Abstract
Catechol-O-methyltransferase (COMT) plays a central role in DNA repair and estrogen-induced carcinogenesis. Many recent epidemiologic studies have investigated the association between the COMT Val158Met polymorphism and cancer risk, but the results are inconclusive. In this study, we performed a meta-analysis to investigate the association between cancer susceptibility and COMT Val158Met in different genetic models. Overall, no significant associations were found between COMT Val158Met polymorphism and cancer risk (homozygote model: odds ratio [OR] =1.05, 95% confidence interval [CI] = [0.98, 1.13]; heterozygote model: OR =1.01, 95% CI = [0.98, 1.04]; dominant model: OR =1.02, 95% CI [0.97, 1.06], and recessive model: OR =1.03, 95% CI [0.97, 1.09]). In the subgroup analysis of cancer type, COMT Val158Met was significantly associated with increased risks of bladder cancer in recessive model, and esophageal cancer in homozygote model, heterozygote model, and dominant model. Subgroup analyses based on ethnicities, COMT Val158Met was significantly associated with increased risk of cancer in homozygote and recessive model among Asians. In addition, homozygote, recessive, and dominant models were significantly associated with increased cancer risk in the subgroup of allele-specific polymerase chain reaction genotyping. Significant associations were not observed when data were stratified by the source of the controls. In summary, this meta-analysis suggested that COMT Val158Met polymorphism might not be a risk factor for overall cancer risk, but it might be involved in cancer development at least in some ethnic groups (Asian) or some specific cancer types (bladder and esophageal cell cancer). Further evaluations of more preclinical and epidemiological studies are required.
Keywords: COMT, polymorphism, cancer, meta-analysis, susceptibility
Introduction
Cancer constitutes an enormous burden on the society in more and less economically developed countries alike.1,2 Based on GLOBOCAN estimates, ~14.1 million new cancer cases and 8.2 million deaths occurred in 2012 worldwide.1 According to the development trend, the new cases in 2030 will reach 22.2 million.2 It is well known that the etiology and development of cancer are as a result of complex interactions between genetic and environmental factors.3 Genes determine the susceptibility of individual to environment, and environmental factors often damage the DNA in turn. Recent studies have shown that host genetic factors are closely related to the pathophysiology of many human cancers.4 The most common form of genetic variation, that is, single-nucleotide polymorphisms, is known to contribute individual susceptibility to cancer.5 Therefore, it is anticipated that the identification of key gene polymorphisms associated with cancer risk is essential for predicting risk of individuals, and that it will greatly assist the global control and therapeutic strategies of this lethal disease.
The catechol-O-methyltransferase (COMT) gene is located on chromosome 22q11.2 and consists of six exons.6 It is an important enzyme involved in the inactivation of endogenous catecholamine and catechol estrogens. Catechol estrogens have been shown to have the ability to damage DNA and carcinogenetic potential.7 Therefore, the loss of or changes in COMT is supposed to contribute to genomic instability and tumor genesis. In line with these considerations, it has been hypothesized that COMT Val158Met might influence the development of all cancers. Up to now, many researches have indicated the link between COMT polymorphism and cancer susceptibility. Several polymorphisms have been identified, including the widely studied polymorphism Val158Met(rs4680).8 This change has been associated with a three- to four-fold decrease in the activity of COMT compared with the wild-type COMT-Val allele.9,10 It is biologically reasonable to hypothesize that women who carry mutant COMT-Met allele may have higher cancer risks.
In recent years, many studies have investigated the relationship between COMT Val158Met polymorphism in different races and different types of cancer, but the results were inconclusive or controversial.11–101 The inconsistent conclusions may be due to a possible minor effect of the polymorphism on cancer or the small sample size in studies with inadequate statistical power of complex traits. Meta-analysis is a powerful statistical tool to pool different studies to overcome deficiencies such as small sample size and to provide more reliable results. Although some previous meta-analyses have reported the association between COMT Val158Met polymorphism and ovarian cancer (up to eight case–control studies included),102,103 breast cancer (up to 56 case–control studies included),65,104–108 endometrial cancer (up to seven case–control studies included),103,109,110 prostate cancer (up to six case–control studies included),111–113 and lung cancer (evidence from six case–control studies),114 only specific cancer types or race populations were included, which led to their limitations. To update the results of previous meta-analyses and to provide a more precise assessment of the association between COMT Val158Met and cancer risk, we performed a comprehensive meta-analysis by including the most recent and relevant articles.
Materials and methods
Identification and eligibility of relevant studies
The meta-analysis was conducted following the criteria of Preferred Reporting Items for Systematic Reviews and Meta Analyses. A comprehensive literature search was performed using the PubMed, Cochrane Library, Chinese National Knowledge Infrastructure, and EMBASE database for relevant articles published (the last search update was February 15, 2015) with keywords “COMT”, “Catechol-O-methyltransferase”, “Val158Met”, “rs4680”, “single nucleotide polymorphism”, “polymorphism”, “Variant”, “Mutation”, “Cancer”, “tumor”, “neoplasm”, “malignancy”, or “Carcinoma”. In addition, studies were identified by a manual search of reviews and retrieved studies. Search results were restricted to human populations, and the articles were written in English or Chinese. We included all the case–control studies and cohort studies that have investigated the association between COMT Val158Met polymorphisms and cancer risk with genotyping data. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. When the same patient population was used in several publications, only the most recent, the largest or the most complete study was included.
Assessment of study quality
The quality of the included studies was assessed by the Newcastle–Ottawa Scale (NOS; http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp),115 including selection of groups, comparability of groups, and ascertainment of exposure. The NOS score ranges from 0 to 10 stars. Studies with NOS score > five stars were included in the final analysis.
Inclusion criteria
All studies were included if they met the following criteria: 1) only the case–control studies or cohort studies were considered, 2) studies that investigated the COMT Val158Met polymorphism and the risk of cancer susceptibility were included, and 3) the genotype distribution of the polymorphism in cases and controls was described in details, and the results were expressed as odds ratio (OR) and corresponding 95% confidence interval (95% CI). Major reasons for exclusion of studies were as follows: 1) not for cancer research, 2) only case population, 3) duplicate of previous publication, and 4) review articles, editorials, case reports, studies with preliminary results not on COMT Val158Met polymorphism or outcome, and investigations of the role of COMT expression related to disease. Ethics approval for the study was granted by the local institute, the People’s Hospital of Three Gorges University Ethics Committee.
Data extraction
Using a standardized form, data from published studies were extracted independently by two reviewers to evaluate their eligibility for inclusion by first screening the title and abstract of each identified reference and then establishing the eligibility of the included papers based on the full text when necessary. For each included study, the following information was collected: first author, year of publication, region, study design, sample size, source of control, geno-typing method, allele or genotype frequencies, and evidence of Hardy–Weinberg equilibrium (HWE). Any discrepancy between the two reviewers was resolved by discussion and consultation with a third reviewer.
Statistical analysis
ORs and their 95% CIs were used to determine the strength of association between the COMT Val158Met polymorphism and cancer risk. The significance of the pooled OR was determined using the Z test, and P<0.05 was considered statistically significant. Homozygote model (AA vs GG), heterozygote model (GA vs GG), dominant model (GA + AA vs GG), and recessive model (AA vs GG + GA) were investigated. Subgroup analysis was performed by ethnicity, cancer type (if one cancer type contained less than two studies, it was defined as “other”), source of controls, and hospital or population controls. Effective modification by a subgroup was assessed by testing the interaction between genotypes and stratification variables by using logistic regression analyses (random-effects estimator). HWE was tested using the chi-square test among controls, and P<0.05 was considered a significant departure from HWE. If the P-value for heterogeneity was >0.05 and I2<50%, indicating an absence of heterogeneity among studies, the fixed-effects model (the Mantel–Haenszel method) was used.116 In contrast, if either the P-value for heterogeneity was ≤0.05 or I2 was ≥50%, indicating heterogeneity among the studies, the more appropriate random-effects model (the DerSimonian and Laird method) was used.117 Sensitivity analyses were performed to assess the stability of the results. Begg’s funnel plots were used to diagnose potential publication bias, and P<0.05 was used to indicate possible publication bias.118 All analyses were performed using RevMan 5.3 (updated in March 2012 by the Cochrane Collaboration). P-values were based on two-sided tests.
Results
Literature search and meta-analysis databases
Following the searching strategy, 337 potentially relevant studies were retrieved. After title and abstract screening, nine of them were ruled out because of repeated data. A total of 202 irrelevance articles were excluded. In addition, after the full texts of the remaining 182 articles were read, 90 articles were excluded for the following reasons: article was a review (n=27), articles had insufficient data (n=13), articles were not related to cancer (n=34), and articles were not related to COMT (n=16). A total of 92 publications with full text were selected and were subjected to further examination. Because seven studies included more than one ethnicity, genotype method, control source, or tumor type and were performed by the same author, we treated them separately in this meta-analysis. Of those, 99 case–control studies with 43,085 cancer cases and 57,882 control subjects were included in our meta-analysis. A flow chart showing the detailed steps of study selection is shown in Figure 1. All studies were case–control studies with the following tumor-type distribution: three were conducted for bladder cancer, two for renal cancer, nine for endometrial cancer, eight for ovarian cancer, 62 for breast cancer, six for lung cancer, three for liver cancer, two for colon cancer, two for esophageal cell cancer, one for thyroid cancer and non-Hodgkin lymphoma, and one for testicular germ cell tumor. Fifty studies investigated the risks in Caucasian populations, 35 studies investigated Asian populations, ten studies investigated mixed populations, and the remaining studies were conducted in African populations. Five main genotyping methods were used such as polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), TaqMan, sequencing, matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF), and allele-specific PCR (AS-PCR). By source of controls, 50 studies were population based, 45 studies were hospital based, and four studies were not clear. The distribution of the genotypes in the control subjects was in agreement with HWE, except for eight studies.34,37,70,72,80,88,95,119 The quality assessment showed that the quality scores ranged from 5 to 9 with a median score of 6, suggesting that all studies were of high quality. The main characteristics of the eligible studies are listed in Table 1.
Figure 1.
Flow chart of publication selection.
Note: A total of 99 studies were included in this meta-analysis and systematically reviewed after a comprehensive study selection.
Abbreviation: COMT, catechol-O-methyltransferase.
Table 1.
Characteristics of studies included in the meta-analysis
| Authors | Year | Country | Ethnicity mixed | Cancer type | Control source | Genotype method | Genotype (cases)
|
Genotype (controls)
|
HWE | NOS score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | |||||||||
| Lavigne et al11 | 1997 | USA | Caucasian | Breast | HB | PCR-RFLP | 35 | 57 | 21 | 31 | 56 | 27 | 0.862 | 6 |
| Millikan et al12 | 1998 | USA | African | Breast | PB | PCR-RFLP | 29 | 106 | 130 | 34 | 118 | 111 | 0.838 | 8 |
| Millikan et al12 | 1998 | USA | Caucasian | Breast | PB | PCR-RFLP | 102 | 184 | 103 | 105 | 188 | 86 | 0.916 | 8 |
| Thompson et al13 | 1998 | USA | Caucasian | Breast | PB | PCR-RFLP | 53 | 159 | 69 | 72 | 139 | 78 | 0.522 | 7 |
| Huang et al14 | 1999 | People’s Republic | Asian | Breast | HB | PCR-RFLP | 13 | 37 | 68 | 4 | 55 | 66 | 0.612 | 5 |
| of China | ||||||||||||||
| Goodman et al15 | 2000 | Germany | Caucasian | Ovarian | HB | PCR-RFLP | 27 | 54 | 27 | 29 | 52 | 25 | 0.905 | 7 |
| Goodman et al16 | 2001 | USA | Mixed | Ovarian | PB | PCR-RFLP | 16 | 57 | 52 | 19 | 57 | 68 | 0.827 | 8 |
| Goodman et al17 | 2001 | USA | Caucasian | Breast | PB | PCR-RFLP | 35 | 57 | 20 | 31 | 55 | 27 | 0.788 | 8 |
| Hamajima et al18 | 2001 | Japan | Asian | Breast | HB | PCR-RFLP | 18 | 72 | 60 | 23 | 63 | 79 | 0.079 | 6 |
| Bergman-Jungestrom and Wingren19 | 2001 | Sweden | Caucasian | Breast | HB | PCR-RFLP | 46 | 64 | 16 | 43 | 61 | 13 | 0.209 | 5 |
| Mitrunen et al20 | 2001 | Finland | Caucasian | Breast | PB | PCR-RFLP | 128 | 238 | 115 | 143 | 237 | 100 | 0.921 | 5 |
| Yim et al21 | 2001 | Korea | Asian | Breast | HB | PCR-RFLP | 3 | 79 | 81 | 16 | 46 | 101 | 0.004 | 6 |
| Garner et al22 | 2002 | USA | Mixed | Ovarian | PB | PCR-RFLP | 48 | 103 | 59 | 54 | 119 | 52 | 0.861 | 6 |
| Kocabas et al23 | 2002 | Turkey | Caucasian | Breast | HB | PCR-RFLP | 14 | 42 | 28 | 13 | 55 | 35 | 0.227 | 7 |
| Comings et al24 | 2003 | USA | Caucasian | Breast | PB | PCR-RFLP | 12 | 24 | 31 | 38 | 78 | 29 | 0.335 | 6 |
| Rossi et al25 | 2003 | Italy | Caucasian | Liver | HB | PCR-RFLP | 15 | 56 | 16 | 23 | 51 | 16 | P>0.05 | 6 |
| Tan et al26 | 2003 | People’s Republic of China | Asian | Breast | HB | PCR-RFLP | 26 | 103 | 121 | 13 | 105 | 132 | 0.174 | 8 |
| Wedrén et al27 | 2003 | Sweden | Caucasian | Breast | PB | DASH | 442 | 767 | 281 | 433 | 662 | 245 | 0.772 | 6 |
| Wu et al28 | 2003 | USA | Asian | Breast | PB | TaqMan | 48 | 213 | 328 | 51 | 229 | 282 | 0.646 | 6 |
| Ahsan et al29 | 2004 | USA | Mixed | Breast | FB | LP | 73 | 156 | 84 | 60 | 144 | 58 | 0.108 | 6 |
| Dunning et al30 | 2004 | UK | Caucasian | Breast | PB | TaqMan | 845 | 1,360 | 645 | 534 | 926 | 448 | 0.232 | 8 |
| Hefler et al31 | 2004 | Austria | Caucasian | Breast | PB | Sequencing | 98 | 192 | 101 | 478 | 835 | 385 | 0.577 | 8 |
| Hung et al32 | 2004 | France | Caucasian | Bladder | HB | PCR-RFLP | 43 | 96 | 62 | 43 | 114 | 57 | P>0.05 | 7 |
| McGrath et al33 | 2004 | USA | Caucasian | Endometrial | HB | PCR-RFLP | 55 | 105 | 55 | 172 | 308 | 161 | 0.874 | 7 |
| Sazci et al34 | 2004 | Turkey | Caucasian | Breast | PB | PCR-RFLP | 28 | 69 | 33 | 16 | 146 | 62 | 0 | 6 |
| Yin et al35 | 2004 | People’s Republic of China | Asian | Liver | HB | PCR-RFLP | 30 | 21 | 3 | 49 | 31 | 6 | NA | 7 |
| Zimarina et al36 | 2004 | Russia | Caucasian | Endometrial | HB | PCR-RFLP | 30 | 65 | 29 | 44 | 73 | 23 | 0.996 | 6 |
| Cheng et al37 | 2005 | People’s Republic of China | Asian | Breast | HB | NR | 35 | 197 | 237 | 58 | 262 | 420 | 0.006 | 6 |
| Doherty et al38 | 2005 | USA | Mixed | Endometrial | PB | PCR-RFLP | 100 | 174 | 97 | 123 | 207 | 90 | 0.953 | 6 |
| Huber et al39 | 2005 | Austria | Caucasian | Colon | PB | PCR-RFLP | 0 | 58a | 18 | 0 | 519a | 203 | NA | 6 |
| Lin et al40 | 2005 | People’s Republic of China | Asian | Breast | PB | PCR-RFLP | 5 | 31 | 51 | 18 | 133 | 190 | 0.393 | 6 |
| Lin et al41 | 2005 | People’s Republic of China | Asian | Breast | PB | PCR-RFLP | 6 | 35 | 58 | 23 | 138 | 205 | 0.972 | 6 |
| Le Marchand et al42 | 2005 | USA | Mixed | Breast | PB | PCR-RFLP | 196 | 624 | 519 | 206 | 614 | 550 | 0.109 | 7 |
| Modugno et al43 | 2005 | USA | Caucasian | Breast | PB | TaqMan | 77 | 124 | 49 | 1,104 | 1,943 | 903 | 0.391 | 8 |
| Sellers et al44 | 2005 | USA | Caucasian | Ovarian | HB | PCR-RFLP | 119 | 224 | 110 | 147 | 269 | 127 | 0.903 | 7 |
| Sellers et al44 | 2005 | USA | African | Ovarian | HB | PCR-RFLP | 0 | 17a | 19 | 0 | 30a | 23 | 0.059 | 6 |
| Skibola et al45 | 2005 | USA | Caucasian | NHL | PB | TaqMan | 77 | 153 | 75 | 163 | 323 | 193 | P>0.05 | 7 |
| Wen et al46 | 2005 | People’s Republic of China | Asian | Breast | PB | PCR-RFLP | 83 | 425 | 612 | 93 | 470 | 628 | 0.698 | 7 |
| Chang et al47 | 2006 | People’s Republic of China | Asian | Breast | HB | PCR-RFLP | 9 | 77 | 103 | 30 | 159 | 132 | 0.068 | 7 |
| Gallicchio et al48 | 2006 | USA | Caucasian | Breast | PB | TaqMan | 24 | 41 | 16 | 371 | 608 | 272 | 0.44 | 9 |
| Gaudet et al49 | 2006 | USA | Caucasian | Breast | PB | MALDI-TOF | 240 | 521 | 287 | 266 | 549 | 277 | 0.853 | 8 |
| Gaudet et al49 | 2006 | Poland | Caucasian | Breast | PB | TaqMan | 439 | 993 | 551 | 539 | 1,123 | 617 | 0.525 | 8 |
| Onay et al50 | 2006 | Canada | Caucasian | Breast | PB | TaqMan | 94 | 202 | 102 | 96 | 196 | 80 | 0.283 | 8 |
| Song et al51 | 2006 | People’s Republic of China | Asian | Breast | NR | PCR-RFLP | 3 | 41 | 66 | 11 | 36 | 65 | 0.09 | 5 |
| Tao et al52 | 2006 | People’s Republic of China | Asian | Endometrial | HB | TaqMan | 85 | 383 | 563 | 67 | 425 | 534 | 0.683 | 6 |
| Akisik and Dalay53 | 2007 | Turkey | Caucasian | Breast | NR | PCR-RFLP | 26 | 59 | 29 | 21 | 53 | 34 | 0.966 | 6 |
| Fan et al99 | 2007 | People’s Republic of China | Asian | Breast | HB | PCR-RFLP | 29 | 75 | 96 | 5 | 44 | 51 | 0.25 | 6 |
| Gemignani et al54 | 2007 | European | Caucasian | Lung | HB | PCR-RFLP | 59 | 144 | 83 | 75 | 146 | 81 | 0.569 | 7 |
| Holt et al55 | 2007 | USA | Caucasian | Ovarian | PB | TaqMan | 79 | 129 | 72 | 137 | 209 | 104 | 0.948 | 8 |
| Holt et al55 | 2007 | USA | African | Ovarian | PB | TaqMan | 10 | 19 | 4 | 16 | 58 | 52 | 0.2 | 8 |
| Hu et al57 | 2007 | People’s Republic of China | Asian | Breast | HB | Sequencing | 11 | 36 | 65 | 3 | 41 | 66 | 0.252 | 6 |
| Liu et al119 | 2007 | People’s Republic of China | Asian | Endometrial | HB | PCR-RFLP | 5 | 33 | 42 | 3 | 46 | 35 | 0.01 | 6 |
| Ralph et al56 | 2007 | USA | Caucasian | Breast | HB | TaqMan | 405 | 825 | 396 | 900 | 1,631 | 755 | 0.758 | 7 |
| Szyllo et al58 | 2007 | Poland | Caucasian | Endometrial | HB | PCR-RFLP | 24 | 81 | 46 | 39 | 110 | 48 | 0.253 | 6 |
| Takata et al59 | 2007 | USA | Mixed | Breast | PB | PCR-RFLP | 89 | 257 | 229 | 47 | 108 | 95 | 0.104 | 8 |
| Tanaka et al60 | 2007 | Japan | Asian | Renal | PB | Sequencing | 10 | 54 | 59 | 11 | 61 | 85 | NA | 8 |
| Zhao et al61 | 2007 | People’s Republic of China | Asian | Endometrial | HB | PCR-RFLP | 16 | 77 | 39 | 8 | 50 | 52 | 0.779 | 6 |
| Delort et al62 | 2008 | France | Caucasian | Ovarian | PB | TaqMan | 18 | 22 | 11 | 283 | 480 | 237 | 0.916 | 7 |
| Hirata et al63 | 2008 | USA | Caucasian | Endometrial | PB | PCR-RFLP | 37 | 81 | 32 | 27 | 90 | 48 | 0.277 | 8 |
| Justenhoven et al64 | 2008 | Germany | Caucasian | Breast | PB | MALDI-TOF | 145 | 298 | 163 | 147 | 305 | 170 | 0.654 | 8 |
| Onay et al65 | 2008 | Canada | Caucasian | Breast | PB | TaqMan | 273 | 642 | 302 | 201 | 353 | 160 | 0.832 | 8 |
| Onay et al65 | 2008 | Finland | Caucasian | Breast | PB | TaqMan | 206 | 361 | 141 | 168 | 267 | 114 | 0.676 | 7 |
| Yuan et al66 | 2008 | People’s Republic of China | Asian | Liver | HB | PCR-RFLP | 18 | 144 | 258 | 32 | 157 | 286 | P>0.05 | 6 |
| Zhu100 | 2008 | People’s Republic of China | Asian | Esophageal | HB | PCR-RFLP | 16 | 51 | 23 | 10 | 37 | 30 | P>0.05 | 5 |
| Zienolddiny et al67 | 2008 | Norway | Caucasian | Lung | PB | Sequencing | 32 | 62 | 163 | 8 | 60 | 202 | 0.182 | 8 |
| Cote et al68 | 2009 | USA | African | Lung | PB | TaqMan | 10 | 46 | 56 | 14 | 47 | 59 | 0.332 | 8 |
| Cote et al68 | 2009 | USA | Caucasian | Lung | PB | PCR-RFLP | 102 | 205 | 78 | 114 | 197 | 92 | 0.696 | 8 |
| Fontana et al69 | 2009 | France | Caucasian | Bladder | HB | TaqMan | 14 | 28 | 9 | 10 | 24 | 11 | NA | 6 |
| He et al71 | 2009 | USA | Caucasian | Breast | HB | TaqMan | 334 | 607 | 271 | 446 | 837 | 400 | 0.85 | 7 |
| Reding et al73 | 2009 | USA | Caucasian | Breast | PB | TaqMan | 240 | 427 | 224 | 236 | 431 | 211 | 0.606 | 8 |
| Sangrajrang et al74 | 2009 | Thailand | Asian | Breast | HB | TaqMan | 42 | 233 | 290 | 30 | 190 | 266 | 0.61 | 7 |
| Shrubsole et al75 | 2009 | People’s Republic of China | Asian | Breast | PB | PCR-RFLP | 0 | 497a | 596 | 0 | 554a | 615 | NA | 7 |
| Yadav et al76 | 2009 | India | Asian | Breast | HB | PCR-RFLP | 28 | 82 | 44 | 29 | 85 | 52 | 0.57 | 7 |
| Zhou98 | 2009 | People’s Republic of China | Asian | Colon | PB | SNPlex | 23 | 121 | 208 | 38 | 262 | 327 | P>0.05 | 7 |
| Delort et al77 | 2010 | France | Caucasian | Breast | PB | TaqMan | 254 | 455 | 201 | 283 | 480 | 237 | 0.23 | 8 |
| Ferlin et al78 | 2010 | Italy | Caucasian | TGCT | HB | PCR-RFLP | 0 | 200 | 34 | 2 | 182 | 34 | P>0.05 | 7 |
| MARIE-GENICA Consortium on Genetic Susceptibility for enopausal Hormone Therapy Related Breast Cancer Risk70 | 2010 | Germany | Caucasian | Breast | PB | MALDI-TOF | 844 | 1,569 | 731 | 1,569 | 2,669 | 1,243 | 0.094 | 8 |
| Jakubowska et al72 | 2010 | Poland | Caucasian | Breast | HB | PCR-RFLP | 84 | 164 | 71 | 54 | 168 | 68 | 0.01 | 8 |
| Li et al97 | 2010 | People’s Republic of China | Asian | Endometrial | HB | PCR-RFLP | 6 | 26 | 90 | 8 | 35 | 71 | 0.22 | 5 |
| Martínez et al101 | 2013 | Mexico | Caucasian | Breast | HB | PCR-RFLP | 32 | 66 | 52 | 23 | 59 | 68 | 0.085 | 7 |
| Moreno-Galvan et al79 | 2010 | Mexico | Caucasian | Breast | HB | PCR-RFLP | 12 | 42 | 37 | 14 | 42 | 38 | 0.669 | 6 |
| Peterson et al80 | 2010 | USA | Caucasian | Breast | PB | TaqMan | 420 | 794 | 370 | 403 | 665 | 348 | 0.026 | 8 |
| Syamala et al81 | 2010 | India | Asian | Breast | HB | PCR-RFLP | 41 | 104 | 74 | 65 | 164 | 138 | 0.183 | 6 |
| Syamala et al81 | 2010 | India | Asian | Breast | FB | PCR-RFLP | 28 | 64 | 48 | 65 | 164 | 138 | 0.183 | 6 |
| Wang et al82 | 2010 | People’s Republic of China | Asian | Breast | PB | AS-PCR | 34 | 62 | 80 | 14 | 66 | 96 | 0.58 | 7 |
| Xu et al96 | 2010 | People’s Republic of China | Asian | Breast | PB | AS-PCR | 38 | 42 | 60 | 10 | 44 | 68 | 0.45 | 7 |
| Cerne et al83 | 2011 | Slovenia | Caucasian | Breast | HB | TaqMan | 144 | 263 | 123 | 67 | 136 | 67 | 0.903 | 7 |
| Cribb et al84 | 2011 | Canada | Caucasian | Breast | HB | PCR-RFLP | 51 | 108 | 48 | 155 | 326 | 140 | 0.208 | 7 |
| Huang et al85 | 2011 | People’s Republic of China | Asian | Esophageal | HB | PCR-RFLP | 25 | 95 | 90 | 30 | 146 | 180 | NA | 6 |
| Lajin et al86 | 2013 | Syria | Mixed | Breast | PB | PCR-RFLP | 31 | 70 | 34 | 30 | 54 | 28 | 0.887 | 7 |
| Naushad et al87 | 2011 | India | Asian | Breast | HB | PCR-RFLP | 66 | 154 | 122 | 26 | 107 | 120 | 0.201 | 6 |
| dos Santos et al88 | 2011 | Brazil | Mixed | Breast | PB | PCR-RFLP | 0 | 41a | 21 | 0 | 26a | 36 | – | 7 |
| Wang et al89 | 2011 | People’s Republic of China c | Asian | Breast | PB | Sequencing | 68 | 145 | 187 | 36 | 156 | 208 | 0.389 | 7 |
| Heck et al90 | 2012 | USA | Mixed | Renal | HB | Sequencing | 0 | 632a | 242 | 0 | 1,496a | 557 | 0.36 | 8 |
| Lim et al91 | 2012 | Singapore | Asian | Lung | HB | PCR-RFLP | 39 | 220 | 284 | 63 | 353 | 549 | 0.539 | 7 |
| Wolpert et al92 | 2012 | Egypt | Mixed | Bladder | PB | TaqMan | 160 | 245 | 110 | 95 | 180 | 114 | P>0.05 | 8 |
| Zhang et al93 | 2013 | People’s Republic of China | Asian | Lung | HB | Sequencing | 11 | 69 | 120 | 19 | 78 | 103 | 0.454 | 8 |
| Ghisari et al94 | 2014 | Denmark | Caucasian | Breast | PB | TaqMan | 13 | 11 | 7 | 41 | 53 | 19 | P>0.05 | 6 |
| Son et al95 | 2015 | Korea | Asian | Breast | HB | Assay | 0 | 423a | 427 | 0 | 212a | 178 | 0.008 | 7 |
Note:
Number of patients with the AA + GA genotype in the case and control groups.
Abbreviations: HWE, Hardy–Weinberg equilibrium; NOS, Newcastle–Ottawa Scale; HB, hospital based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PB, population based; DASH, dynamic allele-specific hybridization; FB, family based; NA, not available; MALDI-TOF, matrix-assisted laser desorption ionization time of flight mass spectrometry; NHL, non-Hodgkin lymphoma; TGCT, testicular germ cell tumor; AS-PCR, allele-specific PCR; LP, Luorescence polarization; NR, not reported.
Quantitative synthesis
Overall, no significant associations between COMT Val158Met and cancer risk were found using homozygote model (OR =1.05, 95% CI [0.98, 1.13]), heterozygote model (OR =1.01, 95% CI [0.98, 1.04]), dominant model (OR =1.02, 95% CI [0.97, 1.06]), or recessive model (OR =1.03, 95% CI [0.97, 1.09]).
Significant heterogeneity was observed among the 99 studies on COMT Val158Met polymorphism. To explore the source of heterogeneity, we performed stratified analyses on ethnicity, cancer type, source of controls, and genotyping method. In the subgroup analysis on cancer type, COMT Val158Met was significantly associated with an increased risk of bladder cancer in recessive model (OR =1.30, 95% CI [1.02, 1.66]), esophageal cell cancer in homozygote model (OR =1.77, 95% CI [1.07, 2.93]), heterozygote model (OR =1.40, 95% CI [1.01, 1.92]), and dominant model (OR =1.46, 95% CI [1.08, 1.98]). However, studies on renal, endometrial, lung, liver, ovarian, colon, and other cancer types have suggested null association (OR =0.70–1.46; Table 2). These studies were further stratified on the basis of ethnicities, and the results showed that COMT Val158-Met polymorphism may be a risk factor for cancer in Asian populations in the homozygote model (OR =1.25, 95% CI [1.03, 1.51]) and recessive model (OR =1.20, 95% CI [1.01, 1.43]). We failed to detect any association between the COMT Val158Met polymorphism and African, Caucasian, and mixed populations. In addition, homozygote models (OR =3.46, 95% CI [2.07, 5.80]), recessive models (OR =3.32, 95% CI [2.02, 5.44]), and dominant models (OR =1.54, 95% CI [1.12, 2.11]) were significantly associated with increased cancer risk in the subgroup of AS-PCR genotyping method, but no significant associations were observed when PCR-RFLP, TaqMan, sequencing, MALDI-TOF, and other genotyping method were used. No significant associations were detected when the studies were stratified on the basis of the source of control subjects.
Table 2.
Meta-analysis of the association between COMT Val158Met and cancer risk
| Variables | No of studies | Homozygote model
|
Heterozygote model
|
Recessive model
|
Dominant model
|
||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% Cl) | I2% | OR (95% Cl) | I2% | OR (95% Cl) | I2% | OR (95% Cl) | I2% | ||
| Total | 99 | 1.05 (0.98, 1.13) | 56 | 1.01 (0.97, 1.05) | 29 | 1.03 (0.97, 1.09) | 51 | 1.02 (0.97, 1.06) | 44 |
| Cancer type | |||||||||
| Bladder | 3 | 1.38 (0.86, 2.21) | 45 | 1.12 (0.71, 1.77) | 57 | 1.30 (1.02, 1.66) | 0 | 1.20 (0.74, 1.94) | 65 |
| Renal | 2 | 1.31 (0.52, 3.28) | – | 1.28 (0.78, 2.09) | – | 1.18 (0.48, 2.86) | – | 1.02 (0.83, 1.25) | 12 |
| Breast | 62 | 1.04 (0.96, 1.13) | 58 | 1.01 (0.96, 1.05) | 21 | 1.03 (0.96, 1.10) | 57 | 1.01 (0.96, 1.06) | 40 |
| Endometrial | 9 | 0.99 (0.73, 1.35) | 55 | 0.90 (0.73, 1.11) | 52 | 1.03 (0.84, 1.26) | 29 | 0.91 (0.73, 1.13) | 61 |
| Lung | 6 | 1.09 (0.68, 1.75) | 76 | 1.11 (0.96, 1.28) | 2 | 1.04 (0.67, 1.57) | 74 | 1.09 (0.87, 1.36) | 60 |
| Liver | 3 | 0.68 (0.42, 1.09) | 0 | 1.03 (0.80, 1.34) | 0 | 0.70 (0.48, 1.03) | 0 | 0.96 (0.75, 1.23) | 0 |
| Ovarian | 8 | 1.05 (0.75, 1.47) | 52 | 1.01 (0.80, 1.28) | 33 | 1.02 (0.84, 1.24) | 20 | 1.00 (0.79, 1.27) | 43 |
| Colon | 2 | 0.95 (0.55, 1.64) | – | 0.73 (0.55, 0.96) | – | 1.08 (0.64, 1.85) | – | 0.92 (0.56, 1.50) | 63 |
| Esophageal | 2 | 1.77 (1.07, 2.93) | 0 | 1.40 (1.01, 1.92) | 0 | 1.46 (0.92, 2.34) | 0 | 1.46 (1.08, 1.98) | 0 |
| Other | 2 | 0.96 (0.29, 3.16) | 24 | 1.18 (0.90, 1.56) | 0 | 0.87 (0.29, 2.62) | 21 | 1.18 (0.91, 1.54) | 0 |
| Ethnicities | |||||||||
| African | 4 | 1.46 (0.43, 4.99) | 83 | 1.23 (0.61, 2.49) | 75 | 1.17 (0.53, 2.56) | 69 | 1.09 (0.60, 1.98) | 73 |
| Caucasian | 50 | 0.98 (0.91, 1.05) | 43 | 1.00 (0.96, 1.05) | 88 | 0.97 (0.92, 1.03) | 38 | 0.99 (0.95, 1.04) | 16 |
| Asian | 35 | 1.25 (1.03, 1.51) | 62 | 1.04 (0.94, 1.14) | 53 | 1.20 (1.01, 1.43) | 60 | 1.06 (0.97, 1.15) | 59 |
| Mixed | 10 | 0.96 (0.78, 1.20) | 49 | 1.00 (0.86, 1.17) | 38 | 0.99 (0.87, 1.13) | 5 | 1.03 (0.88, 1.20) | 58 |
| Controls source | |||||||||
| PB | 50 | 1.03 (0.94, 1.13) | 63 | 0.99 (0.94, 1.04) | 24 | 1.06 (0.95, 1.17) | 58 | 1.01 (0.95, 1.07) | 49 |
| HB | 45 | 1.09 (0.96, 1.24) | 48 | 1.04 (0.96, 1.12) | 36 | 1.02 (0.94, 1.09) | 43 | 1.04 (0.96, 1.11) | 41 |
| Other | 4 | 0.95 (0.59, 1.54) | 48 | 1.00 (0.78, 1.27) | 4 | 1.00 (0.69, 1.46) | 38 | 0.99 (0.78, 1.26) | 7 |
| Genotyping method | |||||||||
| PCR-RFLP | 58 | 1.02 (0.91, 1.15) | 49 | 1.01 (0.94, 1.09) | 36 | 1.01 (0.92, 1.11) | 42 | 1.02 (0.95, 1.09) | 44 |
| TaqMan | 24 | 1.03 (0.94, 1.13) | 46 | 1.02 (0.96, 1.08) | 15 | 1.00 (0.93, 1.07) | 35 | 1.02 (0.95, 1.08) | 34 |
| Sequencing | 6 | 1.55 (0.79, 3.03) | 85 | 0.98 (0.84, 1.14) | 1 | 1.55 (0.84, 2.86) | 84 | 1.09 (0.84, 1.41) | 67 |
| MALDI-TOF | 3 | 0.92 (0.83, 1.02) | 0 | 0.98 (0.90, 1.08) | 0 | 0.93 (0.85, 1.01) | 0 | 0.96 (0.88, 1.05) | 0 |
| AS-PCR | 2 | 3.46 (2.07, 5.80) | 0 | 1.11 (0.78, 1.57) | 0 | 3.32 (2.02, 5.44) | 0 | 1.54 (1.12, 2.11) | 0 |
| Other | 6 | 0.91 (0.77, 1.08) | 0 | 0.94 (0.72, 1.24) | 76 | 0.92 (0.80, 1.05) | 0 | 0.93 (0.81, 1.08) | 57 |
Notes: The bold values indicate that the results are statistically significant.
Abbreviations: COMT, catechol-O-methyltransferase; OR, odds ratio; CI, confidence interval; PB, population based; HB, hospital based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; MALDI-TOF, matrix-assisted laser desorption ionization time of flight mass spectrometry; AS-PCR, allele-specific PCR; I2, variation in OR attributable to heterogeneity.
Test of heterogeneity and sensitivity
Heterogeneity among studies was observed in the overall comparisons as well as in the subgroup analyses. The source of heterogeneity was investigated by cancer ethnicity (European, Asian, African, and mixed; P=0.483), cancer types (bladder, breast, renal, endometrial, lung, liver, ovarian, colon, and other cancer types; P=0.684), control source (population based, hospital based, and family based; P=0.659), and genotyping method (AS-PCR, PCR-RFLP, TaqMan, sequencing, MALDI-TOF, and other genotyping method; P=0.647) using meta-regression, but no covariables were found to contribute to the heterogeneity.
Sensitivity analysis was conducted to verify the effect of each study on the overall OR by repeating the meta-analysis, but one study was omitted each time. When sensitivity analyses were performed without HWE violating studies, all the results were not materially altered. The results showed that the pooled ORs of these three polymorphisms were not materially altered by the contribution of any individual study, thus confirming that the results of this meta-analysis were statistically robust.
Publication bias
Begg’s funnel plot and Egger’s test were performed to evaluate the publication bias of the studies. The shape of the funnel plots showed that the dots were almost symmetrically distributed and were predominantly in 95% confidence limits (dominant model, Figure 2). The results of Egger’s test statistically confirmed the absence of publication bias in the dominant model (t=1.68, P=0.096).
Figure 2.
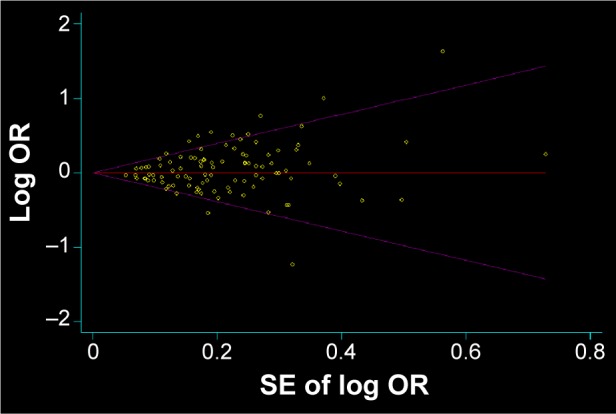
Begg’s funnel plot of the meta-analysis of cancer risk and COMT Val158Met polymorphism (AA + AG vs GG).
Note: Begg’s funnel plot with pseudo 95% confidence limits.
Abbreviations: COMT, catechol-O-methyltransferase; OR, odds ratio; SE, standard error.
Discussion
In the past several years, interest in the genetic susceptibility to cancers has drawn increased attention to the studies on polymorphisms of genes involved in tumor genesis. Genome-wide association study, also known as whole genome association study, is widely used in the study of genetic epidemiology. At present, >1,369 susceptibility loci associated with cancer risk have been identified by genome-wide association study, but none of these studies had reported significant associations between cancer susceptibility and COMT Val158Met polymorphisms. We searched the manufacturers’ websites (http://www.affymetrix.com/index.affx and http://www.illumina.com)120 and the relevant PubMed databases (Probe, Database of Genotypes and Phenotypes, and Gene Expression Omnibus DataSets) and found that the COMT Val158Met polymorphism was not included in the platforms commonly used in genome-wide association studies. But since the identification of COMT Val158Met polymorphism, the role of COMT Val158Met in cancers risk has been reported in an increasing number of studies, but the results remained controversial. Some recent meta-analyses studies reported such an association only for single cancer or specific populations. Importantly, several published studies were not included in the previous meta-analysis, and additional original studies with larger sample sizes have been published since then. Hence, the association between the COMT Val158Met polymorphism and the risk of cancer remains unknown. Therefore, meta-analysis can provide a quantitative summary of the available data supporting the association between COMT Val158Met and cancer risk. Compared with some previous meta-analyses, strengths of our meta-analysis include the large sample size and high statistical power of the analysis based on substantial number of cases and controls from differential studies, which minimized selection bias and led to relatively stable risk estimation.
In the current meta-analysis, 99 case–control studies with 43,085 cancer cases and 57,882 control subjects were considered. The results indicated no significant association between COMT Val158Met polymorphism and overall cancer risk in any genetic comparison model tested. In further subgroup analysis by cancer type, COMT Val158Met was significantly associated with an increased risk of bladder cancer and esophageal cancer in some specific genetic models. However, studies on renal, endometrial, lung, liver, ovarian, colon cancers, and other cancer types have suggested null associations. In line with most previous meta-analyses for single cancer, Zhang et al,111 Du et al102 and Mao et al121 have reported that the COMT Val158Met polymorphism may not contribute to the risk of prostate cancer, ovarian cancer, or breast cancer in any of the assessed genetic model. In the subgroup analysis by ethnicity, no significant associations were found in African, Caucasian, and mixed populations. However, the significant association between the COMT Val158Met polymorphism and cancer risk remains to be determined in Asians. The discrepancy in ethnicity could be attributed to the evident difference in the minor allele frequency of Val158Met polymorphism in Asians and Caucasians in our meta-analysis. This genetic polymorphism variance with ethnicity was consistent with those described in a previous study.8 In addition, stratified analyses by genotyping techniques indicated that studies involving AS-PCR likely acquired significant results in the overall comparison. However, this result should be carefully interpreted because of a relatively small sample size. Moreover, this result should be confirmed by further analysis of additional published studies.
Several limitations should be acknowledged in this meta-analysis. First, only studies in English or Chinese were included in this meta-analysis, which might cause publication bias. Second, the pooled results were based on unadjusted estimates because not all studies had provided adjusted ORs. Even in cases where adjusted ORs were found, they were not adjusted by the same confounders. Hence, a precise analysis should be performed. Third, several factors such as gene–gene or gene–environment interaction may influence gene-disease factor, and the lack of individual data from the included studies limited further evaluation of other potential interactions, as in other genes and environment factors. Finally, cancer is a multifactorial disease resulting from complex interactions among many genetic and environmental factors. Therefore, a single gene or single environmental factor is unlikely to explain cancer susceptibility.
Conclusion
In conclusion, the present meta-analysis suggested that COMT Val158Met polymorphism might not be a risk factor for overall cancer risk, but it might be involved in cancer development at least in some ethnic groups (Asian) or some specific cancer types (bladder and esophageal cancer). Further large-scale and well-designed studies regarding different ethnicities are required to confirm the results of our meta-analysis.
Acknowledgments
This study was supported by National Natural Science Foundation of China (no 81401187).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015 Feb 4; doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human Development Index (2008–2030): a population-based study. The Lancet Oncology. 2012;13(8):790–801. doi: 10.1016/S1470-2045(12)70211-5. [DOI] [PubMed] [Google Scholar]
- 3.Bredberg A. Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer. 2011;117(3):440–445. doi: 10.1002/cncr.25440. [DOI] [PubMed] [Google Scholar]
- 4.Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. American Journal of Epidemiology. 2006;164(1):1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- 5.Foulkes WD. Inherited susceptibility to common cancers. The New England Journal of Medicine. 2008;359(20):2143–2153. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 6.Grossman MH, Emanuel BS, Budarf ML. Chromosomal mapping of the human catechol-O-methyltransferase gene to 22q11.1–q11.2. Genomics. 1992;12(4):822–825. doi: 10.1016/0888-7543(92)90316-k. [DOI] [PubMed] [Google Scholar]
- 7.Cavalieri EL, Stack DE, Devanesan PD, et al. Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(20):10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian C, Liu L, Yang X, Wu H, Ouyang Q. The Val158Met polymorphism in the COMT gene is associated with increased cancer risks in Chinese population. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(4):3003–3008. doi: 10.1007/s13277-013-1387-6. [DOI] [PubMed] [Google Scholar]
- 9.Dawling S, Roodi N, Mernaugh RL, Wang X, Parl FF. Catechol-O-methyltransferase (COMT)-mediated metabolism of catechol estrogens: comparison of wild-type and variant COMT isoforms. Cancer Research. 2001;61(18):6716–6722. [PubMed] [Google Scholar]
- 10.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Lavigne JA, Helzlsouer KJ, Huang HY, et al. An association between the allele coding for a low activity variant of catechol-O-methyltransferase and the risk for breast cancer. Cancer Research. 1997;57(24):5493–5497. [PubMed] [Google Scholar]
- 12.Millikan RC, Pittman GS, Tse CK, et al. Catechol-O-methyltransferase and breast cancer risk. Carcinogenesis. 1998;19(11):1943–1947. doi: 10.1093/carcin/19.11.1943. [DOI] [PubMed] [Google Scholar]
- 13.Thompson PA, Shields PG, Freudenheim JL, et al. Genetic polymorphisms in catechol-O-methyltransferase, menopausal status, and breast cancer risk. Cancer Research. 1998;58(10):2107–2110. [PubMed] [Google Scholar]
- 14.Huang CS, Chern HD, Chang KJ, Cheng CW, Hsu SM, Shen CY. Breast cancer risk associated with genotype polymorphism of the estrogen-metabolizing genes CYP17, CYP1A1, and COMT: a multigenic study on cancer susceptibility. Cancer Research. 1999;59(19):4870–4875. [PubMed] [Google Scholar]
- 15.Goodman JE, Lavigne JA, Hengstler JG, Tanner B, Helzlsouer KJ, Yager JD. Catechol-O-methyltransferase polymorphism is not associated with ovarian cancer risk. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2000;9(12):1373–1376. [PubMed] [Google Scholar]
- 16.Goodman MT, McDuffie K, Kolonel LN, et al. Case-control study of ovarian cancer and polymorphisms in genes involved in catecholestrogen formation and metabolism. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2001;10(3):209–216. [PubMed] [Google Scholar]
- 17.Goodman JE, Lavigne JA, Wu K, et al. COMT genotype, micronutrients in the folate metabolic pathway and breast cancer risk. Carcinogenesis. 2001;22(10):1661–1665. doi: 10.1093/carcin/22.10.1661. [DOI] [PubMed] [Google Scholar]
- 18.Hamajima N, Matsuo K, Tajima K, et al. Limited association between a catechol-O-methyltransferase (COMT) polymorphism and breast cancer risk in Japan. International Journal of Clinical Oncology. 2001;6(1):13–18. doi: 10.1007/pl00012073. [DOI] [PubMed] [Google Scholar]
- 19.Bergman-Jungestrom M, Wingren S. Catechol-O-Methyltransferase (COMT) gene polymorphism and breast cancer risk in young women. British Journal of Cancer. 2001;85(6):859–862. doi: 10.1054/bjoc.2001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitrunen K, Jourenkova N, Kataja V, et al. Polymorphic catechol-O-methyltransferase gene and breast cancer risk. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2001;10(6):635–640. [PubMed] [Google Scholar]
- 21.Yim DS, Parkb SK, Yoo KY, et al. Relationship between the Val158Met polymorphism of catechol O-methyl transferase and breast cancer. Pharmacogenetics. 2001;11(4):279–286. doi: 10.1097/00008571-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Garner EI, Stokes EE, Berkowitz RS, Mok SC, Cramer DW. Polymorphisms of the estrogen-metabolizing genes CYP17 and catechol-O-methyltransferase and risk of epithelial ovarian cancer. Cancer Research. 2002;62(11):3058–3062. [PubMed] [Google Scholar]
- 23.Kocabas NA, Sardas S, Cholerton S, Daly AK, Karakaya AE. Cytochrome P450 CYP1B1 and catechol O-methyltransferase (COMT) genetic polymorphisms and breast cancer susceptibility in a Turkish population. Archives of Toxicology. 2002;76(11):643–649. doi: 10.1007/s00204-002-0387-x. [DOI] [PubMed] [Google Scholar]
- 24.Comings DE, Gade-Andavolu R, Cone LA, Muhleman D, MacMurray JP. A multigene test for the risk of sporadic breast carcinoma. Cancer. 2003;97(9):2160–2170. doi: 10.1002/cncr.11340. [DOI] [PubMed] [Google Scholar]
- 25.Rossi L, Leveri M, Gritti C, et al. Genetic polymorphisms of steroid hormone metabolizing enzymes and risk of liver cancer in hepatitis C- infected patients. Journal of Hepatology. 2003;39(4):564–570. doi: 10.1016/s0168-8278(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 26.Tan W, Qi J, Xing DY, et al. Relation between single nucleotide polymorphism in estrogen-metabolizing genes COMT, CYP17 and breast cancer risk among Chinese women. Zhonghua zhong liu za zhi [Chinese Journal of Oncology] 2003;25(5):453–456. Chinese. [PubMed] [Google Scholar]
- 27.Wedren S, Rudqvist TR, Granath F, et al. Catechol-O-methyltransferase gene polymorphism and post-menopausal breast cancer risk. Carcinogenesis. 2003;24(4):681–687. doi: 10.1093/carcin/bgg022. [DOI] [PubMed] [Google Scholar]
- 28.Wu AH, Tseng CC, Van Den Berg D, Yu MC. Tea intake, COMT genotype, and breast cancer in Asian-American women. Cancer Research. 2003;63(21):7526–7529. [PubMed] [Google Scholar]
- 29.Ahsan H, Chen Y, Whittemore AS, et al. A family-based genetic association study of variants in estrogen-metabolism genes COMT and CYP1B1 and breast cancer risk. Breast Cancer Research and Treatment. 2004;85(2):121–131. doi: 10.1023/B:BREA.0000025401.60794.68. [DOI] [PubMed] [Google Scholar]
- 30.Dunning AM, Dowsett M, Healey CS, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. Journal of the National Cancer Institute. 2004;96(12):936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 31.Hefler LA, Tempfer CB, Grimm C, et al. Estrogen-metabolizing gene polymorphisms in the assessment of breast carcinoma risk and fibroadenoma risk in Caucasian women. Cancer. 2004;101(2):264–269. doi: 10.1002/cncr.20361. [DOI] [PubMed] [Google Scholar]
- 32.Hung RJ, Boffetta P, Brennan P, et al. Genetic polymorphisms of MPO, COMT, MnSOD, NQO1, interactions with environmental exposures and bladder cancer risk. Carcinogenesis. 2004;25(6):973–978. doi: 10.1093/carcin/bgh080. [DOI] [PubMed] [Google Scholar]
- 33.McGrath M, Hankinson SE, Arbeitman L, Colditz GA, Hunter DJ, De Vivo I. Cytochrome P450 1B1 and catechol-O-methyltransferase polymorphisms and endometrial cancer susceptibility. Carcinogenesis. 2004;25(4):559–565. doi: 10.1093/carcin/bgh039. [DOI] [PubMed] [Google Scholar]
- 34.Sazci A, Ergul E, Utkan NZ, Canturk NZ, Kaya G. Catechol-O-methyltransferase Val 108/158 Met polymorphism in premenopausal breast cancer patients. Toxicology. 2004;204(2–3):197–202. doi: 10.1016/j.tox.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 35.Yin PH, Lee HC, Chau GY, et al. Polymorphisms of estrogen-metabolizing genes and risk of hepatocellular carcinoma in Taiwan females. Cancer Letters. 2004;212(2):195–201. doi: 10.1016/j.canlet.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 36.Zimarina TC, Kristensen VN, Imianitov EN, Bershtein LM. Polymorphisms of CYP1B1 and COMT in breast and endometrial cancer. Molekuliarnaia Biologiia. 2004;38(3):386–393. Chinese. [PubMed] [Google Scholar]
- 37.Cheng TC, Chen ST, Huang CS, et al. Breast cancer risk associated with genotype polymorphism of the catechol estrogen-metabolizing genes: a multigenic study on cancer susceptibility. International Journal of Cancer. Journal International Du Cancer. 2005;113(3):345–353. doi: 10.1002/ijc.20630. [DOI] [PubMed] [Google Scholar]
- 38.Doherty JA, Weiss NS, Freeman RJ, et al. Genetic factors in catechol estrogen metabolism in relation to the risk of endometrial cancer. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2005;14(2):357–366. doi: 10.1158/1055-9965.EPI-04-0479. [DOI] [PubMed] [Google Scholar]
- 39.Huber A, Bentz EK, Schneeberger C, Huber JC, Hefler L, Tempfer C. Ten polymorphisms of estrogen-metabolizing genes and a family history of colon cancer – an association study of multiple gene-gene interactions. Journal of the Society for Gynecologic Investigation. 2005;12(7):e51–e54. doi: 10.1016/j.jsgi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Lin WY, Chou YC, Wu MH, et al. Polymorphic catechol-O-methyltransferase gene, duration of estrogen exposure, and breast cancer risk: a nested case-control study in Taiwan. Cancer Detection and Prevention. 2005;29(5):427–432. doi: 10.1016/j.cdp.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Lin SC, Chou YC, Wu MH, et al. Genetic variants of myeloperoxidase and catechol-O-methyltransferase and breast cancer risk. European Journal of Cancer Prevention: The Official Journal of the European Cancer Prevention Organisation. 2005;14(3):257–261. doi: 10.1097/00008469-200506000-00010. [DOI] [PubMed] [Google Scholar]
- 42.Le Marchand L, Donlon T, Kolonel LN, Henderson BE, Wilkens LR. Estrogen metabolism-related genes and breast cancer risk: the multiethnic cohort study. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2005;14(8):1998–2003. doi: 10.1158/1055-9965.EPI-05-0076. [DOI] [PubMed] [Google Scholar]
- 43.Modugno F, Zmuda JM, Potter D, et al. Estrogen metabolizing polymorphisms and breast cancer risk among older white women. Breast Cancer Research and Treatment. 2005;93(3):261–270. doi: 10.1007/s10549-005-5347-8. [DOI] [PubMed] [Google Scholar]
- 44.Sellers TA, Schildkraut JM, Pankratz VS, et al. Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2005;14(11 Pt 1):2536–2543. doi: 10.1158/1055-9965.EPI-05-0142. [DOI] [PubMed] [Google Scholar]
- 45.Skibola CF, Bracci PM, Paynter RA, et al. Polymorphisms and haplotypes in the cytochrome P450 17A1, prolactin, and catechol-O-methyltransferase genes and non-Hodgkin lymphoma risk. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2005;14(10):2391–2401. doi: 10.1158/1055-9965.EPI-05-0343. [DOI] [PubMed] [Google Scholar]
- 46.Wen W, Cai Q, Shu XO, et al. Cytochrome P450 1B1 and catechol-O-methyltransferase genetic polymorphisms and breast cancer risk in Chinese women: results from the shanghai breast cancer study and a meta-analysis. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2005;14(2):329–335. doi: 10.1158/1055-9965.EPI-04-0392. [DOI] [PubMed] [Google Scholar]
- 47.Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ, Huang W. Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast. 2006;15(6):754–761. doi: 10.1016/j.breast.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Gallicchio L, Berndt SI, McSorley MA, et al. Polymorphisms in estrogen-metabolizing and estrogen receptor genes and the risk of developing breast cancer among a cohort of women with benign breast disease. BMC Cancer. 2006;6:173. doi: 10.1186/1471-2407-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaudet MM, Bensen JT, Schroeder J, et al. Catechol-O-methyltransferase haplotypes and breast cancer among women on Long Island, New York. Breast Cancer Research and Treatment. 2006;99(2):235–240. doi: 10.1007/s10549-006-9205-0. [DOI] [PubMed] [Google Scholar]
- 50.Onay VU, Briollais L, Knight JA, et al. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song CG, Hu Z, Yuan WT, et al. Prevalence of Val158Met polymorphism in COMT gene on non-BRCA1/2 hereditary breast cancer. Zhonghua wai ke za zhi [Chinese Journal of Surgery] 2006;44(19):1310–1313. Chinese. [PubMed] [Google Scholar]
- 52.Tao MH, Cai Q, Xu WH, et al. Cytochrome P450 1B1 and catechol-O-methyltransferase genetic polymorphisms and endometrial cancer risk in Chinese women. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2006;15(12):2570–2573. doi: 10.1158/1055-9965.EPI-06-0498. [DOI] [PubMed] [Google Scholar]
- 53.Akisik E, Dalay N. Functional polymorphism of thymidylate synthase, but not of the COMT and IL-1B genes, is associated with breast cancer. Journal of Clinical Laboratory Analysis. 2007;21(2):97–102. doi: 10.1002/jcla.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gemignani F, Landi S, Szeszenia-Dabrowska N, et al. Development of lung cancer before the age of 50: the role of xenobiotic metabolizing genes. Carcinogenesis. 2007;28(6):1287–1293. doi: 10.1093/carcin/bgm021. [DOI] [PubMed] [Google Scholar]
- 55.Holt SK, Rossing MA, Malone KE, Schwartz SM, Weiss NS, Chen C. Ovarian cancer risk and polymorphisms involved in estrogen catabo-lism. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2007;16(3):481–489. doi: 10.1158/1055-9965.EPI-06-0831. [DOI] [PubMed] [Google Scholar]
- 56.Ralph DA, Zhao LP, Aston CE, et al. Age-specific association of steroid hormone pathway gene polymorphisms with breast cancer risk. Cancer. 2007;109(10):1940–1948. doi: 10.1002/cncr.22634. [DOI] [PubMed] [Google Scholar]
- 57.Hu Z, Song CG, Lu JS, et al. A multigenic study on breast cancer risk associated with genetic polymorphisms of ER Alpha, COMT and CYP19 gene in BRCA1/BRCA2 negative Shanghai women with early onset breast cancer or affected relatives. Journal of Cancer Research and Clinical Oncology. 2007;133(12):969–978. doi: 10.1007/s00432-007-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szyllo K, Smolarz B, Romanowicz-Makowska H, Przybylowska K, Lewy J, Kulig B. The risk of endometrial cancer appearance and CYP19 and COMT gene polymorphism. Polski merkuriusz lekarski: organ Polskiego Towarzystwa Lekarskiego. 2007;22(129):208–210. Chinese. [PubMed] [Google Scholar]
- 59.Takata Y, Maskarinec G, Le Marchand L. Breast density and polymorphisms in genes coding for CYP1A2 and COMT: the Multiethnic Cohort. BMC Cancer. 2007;7:30. doi: 10.1186/1471-2407-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka Y, Hirata H, Chen Z, et al. Polymorphisms of catechol-O-methyltransferase in men with renal cell cancer. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2007;16(1):92–97. doi: 10.1158/1055-9965.EPI-06-0605. [DOI] [PubMed] [Google Scholar]
- 61.Zhao XM, Xie MQ, Yang DZ, et al. Polymorphism of catechol-O-methyltransferase gene in relation to the risk of endometrial cancer. Zhonghua fu chan ke za zhi. 2007;42(2):116–119. Chinese. [PubMed] [Google Scholar]
- 62.Delort L, Chalabi N, Satih S, et al. Association between genetic polymorphisms and ovarian cancer risk. Anticancer Research. 2008;28(5B):3079–3081. [PubMed] [Google Scholar]
- 63.Hirata H, Hinoda Y, Okayama N, et al. COMT polymorphisms affecting protein expression are risk factors for endometrial cancer. Molecular Carcinogenesis. 2008;47(10):768–774. doi: 10.1002/mc.20432. [DOI] [PubMed] [Google Scholar]
- 64.Justenhoven C, Hamann U, Schubert F, et al. Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Research and Treatment. 2008;108(1):137–149. doi: 10.1007/s10549-007-9586-8. [DOI] [PubMed] [Google Scholar]
- 65.Onay UV, Aaltonen K, Briollais L, et al. Combined effect of CCND1 and COMT polymorphisms and increased breast cancer risk. BMC Cancer. 2008;8:6. doi: 10.1186/1471-2407-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan X, Zhou G, Zhai Y, et al. Lack of association between the functional polymorphisms in the estrogen-metabolizing genes and risk for hepatocellular carcinoma. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2008;17(12):3621–3627. doi: 10.1158/1055-9965.EPI-08-0742. [DOI] [PubMed] [Google Scholar]
- 67.Zienolddiny S, Campa D, Lind H, et al. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of non-small cell lung cancer in smokers. Carcinogenesis. 2008;29(6):1164–1169. doi: 10.1093/carcin/bgn020. [DOI] [PubMed] [Google Scholar]
- 68.Cote ML, Yoo W, Wenzlaff AS, et al. Tobacco and estrogen metabolic polymorphisms and risk of non-small cell lung cancer in women. Carcinogenesis. 2009;30(4):626–635. doi: 10.1093/carcin/bgp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fontana L, Delort L, Joumard L, et al. Genetic polymorphisms in CYP1A1, CYP1B1, COMT, GSTP1 and NAT2 genes and association with bladder cancer risk in a French cohort. Anticancer Research. 2009;29(5):1631–1635. [PubMed] [Google Scholar]
- 70.Risk M-GCoGSfMHTRBC Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Research and Treatment. 2010;119(2):463–474. doi: 10.1007/s10549-009-0407-0. [DOI] [PubMed] [Google Scholar]
- 71.He C, Tamimi RM, Hankinson SE, Hunter DJ, Han J. A prospective study of genetic polymorphism in MPO, antioxidant status, and breast cancer risk. Breast Cancer Research and Treatment. 2009;113(3):585–594. doi: 10.1007/s10549-008-9962-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jakubowska A, Gronwald J, Menkiszak J, et al. BRCA1-associated breast and ovarian cancer risks in Poland: no association with commonly studied polymorphisms. Breast Cancer Research and Treatment. 2010;119(1):201–211. doi: 10.1007/s10549-009-0390-5. [DOI] [PubMed] [Google Scholar]
- 73.Reding KW, Weiss NS, Chen C, et al. Genetic polymorphisms in the catechol estrogen metabolism pathway and breast cancer risk. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2009;18(5):1461–1467. doi: 10.1158/1055-9965.EPI-08-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sangrajrang S, Sato Y, Sakamoto H, et al. Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. International Journal of Cancer Journal International Du Cancer. 2009;125(4):837–843. doi: 10.1002/ijc.24434. [DOI] [PubMed] [Google Scholar]
- 75.Shrubsole MJ, Lu W, Chen Z, et al. Drinking green tea modestly reduces breast cancer risk. The Journal of Nutrition. 2009;139(2):310–316. doi: 10.3945/jn.108.098699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yadav S, Singhal NK, Singh V, Rastogi N, Srivastava PK, Singh MP. Association of single nucleotide polymorphisms in CYP1B1 and COMT genes with breast cancer susceptibility in Indian women. Disease Markers. 2009;27(5):203–210. doi: 10.3233/DMA-2009-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delort L, Satih S, Kwiatkowski F, Bignon YJ, Bernard-Gallon DJ. Evaluation of breast cancer risk in a multigenic model including low penetrance genes involved in xenobiotic and estrogen metabolisms. Nutrition and Cancer. 2010;62(2):243–251. doi: 10.1080/01635580903305300. [DOI] [PubMed] [Google Scholar]
- 78.Ferlin A, Ganz F, Pengo M, Selice R, Frigo AC, Foresta C. Association of testicular germ cell tumor with polymorphisms in estrogen receptor and steroid metabolism genes. Endocrine-Related Cancer. 2010;17(1):17–25. doi: 10.1677/ERC-09-0176. [DOI] [PubMed] [Google Scholar]
- 79.Moreno-Galvan M, Herrera-Gonzalez NE, Robles-Perez V, Velasco-Rodriguez JC, Tapia-Conyer R, Sarti E. Impact of CYP1A1 and COMT genotypes on breast cancer risk in Mexican women: a pilot study. The International Journal of Biological Markers. 2010;25(3):157–163. doi: 10.1177/172460081002500306. [DOI] [PubMed] [Google Scholar]
- 80.Peterson NB, Trentham-Dietz A, Garcia-Closas M, et al. Association of COMT haplotypes and breast cancer risk in caucasian women. Anticancer Research. 2010;30(1):217–220. [PMC free article] [PubMed] [Google Scholar]
- 81.Syamala VS, Syamala V, Sheeja VR, Kuttan R, Balakrishnan R, Ankathil R. Possible risk modification by polymorphisms of estrogen metabolizing genes in familial breast cancer susceptibility in an Indian population. Cancer Investigation. 2010;28(3):304–311. doi: 10.3109/07357900902744494. [DOI] [PubMed] [Google Scholar]
- 82.Wang Q, Wang YP, Li JY, Yuan P, Yang F, Li H. Polymorphic catechol-O-methyltransferase gene, soy isoflavone intake and breast cancer in postmenopausal women: a case-control study. Chinese Journal of Cancer. 2010;29(7):683–688. doi: 10.5732/cjc.009.10700. [DOI] [PubMed] [Google Scholar]
- 83.Cerne JZ, Pohar-Perme M, Novakovic S, Frkovic-Grazio S, Stegel V, Gersak K. Combined effect of CYP1B1, COMT, GSTP1, and MnSOD genotypes and risk of postmenopausal breast cancer. Journal of Gynecologic Oncology. 2011;22(2):110–119. doi: 10.3802/jgo.2011.22.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cribb AE, Joy Knight M, Guernsey J, et al. CYP17, catechol-o- methyltransferase, and glutathione transferase M1 genetic polymorphisms, lifestyle factors, and breast cancer risk in women on Prince Edward Island. The Breast Journal. 2011;17(1):24–31. doi: 10.1111/j.1524-4741.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 85.Huang CG, Iv GD, Liu T, Liu Q, Feng JG, Lu XM. Polymorphisms of COMT and XPD and risk of esophageal squamous cell carcinoma in a population of Yili Prefecture, in Xinjiang, China. Biomarkers: Biochemical Indicators of Exposure, Response, and Susceptibility to Chemicals. 2011;16(1):37–41. doi: 10.3109/1354750X.2010.522732. [DOI] [PubMed] [Google Scholar]
- 86.Lajin B, Hamzeh AR, Ghabreau L, Mohamed A, Al Moustafa AE, Alachkar A. Catechol-O-methyltransferase Val 108/158 Met polymorphism and breast cancer risk: a case control study in Syria. Breast Cancer. 2013;20(1):62–66. doi: 10.1007/s12282-011-0309-y. [DOI] [PubMed] [Google Scholar]
- 87.Naushad SM, Reddy CA, Rupasree Y, et al. Cross-talk between one-carbon metabolism and xenobiotic metabolism: implications on oxidative DNA damage and susceptibility to breast cancer. Cell Biochemistry and Biophysics. 2011;61(3):715–723. doi: 10.1007/s12013-011-9245-x. [DOI] [PubMed] [Google Scholar]
- 88.dos Santos RA, Teixeira AC, Mayorano MB, Carrara HH, de Andrade J, Takahashi CS. Variability in estrogen-metabolizing genes and their association with genomic instability in untreated breast cancer patients and healthy women. Journal of Biomedicine & Biotechnology. 2011;2011:571784. doi: 10.1155/2011/571784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Q, Li H, Tao P, et al. Soy isoflavones, CYP1A1, CYP1B1, and COMT polymorphisms, and breast cancer: a case-control study in southwestern China. DNA and Cell Biology. 2011;30(8):585–595. doi: 10.1089/dna.2010.1195. [DOI] [PubMed] [Google Scholar]
- 90.Heck JE, Moore LE, Lee YC, et al. Xenobiotic metabolizing gene variants and renal cell cancer: a multicenter study. Frontiers in Oncology. 2012;2:16. doi: 10.3389/fonc.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lim WY, Chen Y, Chuah KL, et al. Female reproductive factors, gene polymorphisms in the estrogen metabolism pathway, and risk of lung cancer in Chinese women. American Journal of Epidemiology. 2012;175(6):492–503. doi: 10.1093/aje/kwr332. [DOI] [PubMed] [Google Scholar]
- 92.Wolpert BJ, Amr S, Saleh DA, et al. Associations differ by sex for catechol-O-methyltransferase genotypes and bladder cancer risk in South Egypt. Urologic Oncology. 2012;30(6):841–847. doi: 10.1016/j.urolonc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, Hua S, Zhang A, et al. Association between polymorphisms in COMT, PLCH1, and CYP17A1, and non-small-cell lung cancer risk in Chinese nonsmokers. Clinical Lung Cancer. 2013;14(1):45–49. doi: 10.1016/j.cllc.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 94.Ghisari M, Eiberg H, Long M, Bonefeld-Jorgensen EC. Polymorphisms in phase I and phase II genes and breast cancer risk and relations to persistent organic pollutant exposure: a case-control study in Inuit women. Environmental Health: A Global Access Science Source. 2014;13(1):19. doi: 10.1186/1476-069X-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Son BH, Kim MK, Yun YM, et al. Genetic polymorphism of ESR1 rs2881766 increases breast cancer risk in Korean women. Journal of Cancer Research and Clinical Oncology. 2014 doi: 10.1007/s00432-014-1849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu YJ, Ge YL, Zhang JY, FN L, Zheng Z. Polymorphism of COMT, p21 and NuMA in sporadic breast cancer. Chin J Clin. 2010;4(5):591–596. [Google Scholar]
- 97.Li L, Li FX, Zhang N, Du YF. Association of gene polymorphism in cytochrome P450 1B1 and COMT, with the expression of mRNA and susceptibility to endometrial cancer in Chinese. Journal of Hebei Medical University. 2010;31(5):1433–1437. [Google Scholar]
- 98.Zhou YH. Association Between Polymorphisms of Estrogen Receptors and Estrogen-Motablic Enzymes and Susceptibility to Colorectal Cancer China: Third Military Medical University. 2009 [Google Scholar]
- 99.Fan Y, Feng Y, Wang L, Wang Y, L F. The relationship between catechol-O-methyltransferase (COMT) polymorphisms and the development of breast cancer. Chin J Clin Oncol. 2007;34(8):430–433. [Google Scholar]
- 100.Zhu MM. The Relationship Between Polymorphisms in Estrogen-metabolizing Genes CYP17, COMT and Risk of Cirrhosis of Hepatocellular Carcinoma in Hepatitis b Infected patients. China: Handong University; 2008. [Google Scholar]
- 101.Martinez-Ramirez OC, Perez-Morales R, Castro C, et al. Polymorphisms of catechol estrogens metabolism pathway genes and breast cancer risk in Mexican women. Breast. 2013;22(3):335–343. doi: 10.1016/j.breast.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 102.Du JZ, Dong YL, Wan GX, et al. Lack of association between the COMT rs4680 polymorphism and ovarian cancer risk: evidence from a meta-analysis of 3,940 individuals. Asian Pacific Journal of Cancer Prevention: APJCP. 2014;15(18):7941–7945. doi: 10.7314/apjcp.2014.15.18.7941. [DOI] [PubMed] [Google Scholar]
- 103.Liu JX, Luo RC, Li R, et al. Lack of associations of the COMT Val158Met polymorphism with risk of endometrial and ovarian cancer: a pooled analysis of case-control studies. Asian Pacific Journal of Cancer Prevention: APJCP. 2014;15(15):6181–6186. doi: 10.7314/apjcp.2014.15.15.6181. [DOI] [PubMed] [Google Scholar]
- 104.Wan GX, Cao YW, Li WQ, Li YC, Li F. The Catechol-O-Methyltransferase Val158Met Polymorphism Contributes to the Risk of Breast Cancer in the Chinese Population: An Updated Meta-Analysis. Journal of Breast Cancer. 2014;17(2):149–156. doi: 10.4048/jbc.2014.17.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qin X, Peng Q, Qin A, et al. Association of COMT Val158Met polymorphism and breast cancer risk: an updated meta-analysis. Diagnostic Pathology. 2012;7:136. doi: 10.1186/1746-1596-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He XF, Wei W, Li SX, et al. Association between the COMT Val158-Met polymorphism and breast cancer risk: a meta-analysis of 30,199 cases and 38,922 controls. Molecular Biology Reports. 2012;39(6):6811–6823. doi: 10.1007/s11033-012-1506-2. [DOI] [PubMed] [Google Scholar]
- 107.McEachin RC, Saccone NL, Saccone SF, et al. Modeling complex genetic and environmental influences on comorbid bipolar disorder with tobacco use disorder. BMC Medical Genetics. 2010;11:14. doi: 10.1186/1471-2350-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dunning AM, Healey CS, Pharoah PD, Teare MD, Ponder BA, Easton DF. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 1999;8(10):843–854. [PubMed] [Google Scholar]
- 109.Lin G, Zhao J, Wu J, et al. Contribution of catechol-O-methyltransferase Val158Met polymorphism to endometrial cancer risk in postmenopausal women: a meta-analysis. Genetics and Molecular Research: GMR. 2013;12(4):6442–6453. doi: 10.4238/2013.December.10.5. [DOI] [PubMed] [Google Scholar]
- 110.Teng Y, He C, Zuo X, Li X. Catechol-O-methyltransferase and cytochrome P-450 1B1 polymorphisms and endometrial cancer risk: a meta-analysis. International Journal of Gynecological Cancer: Official Journal of the International Gynecological Cancer Society. 2013;23(3):422–430. doi: 10.1097/IGC.0b013e3182849e0d. [DOI] [PubMed] [Google Scholar]
- 111.Zhang H, Zhang Z, Wu J, Xu Y, Cheng R, Li L. Lack of association between COMT Val158Met polymorphism and prostate cancer susceptibility. Urologia Internationalis. 2013;91(2):213–219. doi: 10.1159/000345633. [DOI] [PubMed] [Google Scholar]
- 112.Zou LW, Xu XJ, Liu T, et al. No association between COMT Val158-Met polymorphism and prostate cancer risk: a meta-analysis. Genetic Testing and Molecular Biomarkers. 2013;17(1):78–84. doi: 10.1089/gtmb.2012.0216. [DOI] [PubMed] [Google Scholar]
- 113.Xiao L, Tong M, Jin Y, Huang W, Li Z. The l58Val/Met polymorphism of catechol-O-methyl transferase gene and prostate cancer risk: a meta-analysis. Molecular Biology Reports. 2013;40(2):1835–1841. doi: 10.1007/s11033-012-2238-z. [DOI] [PubMed] [Google Scholar]
- 114.Tan X, Chen M. Association between Catechol-O-methyltransferase rs4680 (G>A) polymorphism and lung cancer risk. Diagnostic Pathology. 2014;9:192. doi: 10.1186/s13000-014-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Deeks JJ, Dinnes J, D’Amico R, et al. Evaluating non-randomised intervention studies. Health Technology Assessment. 2003;7(27):iii–x. 1–173. doi: 10.3310/hta7270. [DOI] [PubMed] [Google Scholar]
- 116.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 117.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 118.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 119.Liu J, Yang X, Qu X, Li H. Relation among single nucleotide metabolizing genes CYPl7, COMT and endometrial adenocarcinoma risk in Chinese. J Shandong Univ. 2007;45:18–21. [Google Scholar]
- 120.Han F, Tan Y, Cui W, Dong L, Li W. Novel insights into etiologies of leukemia: a HuGE review and meta-analysis of CYP1A1 polymorphisms and leukemia risk. American Journal of Epidemiology. 2013;178(4):493–507. doi: 10.1093/aje/kwt016. [DOI] [PubMed] [Google Scholar]
- 121.Mao C, Wang XW, Qiu LX, Liao RY, Ding H, Chen Q. Lack of association between catechol-O-methyltransferase Val108/158Met polymorphism and breast cancer risk: a meta-analysis of 25,627 cases and 34,222 controls. Breast Cancer Research and Treatment. 2010;121(3):719–725. doi: 10.1007/s10549-009-0650-4. [DOI] [PubMed] [Google Scholar]