Abstract
In rodents, fibroblast growth factor 21 (FGF21) has emerged as a key metabolic regulator produced by liver. To gather preliminary data on the potential importance of FGF1, co-regulated genes, and upstream metabolic genes, we examined the hepatic mRNA expression in response to nutrition and inflammation in dairy cows. In experiment 1, induction of ketosis through feed restriction on d 5 postpartum upregulated FGF21, its co-receptor KLB, and PPARA but only elicited a numerical increase in serum FGF21 concentration. In experiment 2, cows in control (CON) or receiving 50 g/d of L-carnitine (C50) from -14 through 21 d had increased FGF21, PPARA, and NFIL3 on d 10 compared with d 2 postpartum. In contrast, compared with CON and C50, 100 g/d L-carnitine (C100) resulted in lower FGF21, KLB, ANGPTL4, and ARNTL expression on d 10. In experiment 3, cows were fed during the dry period either a higher-energy (OVE; 1.62 Mcal/kg DM) or lower-energy (CON; 1.34 Mcal/kg DM) diet and received 0 (OVE:N, CON:N) or 200 μg of LPS (OVE:Y, CON:Y) into the mammary gland at d 7 postpartum. For FGF21 mRNA expression in CON, the LPS challenge (CON:Y) prevented a decrease in expression between d 7 and 14 postpartum such that cows in CON:N had a 4-fold lower expression on d 14 compared with d 7. The inflammatory stimulus induced by LPS in CON:Y resulted in upregulation of PPARA on d 14 to a similar level as cows in OVE:N. In OVE:Y, expression of PPARA was lower than CON:N on d 7 and remained unchanged on d 14. On d 7, LPS led to a 4-fold greater serum FGF21 only in OVE but not in CON cows. In fact, OVE:Y reached the same serum FGF21 concentration as CON:N, suggesting a carryover effect of dietary energy level on signaling mechanisms within liver. Overall, results indicate that nutrition, ketosis, and inflammation during the peripartal period can alter hepatic FGF21, co-regulated genes, and upstream metabolic genes to various extents. The functional outcome of these changes merits further study, and in particular the mechanisms regulating transcription in response to changes in energy balance and feed intake.
Introduction
Fibroblast growth factor 21 (FGF21) is a novel metabolic regulator of the FGF family that is produced by the liver and in rodents has an important role in the regulation of glucose and lipid metabolism [1, 2]. In non-ruminants, the induction of mRNA expression of hepatic FGF21 stimulates glucose uptake in adipocytes and skeletal muscle, thus, improving insulin sensitivity and reducing serum triacylglycerol (TAG) concentrations [3, 4]. Hepatic FGF21 also plays a role in regulation of hepatic oxidation of fatty acids and gluconeogenesis in response to fasting and during consumption of high-fat diets [5]. The physiological importance of FGF21 has been partly demonstrated using FGF21-null mice fed a ketogenic diet, which led to higher rates of lypolysis during fasting and greater deposition of liver TAG [6, 7]. Schoenberg et al. [8] working with periparturient cows reported that the onset of negative energy balance (NEB) after calving was associated with increased plasma FGF21 concentration and greater FGF21 mRNA expression in liver.
Carnitine has an important role in various metabolic functions including mitochondrial long-chain fatty acid (LCFA) oxidation, and has been shown to dramatically decrease or prevent liver lipid accumulation in laboratory animals [9, 10] and dairy cows [11]. Recently, Schlegel et al. [12] working with periparturient cows observed a positive correlation between FGF21 mRNA expression and genes involved in carnitine synthesis. Furthermore, in rodents the onset of infection, inflammation, trauma, and malignancy induces the acute-phase response (APR), which leads to a decrease in hepatic oxidation of fatty acids and ketogenesis [13,14]. Feingold et al. [15] proposed that FGF21 is a positive APR protein that could help protect animals from the toxic effects of LPS and sepsis.
The above data led us to hypothesize that FGF21 has a central role in the adaptations to NEB, ketosis, carnitine supplementation, and inflammatory challenge in peripartal dairy cows. Thus, the aim of the present study was to develop a better understanding of the role of hepatic FGF21, co-regulated genes, and upstream metabolic genes related to hepatic metabolism. To achieve this aim we used liver and serum samples from previous experiments dealing with early postpartal ketosis[16], peripartal dietary L-carnitine supplementation[11], and prepartal level of dietary energy and postpartal intramammary LPS challenge [17].
Materials and Methods
Experimental Design and Treatments
The present study was performed using samples from three different experiments, i.e. early postpartal ketosis[16, 18], peripartal dietary L-carnitine supplementation[11], and prepartal dietary energy level and postpartal inflammatory challenge [17]. All these experiments were performed at the University of Illinois Dairy Research Center under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Illinois.
Early Postpartal Ketosis
The details of the animal management and sample collection were presented earlier [16, 18]. Briefly, all Holstein cows were housed in individual tie-stalls, were fed twice daily at ~1000 and 1500 h, and had unlimited access to fresh water. On d 5 postpartum, cows were randomly assigned to control (n = 7) or ketosis-induction (n = 7) based on a thorough physical examination on d 4 postpartum. Cows in the ketosis-induction group were fed at 50% of d 4 intake from d 5 until d 14 postpartum or until they developed signs of clinical ketosis (anorexia, ataxia, or abnormal behavior) while the control group were fed ad libitum throughout the treatment period. The cows with ketosis had higher (P < 0.05) concentrations of serum NEFA, BHBA but lower glucose, as well as greater total lipid and TAG in liver than did control cows [16]. A single liver biopsy for gene expression analysis was performed prior to the morning meal between d 9 and 14 (ketosis induction) or d 14 postpartum (control). The serum samples for FGF21 analysis were from the same day of the biopsy. Energy balance of the cows at the time of liver biopsy was greater (P < 0.05) for the control (93% of estimated requirements) compared with the ketotic cows (53% of estimated requirements) [18].
Dietary Carnitine Supplementation
Detailed information of the animal management and sample collection was reported by[11]. Briefly, all Holstein cows were housed in individual tie-stalls, were fed twice daily at ~1000 and 1500 h, and had unlimited access to fresh water. Cows were assigned to treatments at d −25 relative to expected calving date and remained on experiment until d 56. Treatments were four amounts of supplemental dietary L-carnitine (L-carnitine; Lonza, Inc., Allendale, NJ): control (CON, 0 g/d of L-carnitine; n = 14); low carnitine (6 g/d; n = 11); medium carnitine (C50, 50 g/d; n = 12); and high carnitine (C100, 100 g/d; n = 12). Carnitine supplementation began on d −14 relative to expected calving and continued until d 21. Liver biopsies harvested at d 2 and 10 postpartum before the morning meal were used for gene expression analysis. Blood serum samples from this study were not available for FGF21 determination. Hepatic concentration of free carnitine on d 2 and 10 was greater (P < 0.05) than controls with C50 and C100 [11]. Furthermore, at d 2 and 10 higher (P < 0.05) total lipid and TAG concentrations were observed in liver from CON cows compared with C50 and C100 [11]. Therefore, liver tissue from d 2 and 10 from CON (n = 6), C50 (n = 6), and C100 (n = 6) were used for gene expression analysis. Energy balance of the cows at the time of liver biopsy was greater (P < 0.05) in CON (87% of estimated requirements) and C50 (85% of estimated requirements) compared with C100 (63% of estimated requirements) [11].
Prepartal Dietary Energy and Postpartal Intramammary LPS Challenge
Detailed information of the animal management and sample collection was presented by[17]. Briefly, all Holstein cows were housed in individual tie-stalls, were fed twice daily at ~1000 and 1500 h, and had unlimited access to fresh water. Cows were assigned randomly (n = 20 per diet) to a lower-energy diet (CON, high-fiber; 1.34 Mcal/kg DM), which was fed ad libitum to provide approximately 100% of calculated NEL requirements, or were fed a diet to provide at least 150% of calculated NEL requirements (OVE, overfed group; 1.62 Mcal/kg DM) during the entire 45 days of dry period [19]. After parturition, cows were moved to a tie-stall barn, fed a common lactation diet (NEL = 1.69 Mcal/kg DM), and milked twice daily (0400 and 1600 h).
At d 7 postpartum, each group (i.e. CON and OVE) were further divided into two additional groups (total of 4 groups) based on whether they received an intramammary E. coli LPS challenge (200 μg, strain 0111:B4, cat. # L2630, Sigma Aldrich, St. Louis, MO) or not, i.e. 0 (OVE:N, CON:N) or 200 μg LPS (OVE:Y, CON:Y). Liver tissue harvested on d 7 (2.5 h post-LPS for OVE:Y, CON:Y) and d 14 postpartum (n = 6/dietary group) before the morning meal was used for gene expression analysis. Blood sampled from the coccygeal vein or artery on d 7 (prior to LPS and liver biopsy) and 14 relative to parturition was used for FGF21 determination. Energy balance of the cows at the time of liver biopsy was greater (P < 0.05) in CON (78% of estimated requirements) compared with OVE (65% of estimated requirements) [17].
RNA Extraction, Primer Design, and qPCR Analysis
The complete procedures for RNA extraction and qPCR analysis have been published previously[20,21]. Briefly, approximately 0.2 to 0.3 g of liver tissue was homogenized in 1 to 2 mL ice-cold TRIzol reagent (Invitrogen, Carlsbad, CA, Cat. No. 15596–026) and RNA extraction was performed as described previously[22]. Concentration of RNA was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), while the RNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). The average RNA integrity number value of all samples used was 8 ± 0.4. Total RNA was purified with the RNeasy Mini Kit and residual DNA removed using the RNase-Free DNase Set (Qiagen, Valencia, CA). The extracted and cleaned RNA was re-suspended in RNase free water (Qiagen, Cat No. 74104) and stored at −80°C until qPCR analysis.
The evaluation of direct links among nutrition, ketosis and inflammation were evaluated through the assessment of genes associated with fatty acid oxidation (carnitine palmitoyltransferase 1A, CPT1A; peroxisome proliferator-activated receptor alpha, PPARA), FGF21 signaling (klotho beta, KLB), and the hepatokine angiopoietin-like 4 (ANGPTL4). In addition, recent data from rodent work provided evidence that the BMAL1 (aryl hydrocarbon receptor nuclear translocator-like, ARNTL)-CLOCK complex activates the rodent FGF21 promoter, whereas another circadian gene (nuclear factor, interleukin 3 regulated, NFIL3) suppresses it[23, 24]. Furthermore, the downregulation of FGF21 with insulin in non-ruminants is mediated through NFIL3[23]. Therefore, the expression of selected genes associated with circadian rhythms (ARNTL; clock circadian regulator, CLOCK; NFIL3) and insulin signaling (v-akt murine thymoma viral oncogene homolog 1, AKT1) also were investigated. The final data were normalized using the geometric mean (stability = 0.20) [25] of ubiquitously-expressed transcript (UXT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and ribosomal protein S9 (RPS9). Details of the primers and sequences are presented in supplementary Tables A-D in the S1 File.
Serum Concentration of FGF21
Because FGF21 is a secreted protein, ELISA was performed to determine the serum concentration of FGF21 using a Mouse and Rat FGF-21 ELISA (BioVendor, Laboratorni medicina a.s., Brno, Czech Republic, Cat. No. RD291108200R). The antibody provided with the kit reportedly has cross reactivity with the bovine FGF21 protein, which we previously confirmed[20]. However, it should be kept in mind that the primary antibody also cross-reacts with other bovine proteins. Thus, the data generated should be regarded as qualitative than quantitative. All the procedures were performed according to the manufacturer’s protocol. The minimal detectable concentration of FGF21 with this assay estimated by the manufacturer is 18.4 pg/mL. The lowest concentration detected in cow serum in the present study was 48.88 pg/mL. The intra- and inter-assay coefficients of variation were less than 9.0% and less than 8.0%, respectively.
Statistical Analysis
The MIXED procedure of SAS (version 9.1; SAS Institute Inc., Cary, NC) was used for statistical analysis. In experiment 1, diet (control and ketosis) was included as fixed effect. In experiment 2, the model considered as fixed effects diet (CON, C50, and C100), d (2 and 10) and the interaction between diet and day. A repeated measures analysis was performed using the AR(1) covariate structure. In experiment 3, the model included as fixed effects diet (CON and OVE), LPS (No and Yes), d (7 and 14) and all possible interactions. Data were normalized by logarithmic transformation prior to statistical analysis. All means were compared using the PDIFF statement of SAS. Gene expression data reported in tables were back-transformed after statistical analysis. The 95% confidence intervals were calculated with logarithmic transformed data using a modified version of the Cox method as suggested by Olsson [26].
Results
Hepatic Gene Expression
Early Postpartal Ketosis
A greater FGF21 (P < 0.01) and KLB (P = 0.02) mRNA expression was detected in ketotic cows compared with control cows (Table 1). No significant change (P > 0.05) was observed between control and ketotic cows for the mRNA expression of ARNTL, CLOCK, NFIL3 and AKT1. The mRNA expression of CPT1A and PPARA was not measured in this study because they were reported by Loor et al.[16].
Table 1. Hepatic mRNA expression of FGF21, FGF21 binding, and genes associated with circadian rhythms and insulin signaling in control cows (n = 7) and cows induced to develop ketosis (n = 7) by undernutrition after calving.
Liver biopsy was performed at d 9 to 14 (ketosis induction) or 14 d postpartum (control) before the morning meal. The 95% confidence interval (CI) is reported.
| Gene | Treatment | Relative expression 1 | CI | P value |
|---|---|---|---|---|
| FGF21 | Control | 3.26 | 1.33 to 7.99 | <0.01 |
| Ketotic | 133.4 | 94.61 to 188.20 | ||
| KLB | Control | 7.97 | 5.66 to 11.24 | 0.02 |
| Ketotic | 25.5 | 18.08 to 35.89 | ||
| ARNTL | Control | 1.48 | 1.17 to 1.87 | 0.26 |
| Ketotic | 2.16 | 1.71 to 2.72 | ||
| CLOCK | Control | 0.37 | 0.29 to 0.47 | 0.20 |
| Ketotic | 0.57 | 0.40 to 0.65 | ||
| NFIL3 | Control | 0.55 | 0.43 to 0.70 | 0.39 |
| Ketotic | 0.74 | 0.58 to 0.93 | ||
| AKT1 | Control | 0.42 | 0.37 to 0.49 | 0.86 |
| Ketotic | 0.44 | 0.38 to 0.50 |
1Calculated after normalization with the geometric mean of UXT, GAPDH and RPS9 (see materials and methods).
Dietary L-Carnitine Supplementation
There was an interaction (diet × day; P < 0.05; Table 2) for all genes except AKT1 and a tendency for interaction (P = 0.06) for CLOCK. The mRNA expression of FGF21, CPT1A, and ARNTL was not affected (P > 0.05) by diet or time, but an interaction occurred (diet × day; P < 0.05). No main effects of carnitine (P > 0.05) were observed for AKT1. Supplemental carnitine did not affect FGF21 at d 2, while at d 10 feeding C100 resulted in a marked decrease in FGF21 compared with control. No main effect of carnitine or time were observed for CPT1A expression; however, feeding C50 increased CPT1A expression at d 2 compared with d 10 (diet × day; P < 0.05). At d 2, no change was observed in the expression of ARNTL between the treatments, whereas at d 10 feeding C100 decreased the expression of ARNTL compared with control and C50. Furthermore, between d 2 and 10, ARNTL expression was lower in cows fed C100.
Table 2. Hepatic mRNA expression of FGF21, FGF21 binding, and genes associated with circadian rhythms and insulin signaling in cows fed control (0 L-carnitine; n = 6) and dietary L-carnitine at a rate of 50 g/d (C50; n = 6) or 100 g/d (C100; n = 6) from d −14 through 21 around parturition.
Liver biopsies harvested at d 2 and 10 postpartum before the morning meal. The 95% confidence interval is reported in parentheses.
| Gene | Day | Treatment | P value | ||||
|---|---|---|---|---|---|---|---|
| CON | C50 | C100 | Diet (T) | Day (D) | T × D | ||
| FGF21 | 2 | 0.89 (0.52 to 1.22) | 0.32 (0.19 to 0.53) | 0.62* (0.40 to 0.74) | 0.21 | 0.85 | 0.03 |
| 10 | 1.34 A (0.80 to 1.51) | 0.51 A B (0.30 to 0.84) | 0.23 B * (0.15 to 0.34) | ||||
| PPARA | 2 | 1.16 (1.04 to 1.29) | 1.06* (0.97 to 1.18) | 1.11 (0.99 to 1.24) | 0.08 | 0.68 | 0.02 |
| 10 | 1.23 A (1.14 to 1.37) | 1.33 A * (1.23 to 1.50) | 1.04 B (0.93 to 1.10) | ||||
| KLB | 2 | 1.08 (0.95 to 1.23) | 0.93 (0.82 to 1.05) | 1.06* (0.92 to 1.21) | <0.01 | 0.46 | <0.01 |
| 10 | 1.06 A (0.94 to 1.20) | 1.02 A (0.90 to 1.16) | 0.69 B * (0.60 to 0.80) | ||||
| CPT1A | 2 | 1.01 (0.90 to 1.12) | 0.91* (0.82 to 1.02) | 1.06 (0.94 to 1.19) | 0.81 | 0.39 | 0.05 |
| 10 | 1.13 (1.07 to 1.25) | 1.11* (1.04 to 1.24) | 0.90 (0.80 to 1.02) | ||||
| ANGPTL4 | 2 | 0.73 A * (0.63 to 0.85) | 1.23 B (1.06 to 1.42) | 1.26 B * (1.07 to 1.48) | 0.23 | <0.01 | <0.01 |
| 10 | 1.05 A * (0.91 to 1.21) | 1.01 A (0.87 to 1.16) | 0.53 B * (0.45 to 0.62) | ||||
| ARNTL | 2 | 0.37 (0.28 to 0.48) | 0.24 (0.19 to 0.32) | 0.30* (0.23 to 0.39) | 0.37 | 0.41 | <0.05 |
| 10 | 0.33 A (0.25 to 0.42) | 0.33 A (0.26 to 0.42) | 0.17 B * (0.13 to 0.22) | ||||
| CLOCK | 2 | 0.73 A (0.45 to 1.19) | 0.08 B * (0.05 to 0.14) | 0.93 A (0.53 to 1.60) | 0.06 | 0.02 | 0.06 |
| 10 | 0.76 (0.47 to 1.23) | 0.36* (0.24 to 0.62) | 1.05 (0.59 to 1.82) | ||||
| NFIL3 | 2 | 0.98 A * (0.88 to 1.10) | 1.34 B * (1.20 to 1.50) | 1.21 A B (1.06 to 1.36) | <0.01 | 0.14 | 0.03 |
| 10 | 1.35 A B * (1.22 to 1.56) | 1.72 B * (1.54 to 1.92) | 1.21 A (1.07 to 1.36) | ||||
| AKT1 | 2 | 0.12 (0.10 to 0.14) | 0.13 (0.11 to 0.15) | 0.16 (0.14 to 0.18) | 0.33 | 0.49 | 0.26 |
| 10 | 0.13 (0.12 to 0.15) | 0.14 (0.13 to 0.16) | 0.15 (0.13 to 0.17) | ||||
1Calculated after normalization with the geometric mean of UXT, GAPDH and RPS9 (see materials and methods).
*Means within the treatment group differ (P < 0.05) between d 2 and 10.
ABCWithin time point (d 2 and 10), treatment means (CON, C50 and C100) without a common superscript differ (P < 0.05).
There was an interaction (diet × day; P < 0.05; Table 2) for PPARA, KLB, and NFIL3. The mRNA expression of PPARA, KLB, and NFIL3 was affected by diet (P < 0.01); however, no effect of time (P > 0.05) was observed for these genes. The mRNA expression of PPARA and KLB were not affected by diets at d 2, whereas at d 10 the C100 decreased their expression. Feeding C50 increased expression of PPARA and C100 increased expression of KLB. Furthermore, at d 10 mRNA expression of PPARA and KLB was lower with C100 in comparison with control and C50. At d 2, the mRNA expression of NFIL3 was increased with feeding C50 (P < 0.05) in comparison with control; while at d 10 feeding C100 decreased (P < 0.05) NFIL3 expression compared with C50.
The mRNA expression of ANGPTL4 decreased (diet × day, P < 0.05; Table 2) from d 2 to 10 with the highest dose of carnitine (C100) supplementation, whereas an increase was observed in controls between d 2 and 10 relative to parturition (P < 0.01). However, no significant change was observed in cows fed C50 between d 2 and 10 relative to parturition. Overall, at d 2, the control had lower mRNA expression of ANGPTL4, but at d 10 feeding C100 result in a marked decrease. A tendency for a main effect of diet (P = 0.06) and an interaction (P = 0.06) were observed for CLOCK. At d 2, C50 decreased CLOCK mRNA expression in comparison with control and C100; however, C50 did not induce differences (P > 0.05) compared with other treatments at d 10.
Prepartal Energy and Postpartal Intramammary LPS Challenge
Interactions among diet, time, and LPS (P < 0.05) were observed for FGF21, PPARA, NFIL3, and CLOCK (Table 3). A day × LPS interaction (P < 0.05) was observed for ANGPTL, NFIL3, CLOCK, ARNTL, and AKT1. Except for FGF21 (P = 0.07), no other gene had an interaction between diet and LPS. A diet × day (P < 0.05) interaction was only observed for AKT1.
Table 3. Hepatic mRNA expression of FGF21, FGF21 binding, and genes associated with circadian rhythms and insulin signaling in cows overfed energy (OVE:N; n = 6) or fed to meet energy requirements (CON:N; n = 6) during the entire dry period, and receiving an intramammary LPS challenge at d 7 postpartum (CON:Y, OVE:Y).
Liver tissue harvested at 2.5 h post-LPS on d 7 and 14 postpartum before the morning meal. The 95% confidence interval is reported in parenthesis.
| Gene | Day | Treatment | P value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OVE:N | OVE:Y | CON:N | CON:Y | Diet (T) | Day (D) | LPS (L) | T × D | T × L | D × L | T × D × L | ||
| FGF21 | 7 | 0.81 A B (0.49 to 1.33) | 1.27 A * (1.08 to 1.47) | 0.25 B * (0.16 to 0.52) | 0.82 A B (0.51 to 1.33) | <0.01 | 0.04 | 0.07 | 0.11 | 0.07 | 0.56 | <0.01 |
| 14 | 0.87 A (0.53 to 1.42) | 0.55 A * (0.34 to 0.84) | 0.06 B * (0.03 to 0.10) | 0.66 A (0.41 to 1.07) | ||||||||
| PPARA | 7 | 0.77 A * (0.68 to 0.88) | 0.81 A (0.72 to 0.92) | 1.13 B (0.97 to 1.32) | 0.86 A B * (0.76 to 0.98) | 0.06 | <0.01 | 0.42 | 0.43 | 0.68 | 0.82 | <0.01 |
| 14 | 1.01 A B * (0.90 to 1.15) | 0.87 A (0.77 to 0.98) | 1.26 B (1.08 to 1.47) | 1.20 B * (1.06 to 1.37) | ||||||||
| CPT1A | 7 | 1.10 (1.01 to 1.19) | 1.05 (0.97 to 1.14) | 0.89 (0.83 to 1.06) | 1.01 (0.94 to 1.10) | 0.16 | 0.53 | 0.85 | 0.74 | 0.16 | 0.27 | 0.27 |
| 14 | 1.14 (1.04 to 1.24) | 0.97 (0.89 to 1.09) | 0.88 (0.80 to 1.05) | 1.01 (0.93 to 1.09) | ||||||||
| ANGPTL4 | 7 | 1.27 (0.99 to 1.80) | 2.19 (1.72 to 2.80) | 0.51 (0.38 to 0.70) | 2.27 (1.77 to 2.90) | 0.06 | <0.01 | 0.21 | 0.66 | 0.16 | <0.01 | 0.17 |
| 14 | 0.96 (0.65 to 1.22) | 0.52 (0.41 to 0.67) | 0.46 (0.33 to 0.72) | 0.39 (0.31 to 0.50) | ||||||||
| KLB | 7 | 1.13 (0.65 to 1.47) | 0.24 (0.14 to 0.41) | 1.06 (0.54 to 1.54) | 0.15 (0.09 to 0.24) | 0.82 | 0.32 | <0.01 | 0.32 | 0.77 | 0.30 | 0.83 |
| 14 | 1.03 (0.59 to 1.35) | 0.35 (0.10 to 0.26) | 1.17 (0.60 to 1.75) | 0.13 (0.08 to 0.23) | ||||||||
| ARNTL | 7 | 1.37 (1.04 to 1.51) | 0.03 (0.02 to 0.05) | 0.95 (0.70 to 1.26) | 0.02 (0.01 to 0.04) | 0.71 | 0.67 | <0.01 | 0.19 | 0.73 | <0.01 | 0.26 |
| 14 | 0.83 (0.51 to 1.10) | 0.03 (0.02 to 0.05) | 0.60 (0.44 to 0.87) | 0.05 (0.03 to 0.08) | ||||||||
| CLOCK | 7 | 0.95 A (0.72 to 1.24) | 0.14 B * (0.08 to 0.22) | 1.04 A (0.82 to 1.37) | 0.11 B (0.07 to 0.18) | 0.63 | 0.10 | <0.01 | 0.20 | 0.75 | 0.02 | 0.01 |
| 14 | 0.95 A (0.70 to 1.29) | 0.04 B * (0.02 to 0.06) | 1.02 A (0.78 to 1.32) | 0.11 B (0.06 to 0.19) | ||||||||
| NFIL3 | 7 | 1.09 A (0.93 to 1.29) | 4.03 B * (3.42 to 4.71) | 1.24 A * (1.15 to 1.38) | 4.69 B * (3.99 to 5.14) | 0.25 | <0.01 | 0.38 | 0.10 | 0.26 | <0.01 | 0.02 |
| 14 | 1.08 A (0.92 to 1.28) | 0.47 B * (0.40 to 0.55) | 1.69 C * (1.43 to 2.00) | 0.58 B * (0.49 to 0.68) | ||||||||
| AKT1 | 7 | 1.34 (1.17 to 1.53) | 0.43 (0.37 to 0.49) | 1.23 (1.05 to 1.44) | 0.39 (0.33 to 0.44) | 0.77 | 0.07 | <0.01 | 0.02 | 0.69 | <0.01 | 0.44 |
| 14 | 1.27 (1.11 to 1.45) | 0.28 (0.25 to 0.32) | 1.65 (1.40 to 1.93) | 0.30 (0.27 to 0.35) | ||||||||
1Calculated after normalization with the geometric mean of UXT, GAPDH and RPS9 (see materials and methods).
*Means within the treatment group (OVE:N, OVE:Y, CON:N and CON:Y) differ (P < 0.05) between d 7 and 14.
ABCWithin time point (d 7 and 14), treatment means (OVE:N, OVE:Y, CON:N and CON:Y) without a common superscript differ (P < 0.05).
LPS challenge tended (diet × LPS, P = 0.07) to increase FGF21 mRNA expression at d 7 in OVE:Y and CON:Y compared with the respective controls (Table 3). However, OVE:Y and CON:N resulted in a marked decrease (diet × day × LPS, P < 0.01) in mRNA expression from d 7 to 14 and a nadir in expression was observed in CON:N. The mRNA expression of PPARA was higher (diet × day × LPS, P < 0.05) at d 14 in CON:N and CON:Y groups, whereas no significant change (P > 0.05) was detected for the mRNA expression of CPT1A. The mRNA expression of ANGPTL4 was greater (day, P < 0.05) at d 7, and in OVE:N compared with CON:N (diet, P = 0.06). Furthermore, LPS increased the mRNA expression of ANGPTL4 in both OVE:Y and CON:Y and, similar to controls (OVE:N and CON:N), mRNA expression decreased (LPS × diet, P < 0.05) on d 14. Regardless of prepartal dietary energy, LPS markedly decreased (LPS, P < 0.05) KLB mRNA expression.
The mRNA expression of genes associated with the circadian rhythm was significantly affected by the interaction of diet, day, and LPS challenge (P < 0.05; Table 3); NFIL3 increased while a decrease was observed for ARNTL and CLOCK mRNA expression in cows receiving the LPS challenge (OVE:Y, CON:Y) at d 7. Furthermore, results revealed an increase (diet × LPS, P < 0.05) in NFIL3 mRNA expression due to LPS challenge (OVE:Y, CON:Y) at d 7; however, that was followed by a decrease at d 14. The mRNA expression of ARNTL was greater (day, P < 0.05) in OVE:N at d 7 while it decreased at d 14. The LPS challenge decreased (diet × LPS, P < 0.05) its expression in both groups, but CON:Y increased ARNTL between d 7 and d 14. Furthermore, the LPS treatment decreased (P < 0.05) the mRNA expression of CLOCK irrespective of diet (day × LPS, P > 0.05), and a further decrease (diet × LPS, P < 0.05) was detected in OVE:Y at d 14. The mRNA expression of AKT1 increased (diet × LPS, P > 0.05) at d 14 in CON:N compared with other groups; however, a decrease was observed in the LPS-challenged groups.
Serum Concentration of FGF21
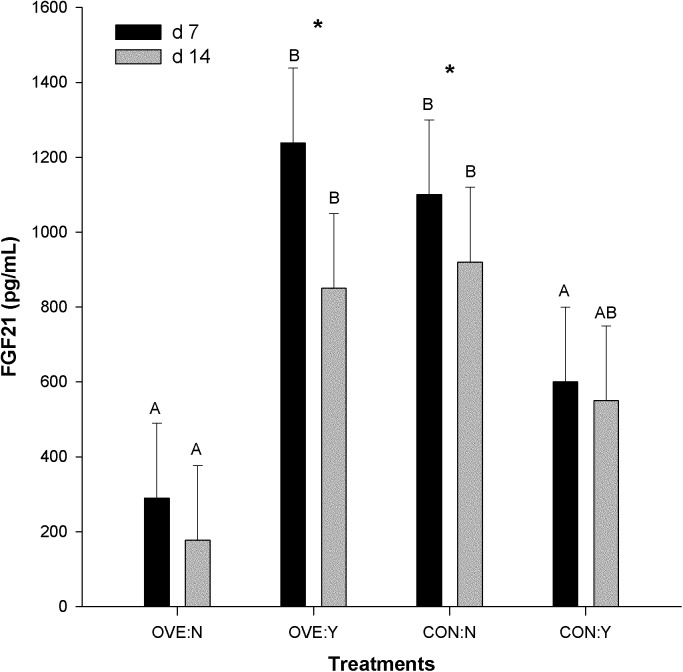
In the absence of serum samples from the dietary carnitine experiment [11], we only measured the blood serum concentration of FGF21 from the ketosis [16]and prepartal higher-energy diet and LPS challenge studies [17]. Although we observed greater mRNA expression of FGF21 in the liver of ketotic cows, the serum concentration of FGF21 was only numerically different (P = 0.25) between diets (Fig 1; 1370 ± 279 vs. 905 ± 275 pg/mL).
Fig 1. Serum concentration of FGF21 in cows overfed energy (OVE:N, n = 6) or fed to meet energy requirements (CON:N, n = 6) during the entire dry period, and receiving an intramammary LPS challenge at d 7 postpartum (OVE:Y, CON:Y).
The error bars represent SEM. The serum contraction of FGF21 tended to have an interaction among diet, time, and LPS (P = 0.08). No main effected of diet (P = 0.48) or LPS (P = 0.37) was observed, but the interaction diet × LPS was significant (P < 0.01). The statistical effects of diet, LPS, days are indicated: *Significant difference at 7 and 14 d postpartum within the same treatment groups; A,BSignificant difference among treatments (diet with or without LPS) at d 7 or 14 postpartum.
The serum contraction of FGF21 tended (P = 0.08) to have an interaction among diet, time, and LPS. No main effected of diet (P = 0.48) or LPS (P = 0.37) was observed, but the interaction diet × LPS was significant (P < 0.01). Intramammary LPS challenge increased (P < 0.05) the blood concentration of FGF21 in OVE:Y but decreased it (diet × LPS, P < 0.05) in CON:Y (Fig 1). Furthermore, OVE:Y and CON:N decreased serum FGF21 concentration between d 7 and 14 (diet × LPS, P < 0.05), whereas OVE:N and CON:Y only had a numerical decrease.
Discussion
Ketosis Induces the mRNA Expression of FGF21
Early lactation in dairy cows is a nutritionally precarious period owing to a combination of increases in energy requeriments for milk production and decreases in voluntary feed intake[27]. During periods of NEB, high-producing cows become more susceptible to developing ketosis, leading to further increases in NEFA, BHBA, hepatic TAG concentration, hepatic mRNA expression of PPARA and angiopoietin-like 4 (ANGPTL4, a hepatokine), and other genes associated with ketogenesis and gluconeogenesis[16]. The 41-fold increase in FGF21 mRNA expression in ketotic cows agrees with previous observations in cows with NEB [8]. Work in rodents revealed that plasma FGF21 increases during fasting and it plays a critical role in metabolic fuel homeostasis during ketosis [28]. It was suggested that increased FGF21 in obese humans may protect against chronic exposure to high concentrations of NEFA, which are toxic in muscle, pancreas, and liver [29]. Schoenberg et al. [8] observed a substantial individual variation in FGF21 serum concentration among cows, particularly at the time of calving and very early in lactation. Therefore, the lack of significant difference in FGF21 serum concentration in the present study can be partly attributed to small sampling size and the high variation among cows. Overall, a period of energy deficit in dairy cows is positively correlated with FGF21 mRNA expression in liver, similar to what is observed in food-deprived rodents and humans [5, 30]. However, it cannot be discerned from our data if the actual reduction of feed intake or the onset of ketosis per se caused the change in FGF21 expression.
The somatotropic axis controls many aspects of growth and lactation; calving and NEB are associated with uncoupling of the somatotropic axis [31]. Calving increases blood concentration of growth hormone (GH), which further augments blood NEFA during NEB [32–34]. In non-ruminants, the NEFA can trigger hepatic FGF21 and ANGPTL4 upregulation and prodution through the activation of PPARA [35,36]. In turn, activation of PPARA and FGF21 are correlated with an increase in the hepatic protein concentration of CPT1A and HMGCS2 via a posttranscriptional mechanism without changes in mRNA[1, 28]. The lack of change in CPT1 activity in hepatic mitochondria [37] and in the mRNA expression of CPT1A with ketosis [16] suggests that CPT1A might not have been affected by ketosis even though FGF21 mRNA expression increased and serum contration was numerically higher.
Yu et al. [38] also observed in beef cattle that GH enhances hepatic FGF21 gene transcription and that this stimulation is mediated, at least in part, by the transcription factor STAT5. They suggested that FGF21 inhibits the JAK-2/STAT5 signaling through a negative loop involving the binding of GH to the hepatic GH receptor (GHR) upon which JAK2-STAT5 signalling starts and can regulate target genes such as FGF21, IGF1, and SOCS2. However, the downregulation of GHR, STAT5, IGF1, and SOCS2 in cows with ketosis in our study [16] does not appear to support the model proposed by Yu et al. [38]. The discrepancy is explained in part by the fact that hepatic GHR decreases around parturition in dairy cattle but not in beef cattle [39]. The decrease in hepatic mRNA expression of STAT5B and SOCS2 coincides with the uncoupling of GH/IGF-1 axis [31].
The specificity of secreted FGF21 action is mediated through FGF receptors (FGFR 1–4) in peripheral tissues, but requires the presence of a co-receptor (KLB) in order to elicit a response [40, 41]. The fact that ketosis increased the hepatic mRNA expression of KLB in a similar fashion to FGF21 at the onset of lactation [8] suggests that FGF21 may exert a paracrine/autocrine action on liver during ketosis. As such, FGF21 may lead to a decrease in the hepatic mRNA expression of IGF1 by increasing the mRNA expression of IGFBP1 [42, 43], which was reported previously [16].
Dietery Carnitine downregulates the mRNA Expression of FGF21
Carlson et al. [11] using the same cows from the present study reported that C50 and C100 resulted in lower liver TAG than control on d 2 and 10, but only feeding C100 led to greater concentration of BHBA compared with control or C50. The lower DMI caused by feeding C100 partly explains the greater BHBA[11]. Although both feeding C50 and C100 increased liver synthesis of acid soluble products (i.e. ketones), the fact that cows fed C100 had lower milk production [11] could partly explain the greater BHBA compared with C50.
During early lactation, adipose tissue lipolysis results in elevated plasma NEFA that can be taken up by liver (~25% of the NEFA flux) to meet its oxidative needs and to produce ketone bodies for extra-hepatic tissues [44]. In non-ruminant liver, hypoinsulinemia due to undernutrition increases NEFA flux into liver where they activate PPARα and forkhead box A2 (FOXA2) resulting in an overall increase in fatty acid oxidation and ketogenesis [45]. Concomitantly, PPARα activation increases FGF21 and its concentration in the circulation, and appears responsible for matching adipose tissue mobilization to oxidative capacity in liver [8] by stimulating hepatic NEFA use while concurrently decreasing lipolysis [6].
The marked decrease between d 2 and 10 in mRNA expression of FGF21 and its co-receptor KLB in the C100 cows agrees with the decrease in metabolism of palmitate to acid-soluble products[11], i.e. despite the lower DMI due to feeding C100 the rates of LCFA oxidation to CO2 did not differ among groups. Furthermore, the greater insulin concentration detected in cows fed C100 compared with controls also could have had a negative effect on LCFA oxidation as in non-ruminants. It is noteworthy that cows fed C50 did not decrease DMI [11] and had an increase in mRNA expression of CPT1A and PPARA between d 2 and 10, suggesting the existence of a threshold of L-carnitine above which there is feedback inhibition of fatty acid oxidation. Furthermore, those data support a role for PPARα activation in the process of LCFA oxidation.
In mice, evidence indicates that the circadian protein NFIL3 alters FGF21 mRNA expression during a circadian cycle and upon food intake [23]. Expression of FGF21 peaks during the post-absorptive phase, but NFIL3 is highest during the fed state; insulin increases NFIL3 mRNA expression and binding to the FGF21 promoter through AKT activation, indicating that NFIL3 is an insulin-responsive repressor of FGF21 mRNA expression [23]. This protein also suppressed the BMAL1 (ARNTL)-CLOCK-activated FGF21 mRNA expression and abolished the PPARα-activated FGF21 mRNA expression [23]. Despite the lack of change in expression of AKT1, the greater blood insulin in C50 compared with controls [11] agrees with the increase in NFIL3 mRNA expression between d 2 and 10 and the concomitant decrease in mRNA expression of FGF21. Together, these data are suggestive that NFIL3 could regulate FGF21 synthesis in ruminants.
A growing amount of evidence indicates that FGF21 and ANGPTL4 are liver-derived biomarkers of energy balance during the peripartal period [20, 22]. Thus, the better energy balance [11] and greater mRNA expression of ANGPTL4 and FGF21 observed in the control group indicate that there are “energy-independent” mechanisms that might control mRNA expression of these genes. For example, the higher ketone body concentration with both carnitine treatments could have promoted pancreatic insulin secretion as reported in rats [46]. As such, ketone bodies could have played a role in mediating the downregulation of FGF21 through NFIL3 [23]. The fact that control cows had lower blood insulin concentrations offers support for this mechanism.
Prepartal Energy and LPS Challenge Alter FGF21 mRNA Expression
Overfeeding dairy cows during the dry period leads to greater BCS before calving, a pronounced decrease in appetite around calving, and as a result more severe NEB [47]. The overfed cows used in this study were in positive energy balance and had greater BCS prepartum than controls, but were in more NEB early postpartum and before the LPS challenge [17]. After the LPS challenge, blood BHBA decreased, NEFA increased, and liver TAG increased only in the OVE:Y group [17]. The observed increase in FGF21 mRNA expression due to feeding OVE without the LPS challenge was associated with higher NEFA concentration, as reported previously [8]. However, the lack of significant effect on the mRNA expression of PPARA due to OVE or LPS suggests the existence of a PPARα-independent mechanism that controls mRNA expression of FGF21 during inflammation or feeding OVE. The lack of a PPARA response to high NEFA in some studies with transition cows [48, 49] supports this idea. Palin and Petit [50] observed no change in hepatic mRNA expression of PPARA in response to feeding above 100% of prepartal energy requirements and different sources of fat. Furthermore, Carriquiry et al. [48] speculated that the lack of increase in PPARA postpartum in response to diets providing 8% more fat than typically fed to dairy cows and enriched with PUFA could be related with a hepatic inflammatory response induced by prepartal high-fat intake. All these observations suggest the existence of a PPARα-independent mode of action on the regulation of FGF21 in dairy cows consuming high-prepartal energy and fat levels and afflicted by inflammatory conditions after calving.
In non-ruminants, recent evidence indicates that the circadian protein NFIL3 regulates the hepatic mRNA expression of FGF21 during a circadian cycle and upon food intake [23]. In ruminants, a recent study provided some evidence for circadian control of FGF21 [51]. The NFIL3 protein suppressed the activation of FGF21 mRNA expression by BMAL1 (ARNTL)-CLOCK and also abolished PPARα activation of FGF21 expression [23]. In the present study, only the marked upregulation of NFIL3 coupled with the downregulation of CLOCK after LPS in both OVE and CON offer the clearest support for an antagonistic role similar to that in non-ruminants. Whether, the CLOCK response had an effect at the protein expression level for PPARA and ARNTL remains to be determined. The observed increase in FGF21 mRNA expression (Table 3) and serum concentration in OVE:Y (Fig 1) support the existence of another regulatory mechanism for FGF21 mRNA expression under inflammatory conditions.
The marked decrease in hepatic mRNA expression of STAT5B and SOCS2 in OVE:Y between d 7 and 14 relative to parturition [17] confirms the uncoupling of GH/IGF-1 axis [31] and argues against the possibility of GH-mediated activation of FGF21 via JAK2/STAT5 signaling [38]. It has been recognized recently in non-ruminants that glucagon increases the hepatic mRNA expression of FGF21 either via PPARα mediated activation or directly via protein kinase A (PKA) [35, 36]. Calving increases the concentration of glucagon [52] as part of the homeorhetic mechanisms to coordinate lactation. The increase in glucagon around calving [52] and the NEB at d 7 [17] might have led to the activation of PKA. This idea is supported by a recent study in rodents and humans that detected increases in both mRNA expression and secretion of FGF21 in response to both native glucagon and glucagon receptor (GcgR) agonists [53]. Those data demonstrated that glucagon achieves its long-term effects on energy and lipid and glucose metabolism at least in part via FGF21-dependent pathways.
We speculate the existence of a glucagon-mediated activation of FGF21 via PKA. For example, both intravenous and intramuscular injection of glucagon reduced NEFA and alleviated fatty liver in dairy cows [54, 55]. If FGF21 increased with glucagon injections, then the protein could exert some inhibition of adipose lipolysis as demonstrated in rodents [56]. Therefore, we suggest that glucagon and consequently NEFA can induce hepatic mRNA expression of FGF21 in order to achieve its lipolysis inhibitory effect. Further studies are required to demonstrate these mechanistic relationships.
The increase in serum concentration of FGF21 with LPS challenge agrees with the greater mRNA expression in the liver, which is the main contributor of blood FGF21 in dairy cows [8]. Glucagon is a potent anti-inflammatory hormone [57], thus, the upregulation of FGF21 with LPS also might be attributed to an increase in glucagon coupled with a decrease in DMI [17]. As reported in non-ruminants [58], in bovine, PPARδ was suggested to regulate the mRNA expression of ANGPTL4 and other hepatokines during acute inflammation[59]. Furthermore, in the cows under study the LPS challenge induced the hepatic mRNA expression of PPARD and genes associated with inflammation and stress [17]. The parallel increase of ANGPTL4 mRNA expression with LPS, which has recently been reported as a positive acute-phase protein in mice challenged with LPS [60], supports the speculation that LPS induced the hepatic mRNA expression of both hepatokines ANGPTL4 and FGF21 via PPARδ.
Our observations of increased serum FGF21 in response to LPS administration agrees with the increase in serum FGF21 with LPS injection in mouse [15]; however, in contrast with our results, Feingold et al. [15] demonstrated that adipose tissue is a major contributor of serum concentration of FGF21 in LPS-challenged mice. Previous work from our group [61] failed to detect FGF21 mRNA expression in subcutaneous adipose tissue around parturition, casting doubt on the contribution of this tissue to peripheral FGF21 concentration. Furthermore, Schoenberg et al. [8] observed little or no mRNA expression of FGF21 in white adipose tissue, skeletal muscle, and mammary gland, suggesting those tissues do not contribute to circulating FGF21. However, as discussed in the previous sections, β-Klotho (KLB; FGF21 co-receptor) determines the target specificity of FGF21 action; down regulation of hepatic KLB with LPS suggests an extra-hepatic role of FGF21 during acute inflammation. We speculate that LPS might have induced FGF21 in adipose tissue to suppress lipolysis in order to reduce the NEFA flow to liver as a way to decrease lipidosis.
Overall, the lack of FGF21 mRNA expression in subcutaneous adipose tissue around parturition [61] and the observations from Schoenberg et al. [8] support the view that under normal conditions the adipose tissue is not a contributor to blood FGF21. However, in dairy cows it appears that adipose tissue is the second major target tissue after liver. In a survey of 15 tissues that included the mammary gland, the mRNA expression of KLB and a subset of interacting FGF receptors was restricted to liver and white adipose tissue and was modestly affected by the transition from late-pregnancy to early lactation in liver but not in adipose [8].
Overall, the data obtained for FGF21 confirm that this gene is a biomarker of negative energy balance as reported previously [8, 20]. Beyond evidencing the important role of FGF21 in peripartal cow biology, the responses across the three experiments suggest different adaptations in hepatic signaling due to onset of ketosis, nutritional management, and inflammation. For instance, the role of KLB in hepatic adaptations to parturition does not seem to be important unless there are marked changes in NEFA and BHBA production as would occur in clinical ketosis. Acute inflammation proved to be a strong chronic inhibitor of signaling via KLB, the clock network, and potentially the insulin pathway. Furthermore, there seems to be a threshold up to which enhancing LCFA influx into the mitochondria via L-carnitine is effective for allowing liver to achieve higher rates of beta-oxidation at least in part through upregulation of PPARA and CPT1A. Under those “ideal” conditions the level of intra-hepatic “stress” is diminished, hence, the lower and stable FGF21. A novel mechanism of insulin action via NFIL3 upregulation at “optimal” rates of LCFA oxidation (cows in C50) was surmised, and suggests a degree of insulin sensitivity in liver despite NEB.
Perspectives
Although the data generated from the studies reported herein provide a preliminary evaluation of the likely roles of liver-derived FGF21 in helping to coordinate the cow’s adaptations to changes in energy balance, onset of ketosis or inflammation, and stimulation of fatty acid oxidation there is a need to conduct more mechanistic studies aimed specifically at deciphering the transcriptional control of FGF21 in bovine liver. Specifically during the immediate period after parturition when most if not all cows are most susceptible to developing ketosis or an inflammatory disorder. To achieve greater mechanistic understanding, techniques such as ChIP assays could be used. Because there was no bovine antibody specific for FGF21 available at the time these experiments were conducted, there would be a need to replicate them. Furthermore, the differences in feed intake induced by the treatments we studied also confound a precise interpretation of the mechanisms driving the changes in hepatic FGF21. Lastly, future work on the regulation of bovine hepatic FGF21 needs to encompass in vitro studies where the roles of specific molecules such as glucagon could be studied in a more controlled manner.
Supporting Information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding for the gene expression work was provided by USDA National Institute of Food and Agriculture (NIFA) and by Hatch funds allocated to JJL by NIFA under project NC-2040 (ILLU-538-914). Partial fellowship support from COMSATS Institute of Information and Technology (Pakistan) to H. Akbar during his doctoral degree is acknowledged.
References
- 1. Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–25. [DOI] [PubMed] [Google Scholar]
- 2. Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci U S A. 2009;106:10853–8. 10.1073/pnas.0904187106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, et al. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115:1627–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mashili FL, Austin RL, Deshmukh AS, Fritz T, Caidahl K, et al. Direct effects of FGF21 on glucose uptake in human skeletal muscle: implications for type 2 diabetes and obesity. Diabetes-Metab Res. 2011;27:286–97. [DOI] [PubMed] [Google Scholar]
- 5. Domouzoglou EM, Maratos-Flier E. Fibroblast growth factor 21 is a metabolic regulator that plays a role in the adaptation to ketosis. Am J Clin Nutr. 2011;93:901s–5s. 10.3945/ajcn.110.001941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Badman MK, Koester A, Flier JS, Kharitonenkov A, Maratos-Flier E. Fibroblast Growth Factor 21-Deficient Mice Demonstrate Impaired Adaptation to Ketosis. Endocrinology. 2009;150:4931–40. 10.1210/en.2009-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hotta Y, Nakamura H, Konishi M, Murata Y, Takagi H, et al. Fibroblast Growth Factor 21 Regulates Lipolysis in White Adipose Tissue But Is Not Required for Ketogenesis and Triglyceride Clearance in Liver. Endocrinology. 2009;150:4625–33. 10.1210/en.2009-0119 [DOI] [PubMed] [Google Scholar]
- 8. Schoenberg KM, Giesy SL, Harvatine KJ, Waldron MR, Cheng C, et al. Plasma FGF21 Is Elevated by the Intense Lipid Mobilization of Lactation. Endocrinology. 2011;152:4652–61. 10.1210/en.2011-1425 [DOI] [PubMed] [Google Scholar]
- 9. Bykov I, Jarvelainen H, Lindros K. L-carnitine alleviates alcohol-induced liver damage in rats: role of tumour necrosis factor-alpha. Alcohol Alcohol. 2003;38:400–6. [DOI] [PubMed] [Google Scholar]
- 10. Spaniol M, Kaufmann P, Beier K, Wuthrich J, Torok M, et al. Mechanisms of liver steatosis in rats with systemic carnitine deficiency due to treatment with trimethylhydraziniumpropionate. J Lipid Res. 2003;44:144–53. [DOI] [PubMed] [Google Scholar]
- 11. Carlson DB, McFadden JW, D'Angelo A, Woodworth JC, Drackley JK. Dietary L-carnitine affects periparturient nutrient metabolism and lactation in multiparous cows. J Dairy Sci. 2007;90:3422–41. [DOI] [PubMed] [Google Scholar]
- 12. Schlegel G, Ringseis R, Keller J, Schwarz FJ, Windisch W, et al. Expression of fibroblast growth factor 21 in the liver of dairy cows in the transition period and during lactation. J Anim Physiol Anim Nutr (Berl). 2012;8:1.28. [DOI] [PubMed] [Google Scholar]
- 13. Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–54. [DOI] [PubMed] [Google Scholar]
- 14. Kim MS, Sweeney TR, Shigenaga JK, Chui LG, Moser A, et al. Tumor necrosis factor and interleukin 1 decrease RXR alpha, PPAR alpha, PPAR gamma, LXR alpha, and the coactivators SRC-1, PGC-1 alpha, and PGC-1 beta in liver cells. Metabolism. 2007;56:267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feingold KR, Grunfeld C, Heuer JG, Gupta A, Cramer M, et al. FGF21 Is Increased by Inflammatory Stimuli and Protects Leptin-Deficient ob/ob Mice from the Toxicity of Sepsis. Endocrinology. 2012;153:2689–700. 10.1210/en.2011-1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loor JJ, Everts RE, Bionaz M, Dann HM, Morin DE, et al. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics. 2007;32:105–16. [DOI] [PubMed] [Google Scholar]
- 17. Graugnard DE, Moyes KM, Trevisi E, Khan MJ, Keisler D, et al. Liver lipid content and inflammometabolic indices in peripartal dairy cows are altered in response to prepartal energy intake and postpartal intramammary inflammatory challenge. J Dairy Sci. 2013;96:918–35. 10.3168/jds.2012-5676 [DOI] [PubMed] [Google Scholar]
- 18. Dann HM, Morin DE, Murphy MR, Bollero GA, Drackley JK. Prepartum intake, postpartum induction of ketosis, and periparturient disorders affect the metabolic status of dairy cows. J. Dairy Sci. 2005;88:3249–3264. [DOI] [PubMed] [Google Scholar]
- 19. NRC-National Research Council. Nutrient requirements of dairy cattle, seventh revised ed. National Academic Press; 2001. Washington, DC, USA. [Google Scholar]
- 20. Khan MJ, Jacometo CB, Graugnard DE, Correa MN, Schmitt E, et al. Overfeeding Dairy Cattle During Late-Pregnancy Alters Hepatic PPARalpha-Regulated Pathways Including Hepatokines: Impact on Metabolism and Peripheral Insulin Sensitivity. Gene regulation and systems biology. 2014;8:97–111. 10.4137/GRSB.S14116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akbar H, Grala TM, Riboni MV, Cardoso FC, Verkerk G, et al. Body condition score at calving affects systemic and hepatic transcriptome indicators of inflammation and nutrient metabolism in grazing dairy cows. J Dairy Sci. 2015;98:1019–32. 10.3168/jds.2014-8584 [DOI] [PubMed] [Google Scholar]
- 22. Loor JJ, Dann HM, Everts RE, Oliveira R, Green CA, et al. Temporal gene expression profiling of liver from periparturient dairy cows reveals complex adaptive mechanisms in hepatic function. Physiol Genomics. 2005;23:217–26. [DOI] [PubMed] [Google Scholar]
- 23. Tong X, Muchnik M, Chen Z, Patel M, Wu N, et al. Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. J Biol Chem. 2010;285:36401–9. 10.1074/jbc.M110.172866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tong X, Zhang D, Buelow K, Guha A, Arthurs B, et al. Recruitment of histone methyltransferase G9a mediates transcriptional repression of Fgf21 gene by E4BP4 protein. J Biol Chem. 2013;288:5417–5425. 10.1074/jbc.M112.433482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vandesompele J, De Preter K, Pattyn F, Poope B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olsson U. Confidence intervals for the mean of a log-normal distribution. Journal of Statistics Education. 2005;13:1–8. [Google Scholar]
- 27. Drackley JK. Biology of dairy cows during the transition period: The final frontier? Journal of Dairy Science. 1999;82:2259–73. [DOI] [PubMed] [Google Scholar]
- 28. Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007; 5:426–37. [DOI] [PubMed] [Google Scholar]
- 29. Sypniewska G. Laboratory assessment of cardiometabolic risk in overweight and obese children. Clinical Bochemistry. 2015; 48:370–376. [DOI] [PubMed] [Google Scholar]
- 30. Galman C, Lundasen T, Kharitonenkov A, Bina HA, Eriksson M, et al. The circulating metabolic regulator FGF21 is induced by prolonged fasting and PPARalpha activation in man. Cell Metab. 2008;8:169–74. 10.1016/j.cmet.2008.06.014 [DOI] [PubMed] [Google Scholar]
- 31. Lucy MC. Functional differences in the growth hormone and insulin-like growth factor axis in cattle and pigs: implications for post-partum nutrition and reproduction. Reprod Domest Anim. 2008;43 Suppl 2:31–9. 10.1111/j.1439-0531.2008.01140.x [DOI] [PubMed] [Google Scholar]
- 32. Adewuyi AA, Gruys E, van Eerdenburg FJ. Non esterified fatty acids (NEFA) in dairy cattle. A review. Vet Q. 2005;27:117–26. [DOI] [PubMed] [Google Scholar]
- 33. Akers RM. Major advances associated with hormone and growth factor regulation of mammary growth and lactation in dairy cows. J Dairy Sci. 2006;89:1222–34. [DOI] [PubMed] [Google Scholar]
- 34. Bradford BJ, Allen MS. Negative energy balance increases periprandial ghrelin and growth hormone concentrations in lactating dairy cows. Domest Anim Endocrin. 2008;34:196–203. [DOI] [PubMed] [Google Scholar]
- 35. Berglund ED, Kang L, Lee-Young RS, Hasenour CM, Lustig DG, et al. Glucagon and lipid interactions in the regulation of hepatic AMPK signaling and expression of PPAR alpha and FGF21 transcripts in vivo. Am J Physiol-Endoc M. 2010;299:E607–E14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Uebanso T, Taketani Y, Yamamoto H, Amo K, Ominami H, et al. Paradoxical Regulation of Human FGF21 by Both Fasting and Feeding Signals: Is FGF21 a Nutritional Adaptation Factor? Plos One. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dann HM, Drackley JK. Carnitine palmityoltransferase I in liver of periparturient dairy cows: effects of prepartum intake, postpartum induction of ketosis, and periparturient disorders. J. Dairy Sci. 2005;88:3851–59. [DOI] [PubMed] [Google Scholar]
- 38. Yu J, Zhao LD, Wang AH, Eleswarapu S, Ge XM, et al. Growth Hormone Stimulates Transcription of the Fibroblast Growth Factor 21 Gene in the Liver through the Signal Transducer and Activator of Transcription 5. Endocrinology. 2012;153:750–8. 10.1210/en.2011-1591 [DOI] [PubMed] [Google Scholar]
- 39. Jiang H, Lucy MC, Crooker BA, Beal WE. Expression of growth hormone receptor 1A mRNA is decreased in dairy cows but not in beef cows at parturition. J Dairy Sci. 2005;88:1370–7. [DOI] [PubMed] [Google Scholar]
- 40. Ogawa Y, Kurosu H, Yamamoto M, Nandi A, Rosenblatt KP, et al. beta Klotho is required for metabolic activity of fibroblast growth factor 21. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kharitonenkov A, Dunbar JD, Bina HA, Bright S, Moyers JS, et al. FGF-21/FGF-21 receptor interaction and activation is determined by beta Klotho. Journal of Cellular Physiology. 2008;215:1–7. [DOI] [PubMed] [Google Scholar]
- 42. Inagaki T, Lin VY, Goetz R, Mohammadi M, Mangelsdorf DJ, et al. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab. 2008;8:77–83. 10.1016/j.cmet.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fisher FM, Estall JL, Adams AC, Antonellis PJ, Bina HA, et al. Integrated regulation of hepatic metabolism by fibroblast growth factor 21 (FGF21) in vivo. Endocrinology. 2011;152:2996–3004. 10.1210/en.2011-0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Drackley JK, Overton TR, Douglas GN. Adaptations of glucose and long-chain fatty acid metabolism in liver of dairy cows during the periparturient period. J Dairy Sci. 2001;84(Elect. Supplement):E100–E12. [Google Scholar]
- 45. Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–32. [DOI] [PubMed] [Google Scholar]
- 46. Biden TJ, Taylor KW. Effects of ketone-bodies on insulin release and islet-cell metabolism in the rat. Biochemical J. 1983;212:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hayirli A, Grummer RR, Nordheim EV, Crump PM. Animal and dietary factors affecting feed intake during the prefresh transition period in Holsteins. J Dairy Sci. 2002;85:3430–43. [DOI] [PubMed] [Google Scholar]
- 48. Carriquiry M, Weber WJ, Fahrenkrug SC, Crooker BA. Hepatic gene expression in multiparous Holstein cows treated with bovine somatotropin and fed n-3 fatty acids in early lactation. J Dairy Sci. 2009;92:4889–900. 10.3168/jds.2008-1676 [DOI] [PubMed] [Google Scholar]
- 49. van Dorland HA, Richter S, Morel I, Doherr MG, Castro N, et al. Variation in hepatic regulation of metabolism during the dry period and in early lactation in dairy cows. J Dairy Sci. 2009;92:1924–40. 10.3168/jds.2008-1454 [DOI] [PubMed] [Google Scholar]
- 50. Palin MF, Petit HV. Effects of polyunsaturated fatty acids on hepatic PPAR alpha mRNA levels in the transition cow. J Anim Feed Sci. 2004;(Polish Acad. Sci. 13(Suppl. 1)):445–8. [Google Scholar]
- 51. Wang M, Zhou Z, Khan MK, Gao J, Loor JJ. Clock circadian regulator (CLOCK) gene network expression patterns in bovine adipose, liver, and mammary gland at 3 time points during the transition from pregnancy into lactation. J. Dairy Sci. 2015;98:1–12. [DOI] [PubMed] [Google Scholar]
- 52. Deboer G, Trenkle A, Young JW. Glucagon, Insulin, Growth-Hormone, and Some Blood Metabolites during Energy Restriction Ketonemia of Lactating Cows. J Dairy Sci. 1985;68:326–37. [DOI] [PubMed] [Google Scholar]
- 53. Habegger KM, Stemmer K, Cheng C, Muller TD, Heppner KM, et al. Fibroblast Growth Factor 21 Mediates Specific Glucagon Actions. Diabetes. 2013;62:1453–63. 10.2337/db12-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hippen AR, She P, Young JW, Beitz DC, Lindberg GL, et al. Alleviation of fatty liver in dairy cows with 14-day intravenous infusions of glucagon. J Dairy Sci. 1999;82:1139–52. [DOI] [PubMed] [Google Scholar]
- 55. Nafikov RA, Ametaj BN, Bobe G, Koehler KJ, Young JW, et al. Prevention of fatty liver in transition dairy cows by subcutaneous injections of glucagon. J Dairy Sci. 2006;89:1533–45. [DOI] [PubMed] [Google Scholar]
- 56. Li X, Ge H, Weiszmann J, Hecht R, Li YS, et al. Inhibition of lipolysis may contribute to the acute regulation of plasma FFA and glucose by FGF21 in ob/ob mice. FEBS Lett. 2009;583:3230–4. 10.1016/j.febslet.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 57. Sigalet DL, Wallace LE, Holst JJ, Martin GR, Kaji T, et al. Enteric neural pathways mediate the anti-inflammatory actions of glucagon-like peptide 2. Am J Physiol Gastrointest Liver Physiol. 2007;293:G211–G21. [DOI] [PubMed] [Google Scholar]
- 58. Staiger H, Haas C, Machann J, Werner R, Weisser M, et al. Muscle-Derived Angiopoietin-Like Protein 4 Is Induced by Fatty Acids via Peroxisome Proliferator-Activated Receptor (PPAR)-delta and Is of Metabolic Relevance in Humans. Diabetes. 2009;58:579–89. 10.2337/db07-1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bionaz M, Chen S, Khan MJ, Loor JJ. Functional Role of PPARs in Ruminants: Potential Targets for Fine-Tuning Metabolism during Growth and Lactation. PPAR Res. 2013;2013:684159 10.1155/2013/684159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lu B, Moser A, Shigenaga JK, Grunfeld C, Feingold KR. The acute phase response stimulates the expression of angiopoietin like protein 4. Biochem Biophys Res Commun. 2010;391:1737–41. 10.1016/j.bbrc.2009.12.145 [DOI] [PubMed] [Google Scholar]
- 61. Ji P, Osorio JS, Drackley JK, Loor JJ. Overfeeding a moderate energy diet prepartum does not impair bovine subcutaneous adipose tissue insulin signal transduction and induces marked changes in peripartal gene network expression. J Dairy Sci. 2012;95:4333–51. 10.3168/jds.2011-5079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.