Abstract
The cancer epigenome exhibits global loss of DNA methylation, which contributes to genomic instability and aberrant gene expression by mechanisms that are yet to be fully elucidated. We previously discovered over 3300 long non-coding (lnc)RNAs in human cells and demonstrated that specific lncRNAs regulate gene expression via interactions with chromatin-modifying complexes. Here, we tested whether lncRNAs could also associate with DNA methyltransferases to regulate DNA methylation and gene expression. Using RIP-seq, we identified a subset of lncRNAs that interact with the DNA methyltransferase DNMT1 in a colon cancer cell line, HCT116. One lncRNA, TCONS_00023265, which we named DACOR1 (DNMT1-associated Colon Cancer Repressed lncRNA 1), shows high, tissue-specific expression in the normal colon (including colon crypts) but was repressed in a panel of colon tumors and patient-derived colon cancer cell lines. We identified the genomic occupancy sites of DACOR1, which we found to significantly overlap with known differentially methylated regions (DMRs) in colon tumors. Induction of DACOR1 in colon cancer cell lines significantly reduced their ability to form colonies in vitro, suggesting a growth suppressor function. Consistent with the observed phenotype, induction of DACOR1 led to the activation of tumor-suppressor pathways and attenuation of cancer-associated metabolic pathways. Notably, DACOR1 induction resulted in down-regulation of Cystathionine β-synthase, which is known to lead to increased levels of S-adenosyl methionine—the key methyl donor for DNA methylation. Collectively, our results demonstrate that deregulation of DNMT1-associated lncRNAs contributes to aberrant DNA methylation and gene expression during colon tumorigenesis.
Introduction
In humans and other mammals, epigenetic modifications play critical roles in orchestrating gene expression patterns in distinct cell types throughout the life of an organism. These epigenetic modifications are regulated by the highly coordinated functions of chromatin-remodeling complexes, histone-modifying enzymes, DNA methyltransferases and chromatin readers (1). However, the process by which these ubiquitous epigenetic modifiers are recruited, assembled and stabilized at specific genomic loci in distinct cell types is yet to be fully elucidated. Increasing experimental evidence suggests potential key roles for long non-coding RNAs in recruiting and scaffolding such complexes to the genome in mammalian cells (2–12).
The mammalian genome encodes thousands of long non-coding RNAs (lncRNAs) (7,13–16), and studies of select lncRNAs have clearly demonstrated their critical roles in various aspects of mammalian biology (6,17–23). For example, some lncRNAs are critical for embryonic development and tissue morphogenesis in vivo, as demonstrated by genetic disruptions of several lncRNAs in mice and zebrafish (3,24,25). The means by which lncRNAs exert their effects have been demonstrated through several mechanisms: (i) transcription co-activators (17), (ii) guidance of epigenetic complexes to the genome (3,5,6,8,12), (iii) competing endogenous RNAs or microRNA ‘sponges’ (26–28), (iv) regulation of mRNA translation and decay (28–30), (v) decoys for transcription factors (TFs) (31) and other potential mechanisms (32).
Previous studies have identified extensive genome-wide interactions between lncRNAs and several chromatin-modifying complexes, including the polycomb repressive complex 2 (PRC2) (7,33). These interactions are required for proper PRC2-mediated gene repression (7,8), suggesting that deregulation of such lncRNAs could impact the ability of epigenetic-modifying complexes to regulate the epigenome and gene expression programs. Indeed, a number of studies have identified numerous lncRNAs that become highly deregulated in various human diseases, including several cancer types (34–38). Thus, deregulation of specific lncRNAs may result in global changes in the epigenome, including global changes in DNA methylation patterns, and deregulation of gene expression (39–42).
DNA methylation is an important epigenetic mark that is typically associated with repressed genes in mammalian cells (43). Three distinct DNA methyltransferases (DNMT1, DNMT3a and DNMT3b) are known to regulate DNA methylation patterns in mammals. Genome-wide studies of DNA methylation in various tumors versus matched normal tissues have demonstrated that although the promoters of some tumor-suppressor genes become hypermethylated, cancer genomes are largely hypomethylated (43,44). Currently, there is a great interest in understanding how DNA methylation patterns become deregulated in human cancers, with the hope that these studies might lead to novel insights into tumorigenesis as well as the development of novel therapeutic strategies (45). We hypothesized that a subset of lncRNAs may interact with DNA methyltransferases and, consequently, affect their genomic occupancies and/or activities. Thus, deregulation of such lncRNAs in human cancers would result in abnormal DNA methylation patterns without any detectable mutations or changes in the expression levels of the genes that encode DNA methyltransferases.
In this article, we identified specific interactions between a subset of human lncRNAs and the DNA methyltransferase DNMT1 using RNA co-immunoprecipitation (RIP) followed by next generation RNA sequencing (RIP-seq) (Fig. 1A). Analysis of one such lncRNA, TCONS_00023265, which we named DACOR1, revealed a critical role of this lncRNA in regulating DNA methylation and gene expression in colon cells. Furthermore, induction of DACOR1 is sufficient to suppress the growth of colon cancer cells by regulating the expression of specific genes and pathways including cellular metabolism. Our results suggest a potential new mechanism by which the human methylome is regulated in human health and disease.
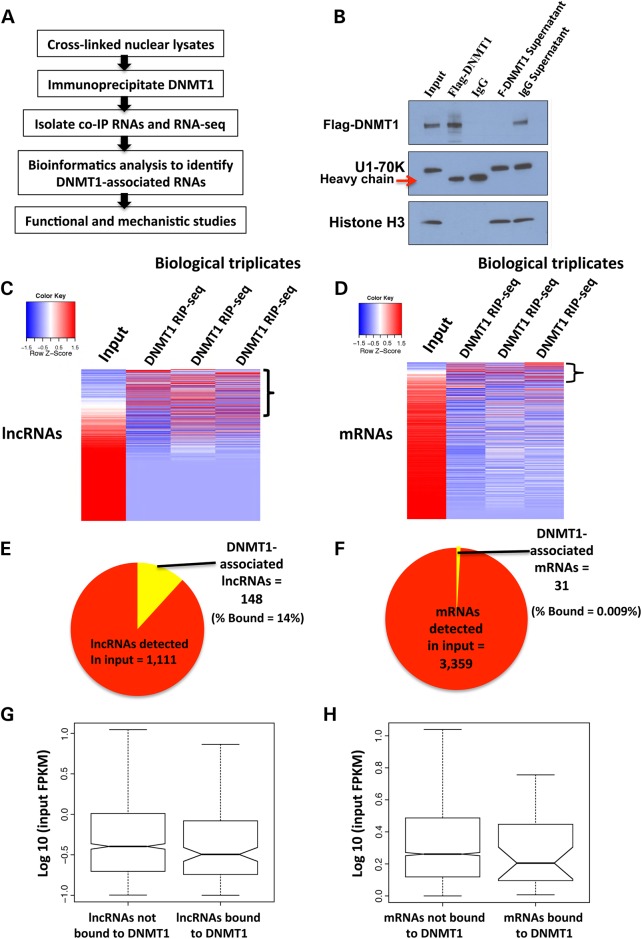
Figure 1.
Numerous long non-coding RNAs (lncRNAs) associate with DNMT1 in human cells. (A) Outline of the experimental strategy utilized to identify DNMT1-associated RNAs. (B) Western blot analysis using an anti-flag-DNMT1 antibody confirms the specific immunoprecipitation (IP) of DNMT1, but not other highly abundant nuclear proteins (histone H3, U1–70K). An IP with anti-IgG antibody demonstrates that there is no detectable background. (C) Heatmap of lncRNAs in input sample versus each of the three biological replicates of DNMT1 RIPs. We observed some variability between the three biological replicates owing to stringent washes to eliminate non-specific RNAs. (D) Heatmap of mRNAs in input versus the three biological replicates of DNMT1 RIPs. (E) Pie chart showing the number of DNMT1-associated lncRNAs versus all lncRNAs expressed in input. Approximately 14% of lncRNAs co-IP with DNMT1. (F) Pie chart showing the number of DNMT1-associated mRNAs versus mRNAs expressed in input, ∼0.009% of mRNAs co-IP with DNMT1. (G–H) Graphs show the expression levels of DNMT1-bound lncRNAs and mRNAs versus non-bound lncRNAs and mRNAs in HCT116 cells. We found that DNMT1-bound lncRNAs and mRNAs show no expression bias over non-bound lncRNAs and mRNAs.
Results
Identification of DNMT1-associated lncRNAs in colon cancer cells
We optimized our RIP protocol in the colon cancer cell line HCT116 (see Material and Methods and Supplementary Material, Appendix S1, Fig. S1A and B) and subsequently utilized it to identify potential interactions between DNMT1 and RNAs. As there are no reliable DNMT1 antibodies that are suitable for RIP applications, we utilized a knock-in DNMT1_3X-flag HCT116 cell line to overcome this limitation (46). First, we confirmed that DNMT1 is specifically immunoprecipitated, but not other abundant nuclear proteins such as U1–70K or histone H3 (Fig. 1B and Supplementary Material, Appendix S1, Fig. S2A). To identify RNAs that potentially interact with DNMT1, we performed triplicate RIPs of DNMT1 using an anti-flag antibody and triplicate RIPs using an anti-IgG antibody as negative controls. We isolated co-immunoprecipitated RNAs and quantified the small amount of DNMT1-bound RNAs (Supplementary Material, Appendix S1, Fig. S2B). We were able to generate RNA-seq libraries from DNMT1 RIPs but not from IgG RIPs (Supplementary Material, Appendix S1, Fig. S2C), owing to depletion of non-specific RNAs by several stringent washes.
Three RNA-seq libraries from three independent biological replicates of DNMT1 RIPs were sequenced and mapped to the human genome (hg19). We also sequenced total nuclear RNA (input) from HCT116 cells as a control for our RIP experiments. We generated fpkm values for mRNAs and lncRNAs detected in the input sample and each of the three biological replicates of DNMT1 RIP-seq (see Material and Methods). The average fpkm of each transcript in the three biological replicates of DNMT1 RIP-seq was divided by the fpkm in the input sample to generate fold changes (Supplementary Material, Files S1 and S2). We identified 148 lncRNAs (14% of lncRNAs detected in the input) and 31 mRNAs (0.009% of mRNAs detected in the input) as DNMT1-associated RNAs based on a 2-fold change or higher above input (Fig. 1C–F). We found the highest fold change of an lncRNA associated with DNMT1 to be ∼41-fold, whereas the highest fold change for an mRNA was only 7-fold, despite mRNAs being expressed at much higher levels than lncRNAs across all cell types (7,13,14,34). To rule out non-specific co-immunoprecipitation of highly abundant RNAs with DNMT1, we compared the expression of all DNMT1-bound versus DNMT1-unbound lncRNAs and mRNAs. We found that there was no expression bias of DNMT1-associated lncRNAs or mRNAs in comparison with unbound lncRNAs and mRNAs (Fig. 1G–H). Lastly, a close examination of DNMT1-associated mRNAs revealed that at least half of these mRNAs are poorly annotated transcripts with predicted open reading frames or miRNA precursors, suggesting that very few mRNAs associate with DNMT1 (Supplementary Material, File S2). In summary, we have identified many lncRNAs and very small number of mRNAs that co-immunoprecipitate with DNMT1 in HCT116 cells by RIP-seq.
The DNMT1-associated lncRNA, DACOR1, is down-regulated in colon cancer cells
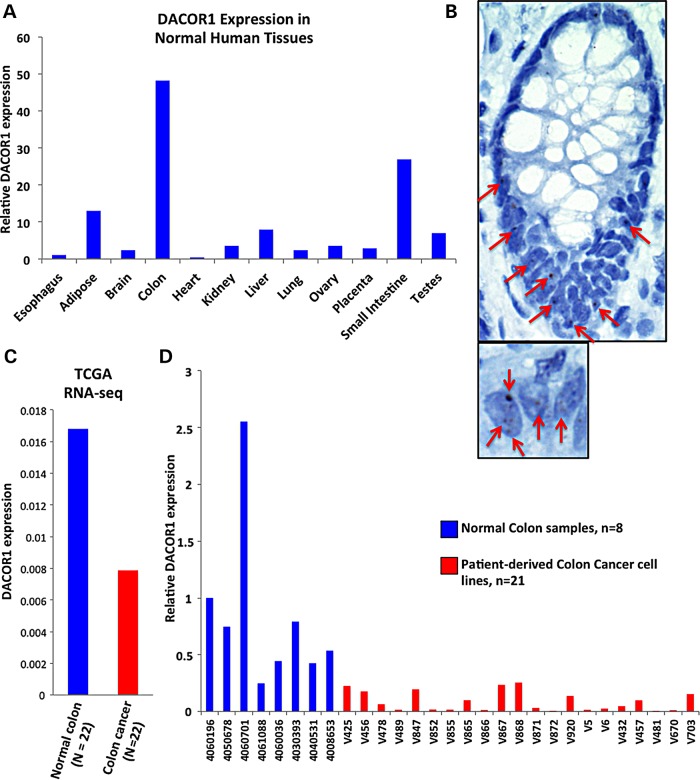
One DNMT1-associated lncRNA, designated TCONS_00023265 (Supplementary Material, Appendix S1, Fig. S3, and File S3), was of interest to us owing to its notable high, tissue-specific expression in normal colon tissues (Fig. 2A and B) and repression in colon tumors and patient-derived colon cancer cell lines (Fig. 2C and D). We therefore named this lncRNA DNMT1-associated Colon Cancer Repressed lncRNA 1 (DACOR1) (see below). In a panel of 12 human normal tissues, DACOR1 shows the highest expression in the colon as measured by qRT-PCR (Fig. 2A). We confirmed the expression of DACOR1 in the normal colon by RNA in situ hybridization and observed DACOR1 expression in the nuclei of colon crypts, the cells from which colon cancer originates (Fig. 2B, large panel). We also observed that DACOR1 occupies several discrete foci in the nucleus (Fig. 2B, small panel). Next, we examined DACOR1 expression in a cohort of 22 colon tumors in comparison with matched normal tissue based on RNA-seq data obtained from The Cancer Genome Atlas (TCGA). This analysis revealed that DACOR1 is down-regulated in colon tumors (Fig. 2C). We also examined the expression of the protein-coding gene SMAD3, the nearest coding gene to DACOR1, in the same TCGA cohort and found that SMAD3 shows variable expression in tumors versus normal colon (Supplementary Material, Appendix S1, Fig. S4). To further confirm that DACOR1 is down-regulated in colon cancer, we examined its expression by qRT-PCR in 8 normal colon samples and 21 patient-derived colon cancer cell lines with limited passage in culture (Fig. 2D, and Supplementary Material, Appendix S1, Fig. S5). Several of the colon cancer cell lines displayed very low expression levels of DACOR1 that were barely detectable by qRT-PCR, further confirming the down-regulation of DACOR1 during colon tumorigenesis (Fig. 2D). These intriguing observations prompted us to further investigate the potential role of DACOR1 in colon cancer biology and its effects on DNA methylation and gene expression.
Figure 2.
The DNMT1-associated lincRNA, DACOR1, is down-regulated in colon cancer. (A) Quantitative real-time PCR (qRT-PCR) of DACOR1 across a panel of human normal tissues demonstrates the high expression of DACOR1 in the colon and small intestine in comparison with other tissues examined. (B) RNA in situ hybridization confirms the expression of DACOR1 in human colon tissues and identifies the colon crypts as one of the major cell types that express it. Red arrows mark DACOR1 signal, which is shown as a chromogenic signal (brown). Close examination of colon cells (small panel) reveals that DACOR1 is retained in the nucleus and potentially interacts with chromatin. (C) Expression analysis of DACOR1 in a cohort of 22 colon cancer tumors versus 22 matched normal tissues in RNA-seq datasets obtained from TCGA demonstrates that DACOR1 is down-regulated in colon tumors. (D) Examining the expression of DACOR1 by qRT-PCR in 8 normal colon samples and 21 patient-derived colon cancer cell lines with limited passage in culture demonstrates that DACOR1 is highly repressed in most colon cancer cells.
DACOR1 affects DNA methylation levels at multiple sites in the human genome
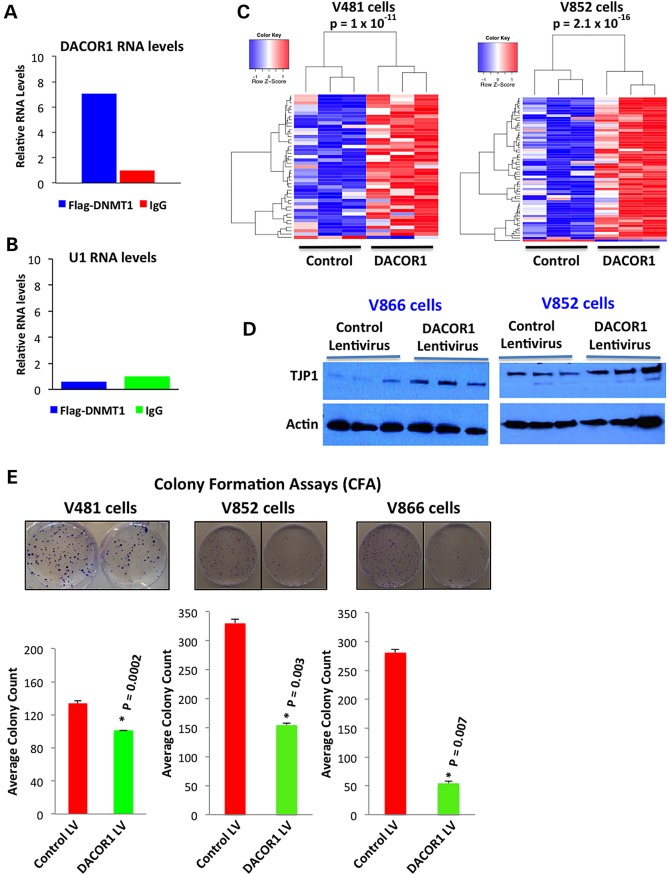
To determine the functional significance of DACOR1 association with DNMT1, we first validated the interaction of DACOR1 with DNMT1 in independent RIP experiments using RIP-qPCR (Fig. 3A). As a negative control, we examined the association of the highly abundant nuclear RNA U1 with DNMT1 and found no association (Fig. 3B). We then tested the effects of DACOR1 induction on DNA methylation in two distinct patient-derived colon cancer cell lines, V481 and V852. We transduced V481 and V852 cells with either a control or DACOR1 lentivirus, and the appropriate expression and nuclear localization of DACOR1 were confirmed by qRT-PCR and RNA in situ, respectively (Supplementary Material, Appendix S1, Fig. S6A and B). We isolated genomic DNA from these cell lines, and equal amount of DNA (1 µg) from each sample (n = 12) was used for DNA methylation analysis using 450K DNA methylation arrays (Illumina). These arrays cover ∼500 000 CpG sites out of the 28 million CpG sites in the human genome. We identified 43 and 59 specific CpG sites in V481 and V852, respectively, which become differentially methylated in response to DACOR1 expression (Fig. 3C, and Supplementary Material, File S4). Of these sites, 42/43 (in V481) and 58/59 (in V852) displayed a gain of DNA methylation (P < 1 × 10−11 and 2.1 × 10 −16, respectively). Next, we determined whether restoration of DACOR1 expression affected DNMT1 protein levels. We performed western blot analyses using a DNMT1 antibody in cells transduced with a control or DACOR1 lentivirus and found that DNMT1 protein levels were unchanged (Supplementary Material, Appendix S1, Fig. S7). In summary, DACOR1 induction appears to enhance DNA methylation at multiple loci without affecting DNMT1 protein levels.
Figure 3.
DACOR1 induction enhances DNA methylation and suppresses growth of colon cancer cells. (A) Validation of the interaction between DNMT1 and DACOR1 by RIP-qPCR. DACOR1 shows a 7-fold enrichment in flag-DNMT1 RIP over IgG RIP. (B) The highly abundant nuclear RNA U1 shows no enrichment in flag-DNMT1 RIP versus IgG RIP, demonstrating the specificity of our method; (C) Induction of DACOR1 expression in two distinct patient-derived colon cancer cell lines (V481 and V852) enhances DNA methylation at multiple genomic loci in trans. Blue color indicates low methylation; red color indicates high methylation. All experiments were performed in triplicates. (D) Induction of DACOR1 expression in patient-derived colon cancer cell lines (V866 and V852) results in up-regulation of tight junction protein 1 (TJP1), suggesting a potential role for DACOR1 in maintaining an epithelial state of colon cells. (E) The colon cancer cell lines V481, V852 and V866 were transduced with either a control or DACOR1 lentivirus. Subsequently, CFAs were carried out (see Material and Methods) in each cell line. Cells with restored DACOR1 expression showed reduced colony formation, suggesting that DACOR1 is potentially a growth suppressor.
DACOR1 may play a role in maintaining the epithelial state of colon crypts
The high expression of DACOR1 in normal colon tissues and the localization of DACOR1 to colon crypts prompted us to examine its potential role in regulating the epithelial state of colon cells. To that end, we examined the effects of DACOR1 induction on the levels of key epithelial markers including Tight Junction Protein 1 (TJP1) and E-cadherin in two distinct colon cancer cell lines. We found that the expression of DACOR1 led to higher levels of TJP1 protein, but not E-Cadherin (Fig. 3D, and Supplementary Material, Appendix S1, Fig. S8). To determine whether the change in TJP1 is at the transcriptional or post-transcriptional level, we measured TJP1 mRNA levels by qRT-PCR in three distinct colon cancer cell lines. We found DACOR1 expression to have no effect on TJP1 mRNA levels (Supplementary Material, Appendix S1, Fig. S9A), suggesting that TJP1 protein levels are regulated post-transcriptionally by DACOR1 in colon cells. We also compared TJP1 mRNA levels in a cohort of 22 colon tumors versus 22 matched normal tissues from TCGA and found that TJP1 mRNA levels are not significantly affected in most patients (Supplementary Material, Appendix S1, Fig. S9B), suggesting that TJP1 protein levels are regulated post-transcriptionally in colon tumors.
DACOR1 induction reduces the clonogenic potential of colon cancer cells
Our studies demonstrated that DACOR1 is down-regulated in colon tumors and patient-derived colon cancer cell lines, but the biological significance of this repression is yet to be determined. Normal colon crypts do not propagate in tissue culture, preventing us from performing knockdown experiments of DACOR1. We, therefore, examined the biological effects of DACOR1 by overexpressing it in several patient-derived colon cancer cell lines. Initially, we utilized three distinct patient-derived colon cancer cell lines (V481, V852 and V866) that we transduced with either a control or a DACOR1 lentivirus. Induction of DACOR1 in these patient-derived colon cancer cell lines resulted in reduced growth of these cells (Supplementary Material, Appendix S1, Fig. S10). To quantify this effect, we performed colony formation assays (CFAs) using all three lines (V481, V852 and V866) and found that the induction of DACOR1 affected colony formation in V481 by ∼25% (P = 0.0002), in V852 cells by ∼53% (P = 0.003) and in V866 by 81% (P = 0.007) (Fig. 3E). The effect of DACOR1 induction, although consistent in reducing colonies, varied among the three lines as each line was derived from a distinct patient tumor and thus has underlying genetic differences.
To test whether the effects we observed on colony formation were due to non-specific effects of overexpressing DACOR1, we performed several control experiments. First, we selected two patient-derived colon cancer cell lines, V703 and V425, that although had reduced levels of DACOR1 relative to normal colon; they still maintained some level of DACOR1 expression (Fig. 2D). Overexpression of DACOR1 in both cell lines had minor effects on colony formation of these cells, when compared with a control lentivirus (Supplementary Material, Appendix S1, Fig. S11A–D). Second, to rule out that the phenotype is due to high expression levels of DACOR1 lentivirus (CMV promoter), we cloned the full length of DACOR1 downstream of a weak Pgk promoter and measured its expression levels in comparison with normal colon and control lentivirus. Using this approach, we are able to bring the overexpression level of DACOR1 closer to the expression levels observed in normal colon (Supplementary Material, Appendix S1, Fig. S12A). We carried out CFAs of control versus DACOR1 lentivirus-transduced cells and also observed significant reduction in colony formation (Supplementary Material, Appendix S1, Fig. S12B). Finally, we cloned the full length of an oncogenic lncRNA, TCON_00011938, which is not associated with DNMT1, downstream of a strong CMV promoter, and found that the overexpression of this distinct lncRNA led to increased colony formation (Supplementary Material, Appendix S1, Fig. S13A and B). Collectively, these results suggest that DACOR1 induction reduces the clonogenic potential of colon cancer cells.
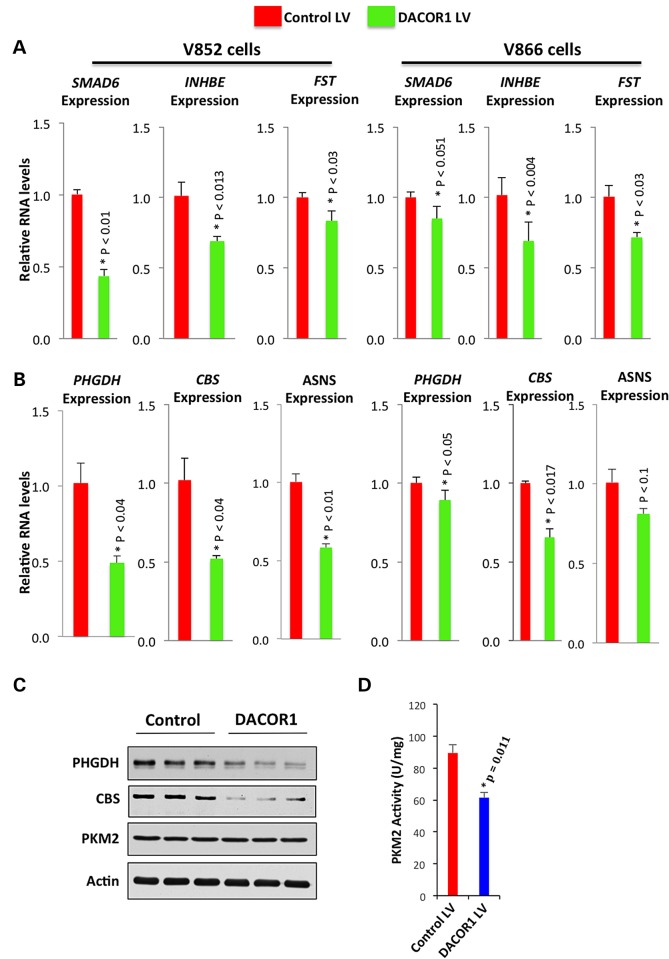
DACOR1 induction affects global gene expression of colon cancer cells
To gain insights into DACOR1 function, we performed RNA-seq using RNA isolated from the colon cancer cell line V852 transduced with either control or DACOR1 lentivirus and identified differentially expressed genes (see Material and Methods) (47). We found that induction of DACOR1 affected the expression of 99 genes (P < 0.05, q < 0.05) (Supplementary Material, File S5). Specifically, we observed that induction of DACOR1 led to the repression of several known inhibitors of TGF-β/BMP signaling, including SMAD6, INHBE (inhibin beta E) and FST (follistatin), which we confirmed by qRT-PCR in two distinct colon cancer cell lines (Fig. 4A) (48). Previous studies have demonstrated that TGF-β/BMP signaling exerts a tumor-suppressor function in the colon, and it becomes inactivated or repressed in a majority of sporadic colorectal cancers (49). SMAD6, which is up-regulated in colon tumors (Supplementary Material, Appendix S1, Fig. S14A) and down-regulated by DACOR1, plays a major role in repressing TGF-β/BMP signaling (48).
Figure 4.
Induction of DACOR1 affects global gene expression in colon cancer cells. (A) qRT-PCR confirmations of RNA-seq data that DACOR1 represses several genes (SMAD6, FST and INHBE) involved in the repression of the TGF-beta/BMP signaling pathway. These observations suggest that induction of DACOR1 enhances the signaling of these pathways, which have tumor-suppressor effects. (B) qRT-PCR validations of RNA-seq data that DACOR1 represses the expression of key genes that are involved in amino acid biosynthesis and metabolism; (C) Western blot analyses demonstrate that DACOR1 induction leads to the repression of PHGDH and CBS but does not affect PKM2 or Actin protein levels in V852 cells; (D) Induction of DACOR1 reduces the activity of PKM2, which is known to be dependent on serine, without affecting overall PKM2 protein levels.
We also found that the induction of DACOR1 led to the down-regulation of several genes involved in amino acid metabolism with known roles in tumorigenesis, including PHGDH, PSAT1, CBS and ASNS (50–52). First, we confirmed that the induction of DACOR1 leads to the repression of these genes in two distinct colon cancer cell lines, V852 and V866, by qRT-PCR (Fig. 4B). We subsequently confirmed the repression of PHGDH at the protein level by western blot analysis (Fig. 4C). PHGDH plays a key role in de novo serine biosynthesis (52–55) and is highly up-regulated in many colon tumors (Supplementary Material, Appendix S1, Fig. S14B). To determine whether the repression of PHGDH by DACOR1 induction affects serine levels, we measured pyruvate kinase M2 (PKM2) activity, which is dependent on serine (55). Indeed, we found that DACOR1 induction leads to reduced PKM2 activity in two independent experiments (three replicates each) (Fig. 4D), without affecting overall PKM2 protein levels (Fig. 4C). Lastly, the repression of Cystathionine β-synthase (CBS) by DACOR1 is intriguing (Fig. 4C), as reduced CBS levels are known to lead to increased methionine, the substrate needed to generate S-adenosyl methionine (SAM). SAM is the key methyl donor utilized by DNA methyltransferases for DNA methylation in mammalian cells. Thus, DNMT1–DACOR1 interaction appears to indirectly regulate cellular SAM levels, and, consequently, genome-wide DNA methylation. Collectively, these findings suggest that DACOR1 plays key roles in regulating DNA methylation and specific tumor-suppressor and metabolic pathways in colon cells to potentially suppress colon tumorigenesis.
DACOR1 interacts directly with chromatin at specific genomic sites
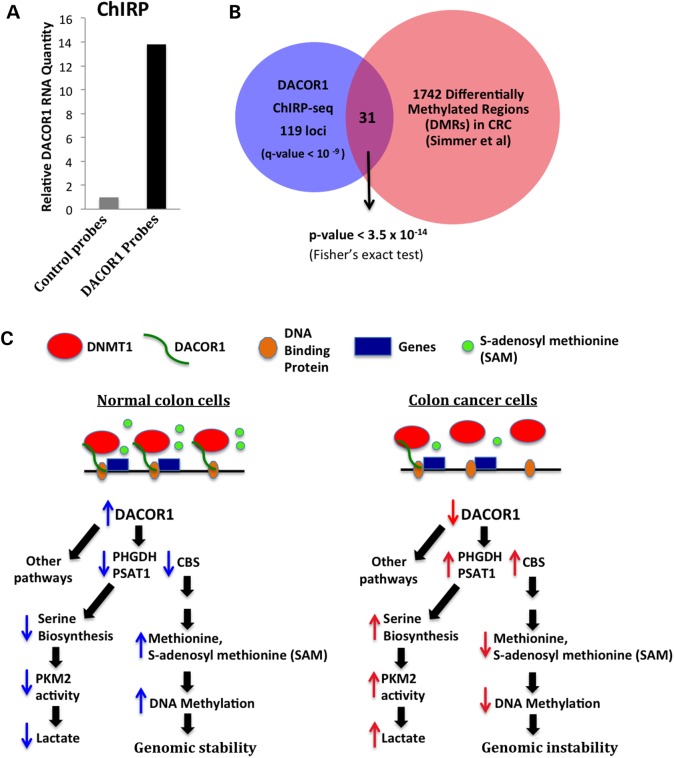
To gain insights into the potential mechanism(s) by which DACOR1 could regulate gene expression and consequently cellular phenotype, we mapped the genomic occupancy of DACOR1 across the entire human genome using ChIRP-seq (56). First, we designed several biotin-modified oligonucleotides complementary to DACOR1 and confirmed that we can specifically isolate DACOR1 from crosslinked cell lysates (Fig. 5A). Subsequent ChIRP-seq and analysis identified 338 DACOR1 genomic occupancy sites, including 161 peaks near 150 annotated genes (multiple peaks per gene in some cases) and 177 sites in intergenic regions (Supplementary Material, File S6). As expected, we observed a peak corresponding to the genomic region of DACOR1 transcription upstream of SMAD3, as we also captured newly synthesized DACOR1 transcripts. We compared the genomic occupancy sites of DACOR1 near annotated genes with differentially methylated regions (DMRs) in a cohort of colon tumors versus matched normal tissues (41). Of the 150 annotated gene loci occupied by DACOR1, 31 sites overlap with these DMRs (P < 3.5 × 10−14) (Fig. 5B). These findings indicate that DACOR1 interacts with both DNMT1 and chromatin and, potentially, recruits and/or assembles the DNMT1 macromolecular protein complex at specific genomic sites to regulate epigenetic modifications and, consequently, the expression of specific genes and pathways (Fig. 5C).
Figure 5.
DACOR1 is associated with chromatin. (A) Confirmation of DACOR1 pull down from crosslinked cell lysates by specific complementary probes, in comparison with non-specific probes (B). Intersection of DACOR1 genome occupancy sites near annotated protein-coding genes identified in this study by ChIRP-seq and differentially methylated regions (DMRs) in colon tumors/normal colon identified by Simmer et al. (41) reveals a significant overlap. This further supports the role of DACOR1, via its interaction with DNMT1, in regulating genome-wide DNA methylation (C). A proposed model of how DNMT1–DACOR1 interactions regulate DNA methylation and gene expression. DACOR1 interacts with specific genomic loci and potentially recruits DNMT1 to establish DNA methylation patterns and/or regulate gene expression. The DNMT1–DACOR1 axis results in modulating the expression of many genes, directly and indirectly, including those involved in amino acid metabolism. When DACOR1 becomes repressed during colon tumorigenesis, several metabolic changes occur. For example, up-regulation of PHGDH levels results in increased serine levels and consequently increased PKM2 activity. Also, up-regulation of CBS levels, which is known to cause a decrease in methionine and SAM levels, could potentially impact the levels of SAM available for DNA methylation causing a global hypomethylation—a hallmark of cancer epigenomes.
Discussion
We previously identified specific interactions between human long non-coding RNAs (lncRNAs) and several chromatin-modifying complexes and demonstrated that these interactions are required for regulating gene expression (7). In this manuscript, we identified specific interactions between the DNA methyltransferase DNMT1 and human lncRNAs, suggesting that in addition to histone modifications, DNA methylation is also indirectly regulated by lncRNAs. DNA methylation is an important epigenetic mark for the regulation of gene expression in mammalian cells from early embryonic development to fully differentiated post-mitotic cells. Our current findings that DNMT1 associates with lncRNAs suggest that these lncRNAs may influence DNMT1 genomic occupancy and/or activities, thereby indirectly regulating the methylome. Thus, deregulation of one or more of DNMT1-associated lncRNAs in human disease would lead to changes in DNA methylation patterns and potentially significant changes in gene expression without any detectable changes in DNMT1 expression levels. Indeed, we found in our studies that the induction of the lncRNA DACOR1 is sufficient to change DNA methylation patterns without affecting DNMT1 protein levels in colon cancer cells. However, the mechanisms of DACOR1-mediated changes in DNA methylation patterns are not yet known. A number of potential mechanisms may be implicated, including DACOR1-mediated recruitment of DNMT1 to specific sites of the genome, similar to what has been observed of lncRNA-mediated recruitment of histone-modifying enzymes (6,8,12). Also, DACOR1 could affect DNMT1 activity at specific CpG sites, potentially by regulating protein components of the DNMT1 macromolecular protein complex.
We currently have several technologies to identify lncRNA–protein interactions; however, it is not currently known how lncRNAs, such as DACOR1, interact with epigenetic-modifying complexes such as histone-modifying enzymes and DNA methyltransferases. It has been proposed that secondary structure of lncRNAs plays a major role in this lncRNA–protein recognition (32,57); however, the experimental evidence for this still requires much needed research. Also, it is not yet clear how epigenetic complexes bind RNAs, as they do not possess canonical RNA-binding domains similar to those found in classic RNA-binding proteins. It is possible that these proteins possess RNA-binding ‘domains’ that are distinct from those identified in classic RNA-binding proteins, or the interaction could be indirect and mediated by RNA-binding proteins. Also, despite the recent development of technologies to map the genome-wide occupancy of nuclear lncRNAs, it is not yet known how lncRNAs can recognize specific genomic loci. Proteins that serve as intermediates between DNA and lncRNAs could also mediate this interaction.
Many lncRNAs, including DACOR1, are poorly conserved, even within mammalian species (58). However, several studies have now demonstrated functionality of lncRNAs despite this lack of sequence conservation (25). The lack of sequence conservation for many lncRNAs makes it challenging to use model organisms to interrogate the function of lncRNAs identified in human systems and diseases. Furthermore, studies of conserved lncRNAs in mouse models do not always recapitulate what has been observed in human cells (2,59). These observations suggest that researchers may have to utilize non-human primates or human organoids to study lncRNAs. As an example, DACOR1 would require a non-human primate such as rhesus macaque for knock-out experiments to delineate its function in vivo.
Gene expression analyses of DACOR1 demonstrated that many colon tumors and colon cancer cell lines dramatically repress DACOR1 expression. Restoring DACOR1 expression into patient-derived colon cancer cells resulted in reduced growth, potentially via the modulation of several pathways. For example, DACOR1 down-regulates the expression of several genes that inhibit TGF-β/BMP signaling and thus potentially enhances TGF-β/BMP signaling, which is known to exert a tumor-suppressor activity in the colon (48,49). DACOR1 also down-regulates several genes involved in metabolism including de novo serine biosynthesis (e.g. PHGDH, PSAT1). Serine is an essential precursor for the synthesis of proteins, nucleic acid and lipids; thus, it is critical for cancer cell growth (52–54). Furthermore, we found that DACOR1 induction is sufficient to attenuate pyruvate kinase M2 (PKM2) activity, which is highly dependent on serine (55). PKM2 has been recently implicated as a key gene in cancer metabolism (55,60,61); thus, the identification of a lncRNA that attenuates its activity, although indirectly, may provide a therapeutic window in cancer biology. Lastly, DACOR1-mediated down-regulation of CBS, the deficiency of which is known to lead to increased levels of methionine and, consequently, SAM, the key methyl donor utilized by DNA methyltransferases to methylate DNA, is also highly significant. These findings suggest that DNMT1, via its interaction with DACOR1, indirectly regulates the cellular levels of SAM and, subsequently, genome-wide DNA methylation. In future studies, we will examine the effect of DACOR1 on DNA methylation at repetitive DNA and transposable elements, which would require whole genome bisulfite sequencing analysis.
In addition to lncRNAs, we also found a small number of mRNAs that co-immunoprecipitate with DNMT1. However, the significance of these interactions is currently unknown. We cannot rule out that some of these mRNAs have dual functions: protein-coding capacity and non-coding function. Previous studies have identified mRNAs that, in addition to their coding potential, function as RNA molecules to regulate a number of biological processes (13,17,26,32). Also, a previous study that utilized a genome-wide computational approach to identify lncRNAs found that many ‘hypothetical’ coding genes are indeed non-coding (62). Thus, future studies may reveal other coding transcripts that also have a non-coding function.
In summary, our current study uncovered a critical role of lncRNAs in regulating the human methylome—these findings could potentially help explain, at least in part, the genome-wide changes in DNA methylation across numerous cancer types. Furthermore, as many lncRNAs have tissue-specific expression patterns, they could serve as biomarkers of disease prognosis as well as for therapeutic strategies with potentially less side-effects than coding genes.
Material and Methods
Sequencing data files
All next-generation RNA- and DNA-sequencing data files are deposited in GEO under GSE58989
Optimization of RIP in HCT116 cells
We have previously utilized RIP in human fibroblasts and HeLa cells to identify interactions between human lncRNAs and several chromatin-modifying complexes (7,63). For this study, we optimized our RIP protocol in HCT116 cells by initially performing control experiments on a well-conserved RNA–protein interaction in the spliceosome: the interaction between U1–70K protein and the small nuclear RNA U1 (64). First, we tested an antibody against U1–70K in immunoprecipitation experiments and confirmed that this antibody specifically immunoprecipitates U1–70K protein from HCT116 cell lysate (Supplementary Material, Appendix S1, Fig. S1A). We also used an IgG antibody that should not recognize any protein as a negative control. Subsequently, we performed three independent biological replicates of U1–70K RIPs from crosslinked HCT116 cell lysate. After several stringent washes, we reversed the formaldehyde crosslinking by heat and isolated associated RNA using Trizol. Quantitative Real-time PCR (qRT-PCR) analysis of U1 RNA using three distinct endogenous controls (GAPDH, 18S rRNA and CLDN3) revealed a specific interaction between U1–70K and U1 RNA (Supplementary Material, Appendix S1, Fig. S1B). These results suggest that our RIP protocol is optimized in HCT116 to detect specific RNA–protein interactions.
Immunoprecipitation (IP) of U1–70K and flag-DNMT1 and western blot analysis
We utilized an antibody against U1–70K (Synaptic Systems, Cat # 203 001) to immunoprecipitate (IP) the U1–70K protein, and anti-flag antibody to IP flag-DNMT1 from HCT116 cell lysates as follows: HCT116 cells were grown in 2 × 15 cm plates before harvesting by trypsin. An equal amount of media was added to quench the reaction, and the cells were collected by centrifugation in a 15-ml conical tube at 500g for 10 min. The pellets were washed twice with PBS prior to fixing in a final concentration of 0.3% formaldehyde for 15 min at room temperature. The reaction was quenched by adding glycine to a final concentration of 0.125 mm and incubated at room temperature for 5 min. The cells were pelleted by spinning at 500g for 10 min and then washed twice with 1× PBS before suspending the pellets in 2.2 ml of RIPA buffer (150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris–HCl (pH 7.4), 1 mm EDTA). The cells were incubated at 37°C for 30 min and vortexed every 5 min at 30-s intervals for the duration of the incubation. Samples were homogenized using a dounce homogenizer to disrupt cellular membranes. The lysate was centrifuged using a microcentrifuge at maximum speed (∼13 300 RPM), and the supernatant was transferred to a new tube. A total of 100 µl of the supernatant was taken as input, and half of remaining supernatant was incubated with an antibody against protein of interest (i.e. U1–70K or flag-DNMT1), and the second half with an IgG antibody (negative control) overnight with rotation at 4°C. Next day, 50 µl of protein A/G magnetic beads was added to each tube and incubated for 30 min at room temperature with rotation. The beads, which now have the antibody and bound protein, were collected using a magnet and washed three times with RIPA buffer and once with 1× PBS. For protein analysis by western blot, we added 100 µl of Laemmli buffer to each tube and incubated the samples at 95°C for 5 min before running the samples on a denaturing SDS–PAGE gel.
RNA co-immunoprecipitation of U1–70K and flag-DNMT1
The same protocol described earlier was utilized for RIP of U1–70K or flag-DNMT1 from HCT116 cells. However, for the isolation of co-immunoprecipitated RNAs, we suspended the magnetic beads + antibody + protein in 100 µl of buffer C (150 mm NaCl, 50 mm Tris–HCl (pH = 7.4), 5 mm EDTA, 10 mm DTT, 1% SDS) and 10 µg of proteinase K. The samples were incubated at 42°C for 30 min for protein digestion, and subsequently at 65°C for 4 h to reverse the formaldehyde crosslinking. RNA was isolated by adding 800 µl of Trizol and 200 µl of chloroform to each sample, mixed and centrifuged at full speed for 10 min, and the upper clear layer (∼600 µl) was transferred to a 1.5-ml tube with 600 µl of 70% ethanol. The mixture was applied to an RNeasy mini kit column (Qiagen) according to the manufacturer's protocol. All samples were treated with DNase prior to final washes and elution with 20 µl of RNase-free water.
Analysis of RNA-seq data from RIP-seq samples
RNA-sequencing libraries were made using a stranded ScriptSeq V2 (Illumina) according to the manufacturer's protocol. Raw RNA-seq fastq files were aligned to UCSC human hg19 using TopHat v2.0.10. Transcript assembly was performed using Cufflinks v2.1.1. Relative transcript abundance for both mRNAs and lncRNAs was reported as fragments per kilobase of exon per million fragments mapped (fpkm). If fpkm values reported in the input sample were less than 1.0 for mRNAs and less than 0.1 for lncRNAs, the transcript was filtered as not expressed in HCT116 cells. Fold changes were then calculated as the average fpkm across RIP samples to the fpkm of the input control sample. Transcripts were identified as binding to DNMT1 if their fold change was greater than 2-fold. Heatmaps were generated using the heatmap.2 function in the gplots package (version 2.12.1) in R [R Core Development Team. (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.]
Quantitative real-time-PCR
RNA was converted to cDNA using RNA to cDNA EcoDry™ Premix Random Hexamers (Clontech). TaqMan assays for GAPDH, 18s rRNA, U1, CLDN3 and DACOR1 were purchased from Life Technologies. Other primer pairs were designed using primer3 software, and most primers used were designed to span exon–exon boundaries. TaqMan Mastermix (Life Technologies) or Maxima SyBr Green/ROX qPCR Master Mix (Thermo Scientific) was used for qRT-PCR. A comparative CT quantitation was performed with a hold stage of 50°C for 2 min and 95°C for 10 min followed by 40× cycle of 95°C for 15 s and 60°C for 1 min and finally melt curve at 95°C for 15 s, 60°C for 1 min and a ramp to 95°C at 0.3°C increments. Analysis was done using the method with GAPDH as the reference gene.
Colony formation assa
The colon cancer cell lines V481, V852, V866, V703 and V425 were transduced with either a control or DACOR1 lentivirus, and non-infected cells were eliminated by puromycin. For CFAs, cells were plated in either 6-well or 10-cm plates in triplicates of each condition (control versus DACOR1 lentivirus). Cells were plated at 1250, 2500, 5000 or 10 000 cells per well/plate and kept under puromycin selection. Colonies were fixed with methanol/acetic acid and subsequently stained with 0.1% crystal violet solution. Plates were scanned, and colonies were counted using the publically available ImageJ software (65). Average colony counts were calculated for control and DACOR1 plates for each cell line, and a paired t-test was used to test for statistical significance.
Illumina 450K DNA methylation arrays
DNA was extracted from V481 and V852 cells using the DNeasy Blood and Tissue kit (Qiagen). DNA methylation profiling was performed at the Genomics Core Facility at Case Western Reserve University using the Illumina 450K HumanMethylation BeadChip (12 samples/chip). Biological triplicates from both the control and DACOR1 lentivirus-transduced cells were tested in order to detect accurate methylation status. Beta values, a ratio of the methylated/un-methylated signal, were reported and ranged from 0 (completely un-methylated) to 1 (completely methylated). In filtering probes, each cell line was analyzed separately. Reported beta values were removed if the P-value for detectable probe signal was >0.05. Targets were then filtered if only a single beta value remained in either condition. The median beta value was calculated for control and DACOR1 samples. Targets were further filtered if the difference in the maximum beta and minimum beta was >0.1 (10% different). Using the median beta, sites were determined as differentially methylated if the absolute value of the delta–beta was >0.1 (>10%) (Supplementary Material, File S4).
Next-generation RNA sequencing
Six RNA samples were isolated from V852 cells transduced with either a control lentivirus (n = 3) or DACOR1 lentivirus (n = 3). RNAs with RNA integrity number of ≥8 (max is 10) were considered high quality and suitable for RNA-seq. Library preparation was performed using Scriptseq™ Complete Gold (Human/Mouse/Rat) (Illumina) and sequenced on an Illumina Hi-Seq2500. All six samples were run on a single flow cell, and 100-bp paired-end strand-specific sequencing reads were generated and mapped to human genome release hg19 using TopHat with two mismatches allowed for full-length reads. The raw reads were mapped to human genes annotated in RefSeq database using Cufflinks V2.0.2, and CuffDiff was used for identifying differentially expressed genes (47). All expression values were calculated as fragment per kilobase of exon per million of mapped fragments (fpkm).
ChIRP-seq of DACOR1
The ChIRP-seq protocol was carried out as previously described by Chu et al. (56). Briefly, 5 × 108 V852 cells with DACOR1 lentivirus were first crosslinked using 1% formaldehyde for 10 min. The cells were spun down, suspended in Buffer A (Hepes 20 mm, KCl 10 mm, MgCl2 1.5 mm, DTT 0.5 mm, 1% Empigen) and dounced before collecting the nuclei by centrifugation. The nuclei were sonicated in nuclei lysis buffer (Tris–HCl pH 7.5, 20 mm, EDTA 10 mm, 1% SDS, 1 mm DTT, protease inhibitor cocktail, RNaseOut 80 U/ml) to produce 100- to 500-bp DNA fragments. LiCl2 was added at 0.5 m to nuclear lysates. Equal amounts of nuclear lysates were incubated with either DACOR1-specific or non-specific DNA probes modified with a TEG linker and Biotin at their 5′ ends and incubated for 24 h at 37°C with rotation. Next day, RiboMinus™ streptavidin-coated magnetic beads (Life Technologies) were blocked with 800 µg/ml yeast tRNA and 800 µg/ml BSA for 1 h at 37°C in hybridization buffer (Tris–HCl pH 7.5 5 mm, EDTA 10 mm, LiCl2 500 mm) before washing and adding to nuclear lysates for 30 min. The beads were then washed three times with nuclear lysis buffer, wash buffer (Tris–HCl pH 7.5 5 mm, EDTA 0.5 mm, NaCl 1 m) and PBS. The beads were suspended in 200 µl of PBS and incubated at 75°C for 5 min; the supernatant was collected from the beads and incubated at 65°C overnight to reverse crosslinking before extracting DNA using DNeasy Blood & Tissue Kit (Qiagen). Paired-end DNA sequencing was performed on a HiSeq2000/2500 at Otogenetics Corporation. DNA reads were mapped against human genome (hg19) using Bowtie 2, and peak calling was performed by using MACS2. Peak annotation was completed using ChIPpeakAnno. Complete list of DACOR1 genomic occupancy is presented in Supplementary Material, File S6.
PKM2 activity assay
Cells were collected by trypsinization, and pellets were washed twice by cold PBS. The pellets were then resuspended in RIPA buffer (150 mm NaCl, 1 mm EDTA, 1 mm DTT, 1% Triton X-100, 25.5 mm deoxycholic acid and 50 mm Tris–HCl, pH 7.5) and sonicated briefly at 4 C. The total extracts were subjected to PKM activity assay as follows: reaction mixtures contain 50 mm Tris–HCl pH 7.5, 100 mm KCl, 5 mm MgCl2, 0.5 mm ADP, 0.2 mm NADH, 8 units LDH (lactate dehydrogenase from sigma) and 1 mm DTT. The lysates (1–10 µg of total protein) were added to the assay mixture to reach 200 of the final volume in 96-well plates. The enzymatic reaction was initiated by the addition of PEP (phosphoenolpyruvic acid, 0.5 mm) as the substrate. The oxidation of NAPH was monitored at 340 nm for 3 min using a Thermo Max microplate reader (Molecular Devices). The number of units of NADH oxidation was calculated using the standard extinction coefficient of NADH (ϵ = 6.22 mm−1 cm−1). This value was then divided by the total amount of protein added in the assay giving units per milligram of protein from the cell extracts. For all analyses, PKM2 activity was calculated using an amount of cell lysate where the reaction rates fell within the linear range of dependence on the concentration of lysate.
Supplementary Material
Funding
This study was supported by a new investigator startup funds from CWRU (A.M.K.), and by NIH awards P50CA150964 (S.M.), UO1 CA152756 (S.M.), RO1CA127590 (Z.W.), R21CA160060 (Z.W.), R37-DK060596 (M.H.) and RO1-DK053307 (M.H.), and by the Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS).
Supplementary Material
Acknowledgements
We thank Ernest Chan, Ph.D. and Sarah McMahon for help with bioinformatic analyses of next-generation sequencing data; Drs. Anthony Wynshaw-Boris, Thomas LaFramboise, Mark Chance, Peter Scacheri, Paul Tesar and Peter Harte for discussion on results. Next-generation RNA-sequencing and DNA methylation analyses were performed at CWRU Genomics Core Facility.
Conflict of Interest statement. None declared.
References
- 1.Chen T., Dent S.Y. (2014) Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet., 15, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K., Ward A.J., Raj A., Lee J.T., Sharp P.A. et al. (2014) LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell., 54, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M. et al. (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell., 24, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature, 472, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene, 30, 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertani S., Sauer S., Bolotin E., Sauer F. (2011) The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell., 43, 1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A. et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA, 106, 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. et al. (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell, 152, 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R., Fraser P. (2008) The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science, 322, 1717–1720. [DOI] [PubMed] [Google Scholar]
- 10.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. (2008) Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell, 32, 232–246. [DOI] [PubMed] [Google Scholar]
- 11.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. (2008) Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature, 451, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science, 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res., 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev., 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H. et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 16.Engstrom P.G., Suzuki H., Ninomiya N., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L. et al. (2006) Complex Loci in human and mouse genomes. PLoS Genet., 2, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanz R.B., McKenna N.J., Onate S.A., Albrecht U., Wong J., Tsai S.Y., Tsai M.J., O'Malley B.W. (1999) A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell, 97, 17–27. [DOI] [PubMed] [Google Scholar]
- 18.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature, 349, 38–44. [DOI] [PubMed] [Google Scholar]
- 19.Brockdorff N., Ashworth A., Kay G.F., Cooper P., Smith S., McCabe V.M., Norris D.P., Penny G.D., Patel D., Rastan S. (1991) Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature, 351, 329–331. [DOI] [PubMed] [Google Scholar]
- 20.Sleutels F., Zwart R., Barlow D.P. (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature, 415, 810–813. [DOI] [PubMed] [Google Scholar]
- 21.Ishii N., Ozaki K., Sato H., Mizuno H., Saito S., Takahashi A., Miyamoto Y., Ikegawa S., Kamatani N., Hori M. et al. (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet., 51, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 22.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. (2009) MEN varepsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res., 19, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C. et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature, 491, 454–457. [DOI] [PubMed] [Google Scholar]
- 24.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M.. et al (2013) Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife, 2, e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell, 147, 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 465, 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell, 147, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnsson P., Ackley A., Vidarsdottir L., Lui W.O., Corcoran M., Grander D., Morris K.V. (2013) A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol., 20, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong C., Maquat L.E. (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature, 470, 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S., Lojek L., Khalil A.M., Baker K.E., Coller J. (2012) Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell, 45, 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal, 3, ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris K.V., Mattick J.S. (2014) The rise of regulatory RNA. Nat. Rev. Genet., 15, 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell, 40, 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet., 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merry C.R., McMahon S., Thompson C.L., Miskimen K.L., Harris L.N., Khalil A.M. (2015) Integrative transcriptome-wide analyses reveal critical HER2-regulated mRNAs and lincRNAs in HER2+ breast cancer. Breast Cancer Res. Treat., 150, 321–334. [DOI] [PubMed] [Google Scholar]
- 36.Hou P., Zhao Y., Li Z., Yao R., Ma M., Gao Y., Zhao L., Zhang Y., Huang B., Lu J. (2014) LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis., 5, e1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yildirim E., Kirby J.E., Brown D.E., Mercier F.E., Sadreyev R.I., Scadden D.T., Lee J.T. (2013) Xist RNA is a potent suppressor of hematologic cancer in mice. Cell, 152, 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morton M.L., Bai X., Merry C.R., Linden P.A., Khalil A.M., Leidner R.S., Thompson C.L. (2014) Identification of mRNAs and lincRNAs associated with lung cancer progression using next-generation RNA sequencing from laser micro-dissected archival FFPE tissue specimens. Lung. Cancer, 85, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Y., Wong C.J., Kaz A.M., Dzieciatkowski S., Carter K.T., Morris S.M., Wang J., Willis J.E., Makar K.W., Ulrich C.M. et al. (2014) Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology, 147, 418–429 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziller M.J., Gu H., Muller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E. et al. (2013) Charting a dynamic DNA methylation landscape of the human genome. Nature, 500, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmer F., Brinkman A.B., Assenov Y., Matarese F., Kaan A., Sabatino L., Villanueva A., Huertas D., Esteller M., Lengauer T. et al. (2012) Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics, 7, 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu H., Bock C., Mikkelsen T.S., Jager N., Smith Z.D., Tomazou E., Gnirke A., Lander E.S., Meissner A. (2010) Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods, 7, 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H., Zhang Y. (2014) Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell, 156, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrlich M. (2009) DNA hypomethylation in cancer cells. Epigenomics, 1, 239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S., Kelly T.K., Jones P.A. (2010) Epigenetics in cancer. Carcinogenesis, 31, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Guo C., Chen Y., Shulha H.P., Schnetz M.P., LaFramboise T., Bartels C.F., Markowitz S., Weng Z., Scacheri P.C. et al. (2008) Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat. Methods, 5, 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol., 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hata A., Lagna G., Massague J., Hemmati-Brivanlou A. (1998) Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev., 12, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kodach L.L., Wiercinska E., de Miranda N.F., Bleuming S.A., Musler A.R., Peppelenbosch M.P., Dekker E., van den Brink G.R., van Noesel C.J., Morreau H. et al. (2008) The bone morphogenetic protein pathway is inactivated in the majority of sporadic colorectal cancers. Gastroenterology, 134, 1332–1341. [DOI] [PubMed] [Google Scholar]
- 50.Bhattacharyya S., Saha S., Giri K., Lanza I.R., Nair K.S., Jennings N.B., Rodriguez-Aguayo C., Lopez-Berestein G., Basal E., Weaver A.L. et al. (2013) Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS One, 8, e79167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balasubramanian M.N., Butterworth E.A., Kilberg M.S. (2013) Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am. J. Physiol. Endocrinol. Metab., 304, E789–E799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.K., Jang H.G., Jha A.K. et al. (2011) Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature, 476, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amelio I., Cutruzzola F., Antonov A., Agostini M., Melino G. (2014) Serine and glycine metabolism in cancer. Trends Biochem. Sci., 39, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Locasale J.W. (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer, 13, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaneton B., Hillmann P., Zheng L., Martin A.C., Maddocks O.D., Chokkathukalam A., Coyle J.E., Jankevics A., Holding F.P., Vousden K.H. et al. (2012) Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature, 491, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell., 44, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khalil A.M., Rinn J.L. (2011) RNA-protein interactions in human health and disease. Semin. Cell Dev. Biol., 22, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grutzner F., Kaessmann H. (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature, 505, 635–640. [DOI] [PubMed] [Google Scholar]
- 59.Schorderet P., Duboule D. (2011) Structural and functional differences in the long non-coding RNA hot air in mouse and human. PLoS Genet., 7, e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong N., De Melo J., Tang D. (2013) PKM2, a central point of regulation in cancer metabolism. Int. J. Cell Biol., 2013, 242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christofk H.R., Vander Heiden M.G., Wu N., Asara J.M., Cantley L.C. (2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature, 452, 181–186. [DOI] [PubMed] [Google Scholar]
- 62.Jia H., Osak M., Bogu G.K., Stanton L.W., Johnson R., Lipovich L. (2010) Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA, 16, 1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moran V.A., Niland C.N., Khalil A.M. (2012) Co-Immunoprecipitation of long noncoding RNAs. Methods Mol. Biol., 925, 219–228. [DOI] [PubMed] [Google Scholar]
- 64.Surowy C.S., van Santen V.L., Scheib-Wixted S.M., Spritz R.A. (1989) Direct, sequence-specific binding of the human U1–70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol. Cell Biol., 9, 4179–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.