Abstract
Unexpected tryptic cleavage has been characterized at modified K48 residues in polyubiquitins. In particular, the tryptic products of all seven of the lysine-linked dimers of ubiquitin and of three trimers—linear Ub–48Ub–48Ub, linear Ub–63Ub–63Ub, and the branched trimer [Ub]2–6,48Ub—have been analyzed. In addition to the peptide products expected under commonly used tryptic conditions, we observe that peptides are formed with an unexpected ε-glycinylglycinyl-Lys carboxyl terminus when the site of linkage is Lys48. Trypsin from three different commercial sources exhibited this aberration. Initial cleavage at R74 is proposed in a distal ubiquitin to produce a glycinylglycinyl-lysine residue which is bound by trypsin.
Proteolysis by trypsin is commonly used in proteomic workflows and other biochemical investigations. It is solidly established that the enzyme cleaves proteins and peptides on the carboxyl side of lysine and arginine residues1,2 employing a well-understood specific binding mechanism that recognizes the basic side chains of those amino acids.3−5 It is generally observed that derivatized lysine and arginine residues are not substrates for the enzyme,1,2,6 and this behavior has been used to good advantage to recognize and localize modifications on Lys and Arg in histones and other proteins.
Ubiquitin is enzymatically conjugated with substrate proteins and other ubiquitin moieties through isopeptide bonds between its terminal Gly76 and ε-amino groups of lysine residues (Figure 1). The extent and nature of such conjugations have profound effects on protein function and disposition in the cell. Trypsin can cleave all the potential sites in these polypeptides in an exhaustive incubation; however, it cleaves ubiquitin at Arg74 very rapidly,7 leaving Gly75 and Gly76 residues on the modified lysine of the substrate peptide.8−11 Likewise, in the case of polyubiquitins, rapid tryptic cleavage of a distal ubiquitin at Arg74 leaves an ε-glycinylglycine tag on lysine at the site of conjugation in the proximal ubiquitin.12,13 This glycinylglycine tag is susceptible to analysis by HPLC, tandem mass spectrometry, and bioinformatics, and its formation and localization have formed the basis for much of our understanding of the biochemistry of ubiquitination.9,10 Neither ubiquitinated lysine nor ε-glycinylglycinyl-Lys is expected to be cleaved by trypsin. However, in a recent study of ubiquitinated conjugates in exosomes shed by myeloid-derived suppressor cells, 15 of 65 ε-glycinylglycinyl-peptide products had been formed by tryptic cleavage directly at the derivatized lysine residues.11 These included products from polyubiquitins as well as protein conjugates. Other laboratories have also recently reported peptides identified from tryptic digests of ubiquitin polymers and conjugates that are terminated in ε-glycinylglycinyl-Lys.10,14,15 The experiments reported here were designed to confirm rigorously or not that this irregular tryptic cleavage occurs in polyubiquitins and, by implication, in proteins conjugated with ubiquitins. Unexpectedly, we have observed that the irregular cleavage occurs in polyubiquitins specifically at linkages located at Lys48.
Figure 1.

Sequence of the ubiquitin dimer Ub–48Ub. Top: The peptide product that results from the expected formation of the glycinylglycinyl tag from the distal chain and conventional cleavages at Arg42 and Arg54 in the proximal chain is highlighted. Bottom: The peptide product that results from formation of the glycinylglycinyl tag from the distal chain and cleavage at Arg42 and the unexpected cleavage at Lys48 in the proximal chain is highlighted. (In a ubiquitin–ubiquitin conjugate, distal refers to the ubiquitin unit that is conjugated through its C-terminal Gly76 to a lysine on another ubiquitin, referred to as proximal. In a branched triubiquitin such as [Ub]2–6,48Ub, two distal ubiquitins are attached to two different lysines, in this case Lys6 and Lys48, on the same proximal ubiquitin.)
Experimental Section
Polyubiquitins
All seven of the lysine-linked ubiquitin dimers were available for this study, as well as two unbranched trimers, Ub–48Ub–48Ub and Ub–63Ub–63Ub, and the branched trimer [Ub]2–6,48Ub. Ubiquitin dimers were assembled either enzymatically (K48, K63) or chemically (K6, K11, K27, K29, K33) using recombinant ubiquitin monomers, as detailed in ref (16). Ubiquitin trimers were assembled enzymatically using linkage-specific ubiquitin-conjugating E2 enzymes.
Tryptic Digestion
All tryptic digestions were carried out under commonly used conditions: 16 h at 37 °C, in 50 mM ammonium bicarbonate, pH 7.8, using a 1:50 (ubiquitin dimers) or 1:20 (ubiquitin trimers) ratio of enzyme to substrate. Briefer incubation times were implemented in kinetic studies. Trypsin Gold, mass spectrometry grade porcine trypsin from Promega, was used in the majority of the 16 h digestions and in kinetic studies. However, TPCK (tosylphenylalanyl chloromethyl ketone)-immobilized bovine pancreatic trypsin (Thermo Pierce) and underivatized bovine pancreatic trypsin free from mycoplasma and extraneous viruses (Worthington Biochemical) were also evaluated.
Mass Spectrometry
The product mixtures were analyzed by LC–MS/MS using Prominence LC20ADnano pumps (Shimadzu Scientific) interfaced to an LTQ–orbitrap (Thermo Fisher) tandem mass spectrometer. This product mixture comprised mostly short peptides, which were separated on a capillary C18 column (150 × 0.15 mm, 300 Å, Grace Davidson Discovery Sciences), increasing the gradient from 0% to 50% solvent B (97.5% acetonitrile, 2.5% water, and 0.1% formic acid) through 23 min, followed by an increase from 50% to 85% solvent B in 2 min. Solvent A was 2.5% acetonitrile and 0.1% formic acid in water. The flow rate was 500 nL/min. Precursor scans of product mixtures were acquired in the orbitrap with a resolution of 30 000 at m/z 400, and product ion scans were acquired at unit resolution in the LTQ. The mixture of tryptic peptides from the K48 dimer was also analyzed at 30 000 resolution at m/z 40 for both precursor ions and fragment ions. In addition to data-dependent analysis, the precursor ions listed in Table 1 and Table 2 were selected for targeted analysis. Product mixtures were analyzed from six replicate digestions of each ubiquitin trimer, and three to six replicate digestions of each dimer were studied.
Table 1. Glycinylglycinyl-Peptides Observeda from Proteolysis of Ubiquitin Dimers Linked at Lys63, Lys48, Lys33, Lys29, Lys27, Lys11, and Lys6.
| cleavage
at unmodified Lys |
cleavage
at modified Lys |
|||||
|---|---|---|---|---|---|---|
| dimer linkage | peptide sequence | precursor mass (Da) | obsd | peptide sequence | precursor mass (Da) | obsd |
| K63 | TLSDYNIQKGGESTLHLVLR | 2244.19 | yes | TLSDYNIQKGG | 1195.59 | no |
| K48 | LIFAGKGGQLEDGR | 1460.78 | yes | LIFAGKGG | 762.45 | yes |
| K33 | IQDKGGEGIPPDQQR | 1637.82 | yes | IQDKGG | 617.32 | no |
| K29 | AKGGIQDK | 816.46 | yes | TITLEVEPSDTIENVKAKGG | 2101.10 | no |
| K27 | TITLEVEPSDTIENVKGGAK | 2101.10 | yes | TITLEVEPSDTIENVKGG | 1901.97 | no |
| K11 | TLTGKGGTITLEVEPSDTIENVK | 2402.27 | yes | TLTGKGG | 633.36 | no |
| K6 | MQIFVKGGTLTGK | 1379.77 | yes | MQIFVKGG | 879.48 | no |
“Yes” indicates that the peptide was observed; “no” indicates that the peptide was not observed.
Table 2. Glycinylglycinyl-Peptides Observeda from Proteolysis of Ubiquitin Trimers Ub–48Ub–48Ub, Ub–63Ub–63Ub, and [Ub]2–6,48Ub.
| cleavage
at unmodified Lys |
cleavage
at modified Lys |
|||||
|---|---|---|---|---|---|---|
| trimer linkage | peptide sequence | precursor mass (Da) | obsd | peptide sequence | precursor mass (Da) | obsd |
| Ub–63Ub–63Ub | TLSDYNIQKGGESTLHLVLR | 2244.19 | yes | TLSDYNIQKGG | 1195.59 | no |
| Ub–48Ub–48Ub | LIFAGKGGQLEDGR | 1460.78 | yes | LIFAGKGG | 762.45 | yes |
| [Ub]2–6,48Ub | MQIFVKGGTLTGK | 1379.77 | yes | MQIFVKGG | 879.48 | no |
| LIFAGKGGQLEDGR | 1460.78 | yes | LIFAGKGG | 762.45 | yes | |
“Yes” indicates that the peptide was observed; “no” indicates that the peptide was not observed.
Bioinformatics
A MASCOT search was performed against the SwissProt human database with an automatic decoy search and allowing one missed cleavage. Precursor and fragment ion tolerances of 5 ppm and 0.6 Da, respectively, were used. Glycinylglycine-modified lysine was allowed as a variable modification. Technical and proteolytic replicates were scored using MASCOT’s “MudPit” function.
Kinetic Studies
Tryptic products of ubiquitin dimers recovered after brief digestions (5, 15, 30, 60, and 90 min) in kinetic studies were fractionated using a capillary C3 column (150 mm × 0.1 mm, 0.5 μm, Agilent) with the same gradient described above and a flow rate of 300 nL/min. Ions were measured in the orbitrap with a resolution of 30 000 at m/z 400. Data sets were deconvoluted, and peak areas were determined in selected ion current chromatograms.
Results and Discussion
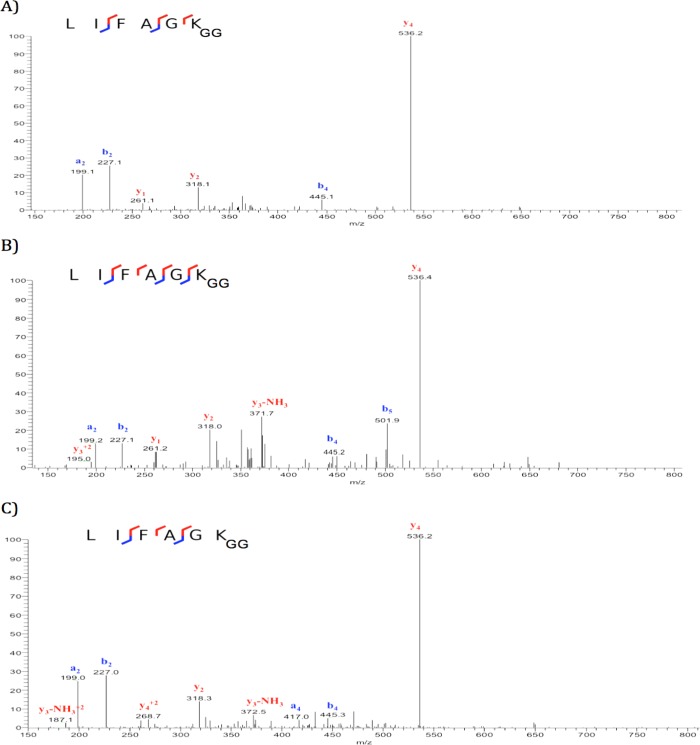
Examination of tryptic products from more than 30 experiments indicates that digestion is exhaustive under the conditions used for the 16 h proteolysis with each of the three samples of trypsin. The tryptic peptide LIFAGKGGQLEDGR [43–54] (Figure 1) was formed from the Ub–48Ub dimer by the expected mechanism with Trypsin Gold. Moreover, an additional glycinylglycinyl-peptide, LIFAGKGG [43–48], was identified, in which tryptic cleavage had occurred at Lys48 to produce a peptide terminated by glycinylglycinyl-Lys48 (Figure 1). As described in the Experimental Section, the peptide was identified by an unbiased search of its tandem mass spectrum (Figure 2A) against the SwissProt human protein database The peptide LIFAGKGG was identified with an expect value of 0.014 (Supplemental Table 1, Supporting Information).
Figure 2.
Tandem mass spectrum of tryptic peptide LIFAGKGG formed in tryptic digestion of (A) ubiquitin dimer Ub–48Ub, (B) ubiquitin trimer Ub–48Ub–48Ub, and (C) ubiquitin trimer [Ub]2–6,48Ub.
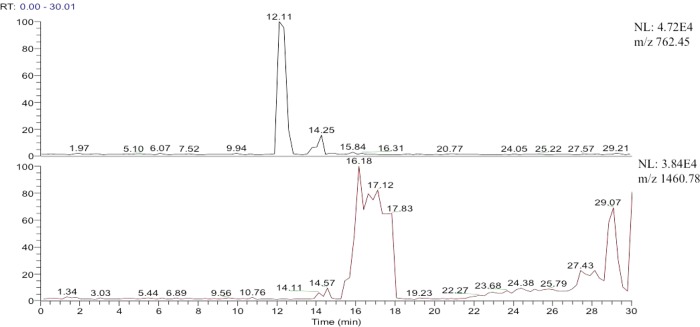
The production of both the expected and aberrant tryptic peptides in the tryptic product mixture is confirmed by the extracted ion chromatograms shown in Figure 3. The ratio of the expected glycinylglycinyl product to the novel peptide is approximately 5:1, estimated by peak areas in the ion chromatograms (Figure 3). These two peptides and the corresponding glycinylglycinyl-peptides that might be formed from dimers with other linkage sites are listed in Table 1. Comments in the table confirm that the expected tryptic products were observed for all the dimers, formed with cleavage of the distal ubiquitin at Arg74, blocked cleavage at the glycinylglycinyl-lysine in the proximal ubiquitin, and cleavage at the next lysine or arginine. The absence to at least the 1% level of any peptides formed by aberrant cleavages in the product mixtures from the other dimers is supported by extracted ion chromatograms and targeted MS/MS experiments. The aberrant or unexpected product was also found to be formed when the Ub–48Ub dimer was incubated with TPCK-immobilized bovine pancreatic trypsin (expected:unexpected = 85:1) and mycoplasma- and extraneous-virus-free underivatized bovine pancreatic trypsin (expected:unexpected = 91:1). The differences in product distribution suggest that bovine trypsin cleaves the Ub–48Ub dimer with more fidelity than porcine trypsin, not surprising in view of the significant differences documented recently in proteolysis of albumin by porcine and bovine trypsin.17
Figure 3.
Extracted ion chromatograms of (top) m/z 762.5, the molecular ion of the peptide formed by tryptic cleavage at glycinylglycine-modified K48, and (bottom) m/z 1460.8, the molecular ion of the peptide formed by tryptic cleavage at K54 with glycinylglycine attached at K48.
Table 2 reports glycinylglycinyl-peptides recovered from the Trypsin Gold digestion of two unbranched ubiquitin trimers, Ub–63Ub–63Ub and Ub–48Ub–48Ub, and the branched trimer [Ub]2–6,48Ub. In the case of Ub–63Ub–63Ub, only the expected cleavage products are observed, with no cleavage of lysine at the sites of conjugation. In the case of Ub–48Ub–48Ub, the expected product [43–54] is formed with cleavage at Arg54, and in addition, peptide [43–48] is identified in which the carboxyl terminus carries the glycinylglycinyl tag (Figure 2B). The third trimer offers a Lys48 modification and also one at Lys6. Digestion with Trypsin Gold produced the expected peptides with missed cleavages at glycinylglycinyl-Lys6 and -Lys48. In addition, a third product was identified, on the basis of accurate mass determination and tandem mass spectra, which terminates in glycinylglycinyl-Lys48 (Figure 2C). Overall, the digestion products detected from these trimers further support the conclusion that proteolysis occurs uniquely at modified Lys48, and not at Lys residues modified at other positions in polyubiquitins.
These observations introduce several questions: What is the substrate for trypsin that produces the unexpected cleavage? Why does the aberrant cleavage take place selectively at substituted K48 in polyubiquitins?
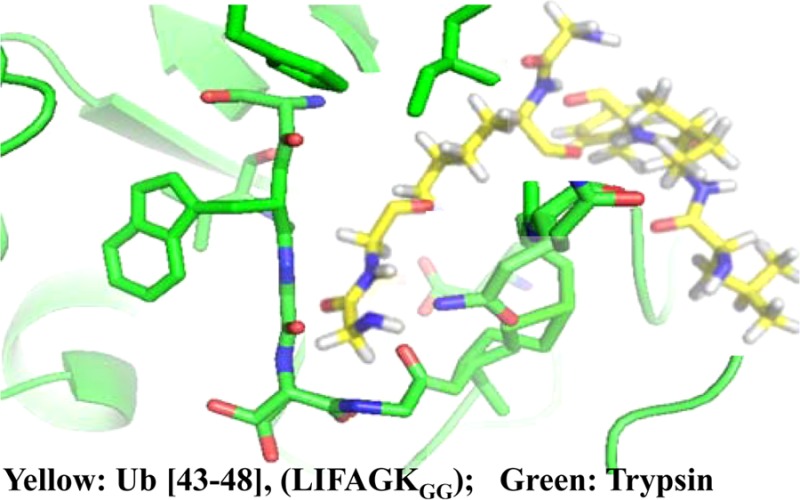
A kinetic study presented in Supplemental Figure 1 (Supporting Information) shows rapid tryptic cleavage at R74 through the first 90 min of digestion of Ub–48Ub and Ub–63Ub dimers and the absence of detectable cleavage at K48 and K63, respectively. This is consistent with earlier reports.7,12,13 We propose that peptides carrying glycinylglycinyl-lysine are bound by trypsin as substrates for subsequent cleavage to produce the unexpected product (Figure 1). This binding would occur in competition with normal substrates. Because the glycinylglycine fragment carries a positive charge at its N-terminus and contains no bulky groups, it is bound in the recognition pocket by which trypsin normally recognizes Lys and Arg side chains. This surrogate binding may not position the polypeptide chain correctly to allow cleavage to occur at the normal rate. The binding pocket of trypsin is 10–12 Å deep, on the basis of crystal structures.18,19 On the basis of a crystal structure available for the K48-linked ubiquitin dimer,20 the length of the extended lysine side chain modified by glycinylglycine is 10.9 Å. It is relevant that S-(β-1-aminoethyl)cysteine, a side chain that also presents a positive terminal charge, was recognized as a substrate for trypsin some years ago.21
The second question is more challenging. If the substrate for the unexpected cleavage is not ubiquitin-modified Lys48, but rather glycinylglycinyl-Lys48-Ub, then we need to ask why trypsin binds and cleaves glycinylglycinyl-Lys at position 48 of diubiquitin, but does not bind and cleave glycinylglycinyl-Lys at other positions. In an initial consideration of the primary sequence, we used the MEROPS tool22 to correlate cleavage frequencies experimentally associated with the three residues N-terminal and four residues C-terminal to the cleavage sites in the various ubiquitin dimers. Although the sequence around K48 scored well, considerations of primary sequence were not definitive and suggest that tertiary structure and topology should be considered. The compact and dynamic structures of all the linkage types of diubiquitin have been modeled recently and found to be distinctive.23,24 We speculate that trypsin has a specific interaction with the surface of ubiquitin around Lys48 and that this interaction promotes binding and/or cleavage of glycinylglycinyl-Lys48 as a substrate. We are currently exploring the possibility of such an interaction using NMR.
In summary, we have confirmed that trypsin does cleave some polyubiquitins to produce peptides that carry glycinylglycinyl-lysine at the carboxyl terminus. We propose that rapid cleavage of a distal ubiquitin chain occurs first in these cases, and that the resulting glycinylglycinyl-lysine side chain is bound as a surrogate to arginine (or lysine) in trypsin’s active site. Subsequent proteolysis can occur if the polypeptide chain is suitably positioned on the enzyme. This hypothesis is extended to proteins conjugated with ubiquitin, consistent with reports of unexpected formation of peptides with glycinylglycinyl-lysine carboxyl termini.10,11,14,15 These observations suggest that qualitative and quantitative studies that rely on analysis only of peptides carrying internal glycinylglycinyl tags may require reinterpretation.
Acknowledgments
This research was supported by grants from the National Institutes of Health (GM021248 and GM065334).
Supporting Information Available
Table listing the peptide identifications from the Ub–48Ub dimer (Excel) and figure showing the peak area as a function of time for the digestion of Ub–48Ub and Ub–63Ub. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.5b01960.
The authors declare no competing financial interest.
Supplementary Material
References
- Olsen J. V.; Ong S.-E.; Mann M. Mol. Cell. Proteomics 2004, 3, 608–614. 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- Rodriguez J.; Gupta N.; Smith R. D.; Pevzner P. A. J. Prot. Res. 2008, 7, 300–305. 10.1021/pr0705035. [DOI] [PubMed] [Google Scholar]
- Sprang S.; Standing T.; Fletterick R. J.; Stroud R. M.; Finer-Moore J.; Xuong N.-H.; Hamlin R.; Rutter W. J.; Craik C. S. Science 1987, 237, 905–909. 10.1126/science.3112942. [DOI] [PubMed] [Google Scholar]
- Craik C. S.; Roczniak S.; Largman C.; Rutter W. J. Science 1987, 237, 909–913. 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- Corey D. R.; Craik C. S. J. Am. Chem. Soc. 1992, 114, 1784–1790. 10.1021/ja00031a037. [DOI] [Google Scholar]
- Ong S.-E.; Mittler G.; Mann M. Nat. Methods 2004, 1, 119–126. 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. D.; Audhya T. K. J. Biol. Chem. 1981, 256, 9235–9241. [PubMed] [Google Scholar]
- Van Nocker S.; Vierstra R. D. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 10297–10301. 10.1073/pnas.88.22.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.; Schwartz D.; Elias J. E.; Thoreen C. C.; Cheng D.; Marsischky G.; Roelofs J.; Finley D.; Gygi S. P. Nat. Biotechnol. 2003, 21, 921–926. 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Denis N. J.; Vasilescu J.; Lambert J. P.; Smith J. C.; Figeys D. Proteomics 2007, 7, 868–874. 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- Burke M. C.; Oei M. S.; Edwards N. J.; Ostrand-Rosenberg S.; Fenselau C. J. Prot. Res. 2014, 13, 5965–5972. 10.1021/pr500854x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P.; Peng J. Anal. Chem. 2008, 80, 3438–3444. 10.1021/ac800016w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkevich E. M.; Sanchez N.; Ge Y.; Strieter E. R. Biochemistry 2014, 53, 4979–4989. 10.1021/bi5006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen J. R.; Sylvestersen K. B.; Bekker-Jensen S.; Szklarczyk D.; Poulsen J. W.; Horn H.; Jensen L. J.; Mailand N.; Nielsen M. L. Mol. Cell. Proteomics 2011, 10, M110.003590. 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.; Paige J. S.; Jaffrey S. R. Nat. Biotechnol. 2010, 28, 868–873. 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. E.; Castaneda C.; Wang Y.; Fushman D.; Fenselau C. J. J. Mass Spectrom. 2014, 49, 1272–1278. 10.1002/jms.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley S. J.; Rudnick P. A.; Liang Y.; Dong Q.; Stein S. E.; Nesvizhskii A. I. J. Prot. Res. 2013, 12, 5666–5680. 10.1021/pr400611h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A.; Hendekson R.; Blow D. M. J. Mol. Biol. 1969, 46, 337–348. 10.1016/0022-2836(69)90426-4. [DOI] [PubMed] [Google Scholar]
- Liebschner D.; Dauter M.; Brzuszkiewicz A.; Dauter Z. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1447–1462. 10.1107/S0907444913009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.-Y.; Zhang N.; LaRonde-LeBlanc N.; Fushman D. Biochim. Biophys. Acta, Mol. Cell Res. 2012, 1823, 2046–2056. 10.1016/j.bbamcr.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindley H. Nature 1956, 178, 647–648. 10.1038/178647a0. [DOI] [PubMed] [Google Scholar]
- Rawlings N. D.; Waller M.; Barrett A. J.; Bateman Nucleic Acids Res. 2014, 42, D503–D509. 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D.; Walker O. J. Mol. Biol. 2010, 395, 803–814. 10.1016/j.jmb.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Tang C.; Wang E.; Wang J. PLoS Comput. Biol. 2014, 10, e1003691. 10.1371/journal.pcbi.1003691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.