Abstract
Background
Heme is an essential iron-containing molecule for cardiovascular physiology, but in excess it may increase oxidative stress. Failing human hearts have increased heme levels, with upregulation of the rate-limiting enzyme in heme synthesis, δ-aminolevulinic acid synthase 2 (ALAS2), which is normally not expressed in cardiomyocytes. We hypothesized that increased heme accumulation (through cardiac overexpression of ALAS2) leads to increased oxidative stress and cell death in the heart.
Methods and Results
We first showed that ALAS2 and heme levels are increased in the hearts of mice subjected to coronary ligation. To determine the causative role of increased heme in the development of heart failure, we generated transgenic mice with cardiac-specific overexpression of ALAS2. While ALAS2 transgenic mice have normal cardiac function at baseline, their hearts display increased heme content, higher oxidative stress, exacerbated cell death, and worsened cardiac function after coronary ligation compared to nontransgenic littermates. We confirmed in cultured cardiomyoblasts that the increased oxidative stress and cell death observed with ALAS2 overexpression is mediated by increased heme accumulation. Furthermore, knockdown of ALAS2 in cultured cardiomyoblasts exposed to hypoxia reversed the increases in heme content and cell death. Administration of the mitochondrial antioxidant MitoTempo to ALAS2-overexpressing cardiomyoblasts normalized the elevated oxidative stress and cell death levels to baseline, indicating that the effects of increased ALAS2 and heme are through elevated mitochondrial oxidative stress. The clinical relevance of these findings was supported by the finding of increased ALAS2 induction and heme accumulation in failing human hearts from patients with ischemic cardiomyopathy compared to nonischemic cardiomyopathy.
Conclusions
Heme accumulation is detrimental to cardiac function under ischemic conditions, and reducing heme in the heart may be a novel approach for protection against the development of heart failure.
Keywords: apoptosis, heart failure, iron, metabolism, myocardial infarction
The high incidence of heart failure in the developed world makes it an important healthcare problem, and its rapidly growing prevalence underpins its urgency.1 Current medical therapies, which are commonly aimed at suppressing neurohormonal activation, are effective in managing heart failure but offer no cure for the disease.2 Therefore, novel approaches are needed to improve cardiac performance and prevent the progressive cardiac dysfunction that characterizes this disease. The failing heart undergoes significant metabolic changes, and therefore modifying metabolic gene expression could improve clinical outcomes in heart failure patients.3–5
Recently, there has been significant interest in the role of iron in heart failure. Iron is essential for the maintenance of cellular viability and function through its roles in oxidative phosphorylation, antioxidant enzyme activities, ribosome biogenesis, and oxygen storage and delivery.6 The majority of iron in the heart exists in the forms of heme and iron–sulfur (Fe/S) cluster proteins. While it has been suggested that intravenous iron may provide clinical benefit in iron-deficient patients with chronic systolic heart failure,7–10 free iron is a highly reactive metal that can catalyze the production of toxic hydroxyl radicals from less reactive species, such as hydrogen peroxide and superoxide anion, via the Fenton reaction.11 Several lines of evidence suggest that cardiac dysfunction is a prominent feature of iron-overload diseases. For example, hereditary hemochromatosis leads to the development of cardiomyopathy,12 and patients with Friedrich’s ataxia develop progressive and lethal cardiomyopathy that is characterized by mitochondrial accumulation of iron and extensive oxidative damage.13,14 Furthermore, cardiomyopathy in thalassemia major patients with iron overload due to regular blood transfusions is attributed to reactive oxygen species (ROS) generation due to intracardiac iron accumulation.15–17 In addition, mice with cardiac-specific overexpression of the alpha subunit of the Gq protein develop cardiomyopathy and significant cardiac accumulation of iron.18 Finally, our lab has shown that mice with genetic deletion of a mitochondrial iron export protein display mitochondrial iron accumulation and cardiomyopathy.19 Therefore, maintenance of iron and iron-containing compounds within the heart is critical for cardiac function.
Heme is an essential iron-containing metabolite for aerobic organisms and serves as a prosthetic group for hemoproteins involved in numerous cardiovascular processes, including oxygen transport (hemoglobin), oxygen storage (myoglobin), oxygen metabolism (oxidases), anti-oxidation (peroxidases, catalases), electron transport (cytochromes), signaling (guanylate cyclase), and enzyme catalysis (cyclo-oxygenase, nitric oxide synthase).20–23 The function of heme-containing proteins strongly depends on the ability of heme to coordinate an iron atom inside its structure and facilitate reduction–oxidation (redox) reactions.24 However, this biochemical property of iron also enables free heme (heme not bound to protein) to generate toxic hydroxyl radicals through the Fenton reaction and cause cellular injury.11,25
Heme production begins in the mitochondria with the condensation of glycine and succinyl coenzyme A to form δ-aminolevulinic acid (ALA) by the rate-limiting enzyme ALA synthase (ALAS). Two isoforms of ALAS exist; ALAS1 is a ubiquitously expressed housekeeping gene, while ALAS2 is traditionally studied in the context of erythrocyte maturation, where it is induced to stimulate hemoglobinization. The next 5 conversions are carried out in the cytosol. Finally, the synthesis is completed in the mitochondria with the insertion of an iron atom into protoporphyrin IX (PPIX) to form heme by ferrochelatase.26 Mature heme can then be incorporated into various mitochondrial proteins or exported into the cytoplasm.27
Recently, a metabolomic analysis revealed that heme is increased in the hearts of mice after permanent coronary ligation, suggesting that myocardial accumulation of free heme may play a role in hypoxic injury and the development of heart failure.28 In addition, our lab has shown that both cytosolic and mitochondrial heme levels are increased in failing human hearts, with feedback inhibition of the heme synthetic enzymes except for ALAS2.29 Upregulation of ALAS2 in failing human hearts is significant because (1) ALAS2 is the rate-limiting enzyme in heme synthesis and could account for the increased heme content in failing hearts, and (2) ALAS2 expression was previously reported to be restricted to the hematopoietic system.30 Unlike other components of the heme synthesis pathway, ALAS2 is not inhibited by high levels of heme and is positively regulated by hypoxia.31 We have also previously shown that ALAS2 overexpression in cultured cardiomyoblasts leads to increased heme content, representing the first heart-specific function for this “noncardiac” isoform of ALAS.29 However, it is not known whether the increased heme in heart failure is an adaptive or maladaptive process.
To further characterize the role of heme in the heart and to determine whether an increase in the rate-limiting enzyme in heme synthesis, ALAS2, is sufficient to induce cardiac injury, we generated a cardiac-specific ALAS2-overexpressing transgenic (TG) mouse model and studied the effects of its expression on cardiac function and heme homeostasis at baseline and in heart failure. We show that while ALAS2 TG mice have normal cardiac function at baseline, their hearts display increased heme content, higher oxidative stress, exacerbated cell death, and worsened cardiac function after coronary ligation compared to control animals. The normal cardiac function of TG mice at baseline can be attributed to a compensatory increase in heme degradation via heme oxygenase-1 (HO-1), which is attenuated in TG mice after coronary ligation. We confirm that heme accumulation is responsible for the increased levels of oxidative stress and cell death seen with ALAS2 overexpression in cultured cardiomyoblasts. Furthermore, knockdown of ALAS2 in cultured cardiomyoblasts exposed to hypoxia normalizes the increases in heme content and cell death. We also show that human hearts from patients with ischemic cardiomyopathy have increased ALAS2 levels and heme accumulation compared to patients with nonischemic cardiomyopathy. Finally, administration of the mitochondrial antioxidant MitoTempo to ALAS2-overexpressing cardiomyoblasts returns the heightened levels of oxidative stress and cell death to baseline, indicating that mitochondrial oxidative stress mediates the detrimental effects of heme accumulation due to increased ALAS2 expression.
Methods
Generation of ALAS2 TG Mice
Human ALAS2 cDNA was obtained from OpenBioSystems. The coding sequence of ALAS2 cDNA was amplified using polymerase chain reaction (PCR) with the addition of a 3′ FLAG tag. The PCR product was first cloned into pCR2.1 TOPO vector (Invitrogen) and the sequence was verified by sequencing. The entire coding region was then excised by SalI and HindIII and subcloned into a transgenic plasmid containing the α-myosin heavy chain (MHC) promoter and human growth hormone poly(A) sequence (provided by Dr Jeffrey Robbins; Cincinnati Children’s Hospital, Cincinnati, OH). The plasmid was purified using the Qiagen EndoFree Plasmid Maxi Kit. The purified plasmid was digested with NotI (which flanks the region containing the transgene), and the fragment was gel isolated. The microinjections of the transgene were performed by the Northwestern Transgenic and Targeted Mutagenesis Core Facility, as described previously.32 Potential founder mice (which grew from the injected zygotes) were screened for insertion of the transgene by PCR analysis after DNA was obtained from their tail tip. A stable TG line expressing a high level of ALAS2 in the heart with intact fertility was selected for subsequent breeding. This founder mouse was backcrossed to C57BL/6 mice. Tissue-specific expression patterns and the level of expression were determined by Western blot analysis and quantitative real-time polymerase chain reaction (qRT-PCR). For all studies, age- and gender-matched littermates were used as controls. A total of ≈25 TG and 25 non-TG mice were used for our studies.
Coronary Ligation
The surgical protocol was performed as previously described.33 Briefly, mice were anesthetized with isoflurane with induction at 3% and maintenance at 1.5% to 2%. The animals were placed in a supine position and ECG leads were attached. The body temperature was monitored using a rectal probe and was maintained at 37°C with heating pads throughout the experiment. A catheter was inserted into the trachea and was then attached to the mouse ventilator via a Y-shaped connector. The mice were ventilated at a tidal volume of 200 μL and a rate of 105 breaths/min using a rodent ventilator. The chest was then opened by an incision of the left fourth intercostal space. The left anterior descending artery was occluded with an 8–0 silk suture. Ischemia was confirmed by pallor of the anterior wall of the left ventricle and by ST-segment elevation and QRS widening on the ECG. After confirmation of ligation, the chest was closed in layers. The mice were kept warm with heating pads and on 100% oxygen via nasal cannula. Animals were given buprenorphine for postoperative pain. All animal procedures were followed in accordance with institutional guidelines.
Echocardiography
Echocardiography was performed using a Vevo 770 High-Resolution Imaging System. Animals were anesthetized with isoflurane and placed in the supine position. The chests were shaved, and the parasternal short- and long-axis views were used to obtain 2-dimensional and M-mode images. At least 10 independent cardiac cycles per each experiment were obtained.
Histochemical Analysis
Heart tissue was excised, rinsed in phosphate-buffered saline (PBS), and perfused with Hanks buffered saline solution using a Langerdorff working heart perfusion system. Following perfusion, tissue was fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. To determine cardiac fiber morphology, sections were stained with hematoxylin and eosin according to standard protocols. Heart fibrosis was assessed by Masson trichrome staining.
For assessment of area at risk (AAR), mice were subjected to coronary ligation for 24 hours and anesthetized with an intraperitoneal injection of avertin. The hearts were excised, cannulated, and washed with PBS, followed by injection of 500 μL of 1% Evans blue into the right ventricle. Left ventricles were excised and frozen at −20°C for 15 minutes to prevent smearing of dye into unstained tissues, sectioned transversely into 5 sections, and incubated in 10% neutral-buffered formaldehyde for 24 hours. Sections were weighed and photographed using a digital camera (Sony), then analyzed using ImageJ (National Institutes of Health, Bethesda, MD). Viable myocardium stained blue while AAR was unstained. The size of AAR was determined by the following equations: weight of AAR=(A1×W1)+(A2×W2)+(A3×W3)+(A4×W4)+(A5×W5), where A is the percent area of AAR by planimetry and W is the weight of each section; AAR as a percentage of left ventricle (LV)=[(weight of AAR)/(weight of LV)]×100.
Lipid Peroxidation Assay
Malondialdehyde and 4-hydroxyalkenals concentrations were determined spectrophotometrically using a commercially available Lipid Peroxidation Microplate Assay Kit (Oxford Biomedical Research). This assay is based on the reaction of a chromogenic reagent, N-methyl-2-phenylindole, with malondialdehyde and 4-hydroxyalkenals, yielding a stable chromophore with maximal absorbance at 586 nm. Freshly isolated and PBS-rinsed heart tissue (≈10 mg) was used for the assay.
Mitochondrial DNA Damage Assay
Genomic DNA was isolated from the cardiac AAR of mice sacrificed 28 days after sham thoracotomy or myocardial infarction using the GeneJet Genomic DNA Purification Kit (Thermo Scientific). Primers were designed using Primer3 (version 0.4.0) software to target a 9-kb mitochondrial DNA sequence (long fragment) and ≈100 base sequence of the mitochondrially encoded Cox2 gene (short fragment). DNA levels were amplified separately on a 7500 Fast Real-Time PCR system with PerfeCTa SYBR Green SuperMix (Quanta). The PCR conditions for the different fragments were optimized to achieve similar amplification efficiencies, with product specificity monitored by dissociation curve analysis. Relative mitochondrial DNA levels of the long fragment were calculated by the comparative threshold cycle method and normalized to the short fragment. Mitochondrial DNA lesion rate is expressed as the inverse of the relative levels of long-to-short mitochondrial DNA levels.
Human Samples
Nonfailing and failing human heart samples were obtained from the Human Heart Tissue Collection at the Cleveland Clinic. Failing human hearts consisted of ischemic and nonischemic (dilated, postpartum) cardiomyopathy samples. A total of 4 nonfailing, 4 ischemic failing, and 8 nonischemic failing human heart samples were used in the study. Informed consent was obtained from all the transplant patients and from the families of the organ donors before tissue collection. Protocols for tissue procurement were approved by the Institutional Review Board of the Cleveland Clinic (Cleveland, OH), which is Association for the Accreditation of Human Research Protection Programs (AAHRPP) accredited, and subjects gave informed consent.
Cell Culture
H9c2 cardiac myoblasts were purchased from ATCC and kept in complete Dulbecco’s Modified Eagle Medium (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (Invitrogen, Grand Island, NY) and 1% penicillin—streptomycin. For hypoxic experiments, cells were maintained in a hypoxic chamber at 37°C and 5% carbon dioxide in the presence of 1% oxygen for up to 4 days. Medium was replaced every 2 days, and cells were collected under hypoxia before analysis. For pharmacological treatments, cells were grown to 80% to 90% confluence and incubated with indicated doses of hemin (Sigma-Aldrich, St. Louis, MO) in complete medium for 6 hours, ALA (Sigma) in complete medium for 8 hours, or 25 μmol/L MitoTempo (Sigma) in serum-free media for 2 hours before induction of cell death by H2O2 or for the last 6 hours of ALAS1/2 modulation before mitochondrial ROS quantification.
Knockdown and Overexpression of ALAS2
For knockdown experiments, ALAS2 or control nontargeting siRNA (Dharmacon) were transfected into H9c2 cells using Dharmafect I reagent (Thermo Scientific, Hanover Park, IL) for 48 hours according to the manufacturer’s protocol. For hypoxia experiments, transfections were repeated every 48 hours to maintain low levels of ALAS2 expression throughout the study. ALAS2 overexpression was achieved through lentiviral transduction. Lentiviral particles coding for ALAS2-GFP (green fluorescent protein) fusion protein or green fluorescent protein-only control were transduced into H9c2 cells at equal multiplicities of infection for 48 hours. Overexpression of ALAS2 was confirmed by Western blot analysis.
Mitochondrial Fractionation
A Mitochondrial Isolation Kit for Tissue or Cultured Cells (Pierce) was used to purify the mitochondrial fraction according to the manufacturer’s protocol. To maintain the integrity of the mitochondria, samples were kept on ice during the isolation process. Following differential centrifugation at 4°C, the supernatant (cytosolic fraction) was collected, and the remaining pellet containing the mitochondria-enriched fraction was dissolved in 1% Triton X-100 (Sigma-Aldrich) in Tris-buffered saline (Cellgro, Manassas, VA).
Nonheme Measurement
Mitochondrial and cytosolic fractions were isolated using the Mitochondrial Isolation Kit for Tissue or Cultured Cells (Pierce) as described above. Nonheme iron was quantified spectrophotometrically as the formation of the Fe(II)-bathophenanthroline disulfonate complex, as previously described.34
Heme Measurement
For determination of total heme levels, ≈5 mg of freshly frozen tissue was homogenized in 1% Triton X-100 in Tris-buffered saline and centrifuged at 5000g for 10 minutes to remove debris. For determination of mitochondrial and cytosolic heme levels, the mitochondrial fraction was isolated using the Mitochondrial Isolation Kit for Tissue (Pierce) as described above. Protein concentration of cytosolic or mitochondrial lysate was quantified by bicinchoninic acid assay (Pierce), and heme was quantified as previously described.35 Briefly, equal amounts of protein were mixed with 2 mol/L oxalic acid and heated to 95°C for 30 minutes to release iron from heme and generate fluorescent PPIX. Samples were then centrifuged for 10 minutes at 1000g at 4°C to remove debris; the fluorescence of the supernatant was assessed at Ex 405 nm/Em 600 nm on a Spectra Max Gemini fluorescence microplate reader and normalized to the protein concentration of each sample. For determination of unsaturated PPIX levels, incubation and heating with oxalic acid steps were omitted. Instead, samples were diluted in PBS, followed by fluorescence measurement and normalization to protein content.
Quantitative Real-Time PCR
RNA was isolated with RNA STAT-60 (TEL-TEST, Inc, Friendswood, TX), reverse transcribed with qScript cDNA SuperMix (Quanta), and amplified on a 7500 Fast Real-Time PCR system with PerfeCTa SYBR Green SuperMix (Quanta). Primers were designed using Primer3 (version 0.4.0) software to target sequences spanning an exon–intron–exon boundary, and their specificity was confirmed by running a dissociation curve. mRNA levels were calculated by the comparative threshold cycle method and normalized to the β-actin and/or 18S genes.
Western Blot
Heart tissue was excised, rinsed in PBS, and perfused with Hanks buffered saline solution using a Langerdorff heart perfusion system. Approximately 10 mg of perfused tissue was homogenized in RIPA buffer in the presence of Protease Arrest protease inhibitors (G-Biosciences), centrifuged at 5000g for 15 minutes to remove debris, and protein concentration of the supernatant determined by bicinchoninic acid assay. Fifty to 100 μg of protein were loaded on sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes (Invitrogen). The membranes were probed with antibodies against HO-1 (Abcam, Cambridge, MA), ALAS1 (Abcam), ALAS2 (Abcam, Novus Biologicals, Littleton, CO), and GAPDH (Santa Cruz, Dallas, TX). Horseradish peroxidase–conjugated donkey anti-rabbit and donkey anti-mouse were used as secondary antibodies (Santa Cruz) and visualized by Pierce SuperSignal Chemiluminescent substrates.
Mitochondrial ROS Quantification
MitoSox Red (Invitrogen) was used to assess mitochondrial O2· production. Cells were visualized by microscopy and ROS levels were quantified by ImageJ software. Multiple fields per each sample were obtained and averaged. Nuclei were counterstained with Hoescht 33342 dye (Invitrogen) and subtracted from the total MitoSox fluorescence to exclude the signal from localization of dye into the nucleus. An accurate overlay between Hoescht and nuclear MitoSox dye was achieved by adjusting the microscopy settings before data collection.
Cell Death Analysis
Following ALAS1/2 overexpression or silencing, cell death was induced with 200 μmol/L H2O2 in serum-free media for 6 hours. H9c2 cells were then resuspended in 1x annexin-binding buffer, double-labeled with propidium iodine (Sigma-Aldrich) and Alexa Fluor 350-conjugated Annexin V (Molecular Probes, Eugene, OR), and analyzed by flow cytometry in a LSRII flow cytometer (BD Biosciences, San Jose, CA). The data are presented as the sum of apoptotic and necrotic cells normalized to the control. Cultured cardiomyoblast viability was also determined by propidium iodine and Hoescht double-staining after H2O2 treatment. Propidium iodine–positive nuclei were counted under light microscopy and expressed as percentage of the total number of cells, as determined by the sum of propidium iodine and Hoescht-positive cells. For determination of myocardial apoptosis, freshly isolated hearts were perfused with PBS, embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, CA), and gradually frozen in liquid nitrogen. Frozen heart sections were permeabilized in 0.1% Triton X-100 in PBS containing 0.1% sodium citrate for 2 minutes and washed, and the fluorescent terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (Roche) mixture was added for 60 minutes at 37°C in the dark, followed by counterstaining with Alexa Flour 568 phalloidin for 20 minutes and 4′,6-diamidino-2-phenylindole (DAPI) for 10 minutes at room temperature. The number of TUNEL-positive cardiomyocytes was randomly counted in high-power fields on the left ventricular wall around the infarct border zone. The percentage of TUNEL-positive cardiomyocytes was calculated by dividing the number of TUNEL-positive cardiomyocytes by the total number of nuclei. Representative TUNEL and DAPI double-stained images are presented in this study.
Statistical Analysis
Data are expressed as mean±SEM. Statistical significance was assessed with the unpaired Student t test or ANOVA with Tukey’s post hoc test when appropriate. The 24, 48, 72, and 96-hour time-points in Figure 7A were each compared to the 0 hour time-point using the unpaired Student t test. A P value of <0.05 was considered statistically significant.
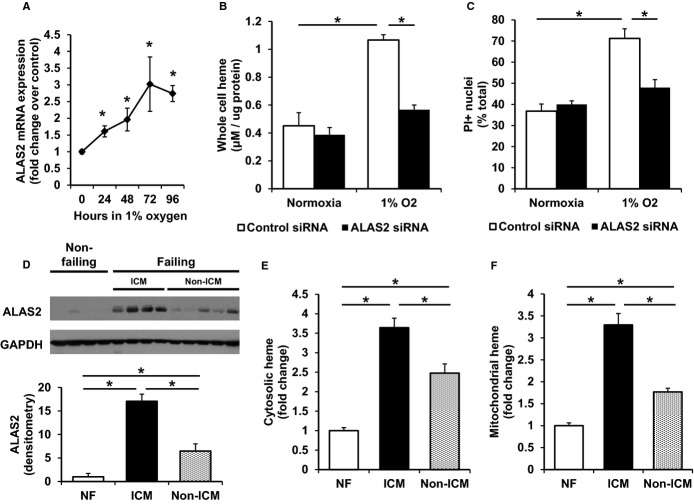
Figure 7.
Knockdown of hypoxia-induced ALAS2 in vitro reverses the increases in heme and cell death. A, ALAS2 mRNA expression is induced in H9c2 cardiac myoblasts subjected to hypoxia (n=4). B, Cellular heme content is increased in H9c2 cardiac myoblasts after 72 hours in hypoxia compared to normoxia, and returns to baseline levels with ALAS2 siRNA treatment (n=5 to 6). C, H2O2-induced cell death is increased in H9c2 cells following 72 hours in hypoxia, and is reduced with ALAS2 silencing RNA (siRNA) treatment, as assessed by PI-positive nuclei (n=3, 3 fields per sample). D, ALAS2 protein is induced in failing human hearts, particularly in patients with ischemic cardiomyopathy (ICM) (n=4 to 5). Densitometry analysis is provided below the Western blot. E, Cytosolic and (F) mitochondrial heme levels are increased in failing human hearts, particularly in patients with ICM (n=4 to 8). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; ICM, ischemic cardiomyopathy; NF, non-failing hearts; PI, propidium iodine.
Results
Genetic Overexpression of ALAS2 in the Heart
While the expression of ALAS2 was previously thought to be restricted to the hematopoietic system, we recently demonstrated its expression in human failing hearts. However, it is not clear whether this expression is a cause or a consequence of heart failure.29 To further characterize the heart-specific functions of this isoform of ALAS and to determine whether the expression of this isoform is sufficient to increase cardiac heme levels and induce cardiac injury, we generated cardiomyocyte-specific ALAS2 TG mice by cloning the human ALAS2 coding sequence downstream of the α-MHC promoter. A founder mouse was selected and backcrossed to C57/B6 mice. The presence of the transgene was confirmed at the level of genomic DNA (Figure 1A), and its expression was verified at the mRNA (Figure 1B) and protein levels (Figure 1C). To minimize detection of ALAS2 from the blood present in the tissue, the hearts were perfused with Hanks buffered saline solution using a Langerdorff heart perfusion system prior to analysis. There was no significant change in ALAS2 expression in other tissues (Figure 1C), and cardiac ALAS1 levels were unchanged between ALAS2 TG and nontransgenic (NTG) mice (Figure S1A).
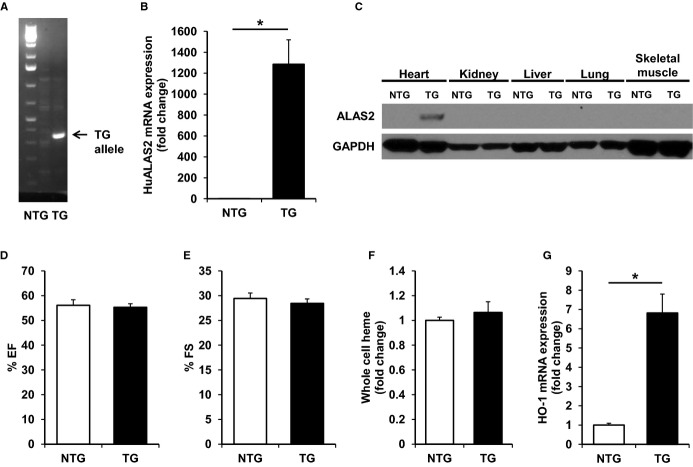
Figure 1.
Cardiac-specific ALAS2 overexpressing mice have normal cardiac function and heme levels at baseline. A, The TG ALAS2 allele is detected in ALAS2 TG mice by DNA genotyping. B, TG human ALAS2 mRNA is expressed in the heart of ALAS2 TG mice as assessed by qRT-PCR (n=5 to 6). C, ALAS2 protein is expressed specifically in the heart (and not other tissues) of ALAS2 TG mice as assessed by Western blot. D, EF and (E) FS are similar in ALAS2 TG and NTG littermates at baseline as assessed by echocardiography (n=4). F, Whole cell heme levels in the hearts of ALAS2 TG and NTG littermates are similar at baseline (n=6). G, The heme degradation enzyme, HO-1, is upregulated in ALAS2 TG mice at the mRNA level compared to control littermates at baseline (n=4). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; EF, ejection fraction; FS, fractional shortening; HO-1, heme oxygenase-1; NTG, nontransgenic; qRT-PCR, quantitative real-time polymerase chain reaction; TG, transgenic.
ALAS2 TG Mice Have Normal Cardiac Function and Heme Levels at Baseline
We first assessed cardiac function in ALAS2 TG and NTG littermates at baseline using echocardiography. ALAS2 TG mice displayed similar ejection fraction and fractional shortening compared to NTG mice (Figure 1D and 1E), suggesting that cardiac-specific ALAS2 overexpression does not alter cardiac function at baseline. There were also no changes in the whole cell (Figure 1F), cytosolic (Figure S1B), or mitochondrial (Figure S1C) levels of heme in hearts from ALAS2 TG and NTG mice. Other than ALAS2, there was no change in the enzymes of heme synthesis in ALAS2 TG and control mice (Figure S1D). However, the mRNA (Figure 1G) and protein levels (Figure S1E) of HO-1, the heme-inducible enzyme that catalyzes the degradation of heme, were significantly upregulated in the hearts of ALAS2 TG compared to NTG mice at baseline. Cardiac mRNA expression of the constitutive isoform HO-2 was unchanged between ALAS2 TG and NTG mice (Figure S1D). These findings suggest that while ALAS2 overexpression may cause increased heme production, it also leads to a compensatory increase in HO-1 expression, which collectively lead to normalized heme levels in the heart and normal cardiac function of ALAS2 TG mice at baseline.
ALAS2 TG Mice Have Reduced Cardiac Function After Coronary Ligation
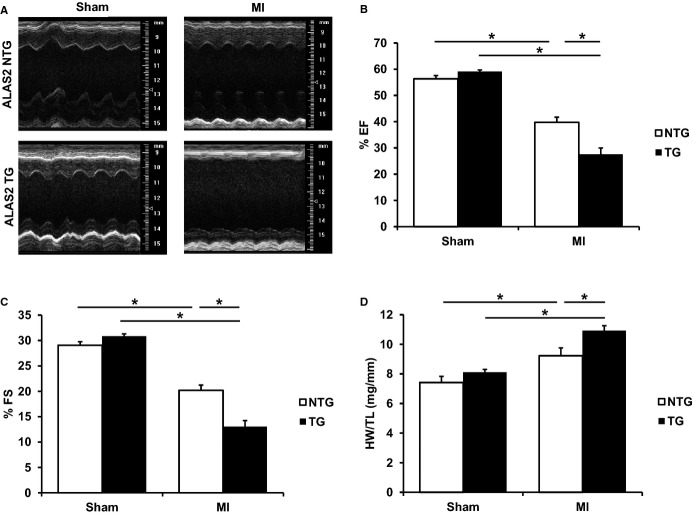
Since we previously reported that ALAS2 expression is increased in failing human hearts, we subjected ALAS2 TG and NTG mice to permanent coronary ligation of the left anterior descending artery to induce heart failure.29 While AAR was similar for ALAS2 TG and NTG mice 24 hours after coronary ligation (Figure S2A), ALAS2 TG mice had significantly reduced cardiac function 28 days after coronary ligation, as assessed by ejection fraction and fractional shortening, compared to NTG mice (Figure 2A through 2C). Murine heart rates were similar among the groups (Figure S2B). Twenty-eight days after coronary ligation, ALAS2 TG mice also demonstrated increased heart weight to tibia length ratio (Figure 2D) and increased lung weight to tibia length ratio (Figure S2C), compared to control mice, which are consistent with more severe heart failure. These data suggest that overexpression of ALAS2 in the heart results in a more prominent reduction in cardiac contractility as compared to control animals in response to coronary ligation.
Figure 2.
ALAS2 TG mice have worsened cardiac function following coronary ligation. A, Representative M-mode echocardiographic images of ALAS2 NTG and TG mice are shown 28 days after sham thoracotomy or permanent coronary ligation (MI). B, EF and (C) FS are worsened in ALAS2 TG mice 28 days after MI compared to NTG littermates (n=4 to 5). D, HW to TL ratio is increased in ALAS2 TG mice 28 days after MI compared to NTG littermates (n=5 to 6). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; EF, ejection fraction; FS, fractional shortening; HW, heart weight; MI, myocardial infarction; NTG, nontransgenic; TG, transgenic; TL, tibia length.
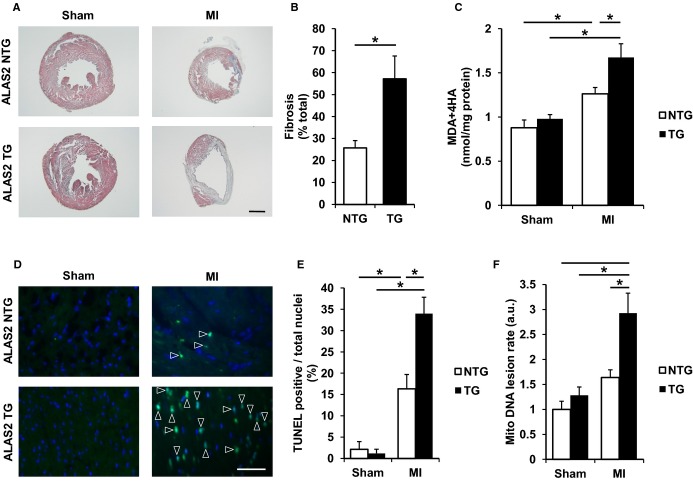
Consistent with exacerbated ischemic injury, histological examination of ALAS2 TG mice revealed increased cardiac fibrosis after 28 days of coronary ligation compared to control mice (Figure 3A and 3B). While the hearts of ALAS2 TG mice displayed normal cellular architecture based on hematoxylin and eosin staining after sham operation (Figure S3A), they showed increased levels of lipid peroxidation products (Figure 3C) 28 days after coronary ligation and increased number of TUNEL-positive nuclei (Figure 3D and 3E) in the infarct border zone after coronary ligation compared to control mice. Mitochondrial DNA lesion rate, a marker of mitochondrial oxidative stress, was also increased in the AAR of ALAS2 TG hearts 28 days after coronary ligation compared to control mice (Figure 3F). Taken together, these results suggest that overexpression of ALAS2 in the heart leads to increased oxidative stress, exacerbated cell death, and worsened cardiac function in response to coronary ligation compared to control animals.
Figure 3.
ALAS2 TG hearts display exacerbated structural abnormalities following coronary ligation. A, Representative images of cellular fibrosis in the hearts of ALAS2 NTG and TG mice are shown 28 days after sham thoracotomy or MI using Masson’s trichrome staining. Scale bar represents 4000 μmol/L. B, Cellular fibrosis is increased in the hearts of ALAS2 TG mice 28 days after MI using Masson’s trichrome staining compared to NTG littermates (n=4 to 5). Cellular fibrosis was quantified by dividing the arc length of the fibrotic scar over the heart circumference. C, Lipid peroxidation products, MDA and HAE, are increased in the hearts of ALAS2 TG mice after MI compared to NTG littermates (n=4 to 6). D, Representative images of cell death in the hearts of ALAS2 NTG and TG mice are shown 48 hours following sham thoracotomy or MI using TUNEL staining. Scale bar represents 50 μmol/L. Apoptotic cells are marked with an arrowhead. E, Cell death is increased in the hearts of ALAS2 TG mice after MI using TUNEL staining compared to NTG littermates (n=3 to 4, 3 fields per sample). Cell death was quantified by dividing the apoptotic cells over the total number of nuclei. F, Mitochondrial DNA lesion rate is increased in ALAS2 TG mice 28 days after MI compared to NTG littermates (n=3). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; a.u., arbitrary units; HAE, 4-hydroxyalkenals; MDA, malondialdehyde; MI, myocardial infarction; NTG, nontransgenic; TG, transgenic; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
ALAS2 TG Mice Have Increased Heme Levels After Coronary Ligation
We next asked whether cardiac-specific ALAS2 overexpression leads to an exaggerated response to ischemic injury through increased heme production and subsequent free heme accumulation in the heart. Heme levels were unchanged in the cytosolic and mitochondrial fractions from ALAS2 TG and control murine hearts 28 days after sham thoracotomy (Figure 4A and 4B). This observation is consistent with the normal cardiac function and heme levels of ALAS2 TG mice at baseline, suggesting that compensatory mechanisms are intact in unstressed ALAS2 TG mice to maintain appropriate levels of cardiac heme.
Figure 4.
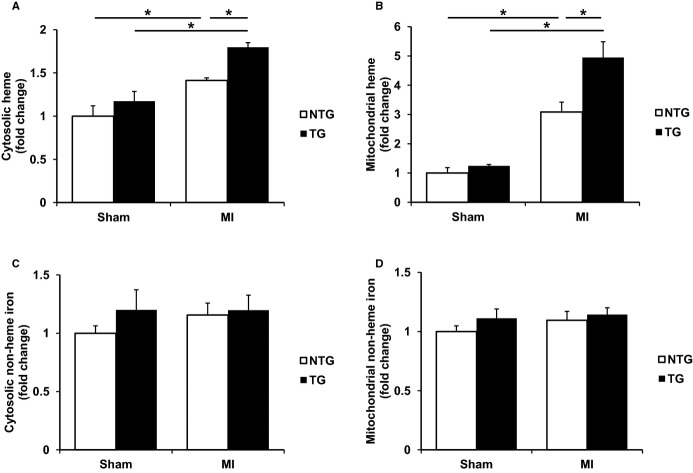
ALAS2 TG hearts display increased heme following coronary ligation. A, Cytosolic and (B) mitochondrial heme levels are increased in the hearts of ALAS2 TG mice 28 days after MI compared to NTG littermates (n=4 to 6). C, Cytosolic and (D) mitochondrial nonheme iron levels are similar in the hearts of ALAS2 NTG and TG mice 28 days after MI (n=5 to 6). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; MI, myocardial infarction; NTG, nontransgenic; TG, transgenic.
However, NTG murine hearts displayed significantly increased cytosolic and mitochondrial heme levels 28 days after coronary ligation compared to sham thoracotomy, and cytosolic and mitochondrial heme levels were further significantly increased in infarcted hearts from ALAS2 TG mice compared to control infarcted hearts (Figure 4A and 4B). To rule out the accumulation of unsaturated PPIX (the last synthetic intermediate in the heme synthesis pathway) contributing to the increased heme content in the above assays, we also measured iron-free PPIX and found no significant difference among the groups in both mitochondrial and cytosolic fractions (Figure S4A). Therefore, hearts overexpressing ALAS2 have physiologically appropriate heme levels at baseline, but exacerbated increases in mitochondrial and cytosolic heme levels after coronary ligation compared to NTG infarcted hearts.
Because the production of heme requires iron, we also measured the levels of nonheme iron in the hearts of ALAS2 TG and NTG mice to rule out the possibility of altered cellular iron homeostasis. As expected, there were no changes in the cytosolic or mitochondrial levels of nonheme iron in hearts from ALAS2 TG and NTG mice after coronary ligation or sham thoracotomy (Figure 4C and 4D), suggesting that delivery of iron to the cytoplasm and mitochondria are intact in ALAS2 TG mice.
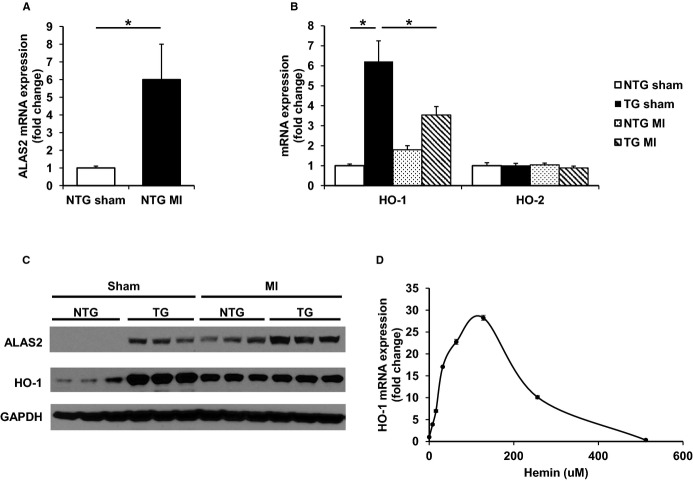
We then assessed the expression of genes directly involved in the synthesis of heme in ALAS2 TG and control mice after coronary ligation or sham thoracotomy. Consistent with the induction of ALAS2 expression in human failing hearts,29 endogenous murine ALAS2 mRNA (Figure 5A) and protein levels (Figure 5C) were significantly induced in infarcted hearts from control mice compared to sham-operated hearts. These observations are also consistent with previous in vitro reports of ALAS2 induction in the setting of hypoxia.29,31
Figure 5.
ALAS2 is upregulated in hearts after coronary ligation, and the increase in HO-1 after coronary ligation is attenuated in the hearts of ALAS2 TG mice. A, ALAS2 mRNA expression is induced in the hearts of ALAS2 NTG mice 28 days after MI compared to sham thoracotomy (n=5). B, The inducible heme degradation enzyme HO-1 is increased at the mRNA level in ALAS2 NTG mice 28 days after MI compared to sham thoracotomy, but reduced in ALAS2 TG mice after MI compared to sham thoracotomy. The mRNA expression of constitutively active HO-2 is similar among ALAS2 NTG and TG mice after sham thoracotomy or MI (n=5). C, Representative protein levels of ALAS2 and HO-1 in hearts of ALAS2 NTG and TG mice 28 days after MI or sham thoracotomy (n=3). D, Dose-curve of hemin treatment in H9c2 cardiac myoblasts demonstrates initial induction of HO-1 mRNA expression, but downregulation with escalating doses of hemin (n=6). Error bars are present but too small to be visualized for some doses. Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; HO-1, heme oxygenase-1; MI, myocardial infarction; NTG, nontransgenic; TG, transgenic.
Other than ALAS2, there was no change in the enzymes of heme synthesis in ALAS2 TG and control mice after coronary ligation or sham thoracotomy (Figure S5A). In fact, several heme synthetic genes showed a downregulated trend in infarcted ALAS2 TG and NTG hearts, suggesting feedback inhibition on the heme synthesis pathway by high levels of heme, as previously reported.36 Combined, these results suggest that ALAS2 induction due to hypoxic myocardium may be responsible for the increased heme levels seen in the infarcted hearts of both ALAS2 TG and control mice.
HO-1 Expression is Upregulated in ALAS2 TG Hearts at Baseline, but Attenuated After Coronary Ligation
In addition to heme synthetic genes, we also examined the expression of genes involved in the degradation of heme in ALAS2 TG and control mice after coronary ligation or sham thoracotomy. These studies were performed to determine whether coronary ligation is associated with different degrees of heme degradation in the hearts of ALAS2 TG and NTG mice. Consistent with previous observations in failing human hearts and animal models of heart failure, the expression of HO-1 was increased in NTG hearts after coronary ligation (Figure 5B).37–39 However, HO-1 mRNA (Figure 5B) and protein levels (Figure 5C) were reduced in ALAS2 TG mice after coronary ligation compared to sham thoracotomy. As ALAS2 TG mice have significantly higher heme levels after coronary ligation compared to NTG mice, our finding suggests that very high levels of heme in the heart may downregulate cardiac HO-1 expression. To confirm this hypothesis, we performed a dose-curve of hemin and measured its effects on HO-1 expression in H9c2 cardiomyoblasts. While induction of HO-1 expression peaked at ≈125 μmol/L hemin, higher concentrations of hemin dramatically reduced HO-1 mRNA expression levels (Figure 5D). Similar results were obtained using a dose-curve of aminolevulinic acid (ALA), the direct product of the ALAS2 reaction (Figure S5B). Thus, further induction of ALAS2 by hypoxia in the ALAS2 TG mice after coronary ligation leads to excess heme production and accumulation, reducing HO-1 expression and contributing to the reduced cardiac function of these mice.
Modulation of ALAS2 In Vitro Alters Heme and Cell Death Levels
The studies thus far suggest that increased ALAS2 is associated with increased ROS, cell death, and heme. However, it is not clear whether this increase in ROS and cell death with ALAS2 overexpression is due to elevated heme production. To confirm that the deleterious effects of ALAS2 on the heart are through upregulation of heme synthesis, we conducted mechanistic in vitro studies in H9c2 cardiomyoblasts using both ALAS2 overexpression and knockdown approaches.
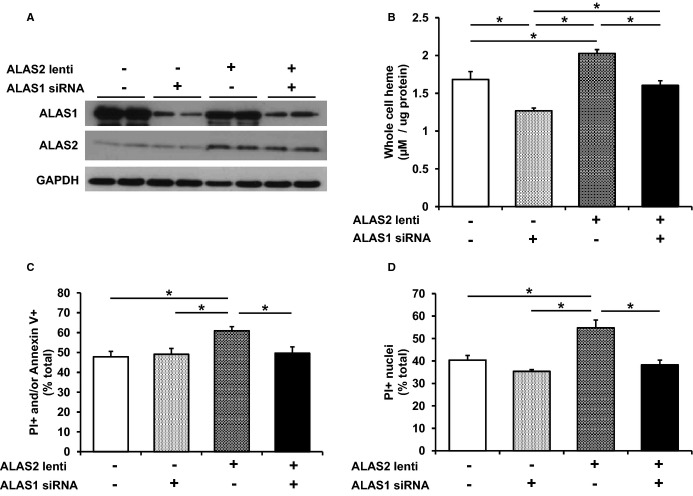
To modulate the levels of heme, we performed siRNA-mediated ALAS1 knockdown (to reduce endogenous heme levels), and/or lentivirus-driven overexpression of ALAS2 (to induce heme production through this isoform) in H9c2 cells (Figure 6A). ALAS1 was chosen to downregulate heme levels because it is endogenously expressed in the heart and serves as the major ALAS isoform responsible for baseline cardiac heme production. As expected, whole cell heme content was elevated in ALAS2-overexpressing cells, reduced in ALAS1 knockdown cells, and returned to baseline levels in cells with both ALAS2 overexpression and ALAS1 knockdown (Figure 6B), with similar changes observed in cytosolic and mitochondrial fractions (Figure S6A and S6B). No change in cytosolic or mitochondrial nonheme iron was observed among the overexpression and knockdown groups (Figure S6C and S6D). After induction of cell death by H2O2 treatment, cell viability was reduced in ALAS2-overexpressing cells, and this was reversed by concurrent downregulation of ALAS1, as assessed by flow cytometry (Figure 6C). Similar results were obtained using fluorescence microscopy after staining for propidium iodide–positive nuclei (Figure 6D). No significant reduction in cell death was observed with ALAS1 knockdown at baseline, possibly because basal heme levels are required to produce essential hemoproteins for cellular viability.
Figure 6.
Modulation of ALAS2 in vitro alters cell death through increased heme. A, Representative protein lentiviral overexpression of ALAS2 and silencing RNA (siRNA) downregulation of ALAS1 in H9c2 cells (n=2). B, Cellular heme content in H9c2 cells is increased with ALAS2 overexpression, reduced with ALAS1 downregulation, and returns to baseline levels with concurrent ALAS2 overexpression and ALAS1 downregulation (n=4 to 6). H2O2-induced cell death is increased in H9c2 cells with ALAS2 overexpression, and returns to baseline levels with concurrent ALAS2 overexpression and ALAS1 downregulation, as assessed by (C) PI/Annexin V double-labeling (n=5 to 8) and (D) PI-positive nuclei (n=3, 3 fields per sample). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; lenti, lentiviral overexpression; PI, propidium iodine.
In addition to ALAS2 overexpression, we also assessed the effects of ALAS2 knockdown on cardiac heme levels and viability. Since ALAS2 is not normally expressed in the heart, we conducted in vitro studies to knockdown ALAS2 under a condition that is associated with ALAS2 induction (ie, hypoxia), followed by ALAS2 downregulation (Figure 7A). Effective knockdown of hypoxia-induced ALAS2 by ALAS2 siRNA was confirmed by RT-PCR (Figure S7A). Cells were then harvested for gene expression and heme measurements, or treated with H2O2 to induce cell death. H9c2 cells incubated in hypoxia displayed increased ALAS2 expression, heme content (Figure 7B), and cell death (Figure 7C). Knockdown of ALAS2 under hypoxic conditions reduced heme levels to baseline (Figure 7B) and protected the cardiomyocytes against cell death (Figure 7C). Thus, our results demonstrate a causative role for ALAS2 in promoting cardiomyocyte death through increased heme content.
Although we have previously shown ALAS2 induction and heme accumulation in failing human hearts, the hearts came from a heterogeneous patient population. To determine whether chronic ischemia is involved in ALAS2 induction in human hearts, we measured ALAS2 levels in failing human hearts, from patients with ischemic cardiomyopathy and nonischemic cardiomyopathy, and nonfailing hearts. ALAS2 mRNA (Figure S7B) and protein (Figure 7A) levels were significantly increased in failing hearts from patients with ischemic cardiomyopathy compared to nonischemic cardiomyopathy. Consistent with increased ALAS2 levels, cytosolic (Figure 7B) and mitochondrial (Figure 7C) heme levels were significantly increased in failing hearts from patients with ischemic cardiomyopathy compared to nonischemic cardiomyopathy. Therefore, cardiac ALAS2 expression is positively regulated by hypoxia treatment in vitro, coronary ligation in vivo, and chronic ischemia in failing human hearts.
MitoTempo Reduces ALAS2-Induced Mitochondrial Oxidative Stress and Cell Death In Vitro
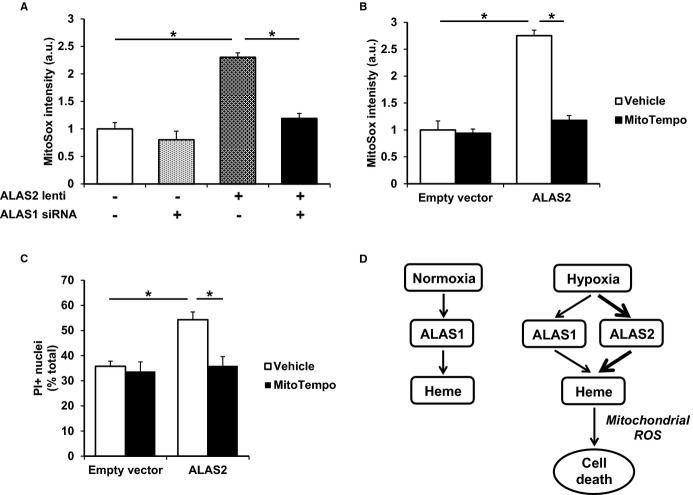
Since heme is predominantly produced in the mitochondria, we next studied whether ALAS2 overexpression contributes to oxidative stress through increased mitochondrial ROS. We first measured mitochondrial ROS levels using MitoSox staining in the setting of ALAS2 overexpression and/or ALAS1 knockdown in H9c2 cardiac myoblasts. Mitochondrial ROS levels were elevated in ALAS2-overexpressing cells, and this was reversed by concurrent ALAS1 downregulation (Figure 8A), confirming the role of heme in ALAS2-mediated mitochondrial ROS production. Addition of a mitochondrial superoxide scavenger, MitoTempo, reversed both mitochondrial ROS production (Figure 8B) and cell death (Figure 8C) in ALAS2-overexpressing H9c2 cells, as assessed by MitoSox and propidium iodide staining, respectively. These results suggest that mitochondrial oxidative stress plays a key role in heme-induced cardiomyocyte death by ALAS2 overexpression.
Figure 8.
ALAS2-induced cell death is associated with increased mitochondrial ROS. A, ImageJ analysis of H9c2 cells stained with the mitochondria-specific ROS-sensitive dye MitoSox demonstrates increased mitochondrial ROS with ALAS2 overexpression, which returns to baseline levels with concurrent ALAS1 knockdown (n=4, 3 fields per sample). B, ImageJ analysis of MitoSox-stained H9c2 cells demonstrates increased mitochondrial ROS with ALAS2 overexpression, which returns to baseline levels with administration of the mitochondrial superoxide scavenger MitoTempo (n=4, 3 fields per sample). C, H2O2-induced cell death is increased in H9c2 cells after ALAS2 overexpression, and is reduced with MitoTempo treatment, as assessed by PI-positive nuclei (n=4, 3 fields per sample). D, Model of cardiac heme production and cell death by ALAS1/2 in normoxic and hypoxic conditions. Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; a.u., arbitrary units; lenti, lentiviral overexpression; PI, propidium iodine; ROS, reactive oxygen species.
Discussion
Although heme is involved in a remarkable array of diverse cardiovascular processes, free heme is also a source of oxidative stress and cellular damage.40 We have previously reported that failing human hearts have increased heme content and ALAS2 levels, a rate-limiting enzyme in heme synthesis whose expression was thought to be restricted to hematopoietic cells.29 In this study, we show that coronary ligation results in ALAS2 induction and heme accumulation in murine hearts. Furthermore, the hearts of cardiac-specific ALAS2-overexpressing mice display increased cytosolic and mitochondrial heme accumulation, higher oxidative stress, exacerbated cell death, and worsened cardiomyopathy after coronary ligation compared to control mice. We also demonstrated in vitro that ALAS2 is induced by hypoxia in cultured cardiomyoblasts, which results in increased heme and cell death, and that these effects are reversed with ALAS2 knockdown. Additionally, we show that failing human hearts from patients with ischemic cardiomyopathy have increased ALAS2 levels and heme accumulation compared to patients with nonischemic cardiomyopathy. Finally, mechanistic studies show that ALAS2 overexpression in cultured cardiomyoblasts results in mitochondrial oxidative stress and cell death through increased heme accumulation, and that levels of mitochondrial oxidative stress and cell death in ALAS2-overexpressing cells are returned to baseline by administering the mitochondrial antioxidant MitoTempo (Figure 8D).
It has been reported that low-dose hemin treatment can be cardioprotective via induction of HO-1, which degrades heme into the antioxidant biliverdin and the anti-inflammatory carbon monoxide.37–39 Indeed, our ALAS2 TG mouse model at baseline demonstrates normal cardiac function and maintains heme at physiologically appropriate levels by balancing increased heme production due to ALAS2 overexpression with upregulation of heme degradation by HO-1. However, we show that upregulation of ALAS2 expression by hypoxia leads to significantly increased heme accumulation, due to insufficient compensatory upregulation of heme degradation by HO-1. Thus, excess heme production may oversaturate HO-1, resulting in incomplete heme degradation and the accumulation of cytotoxic free heme, which may cause cardiac damage.
Despite tight control of iron homeostasis in living organisms, free heme (heme not bound to protein) may accumulate due to increased heme production, excess hemolysis, elevated hemoprotein degradation, compromised integration of heme into hemoproteins, or impaired heme degradation.41 Free heme is a major source of redox-active iron, which causes cellular damage by catalyzing the Fenton reaction to produce toxic free hydroxyl radicals from hydrogen peroxide.11 Heme-driven production of ROS leads to cellular dysfunction by damaging lipid membranes, proteins, and nucleic acids.42–44 To confirm that the deleterious effects of ALAS2 expression on cardiomyocyte viability are through excess heme production, we performed studies in cultured cardiomyoblasts using 2 approaches: (1) ALAS2 knockdown and (2) ALAS2 overexpression with ALAS1 knockdown. We show that ALAS2 knockdown can reverse the increase in heme content and cell death associated with hypoxia. Furthermore, ALAS2 overexpression leads to increased oxidative stress and cell death through increased heme content, as normalization of cellular heme levels in ALAS2 overexpressing cells through concurrent downregulation of ALAS1 reduced levels of heme, oxidative stress, and cell death to baseline. These results suggest that ALAS2 expression in cultured cardiomyoblasts leads to increased oxidative stress and cell death through heme accumulation.
Although ALAS2 is not normally expressed in the heart, there are physiological conditions that lead to its cardiac induction and effects on the heart. Mutations in ALAS2 are most commonly associated with X-linked sideroblastic anemia;45,46 however, multiple case reports of iron overload cardiomyopathy are also associated with mutations in the ALAS2 gene.47,48 Here, we show that ALAS2 is induced in failing human hearts, particularly in patients with ischemic cardiomyopathy. Therefore, we hypothesize that chronic ischemia is involved in the mechanism of cardiac ALAS2 induction. Ischemia is common to failing hearts and our results suggest that, similar to erythroid cells, cardiac ALAS2 expression is positively regulated by hypoxia treatment in vitro, coronary ligation in vivo, and chronic ischemia in failing human hearts.
The treatment of iron deficiency in patients with heart failure has generated significant interest in the past few years. Four randomized controlled trials have suggested symptomatic benefit in treating iron-deficient heart failure patients with intravenous iron.7–10 However, the role of iron and iron-containing proteins in patients with heart failure and ischemic heart disease under normal serum iron levels is not known. Our results suggest that elevated levels of the iron-containing molecule heme lead to increased cardiac damage after ischemia. Thus, our results do not contrast these clinical trials but rather implicate a potential role for reducing heme levels in patients with ischemic heart disease and normal iron levels.
In summary, our results identify ALAS2 induction and heme accumulation as novel features of failing murine hearts after coronary ligation and failing human hearts, particularly in patients with ischemic cardiomyopathy. Furthermore, we show that induction of ALAS2 by hypoxia in cultured cardiomyoblasts leads to increased heme accumulation and reduced cell viability, and that these effects are reversed with ALAS2 knockdown. Additionally, we show that ALAS2 overexpression in cultured cardiomyoblasts leads to increased mitochondrial oxidative stress and cell death through heme accumulation, and that these effects can be reversed by administration of a mitochondrial antioxidant. Finally, the hearts of cardiac-specific ALAS2 overexpressing mice subjected to coronary ligation display increased heme accumulation, heightened oxidative stress, exacerbated cell death, and worsened cardiac function compared to control mice. These results identify heme accumulation as a novel biomarker and therapeutic target in failing hearts, whose levels may be reduced by suppressing cardiac ALAS2 expression.
Acknowledgments
The authors are grateful to all members of the Feinberg Cardiovascular Research Institute (FCVRI) for their insightful comments and support.
Sources of Funding
Dr Ardehali is supported by the NIH grants (K02 HL107448, R01 HL104181, and 1PO1 HL108795). Mr Sawicki is supported by the NIH predoctoral MD/PhD fellowship (F30DK102341).
Disclosures
Dr Ardehali receives speaking honoraria from Merck and is a consultant to Cubist Pharmaceuticals.
Supporting Information
Figure S1.Cardiac-specific ALAS2 overexpressing mice have normal cardiac function and heme levels at baseline. A, ALAS1 protein expression is similar in the hearts of ALAS2 TG and NTG littermates at baseline as assessed by Western blot (n=3). Densitometry analysis is provided below the Western blot. B, Cytosolic and (C) mitochondrial heme levels are similar in the hearts of ALAS2 TG and NTG littermates at baseline (n=5). D, mRNA expression of genes involved in heme synthesis (ALAD, δ-aminolevulinic acid dehydratase; ALAS1, δ-aminolevulinic acid synthase 1; CPOX, coproporphyrinogen oxidase; FECH, ferrochelatase; HMBS, hydroxymethylbilane synthase; PPOX, protoporphyrinogen oxidase; UROS, uroporphyrinogen III synthase; UROD, uroporphyrinogen decarboxylase) and degradation (HO-2, heme oxygenase-2) are similar in the hearts of ALAS2 NTG and TG mice (n=4). E, HO-1 protein expression is increased in the hearts of ALAS2 TG mice at baseline compared to NTG littermates, as assessed by Western blot (n=3). Densitometry analysis is provided below the Western blot. Data are presented as mean±SEM. *P<0.05. NTG indicates nontransgenic; TG, transgenic.
Figure S2. ALAS2 TG mice have worsened cardiac function following coronary ligation. A, Area at risk is similar in ALAS2 TG and NTG mice 24 hours after coronary ligation (n=3 to 4). B, Heart rates are similar in ALAS2 TG and NTG mice 28 days after sham thoracotomy or MI as assessed by ECG during echocardiography (n=4 to 5). C, Lung weight (LW) to tibia length (TL) ratio is increased in ALAS2 TG mice 28 days after MI compared to NTG littermates (n=5 to 6). Data are presented as mean±SEM. *P<0.05. NTG indicates nontransgenic; TG, transgenic.
Figure S3. ALAS2 TG hearts display exacerbated structural abnormalities following coronary ligation. A, Representative images of cellular architecture in the hearts of ALAS2 NTG and TG mice are shown 28 days after sham thoracotomy or MI using hematoxylin and eosin staining. Scale bar represents 4000 μmol/L. ALAS2 indicates δ-aminolevulinic acid synthase 2; NTG indicates nontransgenic; TG, transgenic.
Figure S4. ALAS2 TG hearts display increased heme following coronary ligation. A, Iron-free PPIX levels are similar in the hearts of ALAS2 TG and NTG mice 28 days after MI or sham thoracotomy (n=3 to 6). ALAS2 indicates δ-aminolevulinic acid synthase 2; MI, myocardial infarction; PPIX, protoporphyrin IX; NTG indicates nontransgenic; TG, transgenic.
Figure S5. ALAS2 is upregulated in hearts after coronary ligation, and the increase in HO-1 after coronary ligation is attenuated in the hearts of ALAS2 TG mice. A, mRNA expression of genes involved in heme synthesis (ALAD, δ-aminolevulinic acid dehydratase; PPOX, protoporphyrinogen oxidase; CPOX, coproporphyrinogen oxidase; UROS, uroporphyrinogen III synthase; FECH, ferrochelatase; UROD, uroporphyrinogen decarboxylase; ALAS1, δ-aminolevulinic acid synthase 1; HMBS, hydroxymethylbilane synthase) in the hearts of ALAS2 NTG and TG mice 28 days following sham thoracotomy or MI (n=5 to 6). B, Dose-curve of ALA treatment in H9c2 cardiac myoblasts demonstrates initial induction of HO-1 mRNA expression, but downregulation with escalating doses of ALA (n=4). Error bars are present but too small to be visualized for some doses. Data are presented as mean±SEM. *P<0.05. HO-1 indicates heme oxygenase 1; MI, myocardial infarction; NTG, nontransgenic; TG, transgenic.
Figure S6. Modulation of ALAS2 in vitro alters cell death through increased heme. A, Cytosolic and (B) mitochondrial heme content in H9c2 cells is increased with ALAS2 lentiviral overexpression, reduced with ALAS1 downregulation, and returns to baseline levels with concurrent ALAS2 overexpression and ALAS1 downregulation (n=4 to 6). C, Cytosolic and (D) mitochondrial nonheme iron content are similar in H9c2 cells with ALAS2 overexpression and/or ALAS1 knockdown (n=3 to 6). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; lenti, lentiviral overexpression.
Figure S7. ALAS2 is positively regulated by chronic ischemia in failing human hearts. A, ALAS2 mRNA expression in H9c2 cardiac myoblasts is induced after 72 hours in hypoxia, and reduced to baseline levels with ALAS2 siRNA treatment (n=6). B, ALAS2 mRNA is induced in failing human hearts, particularly in patients with ischemic cardiomyopathy (ICH) (n=4 to 8). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2
References
- Bleumink GS, Knetsch AM, Sturkenboom MC, Straus SM, Hofman A, Deckers JW, Witteman JC, Stricker BH. Quantifying the heart failure epidemic: prevalence, incidence rate, lifetime risk and prognosis of heart failure. The Rotterdam Study. Eur Heart J. 2004;25:1614–1619. doi: 10.1016/j.ehj.2004.06.038. [DOI] [PubMed] [Google Scholar]
- Mehra MR, Uber PA, Francis GS. Heart failure therapy at a crossroad: are there limits to the neurohormonal model? J Am Coll Cardiol. 2003;41:1606–1610. doi: 10.1016/s0735-1097(03)00245-6. [DOI] [PubMed] [Google Scholar]
- Ashrafian H, Frenneaux MP, Opie LH. Metabolic mechanisms in heart failure. Circulation. 2007;116:434–448. doi: 10.1161/CIRCULATIONAHA.107.702795. [DOI] [PubMed] [Google Scholar]
- Fragasso G, Salerno A, Spoladore R, Bassanelli G, Arioli F, Margonato A. Metabolic therapy of heart failure. Curr Pharm Des. 2008;14:2582–2591. doi: 10.2174/138161208786071245. [DOI] [PubMed] [Google Scholar]
- Ardehali H, Sabbah HN, Burke MA, Sarma S, Liu PP, Cleland JG, Maggioni A, Fonarow GC, Abel ED, Campia U, Gheorghiade M. Targeting myocardial substrate metabolism in heart failure: potential for new therapies. Eur J Heart Fail. 2012;14:120–129. doi: 10.1093/eurjhf/hfr173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayeva M, Gheorghiade M, Ardehali H. Mitochondria as a therapeutic target in heart failure. J Am Coll Cardiol. 2013;61:599–610. doi: 10.1016/j.jacc.2012.08.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonko DO, Grzeslo A, Witkowski T, Mandal AK, Slater RM, Roughton M, Foldes G, Thum T, Majda J, Banasiak W, Missouris CG, Poole-Wilson PA, Anker SD, Ponikowski P. Effect of intravenous iron sucrose on exercise tolerance in anemic and nonanemic patients with symptomatic chronic heart failure and iron deficiency FERRIC-HF: a randomized, controlled, observer-blinded trial. J Am Coll Cardiol. 2008;51:103–112. doi: 10.1016/j.jacc.2007.09.036. [DOI] [PubMed] [Google Scholar]
- Toblli JE, Lombrana A, Duarte P, Di Gennaro F. Intravenous iron reduces NT-pro-brain natriuretic peptide in anemic patients with chronic heart failure and renal insufficiency. J Am Coll Cardiol. 2007;50:1657–1665. doi: 10.1016/j.jacc.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Anker SD, Comin Colet J, Filippatos G, Willenheimer R, Dickstein K, Drexler H, Luscher TF, Bart B, Banasiak W, Niegowska J, Kirwan BA, Mori C, von Eisenhart Rothe B, Pocock SJ, Poole-Wilson PA, Ponikowski P Investigators F-HT. Ferric carboxymaltose in patients with heart failure and iron deficiency. N Engl J Med. 2009;361:2436–2448. doi: 10.1056/NEJMoa0908355. [DOI] [PubMed] [Google Scholar]
- Ponikowski P, van Veldhuisen DJ, Comin-Colet J, Ertl G, Komajda M, Mareev V, McDonagh T, Parkhomenko A, Tavazzi L, Levesque V, Mori C, Roubert B, Filippatos G, Ruschitzka F, Anker SD Investigators C-H. Beneficial effects of long-term intravenous iron therapy with ferric carboxymaltose in patients with symptomatic heart failure and iron deficiency. Eur Heart J. 2015;36:657–668. doi: 10.1093/eurheartj/ehu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- Kohgo Y, Ikuta K, Ohtake T, Torimoto Y, Kato J. Body iron metabolism and pathophysiology of iron overload. Int J Hematol. 2008;88:7–15. doi: 10.1007/s12185-008-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael S, Petrocine SV, Qian J, Lamarche JB, Knutson MD, Garrick MD, Koeppen AH. Iron and iron-responsive proteins in the cardiomyopathy of Friedreich’s ataxia. Cerebellum. 2006;5:257–267. doi: 10.1080/14734220600913246. [DOI] [PubMed] [Google Scholar]
- Payne RM. The heart in Friedreich’s ataxia: basic findings and clinical implications. Prog Pediatr Cardiol. 2011;31:103–109. doi: 10.1016/j.ppedcard.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassab-Chekir A, Laradi S, Ferchichi S, Khelil AH, Feki M, Amri F, Selmi H, Bejaoui M, Miled A. Oxidant, antioxidant status and metabolic data in patients with beta-thalassemia. Clin Chim Acta. 2003;338:79–86. doi: 10.1016/j.cccn.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Aessopos A, Farmakis D, Andreopoulos A, Tsironi M. Assessment and treatment of cardiac iron overload in thalassemia. Hemoglobin. 2009;33:S87–S92. doi: 10.3109/03630260903347401. [DOI] [PubMed] [Google Scholar]
- Kremastinos DT, Farmakis D, Aessopos A, Hahalis G, Hamodraka E, Tsiapras D, Keren A. Β-thalassemia cardiomyopathy history, present considerations, and future perspectives. Circ Heart Fail. 2010;3:451–458. doi: 10.1161/CIRCHEARTFAILURE.109.913863. [DOI] [PubMed] [Google Scholar]
- Elas M, Bielanska J, Pustelny K, Plonka PM, Drelicharz L, Skorka T, Tyrankiewicz U, Wozniak M, Heinze-Paluchowska S, Walski M, Wojnar L, Fortin D, Ventura-Clapier R, Chlopicki S. Detection of mitochondrial dysfunction by EPR technique in mouse model of dilated cardiomyopathy. Free Radic Biol Med. 2008;45:321–328. doi: 10.1016/j.freeradbiomed.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Bayeva M, Ghanefar M, Potini V, Sun L, Mutharasan RK, Wu R, Khechaduri A, Jairaj Naik T, Ardehali H. Disruption of ATP-binding cassette B8 in mice leads to cardiomyopathy through a decrease in mitochondrial iron export. Proc Natl Acad Sci USA. 2012;109:4152–4157. doi: 10.1073/pnas.1119338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense SM, Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- Wijayanti N, Katz N, Immenschuh S. Biology of heme in health and disease. Curr Med Chem. 2004;11:981–986. doi: 10.2174/0929867043455521. [DOI] [PubMed] [Google Scholar]
- Tsiftsoglou AS, Tsamadou AI, Papadopoulou LC. Heme as key regulator of major mammalian cellular functions: molecular, cellular, and pharmacological aspects. Pharmacol Ther. 2006;111:327–345. doi: 10.1016/j.pharmthera.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Sawicki KT, Chang HC, Ardehali H. Role of heme in cardiovascular physiology and disease. J Am Heart Assoc. 2015;4:e001138. doi: 10.1161/JAHA.114.001138. doi: 10.1161/JAHA.114.001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos TL. Heme enzyme structure and function. Chem Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol Lett. 2005;157:175–188. doi: 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Khan AA, Quigley JG. Control of intracellular heme levels: heme transporters and heme oxygenases. Biochim Biophys Acta. 2011;1813:668–682. doi: 10.1016/j.bbamcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- Sansbury BE, DeMartino AM, Xie Z, Brooks AC, Brainard RE, Watson LJ, DeFilippis AP, Cummins TD, Harbeson MA, Brittian KR, Prabhu SD, Bhatnagar A, Jones SP, Hill BG. Metabolomic analysis of pressure-overloaded and infarcted mouse hearts. Circ Heart Fail. 2014;7:634–642. doi: 10.1161/CIRCHEARTFAILURE.114.001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khechaduri A, Bayeva M, Chang HC, Ardehali H. Heme levels are increased in human failing hearts. J Am Coll Cardiol. 2013;61:1884–1893. doi: 10.1016/j.jacc.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlon TJ, Dell’Oso T, Surinya KH, May BK. Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis. Int J Biochem Cell Biol. 1999;31:1153–1167. doi: 10.1016/s1357-2725(99)00073-4. [DOI] [PubMed] [Google Scholar]
- Hofer T, Wenger RH, Kramer MF, Ferreira GC, Gassmann M. Hypoxic up-regulation of erythroid 5-aminolevulinate synthase. Blood. 2003;101:348–350. doi: 10.1182/blood-2002-03-0773. [DOI] [PubMed] [Google Scholar]
- Albert Y, Whitehead J, Eldredge L, Carter J, Gao X, Tourtellotte WG. Transcriptional regulation of myotube fate specification and intrafusal muscle fiber morphogenesis. J Cell Biol. 2005;169:257–268. doi: 10.1083/jcb.200501156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wyatt E, Chawla K, Tran M, Ghanefar M, Laakso M, Epting CL, Ardehali H. Hexokinase II knockdown results in exaggerated cardiac hypertrophy via increased ROS production. EMBO Mol Med. 2012;4:633–646. doi: 10.1002/emmm.201200240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa Y, Ghanefar M, Bayeva M, Wu R, Khechaduri A, Naga Prasad SV, Mutharasan RK, Naik TJ, Ardehali H. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J Clin Invest. 2014;124:617–630. doi: 10.1172/JCI72931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH, Jordan I, Kushner JP, Kaplan J. Heme regulation of HeLa cell transferrin receptor number. J Biol Chem. 1984;259:13235–13240. [PubMed] [Google Scholar]
- Ponka P. Tissue-specific regulation of iron metabolism and heme synthesis: distinct control mechanisms in erythroid cells. Blood. 1997;89:1–25. [PubMed] [Google Scholar]
- Wang G, Hamid T, Keith RJ, Zhou G, Partridge CR, Xiang X, Kingery JR, Lewis RK, Li Q, Rokosh DG, Ford R, Spinale FG, Riggs DW, Srivastava S, Bhatnagar A, Bolli R, Prabhu SD. Cardioprotective and antiapoptotic effects of heme oxygenase-1 in the failing heart. Circulation. 2010;121:1912–1925. doi: 10.1161/CIRCULATIONAHA.109.905471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JE, Foresti R, Sarathchandra P, Kaur H, Green CJ, Motterlini R. Heme oxygenase-1-derived bilirubin ameliorates postischemic myocardial dysfunction. Am J Physiol Heart Circ Physiol. 2000;278:H643–H651. doi: 10.1152/ajpheart.2000.278.2.H643. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Maulik N, Ho YS, Alam J, Das DK. H(mox-1) constitutes an adaptive response to effect antioxidant cardioprotection: a study with transgenic mice heterozygous for targeted disruption of the heme oxygenase-1 gene. Circulation. 2001;103:1695–1701. doi: 10.1161/01.cir.103.12.1695. [DOI] [PubMed] [Google Scholar]
- Furuyama K, Kaneko K, Vargas PD. Heme as a magnificent molecule with multiple missions: heme determines its own fate and governs cellular homeostasis. Tohoku J Exp Med. 2007;213:1–16. doi: 10.1620/tjem.213.1. [DOI] [PubMed] [Google Scholar]
- Ryter SW, Tyrrell RM. The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radic Biol Med. 2000;28:289–309. doi: 10.1016/s0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- Meneghini R. Iron homeostasis, oxidative stress, and DNA damage. Free Radic Biol Med. 1997;23:783–792. doi: 10.1016/s0891-5849(97)00016-6. [DOI] [PubMed] [Google Scholar]
- Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- Bayeva M, Sawicki KT, Butler J, Gheorghiade M, Ardehali H. Molecular and cellular basis of viable dysfunctional myocardium. Circ Heart Fail. 2014;7:680–691. doi: 10.1161/CIRCHEARTFAILURE.113.000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Urata G, Takaku F. Aminolevulinic acid synthetase activity in erythroblasts of patients with primary sideroblastic anemia. Nihon Ketsueki Gakkai Zasshi. 1973;36:74–77. [PubMed] [Google Scholar]
- Takaku F, Yano Y, Aoki Y, Nakao K, Wada O. δ-Aminolevulinic acid synthetase activity of human bone marrow erythroid cells in various hematological disorders. Tohoku J Exp Med. 1972;107:217–228. doi: 10.1620/tjem.107.217. [DOI] [PubMed] [Google Scholar]
- Lee P, Rice L, McCarthy JJ, Beutler E. Severe iron overload with a novel aminolevulinate synthase mutation and hepatitis C infection. A case report. Blood Cells Mol Dis. 2009;42:1–4. doi: 10.1016/j.bcmd.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman NL, Lee PL, Dries AM, Schwartz MR, Barton JC. Multi-organ iron overload in an African-American man with ALAS2 R452S and SLC40A1 R561G. Acta Haematol. 2008;120:168–173. doi: 10.1159/000181183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.Cardiac-specific ALAS2 overexpressing mice have normal cardiac function and heme levels at baseline. A, ALAS1 protein expression is similar in the hearts of ALAS2 TG and NTG littermates at baseline as assessed by Western blot (n=3). Densitometry analysis is provided below the Western blot. B, Cytosolic and (C) mitochondrial heme levels are similar in the hearts of ALAS2 TG and NTG littermates at baseline (n=5). D, mRNA expression of genes involved in heme synthesis (ALAD, δ-aminolevulinic acid dehydratase; ALAS1, δ-aminolevulinic acid synthase 1; CPOX, coproporphyrinogen oxidase; FECH, ferrochelatase; HMBS, hydroxymethylbilane synthase; PPOX, protoporphyrinogen oxidase; UROS, uroporphyrinogen III synthase; UROD, uroporphyrinogen decarboxylase) and degradation (HO-2, heme oxygenase-2) are similar in the hearts of ALAS2 NTG and TG mice (n=4). E, HO-1 protein expression is increased in the hearts of ALAS2 TG mice at baseline compared to NTG littermates, as assessed by Western blot (n=3). Densitometry analysis is provided below the Western blot. Data are presented as mean±SEM. *P<0.05. NTG indicates nontransgenic; TG, transgenic.
Figure S2. ALAS2 TG mice have worsened cardiac function following coronary ligation. A, Area at risk is similar in ALAS2 TG and NTG mice 24 hours after coronary ligation (n=3 to 4). B, Heart rates are similar in ALAS2 TG and NTG mice 28 days after sham thoracotomy or MI as assessed by ECG during echocardiography (n=4 to 5). C, Lung weight (LW) to tibia length (TL) ratio is increased in ALAS2 TG mice 28 days after MI compared to NTG littermates (n=5 to 6). Data are presented as mean±SEM. *P<0.05. NTG indicates nontransgenic; TG, transgenic.
Figure S3. ALAS2 TG hearts display exacerbated structural abnormalities following coronary ligation. A, Representative images of cellular architecture in the hearts of ALAS2 NTG and TG mice are shown 28 days after sham thoracotomy or MI using hematoxylin and eosin staining. Scale bar represents 4000 μmol/L. ALAS2 indicates δ-aminolevulinic acid synthase 2; NTG indicates nontransgenic; TG, transgenic.
Figure S4. ALAS2 TG hearts display increased heme following coronary ligation. A, Iron-free PPIX levels are similar in the hearts of ALAS2 TG and NTG mice 28 days after MI or sham thoracotomy (n=3 to 6). ALAS2 indicates δ-aminolevulinic acid synthase 2; MI, myocardial infarction; PPIX, protoporphyrin IX; NTG indicates nontransgenic; TG, transgenic.
Figure S5. ALAS2 is upregulated in hearts after coronary ligation, and the increase in HO-1 after coronary ligation is attenuated in the hearts of ALAS2 TG mice. A, mRNA expression of genes involved in heme synthesis (ALAD, δ-aminolevulinic acid dehydratase; PPOX, protoporphyrinogen oxidase; CPOX, coproporphyrinogen oxidase; UROS, uroporphyrinogen III synthase; FECH, ferrochelatase; UROD, uroporphyrinogen decarboxylase; ALAS1, δ-aminolevulinic acid synthase 1; HMBS, hydroxymethylbilane synthase) in the hearts of ALAS2 NTG and TG mice 28 days following sham thoracotomy or MI (n=5 to 6). B, Dose-curve of ALA treatment in H9c2 cardiac myoblasts demonstrates initial induction of HO-1 mRNA expression, but downregulation with escalating doses of ALA (n=4). Error bars are present but too small to be visualized for some doses. Data are presented as mean±SEM. *P<0.05. HO-1 indicates heme oxygenase 1; MI, myocardial infarction; NTG, nontransgenic; TG, transgenic.
Figure S6. Modulation of ALAS2 in vitro alters cell death through increased heme. A, Cytosolic and (B) mitochondrial heme content in H9c2 cells is increased with ALAS2 lentiviral overexpression, reduced with ALAS1 downregulation, and returns to baseline levels with concurrent ALAS2 overexpression and ALAS1 downregulation (n=4 to 6). C, Cytosolic and (D) mitochondrial nonheme iron content are similar in H9c2 cells with ALAS2 overexpression and/or ALAS1 knockdown (n=3 to 6). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2; lenti, lentiviral overexpression.
Figure S7. ALAS2 is positively regulated by chronic ischemia in failing human hearts. A, ALAS2 mRNA expression in H9c2 cardiac myoblasts is induced after 72 hours in hypoxia, and reduced to baseline levels with ALAS2 siRNA treatment (n=6). B, ALAS2 mRNA is induced in failing human hearts, particularly in patients with ischemic cardiomyopathy (ICH) (n=4 to 8). Data are presented as mean±SEM. *P<0.05. ALAS2 indicates δ-aminolevulinic acid synthase 2