Summary
Higher lipid biosynthesis and accumulation are important to achieve economic viability of biofuel production via microalgae. To enhance lipid content, Chlamydomonas reinhardtii was genetically engineered with a key enzyme diacylglycerol acyltransferase (BnDGAT2) from Brassica napus, responsible for neutral lipid biosynthesis. The transformed colonies harbouring aph7 gene, screened on hygromycin-supplemented medium, achieved transformation frequency of ~120 ± 10 colonies/1 × 106 cells. Transgene integration and expression were confirmed by PCR, Southern blots, staining lipid droplets, proteins and spectro-fluorometric analysis of Nile red-stained cells. The neutral lipid is a major class (over 80% of total lipids) and most significant requirement for biodiesel production; this was remarkably higher in the transformed alga than the untransformed control. The levels of saturated fatty acids in the transformed alga decreased to about 7% while unsaturated fatty acids increased proportionately when compared to wild type cells. Polyunsaturated fatty acids, especially α-linolenic acid, an essential omega-3 fatty acid, were enhanced up to 12% in the transformed line. Nile red staining confirmed formation of a large number of lipid globules in the transformed alga. Evaluation of long-term stability and vitality of the transgenic alga revealed that cryopreservation produced significantly higher quantity of lipid than those maintained continuously over 128 generations on solid medium. The overexpression of BnDGAT2 significantly altered the fatty acids profile in the transformed alga. Results of this study offer a valuable strategy of genetic manipulation for enhancing polyunsaturated fatty acids and neutral lipids for biofuel production in algae.
Keywords: green algae, bioenergy, biodiesel, transgenic algae, polyunsaturated fatty acids, neutral lipids
Introduction
Alternative sources of energy are explored with a focus on economic feasibility and environmental safety considering the challenges associated with coal and petroleum. Microalgae is one of the most promising sources of clean and renewable energy (Chisti, 2008). The production of biodiesel from microalgae is currently under consideration in many countries, to evaluate its potential as a future energy source (Wijffels and Barbosa, 2010), despite higher production cost when compared with petroleum-based fossil fuels.
Microalgae are particularly attractive as biofuel producers as they possess short life cycles, perform photosynthesis, can utilize saline or wastewater for their growth and require nonarable land. Moreover, valuable by-products like animal feed and chemical precursors can be obtained from microalgae-based biofuel generation (Larkum et al., 2012). Nevertheless, there are several barriers that need to be overcome before algal oil-based biofuel or biodiesel can be economically produced at a commercial scale, one of which is the lack of microalgal strains with a high lipid content (Fan et al., 2014). It has been proposed that algal strains that grow faster and have high lipid content can be effectively used to reduce the production cost. However, such a robust strain remains elusive for researchers even after decades of screening natural strains (Vuttipongchaikij, 2012). Most of the strains known to-date possess either one or few of the required characteristics. A few studies have shown that lipid content in algae can be increased by nitrogen or phosphate starvation (James et al., 2011, 2013; Mallick et al., 2012) or by blocking starch production (Li et al., 2010). However, such stresses lower the growth rate, lipid quality, quantity and overall productivity of the system (Chisti, 2007; Hu et al., 2008), which is a major bottleneck for producing oleaginous biofuels at commercial scale. Therefore, genetic engineering could offer potential solutions for generating an improved strain with several desired characteristics (Mata et al., 2010; Radakovits et al., 2010; Wijffels and Barbosa, 2010).
The TAG biosynthetic pathway is relatively well understood in plants, but poorly known in algae. Type-2 DGAT proteins are localized in different subdomains of the endoplasmic reticulum (Shockey et al., 2006), which indicates that enzymatic machinery for the formation of TAGs is located in the endoplasmic reticulum (Turchetto-Zolet et al., 2011). In the de novo triacylglycerol (TAG) biosynthetic pathway, diacylglycerol acyltransferase (DGAT, EC 2.3.1.20) has been reported to be mainly responsible for lipid accumulation in the green alga (Cases et al., 1998). DGAT has also been suggested as a target gene for enhancing lipid content via genetic engineering (Fan et al., 2014) to generate more precursors for triacylglycerol (TAG) accumulation (Moellering and Benning, 2010; Radakovits et al., 2010). Several types of biofuels can be produced from triacylglycerol-containing feedstocks such as biodiesel (mono-alkyl esters) production by transesterification of the oil or ‘renewable diesel’ production by hydrodeoxygenation of fat or oil (Knothe, 2010). Thus, increasing the lipid feedstock is the first step towards making algae an economically feasible source for biofuel production.
The reaction catalysed by DGAT enzyme is considered as a terminal and the only committed step in triglyceride biosynthesis; this is a key enzyme in neutral lipid biosynthesis (Turchetto-Zolet et al., 2011). DGAT catalyses the formation of triacylglycerides from diacylglycerol and Acyl-CoA. So far, many attempts of overexpressing the isoforms of DGATs of same species have been unsuccessful (Vuttipongchaikij, 2012). For instance, overexpression of the endogenous orthologs of DGAT encoding genes in TAG biosynthesis, CrDGAT2a, CrDGAT2b and CrDGAT2c in C. reinhardtii have not shown any significant difference in TAG composition or accumulation despite the higher levels of transcripts observed (La Russa et al., 2012). On the other hand, when an isoform of type 2 (DGAT2) was heterogeneously overexpressed in a different organism, it surprisingly produced more oil bodies along with higher TAG levels and enhanced neutral lipids (Hung et al., 2013; Niu et al., 2013; Sanjaya et al., 2013).
In C. reinhardtii, there are two types of DGAT homologues, and transcripts encoding DGTT2 are reportedly present at consistently low levels under all tested conditions, including N deprivation. On the other hand, mRNA of the other homologue DGTT3 is present at low levels and only increases slightly under N deprivation (Miller et al., 2010), thus, not playing any major role in lipid accumulation.
The single-celled green alga C. reinhardtii has been an excellent photosynthetic model organism to study transgene expression and several foreign genes have been expressed in Chlamydomonas (Hannon et al., 2010). However, heterologous lipid biosynthetic genes have not been explored so far in this system. Therefore, this study is aimed at understanding the function of heterogeneous key enzyme involved in lipid biosynthesis. DGAT type 2 of Brassica napus (rapeseed) was introduced into C. reinhardtii. Results show altered fatty acid composition and higher accumulation of neutral lipid content. Significantly, higher number of oil bodies was observed in the transformed cells under fluorescent microscope. Transgene integration and expression of BnDGAT2 was confirmed by PCR/Southern blots, and eGFP expression was observed under fluorescent microscope. Results of BnDGAT2 expression in transformed alga, its maintenance for longer duration and implications of solid versus liquid medium have been investigated for various biofuel applications.
Results
Comparative homologue of DGATs
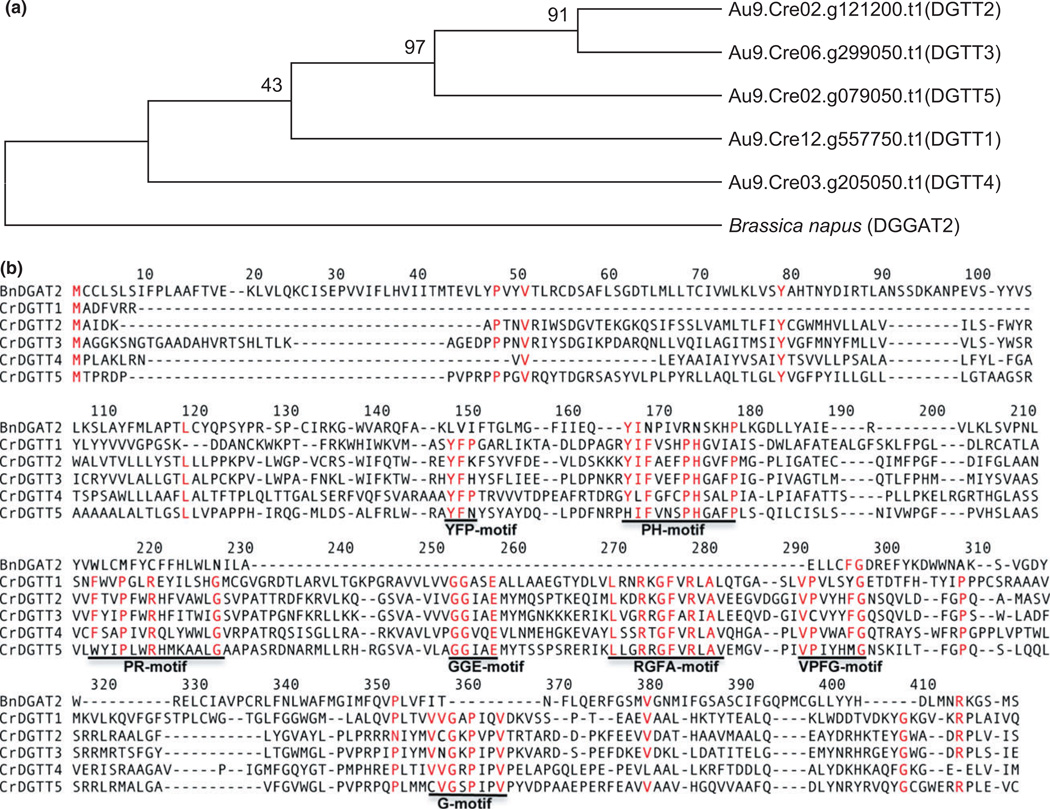
The amino acid sequence of B. napus BnDGAT2 was compared with that of the C. reinhardtii DGAT2 isoforms, namely DGTT1–5 (Boyle et al., 2012), and a phylogenetic tree was constructed (Figure 1a–b). The type 2 DGAT subfamilies of C. reinhardtii (DGTT1, DGTT2 and DGTT3, DGTT4 and DGTT5) were detected to be quite different from each other as well as from BnDGAT2. The amino acid sequence of B. napus BnDGAT2, when compared with C. reinhardtii DGAT type isoforms, Au9.Cre12.g557750.t1 (DGTT1), Au9.Cre03.g205050.t1 (DGTT4), Au9.Cre02. g079050.t1 (DGTT5), Au9.Cre02.g121200.t1 (DGTT2) and Au9.Cre06.g299050.t1 (DGTT3) showed about 19–25% identity (Figure 1b). The maximum per cent identity of BnDGAT2 was observed with CrDGTT4 (~25%), suggested plant-like DGAT2 (Hung et al., 2013). Seven conserved domains of amino acids are observed in the protein sequence alignment in type 2 family of diacylglycerol acyltransferase, as previously reported (Cao, 2011; Liu et al., 2012). These motifs are named PH, PR, GGE, RGFA, VPFG and G block (Figure 1b). The first domain YFP is highly conserved in CrDGTT1 and CrDGTT4 and is replaced by LVI in BnDGAT2, similar to several higher plant species (Arabidopsis thaliana, Olea europaea, Citrus sinensis, Jatropha curcas, Nicotiana tabacum, Solanum tuberosum, Solanum lycopersicum, Glycine max etc.). The second conserved domain, named PH block (also known as HPHG), has a core PH motif in all CrDGTTs, which is replaced by RN in BnDGAT and several higher plant species. The PR motif is conserved in all CrDGTTs, but is completely modified in BnDGAT. The motifs GGE and RGFA are highly conserved among CrDGTTs, but are absent in the BnDGAT2 and higher plant species. In the VPFG motif, FG is conserved in BnDGAT2 and CrDGTT 2, 3 and 4. The G block is conserved in the CrDGTTs family but is absent in BnDGAT2.
Figure 1.
(a–b) Phylogenetic tree and alignment of diacylglycerol acyltransferase type 2 of Brassica napus DGAT2 amino acids with DGATs isoforms (1–5) of Chlamydomonas reinhardtii. (a) Phylogenetic tree showing relationship of C. reinhardtii DGTT 1–5 with DGAT2 of B. napus. The whole-DGAT amino acid sequences were used for calculating the per cent identity. Phylogenetic tree was generated using the MEGA5.0 software and statistical support for tree branches was evaluated by bootstrap analysis (1000 replicates) mentioned on the nodes. (b) Multiple alignment of BnDGAT2 amino acid sequence with CrDGTTs isoforms (1–5) using Tcoffee (http://tcoffee.crg.cat). The red fonts and underlined motifs indicate the seven conserved domains among diacylglycerol acyltransferase type 2 family. The per cent identity of C. reinhardtii DGTTs was observed to be 19–25% with BnDGAT2.
Algal transformation
The unicellular alga C. reinhardtii (CC-125) was transformed with the vector pAlgaeDGAT-eGFP (Figure 2) containing type 2 gene of rapeseed (BnDGAT2) via electroporation. The transformed colonies harbouring aph7 gene were screened on hygromycin-supplemented medium with a transformation frequency of about 120 ± 10 colonies/1 × 106 cells. Initially 15 transformed colonies were tested by PCR, selected on hygromycin, and were further confirmed by GFP analysis under the fluorescence microscope. On the basis of high fluorescence intensity (using Nile red stain), five independent transformed cell lines were selected, and each one was divided into three replicates for evaluation of lipid and protein content. Transformed cells were confirmed for transgene integration and expression using standard molecular biology techniques as described in the Material and Methods section. The transformed cell line number 2, with a single copy of transgene integration, showing maximum fluorescence and growth rate of cells similar to wild type was maintained over a year by subculturing on semisolid medium (up to the 128th generation) and in a cryopreserved phase, as well. Some of the cells from the cryopreserved line were checked every 3 months for DGAT expression in a liquid medium. The average doubling time in control and transformed cells was observed to be about 11 and 12 h, respectively. The average biomass of the transformed alga (0.64 gm/L) was comparable to that of the wild type cells (about 0.73 gm/L) on a dry weight basis, as observed on the sixth day.
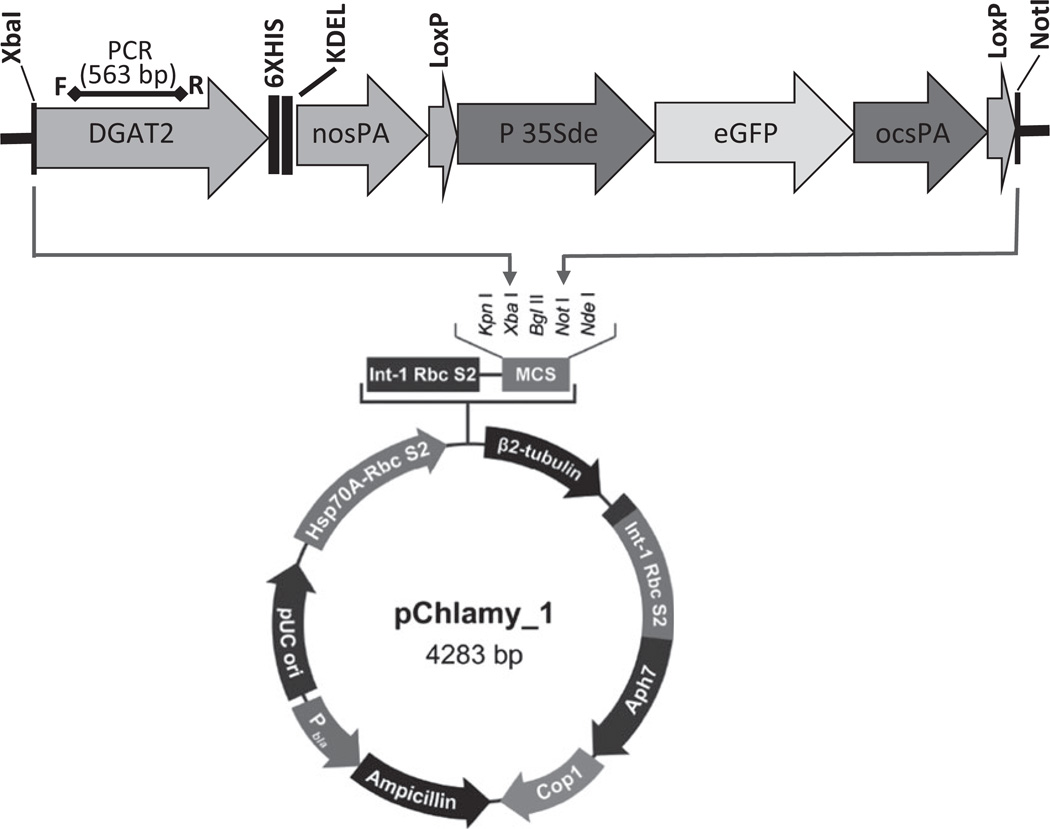
Figure 2.
Construction of pAlgaeDGAT-eGFP vector to transform the Chlamydomonas reinhardtii cells. The synthetic cassette containing BnDGAT2-6XHIS-Tag-KDEL-NOS-PolyA-35Sde-eGFP was cloned in pChlamy_1 at restriction sites XbaI and NotI. BnDGAT2 (Accession No. AF155224) was expressed by Hsp70-RbcS2 promoter and was tagged with 6XHis and KDEL sequences at the C-terminal. Enhanced GFP (Accession No. JN596101) was expressed by a double enhancer 35Sde promoter (Accession No. V00140.1). The letters F and R above DGAT2 gene denote the forward and reverse PCR primers used in the preliminary assessment of transgene integration in algal cells.
Evaluation of transgene integration
Primary transformation events in C. reinhardtii were confirmed by PCR using a BnDGAT2-gene specific primer pair. The genomic DNA was isolated from the transformed cell lines T1–T5. The genomic DNA was amplified by PCR, yielding about 600 bp product in transformed cells while no amplicon was observed in the wild type (Figure 3a). The PCR positive transformed cell lines were further analysed for Green Fluorescent Protein (GFP) expression and lipid accumulation.
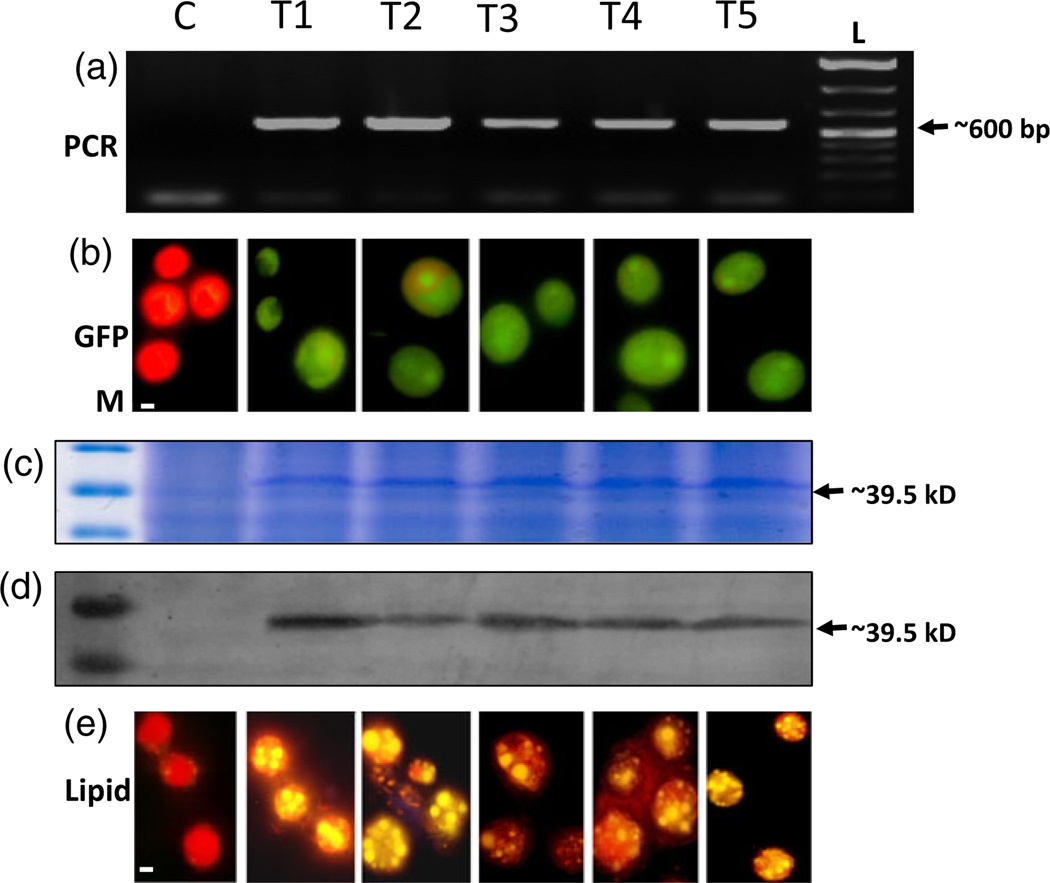
Figure 3.
(a–e) Expression of BnDGAT2 of Brassica napus into C. reinhardtii cells. (a) Transformed lines (T1–T5) versus control nontransformed C. reinhardtii were amplified by PCR primers, which specifically bind to BnDGAT2 as shown in Figure 2 and produced amplicon of ~600bp in transformed cell lines. (b) Fluorescence micrograph of eGFP expression in transformed cell lines T1–T5. Control line appeared red due to autofluorescence of chlorophyll. (c) SDS-PAGE 12.5% gel showing ~39.5 kD protein in lines T1–T5, stained with Coomassie dye G-250. (d) Western blot analysis in lines T1–T5 developed by ECL. (e) Nile red staining of T1–T5 line. Lipid droplets visualized in golden colour. Control cells observed with the least number of lipid droplets. In c–d, no BnDGAT2 signal was detected in control cells. The bar size in b and e is 10 µm, denoted by a white line.
Evaluation of GFP expression
Expression of the eGFP gene was driven by the CaMV35S double enhancer promoter (Kay et al., 1987) to visually identify transformed cell lines. An enhanced GFP expression at 488 nm was observed in all the tested transformed cell lines T1–T5 whereas untransformed cells only produced red colour due to chlorophyll autofluorescence (Figure 3b).
Lipid analysis by Nile red staining
To visualize the intracellular lipid globule accumulation in the transformed alga, cells from the transformed cell lines (T1–T5) and wild type were stained with Nile red and observed under the microscope. The expression of B. napus BnDGAT2 in Chlamydomonas led to the accumulation of a high number of lipid bodies, visualized as golden colour in the transformed cell lines (T1–T5). The untransformed cells showed an infinitesimally small number of lipid droplets and cells appeared mostly red due to chlorophyll autofluorescence (Figure 3e).
Evaluation of BnDGAT2 expression
Cells from all five transformed lines expressing BnDGAT2 under the HSP70A-RBCS2 promoter were analysed by SDS-PAGE, Coomassie blue staining and Western blots. The BnDGAT2 protein bands (~39 kD) appeared in all the transformed cell lines (T1–T5) whereas no specific signal was observed in the control cells (Figure 3c). Similar results were obtained with Western blot analysis. The sample proteins from transformed and wild type cells were transferred to the membrane and hybridized with the 6XHIS monoclonal antibodies, and signal was produced in all the transformed cell lines whereas no signal was noticed in the control cells (Figure 3d).
Evaluation of lipid content
To evaluate lipid content, relative fluorescence intensity was analysed in the transformed cell lines T1–T5 from day 2–7 using the Nile red stain. The maximum intensity was observed in transformed cell lines from day 3–6. On the other hand, wild type cells produced the same intensity during 0–6 days of the growth period. The transformed cell lines were observed with reduced Transformation of alga with brassica DGAT2 3 intensity after sixth day due to lipid catabolism as cells reached their stationary phase (Figure 4). Therefore, cells were harvested on the sixth day for lipid and FAME content studies. Among all the five transformed lines used for different comparative studies, line number T2, which possessed a single copy transgene (Figure 6c), showed optimal growth. It showed about twofold higher fluorescence intensity when compared with wild type cells (Figure 4). Therefore, T2 line was used to analyse long-term stability of transformed alga in the cryopreserved as well as solid phase.
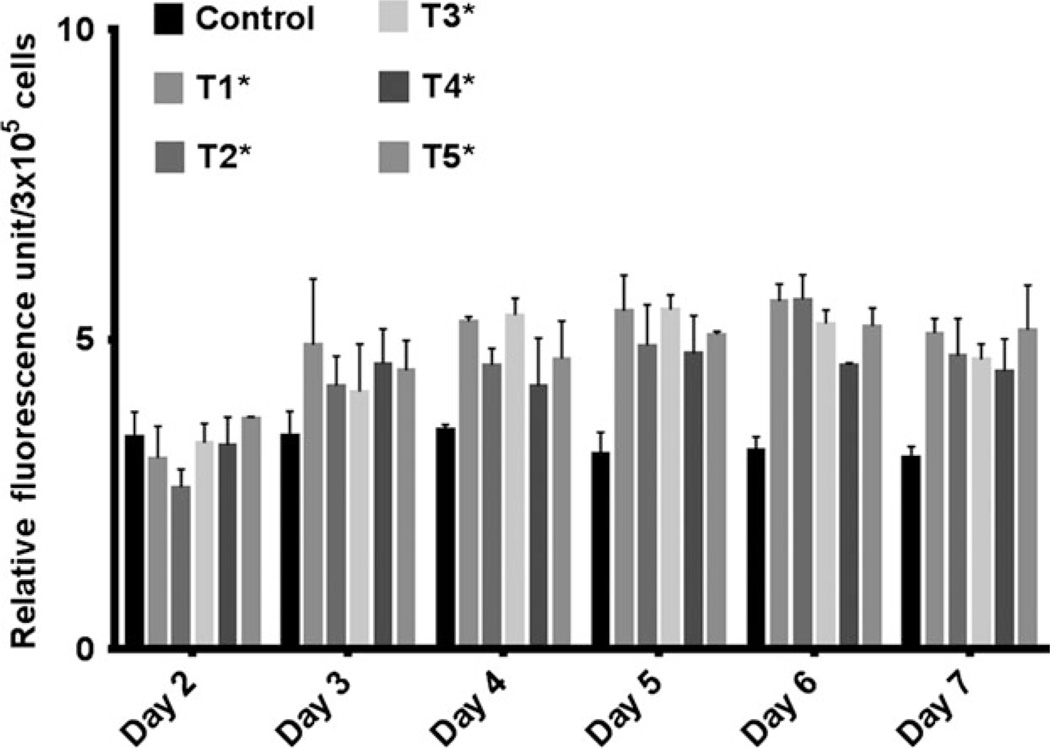
Figure 4.
Relative fluorescence intensity of C. reinhardtii cell lines T1–T5 transformed with BnDGAT2 of B. napus. Triplicate samples of C. reinhardtii cells were stained with Nile Red from days 2–7. Maximum intensity was observed on the sixth day in T1–T5 lines. Transformed cell line T2 was selected for further analysis due to its capacity for accumulating highest amount lipid and maximum growth rate. Relative fluorescent intensity was found significant at *P < 0.05 when compared the mean of relative fluorescent intensity of all transgenics (T1–T5) with the mean of relative fluorescent intensity of wild type samples by one-way anova with Dunnett’s multiple comparisons test (n = 3; average ± SD).
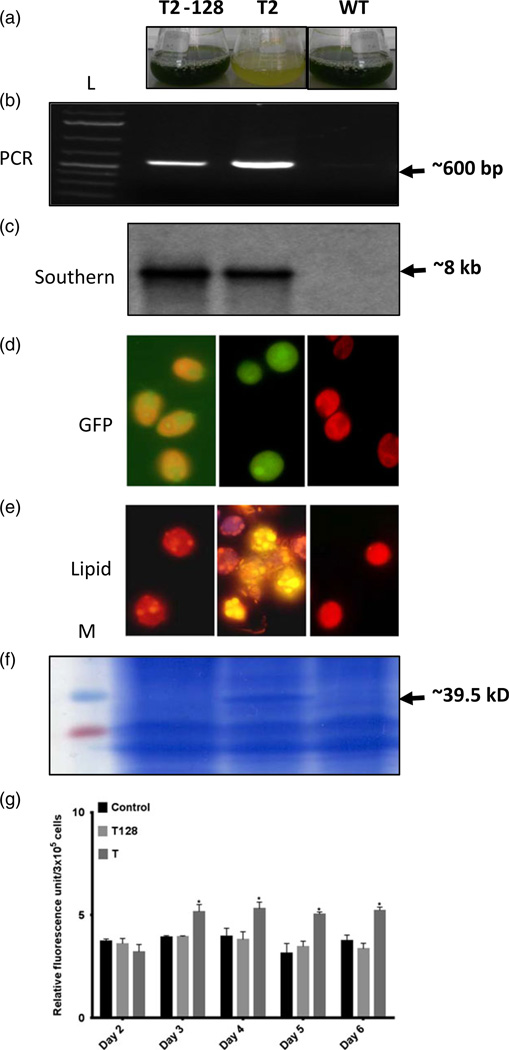
Figure 6.
Comparison of T2 cryopreserved versus T2 derived cell line maintained up to 128 generation on solid medium. (a) Cultures of T2–128 after 128th generation, T2 cell line from cryopreservation and wild type cells (WT). (b) PCR analysis for confirming the transgene presence and (c) Southern Blot confirmation of the stability of integrated DGAT2 transgene in T2–128. (d) Fading GFP expression in T2–128 cultures compared to T2 cells. (e) T2–128 cells stained with Nile red with the least number of lipid droplets. Cells are mostly producing red autofluorescence due to chlorophyll presence as WT cells. (f) Absence of BnDGAT2 protein analysed by Coomassie blue gel. (g). Spectro-fluorometry analysis of Nile red stained T128 (T2–128) algal cells compared with T (T2) cryopreserved line and wild type cells. Significant at *P < 0.05 compared with the wild type by one-way ANOVA with Dunnett’s multiple comparison test (n = 3; average ± SD).
Lipid and FAME analysis in transformed alga
The dry biomass of the transformed line T2 and untransformed alga was used to estimate the total lipid, FAME and neutral lipids. To quantify the total neutral lipids, we performed base-catalysed transesterification reaction from total lipids for conversion into FAME and quantified only those FAMEs that were derived from neutral lipids. We standardized the transesterification reaction using soybean oil with freshly prepared base catalyst and obtained about 47% conversion into FAMEs and a conversion factor of 2.09, analysed by gas chromatography. To calculate the total neutral lipid in alga, the FAME value was multiplied with the conversion factor 2.09. FAME in the transformed cell line T2 and wild type cells (WT) was analysed by gas chromatography (GC), and the peaks were confirmed by GC-Mass Spectrometry (GC-MS). All results were predicted based on the retention time of various saturated and unsaturated fatty acids in the chain length range of C16–C18. The neutral lipids and FAME in T2 were observed to be double the amount when compared to wild type cells (Table 1). When individual fatty acid methyl ester of transformed cells was compared with control cells, a significant alteration in the lipid composition was observed in the FAME of T2. The total saturated fatty acid content (palmitic acid and stearic acid) of chain length C16 and C18 was reduced to about 24% from 31% in the transformed cells when compared with the wild type cells (Figure 5). The monounsaturated fatty acids (MUFA) like oleic acid and palmitoleic acid of carbon chain length C16:1 and C18:1 increased by about 6% in the transformed cells from 5% of the wild type. However, omega (ω) α-linolenic polyunsaturated fatty acid (PUFA) was enhanced significantly from 32% (in wild type cells) to 45% in transformed cells (Figure 5). The overall PUFA increased in the FAME to about 7% in transformed cells when compared to wild type cells whereas saturated fatty acid content was reduced to about 7% in the transformed alga (Figure 6).
Table 1.
Lipid accumulation in transformed algal cell line (T2) expressing rapeseed BnDGAT2 compared with control (wild type) cells. The total lipid was extracted using Bligh and Dyer (1959) method from dry biomass
| Algal cells | Dry biomass (mg) | Total lipid (mg) | Total lipid (%) | Total FAME (mg) | Neutral lipid (mg) | Neutral lipid (%) |
|---|---|---|---|---|---|---|
| Control | 100 | 12.33 ± 2.08 | 12.33 | 1.42 ± 0.18 | 2.98 ± 0.37 | 2.98 |
| Transformed | 100 | 18.76 ± 1.96 | 18.76 | 3.68 ± 0.16 | 7.69 ± 0.34 | 7.69 |
The total lipid, Fatty Acid Methyl Esters (FAME) and neutral lipid, was estimated using three independent replicates and standard deviation is mentioned as ±.
Figure 5.
The per cent Fatty Acid Methyl Esters (FAME) in transformed cell line (T2) and control untransformed cells (C) was analysed with the help of GC-MS. The composition of fatty acids in the predominant region of carbon chain length C16–C18 was altered in the transformed cells. Saturated fatty acids C16:0 and C18:0 were reduced while polyunsaturated acids C18:1 and C18:3 increased in transformed algal cells compared to control cells. SFA, MUFA and PUFA are Saturated, Monounsaturated and Polyunsaturated fatty acids. Significant at ****P < 0.0001 or **P < 0.001 compared with the wild type by two-way ANOVA with Sidaks’s multiple comparisons test (n = 3; average ± SD).
Long-term maintenance and stability
Stability of the transformed algal line T2 (with a single copy of the transgene) was studied for a longer duration on solid media as well as in a cryopreserved state. After 10th generation, cells were either maintained on semisolid medium by subculturing (every 3 days) up to the 128th generation or maintained in a cryopreserved form over a year. The progeny of the T2 line maintained on solid plates displayed conversion of lipid properties similar to wild type cells (Figure 6a) while the cryopreserved cells, which were periodically checked for their viability by transferring in the liquid medium, maintained high lipid accumulation (Table 1). In a comparative study, total lipids in T2–128 line on solid medium were reduced to 13.5% from 18.76% of the T2 line. Neutral lipids were also reduced in the T2–128 line on solid medium to about 3.35% of the T2 line from the initial level of 7.69%. Similarly, composition of FAME analysed by GC-MS and total SFA, MUFA and PUFA was observed to be about 29.86%, 7.79% and 62.35%, respectively, which is closer to wild type cells. The stability of transgene integration in both cells maintained in solid, liquid and cryopreserved states were reconfirmed by PCR and Southern blot (Figure 6b–c). The T2 cell line grown on solid medium expressed GFP poorly when compared to cryopreserved cells (Figure 6d). Thus, on solid medium, T2 lines accumulated fewer lipids like the wild type cells when compared to cryopreserved cells (Figure 6e). These results were also supported by SDS-PAGE data and spectro-fluorometric analysis of Nile red stained cells (Figure 6f–g). These results indicate that if transformed cells were maintained for a longer duration on solid medium, may lose the introduced trait.
Discussion
This study is based on the expression of diacylglycerol acyltransferase type 2 (BnDGAT2) of Brassica napus (rapeseed) in Chlamydomonas reinhardtii for evaluating the triacylglycerol accumulation in transformed alga, which is of increasing interest for potential biodiesel production. Using microalgae as an alternative and renewable source of lipid-rich biomass feedstock for biofuel production has been explored during the past few years. However, a scalable, commercially viable system is yet to emerge. Chlamydomonas is an excellent reference organism to test basic principles of lipid metabolism in microalgae, in particular of the ‘green algal’ lineage (Liu and Benning, 2013). Besides, there are several gaps in the current scientific knowledge regarding DGAT and lipid biosynthesis in C. reinhardtii.
Several DGAT genes have been identified in a wide range of organisms that are involved in lipid biosynthesis. However, in Chlamydomonas, six genes predicted to encode acyl-CoA: diacylglycerol acyltransferases (DGAT) were compared to two genes known for similar activities in plants, suggesting the complexity of TAG biosynthesis in algal cells (Liu and Benning, 2013). Further, the process of lipid biosynthesis in microalgae might be subjected to rate-limiting steps with unknown feedback regulations, which is supported by the overexpression of three type-2 DGAT homologue genes in C. reinhardtii that neither boosted any intracellular TAG accumulation nor altered the fatty acid profiles during nitrogen or sulphur stress conditions (La Russa et al., 2012).
Therefore, it was assumed that overexpressing a homologous gene for lipid biosynthesis is unlikely to increase the lipid yield in C. reinhardtii due to unknown negative feedback inhibition. However, it was hypothesized that such inhibition may not act efficiently when a heterogeneous gene is introduced into an organism due to variation in the enzyme’s active site. The results of this study supported the above hypothesis as BnDGAT2 of B. napus assisted in the accumulation of twofold higher content of TAG in the transformed alga. Similarly, expression of heterologous type 2 DGTT of Chlamydomonas increased the lipid content of Arabidopsis thaliana leaves (Sanjaya et al., 2013). Also, in an yeast mutant (defective in triacylglycerol biosynthesis), the lipid content increased ninefold (Hung et al., 2013). Likewise, type 2 PtDGAT2B of diatom reportedly helped in restoring the complete lipid body formation in a TAG-deficient mutant strain of Saccharomyces cerevisiae (Gong et al., 2013). To ascertain the mechanism, subcellular localization of the heterogeneous proteins and biochemical analysis of purified individual DGAT proteins for their substrate preference may be of interest for further investigation (Liu and Benning, 2013).
In addition to choosing a heterologous lipid biosynthesis gene, suitable regulatory sequences were identified to enhance expression. The hybrid promoter HSP70A fused with RBCS2 has been reported to dramatically overexpress transgenes in C. reinhardtii when compared with HSP70A promoter alone (Schroda et al., 2000). The hybrid promoter was, therefore, used in this study, resulting in significant overexpression of the rapeseed BnDGAT2 in C. reinhardtii.
For an economically viable process to produce biodiesel, microalgae cultures should have both high lipid content and enhanced biomass. In our study, biomass of the transformed alga harvested on the sixth day (0.64 gm/L) was similar to that of the wild type cells (about 0.73 gm/L) based on dry weight. The nitrogen starvation stress studies on transformed cells for analysing the lipid accumulation in previous works have shown the loss of biomass when green alga, Botrycoccus sp. was exposed to nitrogen-deficient conditions (Yeesang and Cheirsilp, 2011). Decreased overall growth of Scenedesmus obliquus under nitrogen-deficient conditions (Gouveia and Oliveira, 2009) and decreased biomass in Nannochloropsis sp. under low nitrate concentration (Hu and Gao, 2006) has also been reported. Therefore, stress conditions were omitted in this study, considering that such conditions diminish the growth rate, lipid quality and quantity as well as productivity of the whole system (Chisti, 2007; Hu et al., 2008).
In the present study, neutral lipids, which are the main components for biodiesel production, doubled (Table 1). In addition, oil quality in most of the algal species is not good enough to be directly used as a substrate for producing good quality biodiesel, if saturated and unsaturated fatty acids are not present in an appropriate proportion. For example, biodiesel containing a higher amount of saturated short or medium chain fatty acids may become resistant to oxidation, but will probably have a lower cloud point and form wax at lower temperature. This concern was successfully addressed in the present study wherein saturated fatty acids like palmitic and stearic (C16:0 and C18:0) were reduced by 7% while monounsaturated fatty acids such as palmitic acid (C16:1) and oleic (C18:1) were increased to 1%. The α-linolenic acid (ALA; omega-3 fatty acids; C18:3n3), an essential fatty acid also known as PUFA, was increased by about 12% in the transformed alga (Figure 5), which has an enormous significance for improving the biofuel values. Unsaturated fats are excellent for producing biodiesel for cold weather, but may require an oxidative stabilizer to avoid the oxidation and rancidification. Thus, in this study, there was a considerable increase in neutral lipid content and alteration of fatty acid composition with a significant enhancement in the proportion of polyunsaturated fatty acids.
Similar phenomenon of fatty acid profile alteration has been reported in earlier studies. The overexpression of Thraustochytrids DGAT2 in Arabidopsis increased oleic acid (monounsaturated fatty acid) content in seeds (Zhang et al., 2013). Yeast transformed with PtDGAT2B of diatom showed alteration in TAG proportion of unsaturated C16 and C18 fatty acids (Gong et al., 2013). Expression of Chlamydomonas DGTT2 in Arabidopsis altered the leaf fatty acid chain length (Sanjaya et al., 2013). The acyl compositions of sphingolipids and surface waxes changed, and cutin levels decreased.
In addition to utilization in biofuels, the PUFA is emerging as a significant dietary supplement. Enhancement of oleic acid in vegetable oils has been a major target for biotechnologists nowadays as such oils have improved oxidative stability for food processing and other bio-based products. These oils are also in increasing demand for use in foods with low trans fat containing hydrogenated oils (Damude and Kinney, 2008).
The algal strain engineered with BnDGAT2 under HSP70A promoter may produce more lipids at high temperature and may be explored for carbon dioxide mitigation generated due to flue gases. The chloroplast-targeted heat shock protein 70 (HSP70) in Chlamydomonas has exhibited a greater photosynthetic efficiency because of the protection of photosystem II (Schroda et al., 1999). Presently, studies are in progress in this laboratory to analyse the impact of different light intensities and heat shock treatment on lipid accumulation in C. reinhardtii, transformed with rapeseed BnDGAT2.
Overcoming nuclear gene silencing in C. reinhardtii has been an ongoing challenge in C. reinhardtii. Previously gene silencing in Chlamydomonas has been observed by Mayfield group by expressing the heterologous gene (Hannon et al., 2010). The C. reinhardtii often silenced or down-regulated the non required heterologous genes when expressed at high levels. However, not all green algae of Chlorophyceae family tend to show active silencing of an introduced heterologous gene, which reaffirmed that transgene stability and reproducibility should not be a concern with other algal candidate for producing biofuels and bioproducts on commercial scale (Hannon et al., 2010). The losses of trait or spontaneous mutation in Chlamydomonas cultures have been reported in solid medium. Chlamydomonas cultures are most commonly maintained as vegetative cells on agar-containing medium for short periods of time to avoid the genetic changes in cultures. To maintain the quality, long-term storage of Chlamydomonas cultures was achieved by cryopreservation under liquid nitrogen (Gonzalez-Ballester et al., 2011; González-Ballester et al., 2005). Thus, such concern can be avoided by freezing the transgenic cells in liquid nitrogen (León and Fernández, 2007). It was observed during the present study that when the same T2 line was maintained in liquid nitrogen, no loss in the transgenic trait was observed for a period of over a year. However, when cells of same line were maintained for a long time on solid medium (i.e. one cycle on antibiotic selection and another without antibiotic up to the 128th generation for a year) as reported previously by Guo et al. (2013), transgenic trait of BnDGAT2 and eGFP seemed to diminish.
Thus, introduction of rapeseed BnDGAT2 in alga helped in producing higher neutral lipid content in C. reinhardtii when compared to untransformed cells. These neutral lipids, main constituents of FAME, were deciphered with higher amount of unsaturated long-chain fatty acids and lower amount of saturated fatty acids. The longer chain length fatty acid molecules reportedly increase the cetane number, heat of combustion and reduce the NOx emissions while reduced saturated fatty acid content is likely to help in diminishing the viscosity and increase the lubricity of the biodiesel. Thus, results derived from the current study can be applied to commercially important algal strains for accumulating higher neutral lipid content and improving the oil quality by introducing the rapeseed BnDGAT2. The HSP70A promoter, which can be induced by heat shock and light, is recommended as a tool to further enhance expression level. This additional advantage should produce more lipids in the later growth phase as very high lipid accumulation in initial phase may retard the growth of the transformed alga. Another significant observation was the phenomenon of gene silencing. Important precautions need to be taken in handling the transformed alga, and based on our study, it is advised that seed cultures should be maintained in liquid or cryopreserved form for long-term stability of the transgenes.
Materials and methods
Strain and growth conditions
Chlamydomonas reinhardtii (wild type strain CC-125) with intact cell wall was obtained from the Chlamydomonas Genetics Centre, Duke University, Durham, North Carolina, USA. Stocks were kept frozen in liquid nitrogen. For experimental purposes, the strain was grown in Tris–acetate–phosphate (TAP) liquid or agar-solidified medium (Harris, 1989) containing NH4Cl (375 mg/L), MgSO4․7H20 (100 mg/L), CaCl2․2H2O (50 mg/L), K2HPO4 (108 mg/L), KH2PO4 (54 mg/L), Tris (hydroxymethyl) aminomethane (2.42 g/L) (pH 7.0), glacial acetic acid (1 mL/L) and trace metal solution (1 mL/L). Stock solution of trace metals was prepared using the following ingredients: EDTA disodium salt (50 g/L), ZnSO4․7H2O (22 g/L), H3BO3 (11.4 g/L), MnCl2․4H2O (5.06 g/L), CoCl2․6H2O (1.61 g/L), Cu.SO4․5H2O (1.57 g/L), (NH4)6Mo7O24․4H2O (1.10 g/L) and KOH (17 g/L). Culture was maintained at 25 °C, under solid-state lamps with light intensity 12000 lux (for 16 : 8 h light and dark condition) on solid plates or shaken at 200 rpm.
Construction of the transformation vector
The pCHLAMY_1 expression vector of size 4283 bp was procured from Life technologies, Invitrogen, USA. It contained hygromycin resistance gene (aph7) driven under the β-tubulin promoter. The Hsp70-RbcS2 promoter was used to drive the expression of BnDGAT2, which was synthesized based on the sequence of Brassica napus (Accession No. AF155224). The six restriction sites namely BclI, HindIII, EcoRI, PstI, NcoI and BamHI were removed by changing codons from an original gene of BnDGAT2 (AF155224). The complete synthetic DNA cassette (DGAT-eGFP) contained the BnDGAT2 gene tagged with 6XHIS and fused with KDEL-NOS-PolyA sequences at the 3′terminal. The addition of carboxy-terminal amino acid sequence KDEL (Lys-Asp-Glu-Leu) at 3′end should help in the retention of DGAT2 protein in the endoplasmic reticulum (Ellgaard et al., 1999; Pelham, 1988). Promoter 35Sde was used to drive the eGFP reporter gene (Accession No. JN596101). The origin of green fluorescent protein (GFP) is the jellyfish Aequorea Victoria and eGFP is codon optimized for increasing the sensitivity of the reporter protein (Zhang et al., 1996). Thus, complete cassette containing ‘BnDGAT2-6XHIS-KDEL-OS-PolyA-35Sde-eGFP-ocspolyA’ was synthesized (BioBasic Inc. Markham, Ontario, Canada) and, cloned at Xba1 and Not1 restriction sites in the pCHLAMY_1 vector and named as ‘pAlgae-DGAT-eGFP’ (Figure 2); it was bulked up by transforming into DH5α cells. The positive clones were selected on LB plates containing 100 µg/mL ampicillin.
Transformation of C. reinhardtii
The plasmid (pAlgae-DGAT-eGFP) was linearized with enzyme PvuI (located in the backbone of the vector). C. reinhardtii cells (15 mL) were collected at 0.6 OD750 and centrifuged at 804 g (2500 rpm) (500 g). Pellet was re-suspended in 250 µL of TAP medium containing 40 mm sucrose solution and 2 µg of the linearized plasmid. The mixture was gently mixed by pipetting and transferred to a 4 mm electrocuvette (Bio-Rad Laboratoreis, Hercules, CA, USA). Cells were electroporated with the help of Gene Pulser (Bio Rad) using voltage-600V, capacitance-50 µF and infinite resistance. Electroporated cells were split into two aliquots of 125 µL each and augmented with 5 mL of TAP medium containing 40 mm sucrose solution. The suspension was poured into a 6-well plate and incubated for 24 h at 25 °C in an orbital shaker (150 rpm). After that, cells were harvested at 804 g (2500 rpm) for 10 min, and the pellet was suspended in TAP medium (150 µL) containing 40 mm sucrose. The resultant mixture was platted on solid TAP medium containing 10 mg/L hygromycin. Plates were incubated in light (16 : 8) at 25 °C for 10–12 days until green colonies appeared on the selection medium.
Comparative analysis of Brassica napus DGAT2 with Chlamydomonas reinhardtii DGATs Isoforms
The protein sequences of the C. reinhardtii DGTT1–5 genes were retrieved by sequence comparison with B. napus DGAT2 gene (Accession No. AF155224) using BLAST (Altschul et al., 1997). Sequences of CrDGTT1–5 and BnDGAT2, proteins were aligned using the CLUSTALW software in MEGA5 (Tamura et al., 2011). In CLUSTALW multiple sequence alignment (Figure 1b), 79 amino acids (position number 205–284) were deleted from CrDGTT5 due to lack of similarity with other diacylglycerol acyltransferase type 2 family. The alignment was then used to construct a phylogenetic tree using the neighbour-joining method in MEGA5 by bootstrapping with 1000 replicates.
Polymerase Chain Reaction
Primary transformation events were confirmed by PCR using a specific primer pair based on BnDGAT2 DNA sequence of B. napus, F-DGAT2, GATTCTGCCTTCTTATCAGGTGACAC and R-DGAT2, CGAACCATCCATTTGTGAACAGG. The genomic DNA from transformed and untransformed cells was extracted by the DNeasy plant mini kit (Qiagen GmbH, Hilden, Germany). PCR reactions were performed in a 25-µL reaction volume containing five picomole each of both primers, 10 mm dNTP, 25 ng template DNA and 0.5 U Taq polymerase (Himedia Labs, Mumbai, India). The PCR amplification was carried out for 35 cycles (denaturation for 30 s at 95 °C, annealing for 30 s at 58 °C and extension for 1 min at 72 °C), including initial denaturation for 2 min at 95 °C and final extension for 10 min at 72 °C. The PCR product was electrophoresed on a 0.8% agarose gel.
Southern blot
Genomic DNA isolated in bulk from transformed and untransformed cells using the DNeasy plant mini kit (Qiagen) was digested with the restriction enzyme NdeI. Digested genomic DNA was electrophoresed on 1% agarose gel, denatured and blotted onto Hybond positive nylon membrane as described (Kumar et al., 2004). The probe was prepared as per the AlkPhos Direct Labeling Reagents kit (GE Healthcare) using BnDGAT2 gene (amplified with primers, F-DGAT2-SB and R-DGAT2-SB). Southern hybridization was carried out according to AlkPhos kit instructions (GE Healthcare, Little Chalfont, United Kingdom).
Evaluation of GFP
Transformed cells were analysed for GFP expression using fluorescent microscope Nikon TS Eclipse. The GFP expression was observed at 488 nm wavelength.
Fluorescence microscopy
Nile red, a fluorescent lipophilic stain (Greenspan et al., 1985), was used to observe the lipid bodies in transformed alga. Prior to staining, 1 mL algal cells grown until the sixth day (mid logarithmic phase) and then harvested (Kightlinger et al., 2014; Neupert et al., 2009) by centrifugation at 12857 g for 10 min, and the pellet was suspended in 1 mL 20% DMSO. Mixture was thoroughly vortexed for 10 min at room temperature. Cells were re-centrifuged at 12857 g for 10 min, pellet was suspended in 1 mL water and vortexed again. Thereafter, 5 µL (1 mg/mL stock, prepared in isopropanol) of Nile Red stain (9-diethylamino-5H-benzo[a]-phenoxazine-5-one) was added to visualize the intracellular lipid bodies of algal cells. The mixture was incubated for 5 min in dark at room temperature, and cells were observed under a fluorescence microscope (Nikon TE2000-U, Tokyo, Japan) using UV light with excitation and emission at 530 and 575 nm, respectively.
Protein extraction
One hundred millilitre of each of transformed and untransformed algal culture grown for 6 days were harvested through centrifugation at 6000 g and grounded in liquid nitrogen. Cells were suspended in protein extraction buffer [2% (w/v) SDS, 56 mm NaCO3, 12% (w/v) Sucrose, 56 mm DTT and 2 mm EDTA, pH 8.0] (Richter et al., 2013). The protein was denatured at 70 °C for 20 min. After centrifugation at 8000 g for 10 min, concentration of protein in the supernatant was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific India Pvt. Ltd., Mumbai, India). The Coomassie Brilliant Blue G-250 dye was used to visualize polypeptides in 12.5% polyacrylamide gels using a solution containing 40% methanol, 10% acetic acid and 0.025% Coomassie Brilliant Blue R-250 (Figure 3c).
Western blot analysis
The purified mouse histidine tag (monoclonal mouse Anti-His Antibodies; IgG1) was used to detect recombinant proteins carrying His tags at the C terminus of BnDGAT2 in the transformed algal cells. About 30 µg reduced protein extract was resolved by 12.5% SDS-PAGE gel and blotted onto Hybond-C membranes (GE Healthcare). Membrane was hybridized with monoclonal anti-His antibody (Sigma, St. Louis, MO) antibodies raised in mouse. The membrane, along with anti-His antibody at dilution 1 : 10000, was incubated for 2 h at 4 °C. The blot was further exposed to an appropriate secondary antibody (raised in goat and used at dilution 1 : 1000) (Sigma) coupled to Horse Radish Peroxidase (HRP). To observe the His signal fused at C terminus of the DGAT2 (Figure 3d), blot was developed using ECL Kit (Genetix) following manufacturer’s instructions, and signals were detected using the FluorChem Imager (ProteinSimple, Santa Clara, CA, USA).
Fluorometric analysis of lipid droplets
Algal cells at various stages of growth were counted using a hemocytometer. Lipid quantification was performed in a 96-well micro titre plate in triplicates. ~3 × 105 cells were re-suspended in 297 µL of 25% aqueous DMSO solution and 3 µL of Nile red stain solution (50 µg/mL) while control unstained cells were suspended in 300 µL of DMSO. The control and stained cells were incubated at 40 °C for 10 min and placed in a shaker at 120 rpm. Fluorescence was recorded using a spectro-fluorometer (Spectra-Max M3, Sunnyvale, CA, USA) at a wavelength of 485-nm excitation and 552-nm emission (Kimura et al., 2004). The observed intensities were corrected by subtracting the fluorescence value difference in Nile red stained and unstained cells.
Lipid extraction
Lipid was extracted as reported (Bligh and Dyer, 1959). The dry biomass was crushed to a fine powder by mortar pestle, and 100 mg powder was transferred to a glass vial. The vial was added with 1 mL chloroform and 2 mL methanol and shaken at 200 rpm for 12 h. Mixture was further augmented with 1 mL chloroform and 1.6 mL distilled water. After vortexing, the mixture was incubated for 2 h until water and chloroform layers separated. Lower layer of chloroform containing lipids was separated by GF/A filter paper in a preweighed glass vial. The filter paper was washed by chloroform and pooled into the glass vial. Pooled solvent was evaporated in a hot air oven at 75 °C. Weight of lipid was measured gravimetrically by subtracting the empty vial’s weight from the lipid-containing vial.
Transesterification of lipid into Fatty acid methyl esters (FAME)
The algal lipid was transesterified into fatty acid methyl esters (FAME) as reported (Ichihara et al., 1996). Each 10 mg of lipid was dissolved in 2 mL hexane and 200 µL of freshly prepared 2-M methanolic KOH (used as a catalyst). The mixture was vortexed for 2–5 min followed by a brief centrifugation, and the upper hexane layer was collected for FAME analysis.
FAME Analysis by GC-MS and GC-FID
Quantification of FAME was carried out using gas chromatography (Agilent) equipped with Omega Wax 250 column (30 m × 0.25 mm × 0.25 µm) and flame ionization detector (FID). The operating conditions were as follows: split ratio 1 : 10, injection volume 1 µL, nitrogen carrier gas with constant linear velocity 33.9 cm/s, H2 at 40 mL/min, air at 400 mL/min, make-up gas (nitrogen) at 30 mL/min, injector temperature 250 °C, detector temperature 260 °C, oven temperature started at 50 °C for 2 min and increased at the rate of 4 °C/min to 240 °C, and holding time of 20 min at 240 °C. Supelco 37-component FAME mix was used as the external standard to identify retention time for specific FAME and quantification (Koberg et al., 2011; Wang et al., 2010). The FAME C16:2 and C16:3 (absent from external standard) were quantified based on the area of C16:1 isomer of palmitic acid. The peak corresponding to specific FAME was further confirmed using GC–MS.
Statistical analysis
For fluorometric analysis, one-way anova with Dunnett’s multiple comparison test was performed to determine the statistical differences between the control and transgenic samples. The statistical equivalence was determined for each day based on reflective fluorescence using n = 3 samples for each group as described in Figures 4 and 6(g). For FAME comparison, two-way anova was conducted with Sidak’s multiple comparisons test. Sample means are considered equivalent if |µ1−µ2| ≤ θ with a 95% confidence interval. Statistical parameters were set at: θ = 0.5, α = 0.05 as described (http://www.graphpad.com).
Acknowledgements
Thanks to Department of Biotechnology, Government of India, for providing the financial support to carry out this research work.
Footnotes
Competing interests
Authors in manuscript have no competing interests.
Authors’ contributions
IA performed all transformation experiments and molecular analysis. SK along with HD conceived the concept, designed the study. SK with the help of IA coordinated the experiments. IA, AKS, HD and SK wrote and edited the manuscript.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012;287:15811–15825. doi: 10.1074/jbc.M111.334052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H. Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Res. Notes. 2011;4:249. doi: 10.1186/1756-0500-4-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Smith SJ, Zheng Y-W, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ. Identification of a gene encoding an acyl CoA: diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl Acad. Sci. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnol. Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008;26:126–131. doi: 10.1016/j.tibtech.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Damude HG, Kinney AJ. Enhancing plant seed oils for human nutrition. Plant Physiol. 2008;147:962–968. doi: 10.1104/pp.108.121681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Fan J, Cui Y, Wan M, Wang W, Li Y. Lipid accumulation and biosynthesis genes response of the oleaginous Chlorella pyrenoidosa under three nutrition stressors. Biotechnol. Biofuels. 2014;7:17. doi: 10.1186/1754-6834-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Zhang J, Guo X, Wan X, Liang Z, Hu CJ, Jiang M. Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-coenzyme A: diacylglycerol acyltransferase family in the diatom Phaeodactylum tricornutum. FEBS Lett. 2013;587:481–487. doi: 10.1016/j.febslet.2013.01.015. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, Pootakham W, Mus F, Yang W, Catalanotti C, Magneschi L, de Montaigu A, Higuera JJ, Prior M, Galván A. Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods. 2011;7:1–13. doi: 10.1186/1746-4811-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, de Montaigu A, Higuera JJ, Galván A, Fernández E. Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 2005;137:522–533. doi: 10.1104/pp.104.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia L, Oliveira AC. Microalgae as a raw material for biofuels production. J. Ind. Microbiol. Biotechnol. 2009;36:269–274. doi: 10.1007/s10295-008-0495-6. [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S-L, Zhao X-Q, Tang Y, Wan C, Alam MA, Ho S-H, Bai F-W, Chang J-S. Establishment of an efficient genetic transformation system in Scenedesmus obliquus. J. Biotechnol. 2013;163:61–68. doi: 10.1016/j.jbiotec.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Hannon M, Gimpel J, Tran M, Rasala B, Mayfield S. Biofuels from algae: challenges and potential. Biofuels. 2010;1:763–784. doi: 10.4155/bfs.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. The Chlamydomonas Sourcebook; p. 780. [DOI] [PubMed] [Google Scholar]
- Hu H, Gao K. Response of growth and fatty acid compositions of Nannochloropsis sp. to environmental factors under elevated CO2 concentration. Biotechnol. Lett. 2006;28:987–992. doi: 10.1007/s10529-006-9026-6. [DOI] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- Hung C-H, Ho M-Y, Kanehara K, Nakamura Y. Functional study of diacylglycerol acyltransferase type 2 family in Chlamydomonas reinhardtii. FEBS Lett. 2013;587:2364–2370. doi: 10.1016/j.febslet.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Ichihara Ki, Shibahara A, Yamamoto K, Nakayama T. An improved method for rapid analysis of the fatty acids of glycerolipids. Lipids. 1996;31:535–539. doi: 10.1007/BF02522648. [DOI] [PubMed] [Google Scholar]
- James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price GD, Djordjevic MA. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour. Technol. 2011;102:3343–3351. doi: 10.1016/j.biortech.2010.11.051. [DOI] [PubMed] [Google Scholar]
- James GO, Hocart CH, Hillier W, Price GD, Djordjevic MA. Temperature modulation of fatty acid profiles for biofuel production in nitrogen deprived Chlamydomonasreinhardtii. Bioresour. Technol. 2013;127:441–447. doi: 10.1016/j.biortech.2012.09.090. [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Kightlinger W, Chen K, Pourmir A, Crunkleton DW, Price GL, Johannes TW. Production and characterization of algae extract from Chlamydomonas reinhardtii. Electron. J. Biotechnol. 2014;17:3–3. [Google Scholar]
- Kimura K, Yamaoka M, Kamisaka Y. Rapid estimation of lipids in oleaginous fungi and yeasts using Nile red fluorescence. J. Microbiol. Methods. 2004;56:331–338. doi: 10.1016/j.mimet.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Knothe G. Biodiesel and renewable diesel: a comparison. Prog. Energy Combust. Sci. 2010;36:364–373. [Google Scholar]
- Koberg M, Cohen M, Ben-Amotz A, Gedanken A. Bio-diesel production directly from the microalgae biomass of Nannochloropsis by microwave and ultrasound radiation. Bioresour. Technol. 2011;102:4265–4269. doi: 10.1016/j.biortech.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol. Biol. 2004;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Russa M, Bogen C, Uhmeyer A, Doebbe A, Filippone E, Kruse O, Mussgnug JH. Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 2012;162:13–20. doi: 10.1016/j.jbiotec.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Larkum AW, Ross IL, Kruse O, Hankamer B. Selection, breeding and engineering of microalgae for bioenergy and biofuel production. Trends Biotechnol. 2012;30:198–205. doi: 10.1016/j.tibtech.2011.11.003. [DOI] [PubMed] [Google Scholar]
- León R, Fernández E. Nuclear Transformation of Eukaryotic Microalgae. Historical Overview, Achievements and Problems. In: León R, Galván-Cejudo A, Fernández E, et al., editors. TransgenicMicroalgae as Green Cell Factories. New York: Springer Science + Business Media; 2007. pp. 1–14. [DOI] [PubMed] [Google Scholar]
- Li Y, Han D, Hu G, Sommerfeld M, Hu Q. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2010;107:258–268. doi: 10.1002/bit.22807. [DOI] [PubMed] [Google Scholar]
- Liu B, Benning C. Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 2013;24:300–309. doi: 10.1016/j.copbio.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ. Acyl-CoA: diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012;51:350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Mallick N, Mandal S, Singh AK, Bishai M, Dash A. Green microalga Chlorella vulgaris as a potential feedstock for biodiesel. J. Chem. Technol. Biotechnol. 2012;87:137–145. [Google Scholar]
- Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: a review. Renew. Sustain. Energy Rev. 2010;14:217–232. [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 2010;154:1737–1752. doi: 10.1104/pp.110.165159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell. 2010;9:97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- Niu Y-F, Zhang M-H, Li D-W, Yang W-D, Liu J-S, Bai W-B, Li H-Y. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs. 2013;11:4558–4569. doi: 10.3390/md11114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988;7:913. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell. 2010;9:486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamaki E, Grimm B. Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol. 2013;162:63–73. doi: 10.1104/pp.113.217141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya, Miller R, Durrett TP, Kosma DK, Lydic TA, Muthan B, Koo AJ, Bukhman YV, Reid GE, Howe GA, Ohlrogge J, Benning C. Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell. 2013;25:677–693. doi: 10.1105/tpc.112.104752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman F-A, Beck CF. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell. 1999;11:1165–1178. doi: 10.1105/tpc.11.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Blöcker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan J-C, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. The Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetto-Zolet AC, Maraschin FS, de Morais GL, Cagliari A, Andrade CM, Margis-Pinheiro M, Margis R. Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol. Biol. 2011;11:263. doi: 10.1186/1471-2148-11-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuttipongchaikij S. Genetic manipulation of microalgae for improvement of biodiesel production. Thai J. Genet. 2012;5:130–148. [Google Scholar]
- Wang L, Li Y, Chen P, Min M, Chen Y, Zhu J, Ruan RR. Anaerobic digested dairy manure as a nutrient supplement for cultivation of oil-rich green microalgae Chlorella sp. Bioresour. Technol. 2010;101:2623–2628. doi: 10.1016/j.biortech.2009.10.062. [DOI] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329:796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- Yeesang C, Cheirsilp B. Effect of nitrogen, salt, and iron content in the growth medium and light intensity on lipid production by microalgae isolated from freshwater sources in Thailand. Bioresour. Technol. 2011;102:3034–3040. doi: 10.1016/j.biortech.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Zhang G, Gurtu V, Kain SR. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem. Biophys. Res. Commun. 1996;227:707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- Zhang C, Iskandarov U, Klotz ET, Stevens RL, Cahoon RE, Nazarenus TJ, Pereira SL, Cahoon EB. A thraustochytrid diacylglycerol acyltransferase 2 with broad substrate specificity strongly increases oleic acid content in engineered Arabidopsis thaliana seeds. J. Exp. Bot. 2013;64:3189–3200. doi: 10.1093/jxb/ert156. [DOI] [PMC free article] [PubMed] [Google Scholar]