Abstract
The c-Myc oncoprotein promotes proliferation and apoptosis, such that mutations that disable apoptotic programmes often cooperate with MYC during tumorigenesis. Here we report that two common mutant MYC alleles derived from human Burkitt’s lymphoma uncouple proliferation from apoptosis and, as a result, are more effective than wild-type MYC at promoting B cell lymphomagenesis in mice. Mutant MYC proteins retain their ability to stimulate proliferation and activate p53, but are defective at promoting apoptosis due to a failure to induce the BH3-only protein Bim (a member of the B cell lymphoma 2 (Bcl2) family) and effectively inhibit Bcl2. Disruption of apoptosis through enforced expression of Bcl2, or loss of either Bim or p53 function, enables wild-type MYC to produce lymphomas as efficiently as mutant MYC. These data show how parallel apoptotic pathways act together to suppress MYC-induced transformation, and how mutant MYC proteins, by selectively disabling a p53-independent pathway, enable tumour cells to evade p53 action during lymphomagenesis.
Human tumours frequently show deregulated expression of the c-Myc proto-oncogene1–3. In Burkitt’s lymphoma, this deregulation occurs through reciprocal translocations that juxtapose c-Myc with an immunoglobulin (Ig) promoter, leading to gross overexpression of c-Myc messenger RNA in the B cell lineage4,5. In addition, point mutations are often found in the translocated MYC alleles, clustering in a conserved region known asMYC box I (refs 6–8). Although some mutations can increase MYC stability and transforming activity in vitro, their impact on the pathogenesis of Burkitt’s lymphoma is unclear9–16. In fact, translocated c-Myc genes are subject to hypermutation in vivo that can also alter non-coding sequences, raising the possibility that these mutations are a consequence and not a cause of tumour development17,18.
To examine the effects of MYC mutation on lymphoma development in vivo, we used a system for rapidly generating tissue-specific transgenic mice (see Supplementary Fig. 1). Two mutant MYC alleles commonly observed in Burkitt’s lymphoma (P57S and T58A)13 were cloned into a murine stem cell virus (MSCV)-based vector that co-expresses green fluorescent protein (GFP). Haematopoietic stem cells (HSCs) derived from normal fetal livers were transduced with retroviruses expressing either wild-type or mutant MYC, and the genetically modified stem cells were then used to reconstitute the haematopoietic system of lethally irradiated recipient animals. This adoptive transfer strategy has several advantages over traditional transgenic approaches, such as: (1) transgene expression is limited to the haematopoietic system; (2) various combinations of transgenes or stem cell genotypes can be rapidly analysed; and (3) only a fraction of the injected stemcells are infected, allowing tumorigenesis to occur in the context of an essentially normal haematopoietic compartment. Also, the strong MSCV promoter produces high levels of MYC expression that recapitulate that observed in Burkitt’s lymphoma (data not shown), and the co-expressed GFP enables assessment of infection efficiency and visualization of tumorigenesis using fluorescence imaging.
MYC mutants show enhanced oncogenicity in vivo
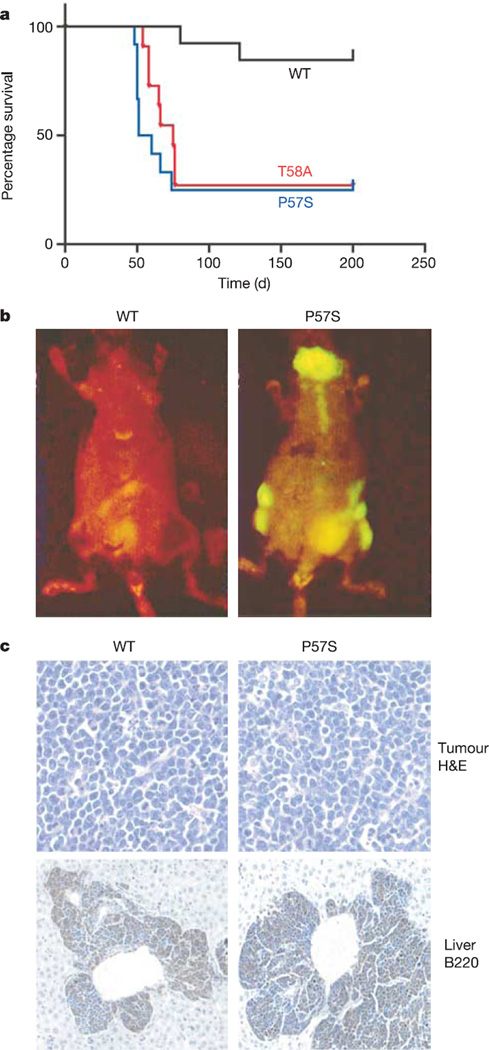
Although the control MSCV vector was not oncogenic (data not shown), recipients of stem cells infected with wild-type MYC developed tumours at low penetrance after a long latency (Fig. 1a; 2 out of 13 recipient mice at >100 days). The tumours that arose were aggressive pre-B cell lymphomas (Fig. 1b, c; see also Supplementary Fig. 2a), similar to those produced by the tissue-specific overexpression of MYC in transgenic mice19. Recipients of stem cells infected with the tumour-derived MYC point mutants also developed pre-B cell lymphomas, but at a significantly higher penetrance (Fig. 1a; 9 out of 12 for P57S and 8 out of 11 for T58A; P < 0.005 for each mutant versus wild-type MYC) and a significantly reduced latency (56 ± 9 days for P57S and 66 ± 9 days for T58A). Flow cytometry for GFP fluorescence showed that the increased tumorigenicity of these mutants was not due to differences in stem cell infection efficiency or transgene expression (see Supplementary Fig. 2b).
Figure 1. Tumour-derived MYC mutants show enhanced oncogenicity in vivo.
a, Kaplan–Meier curve showing survival at various times after adoptive transfer. b, In vivo GFP imaging shows disseminated lymphomas in a mouse 60 days after reconstitution with HSCs transduced with P57S, but not wild-type MYC. c, Haematoxylin/eosin staining and immunohistochemical staining for B220 (a B cell lineage marker) of lymph node and liver sections of animals harbouring wild-type MYC and P57S lymphomas, showing an aggressive B cell disease with perivascular infiltration of B220-positive tumour cells into the liver.
As deregulated MYC expression coordinately induces proliferation and apoptosis20, the increased oncogenicity of the mutant MYC proteins might reflect the altered activation of one or both of these processes. To investigate the proliferation of cells expressing wild-type and mutant MYC, we assessed BrdU incorporation in the bone marrow of pre-malignant mice after stem cell reconstitution. Although all MYC constructs induced an increase in proliferation relative to uninfected controls (data not shown), wild-type and mutant MYC showed indistinguishable BrdU incorporation profiles (Fig. 2a; 25 ± 3% for wild-type MYC, 24 ± 6% for P57S and 27 ± 5% for T58A BrdU-positive cells). To examine the impact of MYC expression on apoptosis, we compared the ability of wild-type and mutant MYC to induce tumours in the presence of a strong apoptotic block. Recipient mice were reconstituted with stem cells co-infected with a retrovirus expressing MYC constructs and a retrovirus expressing the anti-apoptotic protein Bcl2. Notably, when co-expressed with Bcl2, wild-type and mutant MYC produced aggressive B cell lymphomas with indistinguishable latency (34 ± 2 days for wild-type MYC, 35 ± 2 days for T58A and 33 ± 2 days for P57S) and penetrance (Fig. 2b, c). In addition, propidium iodide staining for DNA content revealed no differences in cell cycle profiles between lymphomas expressing wild-type and mutant MYC (Fig. 2d). Thus, in the absence of apoptosis, wild-type and mutant MYC are equally oncogenic.
Figure 2. Wild-type and mutantMYC show apoptotic, but not proliferative, differences in vivo.
a, Flow cytometry showing BrdU incorporation in GFP-positive bone marrow cells 24 days after reconstitution. Data are representative of three independent experiments. b, Kaplan–Meier curve showing survival following adoptive transfer with HSCs co-infected with the indicated MYC-expressing retroviruses and a Bcl2-expressing retrovirus. c, In vivo GFP imaging showing disseminated lymphomas in mice 35 days after adoptive transfer with HSCs co-transduced with wild-type or mutant MYC and Bcl2. d, Representative histograms of DNA content of wild-type MYC/Bcl2 and mutant MYC/Bcl2 lymphomas.
Mutant MYC induces p19ARF and p53, but not Bim
Oncogene-induced apoptosis is often mediated by the induction of p19ARF and subsequent stabilization of p53, which is thought to sense hyperproliferative signals and prevent aberrant proliferation21. To investigate the basis for the apoptotic defect of mutant MYC, we examined p19ARF and p53 expression in murine embryonic fibroblasts (MEFs) and HSC populations infected with wild-type and mutant MYC retroviruses. Interestingly, both P57S and T58A induced p19ARF and p53 as well as (or better than) wild-type MYC (Fig. 3a, c). This p53 was transcriptionally active, as cells expressing either wild-type or mutant MYC showed a similar increase in p53 phosphorylation and expression of the p53 transcriptional targets Bax, Puma and Noxa (Fig. 3c and data not shown). Furthermore, the apoptotic defect was unrelated to the established ability of MYC to repress Bcl2 and Bcl-XL22, because no consistent effect of MYC on Bcl2 expression was observed and both wild-type and mutant MYC were capable of repressing Bcl-XL. Although p21Cip1 levels were reduced in HSCs expressing wild-type MYC, they were substantially higher in cells expressing mutant MYC. However, this effect was not consistently seen inMEFs (Fig. 3c and data not shown), suggesting that the impact of p21Cip1 repression on MYC-induced apoptosis23 might contribute to, but is not essential for, the mutant MYC phenotype.
Figure 3. Impaired Bim induction contributes to the increased oncogenicity of mutant MYC alleles.
a, b, Western blot analysis showing ARF and p53 levels (a) or Bim levels (b) in MEFs stably infected with control (labelled C), wild-type MYC or mutant MYC vectors. The long (BimL) and extra long (BimEL) isoforms of Bim are shown. c,Western blot analysis of HSCs using antibodies against the indicated proteins. d, Kaplan–Meier curve showing free–free survival following adoptive transfer with Bim−/− HSCs infected with the indicated MYC-expressing retroviruses. e, In vivo GFP imaging showing disseminated lymphomas in mice 34 days after adoptive transfer with Bim−/− HSCs transduced with mutant or wild-type MYC. f,Wild type (Bcl2+/+) and Bcl2−/− MEFs transduced with the indicated retroviruses were incubated in low serum and viability was assessed by trypan blue exclusion. Data represent the mean ± standard deviation of three independent experiments.
The pro-apoptotic, BH3-only protein Bim has recently been shown to inhibit MYC-induced lymphomagenesis, and Bim expression is elevated in B cells overexpressing MYC (ref. 24). Accordingly, Bim was acutely induced in response to wild-type MYC in MEFs, FL5.12 cells and HSCs (Fig. 3b, c; see also Supplementary Fig. 3a). This effect was not dependent on MYC induction of p53 because Bim was induced in p53−/− MEFs (data not shown). However, both tumour-derived MYC mutants were unable to induce Bim efficiently. Furthermore, mice reconstituted with Bim−/− stem cells expressing wild-type MYC produced lymphomas at a latency and penetrance indistinguishable from those reconstituted with Bim−/− stem cells expressing mutant MYC (Fig. 3d, e; 40 ± 5 days for wild-type MYC, 36 ± 6 days for T58A and 38 ± 6 days for P57S), implying that the failure of these mutant MYC alleles to induce Bim contributes to their increased oncogenicity. Thus, MYC mutations do not affect the ‘sensing’ of MYC induced hyperproliferative signals by the p19ARF–p53 pathway, but promote lymphomagenesis by abrogating a parallel apoptotic signal transmitted from MYC to Bim.
Bim was initially identified by virtue of its ability to bind Bcl2, and it acts as a potent inhibitor of the pro-survival members of the Bcl2 family25. Thus, failure of mutant MYC to induce Bim should result in a defective ability to suppress Bcl2 activity. Such a model predicts that suppression of Bcl2 activity might rescue the apoptotic defect of mutant MYC. To test this, we assessed the ability of wild-type and mutant MYC to promote apoptosis in cells derived from Bcl2 knockout mice. Wild-type (Bcl2+/+) and Bcl2-deficient (Bcl2−/−) MEFs were infected with retroviruses expressing each MYC allele and then subjected to serum starvation to trigger apoptosis. In the presence of Bcl2, wild-type MYC consistently induced higher levels of apoptosis than either of the MYC point mutants (Fig. 3f). However, in Bcl2−/− MEFs, P57S and T58A were as capable of inducing apoptosis as wild-type MYC. Interestingly, apoptosis induced by wild-type MYC was not affected by Bcl2 ablation, perhaps because wild-type MYC already efficiently inhibits Bcl2. Similarly, recipients of Bcl2+/+ HSCs infected with wild-type MYC showed significantly less GFP-positive bone marrow after reconstitution compared with those receiving mutant MYC-infected stem cells; however, all MYC-expressing cells were strongly selected against during haematopoietic reconstitution with Bcl2−/− HSCs (see Supplementary Fig. 3b). Thus, in the absence of Bcl2, wild-type and mutant MYC were equally capable of promoting cell death, implying that MYC mutants—through reduced Bim induction and perhaps other additional effects—are ultimately deficient in their ability to inhibit Bcl2.
Evasion of p53 tumour suppressor function
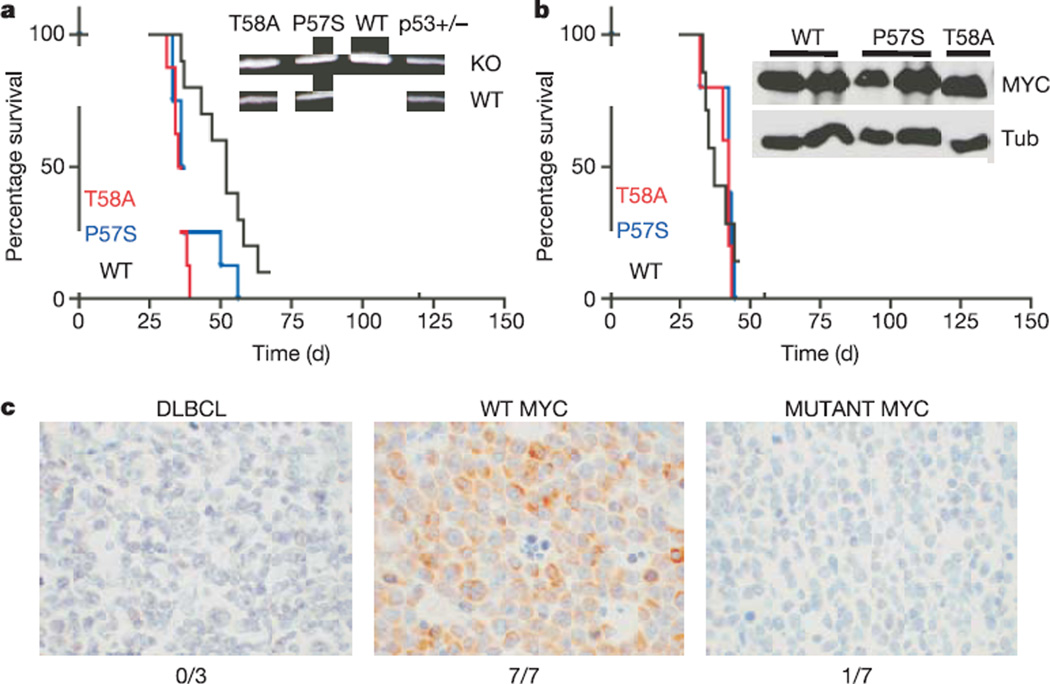
These data suggest that the increased oncogenicity of tumour-derived MYC point mutants results from their inability to efficiently couple proliferation to cell death. Certain targeted apoptotic defects can obviate the requirement for tumour suppressor inactivation during tumorigenesis. For example, both Bcl2 overexpression and Bim deficiency facilitate MYC-induced lymphomagenesis in the absence of p19ARF or p53 mutations24,26. To determine whether MYC point mutations could also bypass the requirement for p53 loss, we examined tumour onset and p53 status in lymphomas derived from p53+/− stem cells. In this context, both wild-type and mutant MYC rapidly produced lymphomas with complete penetrance (Fig. 4a), although those produced by wild-type MYC arose with slightly longer latency (56 ± 24 days for wild-type MYC, 35 ± 3 days for T58A and 40 ± 9 days for P57S; P < 0.001 for both mutants compared with wild type). However, although tumours induced by wild-type MYC invariably lost the wild-type p53 allele during tumour development (0 out of 9 with wild-type p53 allele), those induced by the MYC mutants typically retained the wild-type p53 allele (7 out of 9 with wild-type p53 allele for both P57S and T58A; Fig. 4a, inset).
Figure 4. Impact of mutations in MYC on p53 tumour suppressor action and Bim induction in human Burkitt’s lymphomas.
a, Kaplan–Meier curve showing mouse survival after adoptive transfer with p53+/− HSCs infected with the indicated MYC retroviral constructs. Inset, Representative allele-specific PCR assay showing retention of the wild-type p53 allele in lymphomas produced by mutant but not wild-type MYC. b, Kaplan–Meier curve showing mouse survival after adoptive transfer with p53−/− HSCs infected as in a. Inset, western blot analysis of representative lymphomas harbouring the indicated MYC allele. c, Bim expression in DLBCL and wild-type MYC-expressing and mutant MYC-expressing Burkitt’s lymphomas as assessed by immunohistochemisty using an anti-Bim antibody. The frequency of Bim-positive tumours in each category is indicated.
The requirement to lose the wild-type p53 allele in tumours initiated by wild-type MYC may account for their delayed onset relative to tumours in recipients of mutant MYC-infected stem cells. Indeed, mice reconstituted with p53−/− HSCs expressing either the wild-type or mutant MYC formed tumours rapidly, with an identical penetrance and latency (Fig. 4b; 40 ± 8 days for wild-type MYC, 40 ± 4 days for T58A and 41 ± 5 days for P57S). All of the p53−/− lymphomas were oligoclonal (see Supplementary Fig. 3b), and showed no obvious differences between wild-type and mutant MYC expression (Fig. 4b, inset). Thus MYC mutations can facilitate tumour development in the absence of p53 inactivation.
MYC, p53 and Bim status in Burkitt’s lymphoma
These data predict that Bim expression should be reduced in Burkitt’s lymphomas expressing MYC mutations, and that p53 mutations should be under-represented in Burkitt’s lymphomas harbouring the P57S and T58A mutant MYC alleles. Indeed, seven out of seven Burkitt’s lymphomas expressing wild-type MYC were positive for Bim expression by immunohistochemistry, displaying substantially higher Bim levels than several diffuse, large B cell lymphomas (DLBCL) without MYC translocations (three out of three; Fig. 4c). In contrast, six out of seven Burkitt’s lymphomas harbouring MYC alleles with MYC box I mutations (including four out of five tumours with P57 and T58 mutations) were scored as Bim negative (Fig. 4c). Furthermore, none of the p53 mutant Burkitt’s lymphomas (0 out of 17) that we analysed or identified from the literature6,27,28 had coincident mutations at MYC codons 57 and 58, despite their common occurrence in our data set (12 out of 71) (see Supplementary Table; and data not shown). Of note, two out of two lymphomas harbouring a Pro to Ser mutation at codon 60 of MYC had a p53 mutation at codon 248, suggesting that not all MYC mutations are equivalent. Nevertheless, our data demonstrate that two mutant MYC alleles commonly observed in Burkitt’s lymphoma can bypass p53 action during lymphomagenesis in mice, and provide evidence to suggest that this may also occur in humans.
Discussion
Our results reveal new insights into the biology of the MYC oncoprotein. For example, they suggest that tumour-derived MYC mutants are relevant to the pathology of Burkitt’s lymphoma because they are unable to upregulate Bim and efficiently inhibit Bcl2. Furthermore, they show that the MYC mutants we examined retain their ability to activate the p19ARF–p53 pathway and efficiently promote proliferation. Thus, MYC mutations alter MYC activity in both quantitative12,14 and, as shown here, qualitative ways. These qualitative changes may be particularly important for the development of tumours such as Burkitt’s lymphoma, which acquire independent alterations that quantitatively upregulate MYC. Such qualitative changes to MYC may also be important for normal cell growth; in this regard, some tumour-derived MYC mutations lead to constitutive phosphorylation of MYC at Ser 62, a target of mitogen-stimulated kinases14. Perhaps MYC mutations ‘lock in’ a qualitative state that normally uncouples proliferation from apoptosis in response to mitogens, allowing tissue expansion.
Mutations that are selected for in malignant tumours pinpoint genes and processes that must be altered during tumour evolution. Thus, the fact that certain mutant MYC alleles selected for in human lymphomas are specifically impaired in their ability to promote apoptosis in vivo provides compelling genetic evidence that the phenomenon of oncogene-induced apoptosis is relevant to human tumorigenesis. Furthermore, our results demonstrate that the MYC-induced apoptotic programme is not linear, but instead involves p53-dependent and -independent signals that act in parallel to promote cell death and suppress MYC-induced tumorigenesis. That a cell expressing mutant MYC can activate p53 in response to hyperproliferative signalling but evade p53 action implies that MYC-induced apoptosis involves a threshold phenomenon, such that inactivation of any one of several MYC effectors can cause apoptotic firing to drop below the threshold level and allow unabated proliferation. Consequently, certain mutations in MYC alleviate the requirement for a cooperating gene mutation during tumorigenesis.
METHODS
Retroviral infection and cell culture
HAM (a methionine-rich variant of the widely used haemagglutinin tag)-tagged wild-type MYC and mutants were subcloned into MSCV-IRES-GFP26 for in vivo studies and MSCVpuro (Clon-tech) for cell culture studies. Primary MEFs were generated from embryonic 13.5 (E13.5) day embryos from a wild-type C57BL/6J background or from a Bcl2+/− to Bcl2+/− cross. Retroviral-mediated gene transfer was performed using Phoenix packaging cells as previously described29. Cells for in vivo studies were infected with MSCV-GFP, MSCV-GFP-WT MYC, MSCV-GFP-P57S or MSCV-GFP-T58A. Infected cell populations of FL5.12 cells and MEFs used for in vitro studies were selected in puromycin (2 mg ml−1 for 2 days). Infected HSCs used for in vitro studies were diluted to equal infection efficiency (assessed by GFP) then sorted using magnetic beads to 70–80% purity with a lineage cell depletion kit (Miltenyi Biotec). For cell viability assays, MEFs were infected with MYC-expressing retroviruses as described, and were incubated for 24 h in either 10% fetal bovine serum (high serum) or 0.1% fetal bovine serum (low serum) and analysed for viability by trypan blue exclusion.
Stem cell isolation and adoptive transfer
Pregnant E14.5 C57BL/6 mice from wild-type, p53+/−, Bim+/− or Bcl2+/− crosses were killed to obtain fetal livers. Stem cell infection and transplantation were performed as described26. For stem cell reconstitution and BrdU analysis, total bone marrow was isolated 21–28 days after tail vein injection. In vivo BrdU incorporation was assessed using an APC BrdU Flow Kit (BD Pharmingen). Bone marrow was harvested 2 h after an intraperitoneal injection of 200 ml 10 mg ml−1 BrdU, and BrdU incorporation was assessed for a population of GFP-positive cells at a defined GFP intensity. Flow cytometry analysis was performed on a Becton Dickinson LSRII cell analyser, with FACSVantage DiVa software.
Lymphoma monitoring and analysis
Reconstituted animals were monitored for illness by lymph node palpation, monitoring overall morbidity, and, in some cases, whole-body fluorescence imaging26. Overall survival was defined as the time from stem cell reconstitution until the animal reached a terminal stage and was killed. Statistical analysis was performed using a one-way ANOVA (analysis of variance) test using Graph Pad Prism version 3.0 (Graph Pad Software). Immunohistochemistry was performed using CD45R/B220–clone RA3-6B2 (BD Biosciences, Pharmingen) and rabbit anti-Bim (Stressgen) antibodies. Tumour cell DNA content was determined by FACS analysis with propidium iodide staining of ethanol-fixed cells26. Nested PCR analysis of variable, diversity and joining (V(D)J) recombination was performed as described30.
Protein analysis
Immunoblots were performed from whole-cell lysates obtained by boiling cell pellets that were solubilized in Laemmli sample buffer. Twenty micrograms of protein samples (Bio-Rad protein assay) were separated by SDS polyacylamide electrophoresis (SDS–PAGE) and transferred to Immobilon-P membranes (Millipore). The antibodies used were anti-MYC (Ab1, Oncogene; 1:250), anti-p19ARF (ab80-100, Abcam; 1:1,000), anti-p21 (C-19, Santa Cruz; 1:500), anti-p53 (NCL-p53-505, Novocastra; 1:500), anti-phosphop53 (Ser 15) (16G8, Cell Signaling; 1:1,000), anti-Bim/BOD (AAP-330, Stressgen; 1:1,000), anti-Bcl2 (N-19, Santa Cruz; 1:1,000) and anti-Bcl-XL (S-18, Santa Cruz; 1:1,000). Proteins were visualized using ECL (Amersham) or Lumi-light (Roche).
Supplementary Material
Acknowledgements
We thank M. S. Jiao, C. Rosenthal, M. Yang, S. Ray, C. Yang and I. Linkov for technical assistance and J. Zilfou, R. Dickins, E. Cepero, M. Spector, J. Fridman and other members of the Lowe laboratory for advice and/or critical reading of the manuscript. We also thank A. Hunt and G. Evan for helpful advice, and T. Mak and A. Strasser for mice. This work was supported by a postdoctoral fellowship from the Helen Hay Whitney Foundation (M.T.H.), an NCI postdoctoral training grant (A.B.), a program project grant from the National Cancer Institute (W.P.T and S.W.L.) and a grant from the Irving Hansen Memorial Foundation (W.P.T). W.P.T. is a Leukemia and Lymphoma Society Scholar and S.W.L. is an AACR-NCRF Research Professor.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole MD. Activation of the c-myc oncogene. Basic Life Sci. 1986;38:399–406. doi: 10.1007/978-1-4615-9462-8_43. [DOI] [PubMed] [Google Scholar]
- 3.Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 4.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl Acad. Sci. USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis M, Malcolm S, Rabbitts TH. Chromosome translocation can occur on either side of the c-myc oncogene in Burkitt lymphoma cells. Nature. 1984;308:286–288. doi: 10.1038/308286a0. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia K, et al. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nature Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 7.Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt’s lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 8.Clark HM, et al. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- 9.Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 10.Hoang AT, et al. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol. Cell. Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westaway D, Payne G, Varmus HE. Proviral deletions and oncogene base-substitutions in insertionally mutagenized c-myc alleles may contribute to the progression of avian bursal tumors. Proc. Natl Acad. Sci. USA. 1984;81:843–847. doi: 10.1073/pnas.81.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 2000;20:4309–4319. doi: 10.1128/mcb.20.12.4309-4319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sears RC. The life cycle of c-Myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 15.Frykberg L, Graf T, Vennstrom B. The transforming activity of the chicken c-myc gene can be potentiated by mutations. Oncogene. 1987;1:415–422. [PubMed] [Google Scholar]
- 16.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nature Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 17.Rabbitts TH, Hamlyn PH, Baer R. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature. 1983;306:760–765. doi: 10.1038/306760a0. [DOI] [PubMed] [Google Scholar]
- 18.Bemark M, Neuberger MS. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt’s lymphoma line. Oncogene. 2000;19:3404–3410. doi: 10.1038/sj.onc.1203686. [DOI] [PubMed] [Google Scholar]
- 19.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 20.Evan GI, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 21.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 22.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol. Cell. Biol. 2003;23:7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 24.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl Acad. Sci. USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connor L, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt CA, et al. Dissecting p53 tumour suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 27.Gaidano G, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 1991;88:5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia KG, Gutierrez MI, Huppi K, Siwarski D, Magrath IT. The pattern of p53 mutations in Burkitt’s lymphoma differs from that of solid tumors. Cancer Res. 1992;52:4273–4276. [PubMed] [Google Scholar]
- 29.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 30.Yu D, Thomas-Tikhonenko A. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene. 2002;21:1922–1927. doi: 10.1038/sj.onc.1205244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.