn | ||
| ENH−/−/CypherL+/+ | 28.6% (n=8) | 27.3% (n=6) | 24% (n=6) | 29% (n=9) | 31.8% (n=7) | 29.4% (n=5) | 31% (n=18) | 25% |
| ENH−/−/CypherL+/− | 53.6% (n=15) | 45.4% (n=10) | 56% (n=14) | 48.3% (n=15) | 68.2% (n=15) | 70.6% (n=12) | 69% (n=40) | 50% |
| ENH−/−/CypherL−/− | 17.8% (n=5) | 27.3% (n=6) | 20% (n=5) | 22.5% (n=7) | 0% (n=0) | 0% (n=0) | 0% (n=0) | 25% |
dpn indicates days postnatal; ENH, Enigma homolog protein.
Discussion
The dramatically dilated heart at E9.5, and severe pericardial effusion at E10.5 indicated that ENH−/−/Cypher−/− embryos died of heart failure. Since Cypher and ENH are highly, and almost exclusively, expressed in myocardium at and before dKO embryos displayed abnormal cardiac morphogenesis phenotypes, we concluded that lethality in dKO embryos resulted from loss of both Cypher and ENH in cardiomyocytes.
We have shown previously that neither Cypher−/− nor ENH−/− mice display obvious cardiac developmental defects. Data presented in this report clearly demonstrated that deletion of both Cypher and ENH leads to embryonic lethality with severe cardiac defects, demonstrating that Cypher and ENH play redundant roles in mouse heart development. Our data also demonstrated that only Cypher long isoforms, but not short isoforms, are redundant with ENH to effect normal cardiac development. However, Cypher short isoforms can partially compensate for the loss of the long isoforms, as ENH−/−/CypherL−/− embryos die 1 to 2 days later than ENH−/−/Cypher−/− embryos.
We have shown previously, by analyzing Cypher−/− mice, that Cypher does not play a role in recruiting α-actinin to the Z-line, or striated muscle sarcomerogenesis, but is required for maintaining Z-line structure during muscle contraction.14 It has been reported by 1 group that dZasp, the only member of ALP/Enigma family in Drosophila, is required for recruitment of α-actinin to the Z-line and sarcomere assembly.31 However, 2 other groups reported that dZASP is not essential for recruiting α-actinin to the Z-line, or for sarcomere assembly.32,33 Consistent with the latter observations, our immunostaining data from E9.0 to 9.5 dKO embryos clearly demonstrated normal M-line structure in dKO hearts when compared with control littermate hearts and that α-actinin was still present at the Z-line, albeit not as well organized as in controls. Collectively, our data demonstrated that Cypher and ENH are not required for murine sarcomerogenesis. Cypher and ENH, however, redundantly play an essential role in sustaining Z-line structure from the earliest stages of cardiac function, and are redundantly required to maintain normal embryonic heart function and embryonic viability.
Sources of Funding
Chen is funded by grants from the National Institute of Arthritis and Musculoskeletal and Skin (R01AR059334) and the National Heart, Lung, and Blood Institute. Peter is funded by the National Institutes of Health Training in Cardio-vascular Physiology and Pharmacology Training Grant (5T32HL007444-27). Lange is funded by a National Institutes of Health grant (K99HL107744-01). Ouyang is supported by the National Key Basic Research Program of China (2013CB531200), and the National Science Foundation of China (31370823, 91439130).
Disclosures
None.
Supporting Information
ENH expression at E8.5 and E10.5. Whole-mount RNA in situ hybridization analysis of ENH expression in a wild-type embryo (right and left lateral views) at (A and B) E8.5 and (C and D) E10.5 using an ENH-specific probe.
Figure S2. Cypher ablation does not affect early heart development. Whole-mount microscopic assessment of wild-type and Cypher-null (Cypher−/−) embryos (left lateral view) at different developmental stages. A, Wild-type embryo and (B) somite-matched Cypher−/− embryo at E9.5. C, Wild-type embryo and (D) somite-matched Cypher−/− embryo at E10.5.
Figure S3. ENH−/−/Cypher−/− hearts display no abnormalities in cardiomyocyte apoptosis or proliferation. Immunofluorescence analysis of a transverse cryosection of the heart of (A and C) control and (B and D) somite-matched ENH−/−/Cypher−/− double knockout (dKO) embryos at E9.0 to E9.5. Apoptosis and proliferation were stained using antibodies against (A and B) cleaved caspase 3, cCASP3 (green), and (C and D) phospho-histone H3, pHH3 (green), respectively. Sarcomeres were stained with anti-α-actinin (red); DNA is stained with DAPI (blue). Note: due to thinning of the myocardium and severe dilation of the heart chamber, dKO hearts shrank during sucrose processing.
References
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Frank D, Kuhn C, Katus HA, Frey N. Role of the sarcomeric Z-disc in the pathogenesis of cardiomyopathy. Future Cardiol. 2007;3:611–622. doi: 10.2217/14796678.3.6.611. [DOI] [PubMed] [Google Scholar]
- Chen J, Chien KR. Complexity in simplicity: monogenic disorders and complex cardiomyopathies. J Clin Invest. 1999;103:1483–1485. doi: 10.1172/JCI7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- Kruger M, Linke WA. Titin-based mechanical signalling in normal and failing myocardium. J Mol Cell Cardiol. 2009;46:490–498. doi: 10.1016/j.yjmcc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Sanger JM, Sanger JW. The dynamic Z bands of striated muscle cells. Sci Signal. 2008;1:pe37. doi: 10.1126/scisignal.132pe37. [DOI] [PubMed] [Google Scholar]
- Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290:H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Bang ML, Lange S, Chen J. “Z”eroing in on the role of Cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc Med. 2007;17:258–262. doi: 10.1016/j.tcm.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Cheng H, Banerjee I, Chen J. ALP/Enigma PDZ-LIM domain proteins in the heart. J Mol Cell Biol. 2010;2:96–102. doi: 10.1093/jmcb/mjp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ruiz-Lozano P, Martone ME, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, Valle G, Lanfranchi G. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999;146:465–475. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Cheng H, Li X, Zhang J, Cui L, Ouyang K, Han L, Zhao T, Gu Y, Dalton ND, Bang ML, Peterson KL, Chen J. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum Mol Genet. 2009;18:701–713. doi: 10.1093/hmg/ddn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, Knoll G, Shelton GD, Evans S, Chen J. Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J Cell Biol. 2001;155:605–612. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer DL, Marques IJ, Leito JT, Besser J, Bakkers J, Schoonheere E, Bagowski CP. Zebrafish Cypher is important for somite formation and heart development. Dev Biol. 2006;299:356–372. doi: 10.1016/j.ydbio.2006.07.032. [DOI] [PubMed] [Google Scholar]
- Griggs R, Vihola A, Hackman P, Talvinen K, Haravuori H, Faulkner G, Eymard B, Richard I, Selcen D, Engel A, Carpen O, Udd B. Zaspopathy in a large classic late-onset distal myopathy family. Brain. 2007;130:1477–1484. doi: 10.1093/brain/awm006. [DOI] [PubMed] [Google Scholar]
- Strach K, Reimann J, Thomas D, Naehle CP, Kress W, Kornblum C. ZASPopathy with childhood-onset distal myopathy. J Neurol. 2012;259:1494–1496. doi: 10.1007/s00415-012-6543-1. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- Ueki N, Seki N, Yano K, Masuho Y, Saito T, Muramatsu M. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat enigma homologue (ENH) J Hum Genet. 1999;44:256–260. doi: 10.1007/s100380050155. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Walchli S, Fujita T, Ryser S, Hoshijima M, Schlegel W, Kuroda S, Maturana AD. Splice variants of enigma homolog, differentially expressed during heart development, promote or prevent hypertrophy. Cardiovasc Res. 2010;86:374–382. doi: 10.1093/cvr/cvq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Kimura K, Peter AK, Cui L, Ouyang K, Shen T, Liu Y, Gu Y, Dalton ND, Evans SM, Knowlton KU, Peterson KL, Chen J. Loss of enigma homolog protein results in dilated cardiomyopathy. Circ Res. 2010;107:348–356. doi: 10.1161/CIRCRESAHA.110.218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Zheng M, Peter AK, Kimura K, Li X, Ouyang K, Shen T, Cui L, Frank D, Dalton ND, Gu Y, Frey N, Peterson KL, Evans SM, Knowlton KU, Sheikh F, Chen J. Selective deletion of long but not short Cypher isoforms leads to late-onset dilated cardiomyopathy. Hum Mol Genet. 2011;20:1751–1762. doi: 10.1093/hmg/ddr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci USA. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhou Q, Liang P, Hollander MS, Sheikh F, Li X, Greaser M, Shelton GD, Evans S, Chen J. Characterization and in vivo functional analysis of splice variants of Cypher. J Biol Chem. 2003;278:7360–7365. doi: 10.1074/jbc.M211875200. [DOI] [PubMed] [Google Scholar]
- Liang X, Sun Y, Schneider J, Ding JH, Cheng H, Ye M, Bhattacharya S, Rearden A, Evans S, Chen J. Pinch1 is required for normal development of cranial and cardiac neural crest-derived structures. Circ Res. 2007;100:527–535. doi: 10.1161/01.RES.0000259041.37059.8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou Q, Li X, Sun Y, Lu M, Dalton N, Ross J, Jr, Chen J. PINCH1 plays an essential role in early murine embryonic development but is dispensable in ventricular cardiomyocytes. Mol Cell Biol. 2005;25:3056–3062. doi: 10.1128/MCB.25.8.3056-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun. 2000;272:505–512. doi: 10.1006/bbrc.2000.2787. [DOI] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, Ross J, Jr, Chien KR. Selective requirement of myosin light chain 2v in embryonic heart function. J Biol Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- Fritz-Six KL, Cox PR, Fischer RS, Xu B, Gregorio CC, Zoghbi HY, Fowler VM. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. J Cell Biol. 2003;163:1033–1044. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert S, Bergmann N, Luo X, Erdmann B, Gotthardt M. M line-deficient titin causes cardiac lethality through impaired maturation of the sarcomere. J Cell Biol. 2006;173:559–570. doi: 10.1083/jcb.200601014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani K, Schock F. Zasp is required for the assembly of functional integrin adhesion sites. J Cell Biol. 2007;179:1583–1597. doi: 10.1083/jcb.200707045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benna C, Peron S, Rizzo G, Faulkner G, Megighian A, Perini G, Tognon G, Valle G, Reggiani C, Costa R, Zordan MA. Post-transcriptional silencing of the Drosophila homolog of human ZASP: a molecular and functional analysis. Cell Tissue Res. 2009;337:463–476. doi: 10.1007/s00441-009-0813-y. [DOI] [PubMed] [Google Scholar]
- Rui Y, Bai J, Perrimon N. Sarcomere formation occurs by the assembly of multiple latent protein complexes. PLoS Genet. 2010;6:e1001208. doi: 10.1371/journal.pgen.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ENH expression at E8.5 and E10.5. Whole-mount RNA in situ hybridization analysis of ENH expression in a wild-type embryo (right and left lateral views) at (A and B) E8.5 and (C and D) E10.5 using an ENH-specific probe.
Figure S2. Cypher ablation does not affect early heart development. Whole-mount microscopic assessment of wild-type and Cypher-null (Cypher−/−) embryos (left lateral view) at different developmental stages. A, Wild-type embryo and (B) somite-matched Cypher−/− embryo at E9.5. C, Wild-type embryo and (D) somite-matched Cypher−/− embryo at E10.5.
Figure S3. ENH−/−/Cypher−/− hearts display no abnormalities in cardiomyocyte apoptosis or proliferation. Immunofluorescence analysis of a transverse cryosection of the heart of (A and C) control and (B and D) somite-matched ENH−/−/Cypher−/− double knockout (dKO) embryos at E9.0 to E9.5. Apoptosis and proliferation were stained using antibodies against (A and B) cleaved caspase 3, cCASP3 (green), and (C and D) phospho-histone H3, pHH3 (green), respectively. Sarcomeres were stained with anti-α-actinin (red); DNA is stained with DAPI (blue). Note: due to thinning of the myocardium and severe dilation of the heart chamber, dKO hearts shrank during sucrose processing.


