Abstract
Targeted knockout of genes in primary human cells using CRISPR-Cas9 mediated genome-editing represents a powerful approach to study gene function and to discern molecular mechanisms underlying complex human diseases. We used lentiviral delivery of CRISPR-Cas9 machinery and conditional reprogramming culture methods to knockout the MUC18 gene in human primary nasal airway epithelial cells (AECs). Massively parallel sequencing technology was used to confirm that the genome of essentially all cells in the edited AEC populations contained coding region insertions and deletions (indels). Correspondingly, we found mRNA expression of MUC18 was greatly reduced and protein expression was absent. Characterization of MUC18 knockout cell populations stimulated with TLR2, 3 and 4 agonists revealed that IL-8 (a pro-inflammatory chemokine) responses of AECs were greatly reduced in the absence of functional MUC18 protein. Our results show the feasibility of CRISPR-Cas9 mediated gene knockouts in AEC culture (both submerged and polarized), and suggest a pro-inflammatory role for MUC18 in airway epithelial response to bacterial and viral stimuli.
Introduction
Functional studies of a gene or multiple genes in primary human cells are critical to elucidate the pathological mechanisms underlying complex human diseases. In the past decade, many studies have utilized RNA interference (RNAi) technology to effectively knockdown genes of interest.1 However, this knockdown approach does not result in complete loss of gene/protein expression (knockout) and can often result in off-target effects.2 Thus, methods for complete knockout of a gene in human cells, especially in primary cells, are urgently needed.
By using clustered regularly interspaced short palindrome repeats associated Cas9 nuclease (CRISPR-Cas9) technology, several groups of investigators have successfully generated gene knockouts and made sequence level nucleotide changes in both human transformed and induced pluripotent stem cells (iPS).3-5 Moreover, CRISPR-Cas9 machinery has recently been used successfully to edit the genome of primary mouse cells in vivo.6-8 However, this technology has only been applied to a few primary human cell types currently used in culture to study specific disease processes.9 Application of this technology to primary airway epithelial and other primary cell types is limited by low transfection efficiencies, a requirement for multiple passages/proliferation, and harsh selection conditions required to achieve an edited primary cell population.
Human primary airway epithelial cells (AECs) are the first line of host defense against hazardous inhaled environmental factors such as pathogens and pollutants. The ability to knockout genes in AECs would allow functional analysis of single genes in the epithelial response to these environmental risk factors.
MUC18, also known as CD146 or melanoma cell adhesion molecule (MCAM), is a 113 kD transmembrane glycoprotein and is a member of the immunoglobulin superfamily.10, 11 MUC18 is composed of an extracellular domain, a single transmembrane domain and a short (63 amino acids) cytoplasmic tail. MUC18 over expression was initially identified in human malignant melanoma cells and thought to promote tumor metastasis.12-14 Our recent publications 15, 16 demonstrated that MUC18 is upregulated in asthmatic and COPD patient airway epithelial cells. MUC18 protein is expressed by basal and ciliated airway epithelial cells.15 Our findings further suggest that MUC18 is critical to bacteria-induced murine lung inflammation.15 However, whether MUC18 promotes airway epithelial inflammatory responses to pathogens or Toll-like receptor (TLR) agonists, mimicking pathogen infections, remains unclear.
In the current study, we detail for the first time, generation of primary human nasal airway epithelial cells knocked out for a gene (here MUC18), using CRISPR-Cas9 technology. We use the MUC18 knockout cells to demonstrate a pro-inflammatory function of MUC18 in response to stimulation with various TLR agonists. Our workflow provides a strategy to produce gene knockouts in primary airway epithelial cells, and our results reveal a function of MUC18 in the airway epithelium that may be important to multiple airway diseases.
Results
MUC18 Targeted Knockout Strategy
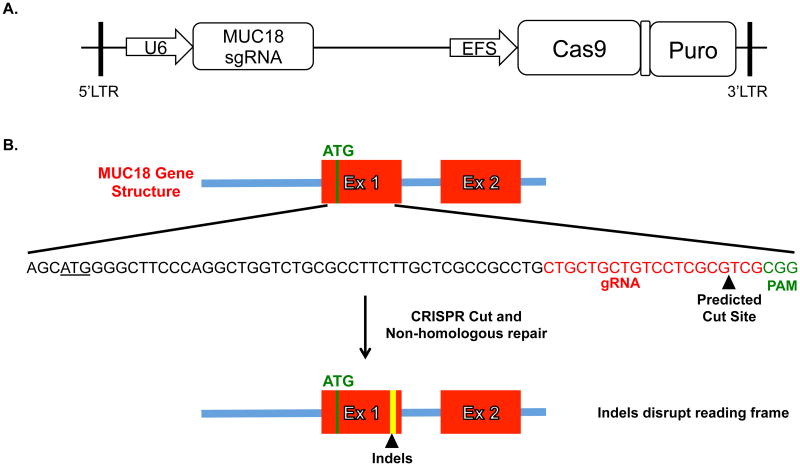
Preliminary studies indicated that primary airway epithelial cells are difficult to transfect at high efficiency and low toxicity. Consequently, a lentiviral transduction strategy was used to introduce the CRISPR-Cas9 machinery. A recently developed lentiviral vector, which expresses the sgRNA, Cas-9 nuclease, and puromycin resistance gene was used (Fig. 1).4 The gRNA was designed to target Cas9 machinery immediately downstream of the MUC18 start codon. Targeting at this site will create double-stranded breaks repaired by non-homologous end joining that will result in frame shift insertions and deletions (indels), and thus “knockout” functional MUC18 protein (Fig. 1). Random integration of the lentiviral expression cassette ensures stable expression of the MUC18 targeting CRISPR-Cas9 and puromycin selection machinery. The application of puromycin allows the selection of cells with successful integration and has previously been shown to eventually lead to a mixed population (with respect to a specific indel) of bi-allelically edited cells.4
Figure 1. Lentiviral vector and MUC18 Knockout Targeting Strategy.
A. Simplified schematic of the published plentiCRISPRv1 vector we used to target MUC18. We designed the MUC18 gRNA and ligated into the plentiCRISPRv1 vector. The plentiCRISPR vector co-expresses both the MUC18 gRNA and the Cas9 protein. B. The gRNA and thus cutting site was chosen to be immediately downstream of the start codon for MUC18 so indel's introduced by non-homologous repair of the double-stranded break would likely disrupt the protein reading frame.
Initial Generation of MUC18 KO Nasal Airway Epithelial Cells
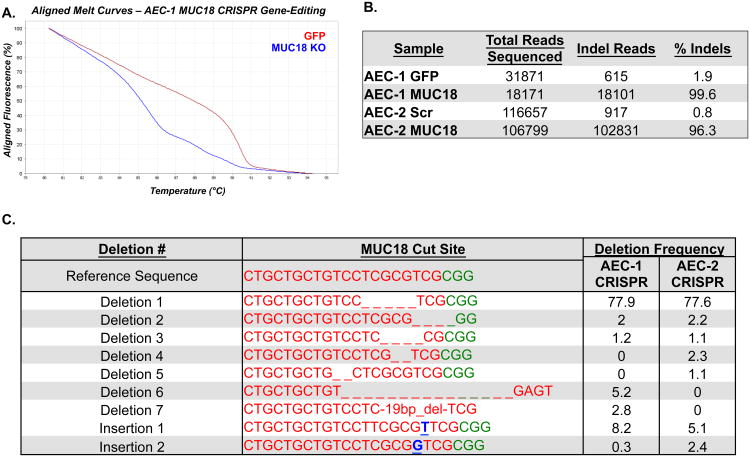
Passage 3 primary human nasal AECs (AEC-1) were infected in standard growth conditions, with addition of a ROCK inhibitor. Transduction efficiencies were determined using a GFP-expressing control virus and were near 100%. Due to the limited proliferative capacity of primary AECs and the need to select the infected cell population, the cells were transitioned into modified Schlegel culture conditions.17 This culture method involves the growth of epithelial cells on an irradiated fibroblast feeder layer, with specialized media additives, and a ROCK inhibitor. Several studies have revealed this method allows near unlimited proliferation and passage of several epithelial cell types without transformation and loss of primary characteristics.17, 18 We generated puromycin-resistant fibroblasts to allow puromycin selection of AECs without disruption of the feeder layer. Cells were seeded at low density (1-3.2× 104/100mm dish) throughout culture to aide in selection. In total, puromycin selection was applied across 5 AEC passages (39 days in this experiment), which based on experiments using this system in cell lines4 should have been adequate time for selection and CRISPR-mediated cutting of the MUC18 target site (Supplemental Fig. 1). A PCR amplicon was designed across the MUC18 cut site to examine generation of indels. High resolution melt (HRM) analysis of PCR products from P8 GFP control and MUC18 CRISPR-Cas9 treated cells revealed significant differences in the melting behavior of CRISPR-Cas9 treated cell amplicons, characteristic of sequence differences in the CRISPR treated cells (Fig. 2A). In contrast melting behavior was highly similar between GFP control and MUC18 CRISPR-Cas9 treated cells for amplicons designed across the top four predicted off-target sites, indicating the gRNA was targeting specifically to the MUC18 exon (Supplemental Fig. 2). To determine the base composition of the sequence changes and quantify their occurrence in the selected cell populations, we generated sequencing libraries from the PCR products, which were examined by massively parallel sequencing technology. Strikingly, we found 99.6% of sequencing reads (from over 18, 101) examined from the selected MUC18 CRISPR-Cas9 cell population contained indels, versus only 1.9% of the selected GFP-control cell population reads (Fig. 2). These results indicate that only integrated cells survived selection, the CRISPR-Cas9 machinery had mediated cutting, and subsequent repair had occurred in nearly all cells in the batch-selected population. Consistent with the batch selection of cells and random nature of indel formation in non-homologous repair, we observed 5 different deletions and one insertion with a frequency of greater than 1% in the MUC18 CRISPR-Cas9 treated cells (Fig. 2). However, we did observe the majority of reads contained a 5-bp deletion, suggesting a bias in the repair process to favor generation of this deletion (Fig. 2).
Figure 2. Genomic characterization of MUC18 CRISPR-Cas9 treated and selected AECs.
PCR was used to amplify a sequence crossing the MUC18 cut site using genomic DNA isolated from GFP-virus infected, scrambled MUC18 CRISPR-Cas9 virus treated, and MUC18 CRISPR-Cas9 virus treated airway cells in two experiments (different donors). A. High resolution melt (HRM) analysis of PCR amplicons, revealing a significant melting curve shift for the amplicons derived from MUC18 CRISPR-Cas9 treated versus GFP-virus treated AEC-1 cells. B. Results of Next-Generation sequencing analysis of the same PCR amplicons for indels over the MUC18 cut site, revealing the presence of indels in nearly all reads originating from MUC18 CRISPR-Cas9 treated AECs from the two experiments. C. We list the most common indels detected across the MUC18 CRISPR-Cas9 cut site from the two experiments.
Optimized Generation of MUC18 KO Nasal Airway Epithelial Cells
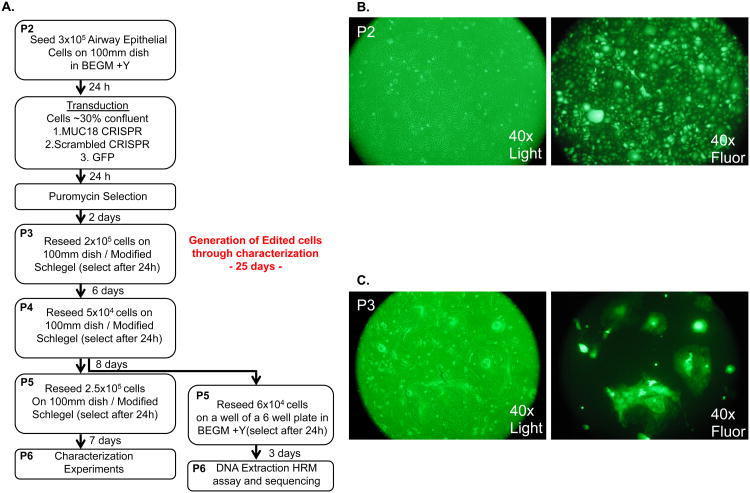
We attempted to repeat MUC18 gene editing in nasal epithelial cells from a new donor (AEC-2) and also to reduce the time and passages necessary to generate the edited cell population (Fig. 3A). In this experiment we also infected cells using a lentivirus carrying the CRISPR machinery with a scrambled MUC18 gRNA sequence, to generate a suitable control cell population for comparison in the characterization experiments of the MUC18 KO cells. Cells from this donor were infected at passage 2, and during selection passages cells were seeded at increased density (0.5-2.5× 105/100mm dish) relative to the first experiment to decrease the time between passages (Fig. 3A). Again high efficiency of transduction was observed by infection of cells with GFP expressing lentivirus (Fig. 3B-C). Editing frequency was determined after each passage to establish when maximum editing frequency was reached. After a single passage MUC18 CRISPR-Cas9 treated cells displayed indels in 62% of sequenced reads (P3 cells), which increased to 95.6% of reads after an additional passage (P4 cells). The frequency of indels detected was 96.9% and 96.2%for P5 and P6 cells, respectively (Fig. 2). Based on this stabilization of edited reads at 96% we suggest that all cells are likely edited at this point and the remaining % of unedited reads may derive from low-level feeder cell contamination. Comparison of HRM amplicons from scrambled gRNA control and the MUC18 gRNA treated cells indicated no editing of off-target sites (Supplementary Fig. 3). Similar to the first donor we observed a mosaic of indels generated at the MUC18 target site, with 77% of edited reads containing the same 5bp deletion favored in the AEC-1 donor (Fig. 2). These results show batch population editing of nearly all cells can be achieved with these methods using only 2 passages and 8 days of selection (Fig. 3).
Figure 3. Optimized experimental workflow to generate MUC18 knockout primary airway epithelial cells through lentiviral CRISPR-Cas9 treatment.
A. We show the time course and culture conditions for generation of AEC-2 MUC18 knockout cells. Reseeding time refers to the MUC18 CRISPR treated cultures, but approximates growth times in scrambled MUC18 CRISPR-Cas9 control cells as well. Note in an extra effort to eliminate potential fibroblast contamination from altering sequencing results, the cells were passed a last time on collagen coated plates in BEGM before DNA isolation. B. Microscopy pictures of P2 cells immediately before passage, revealing high transduction efficiency. C. Microscopy pictures of P3 cells immediately before passage, revealing all epithelial colonies are GFP positive. Note cells are present in colonies because they are being grown with Schlegel culture methods. Also note green tint in the light microscopy picture is due to the GFP filter.
Characterization of MUC18 mRNA / Protein Expression in the MUC18 edited AECs
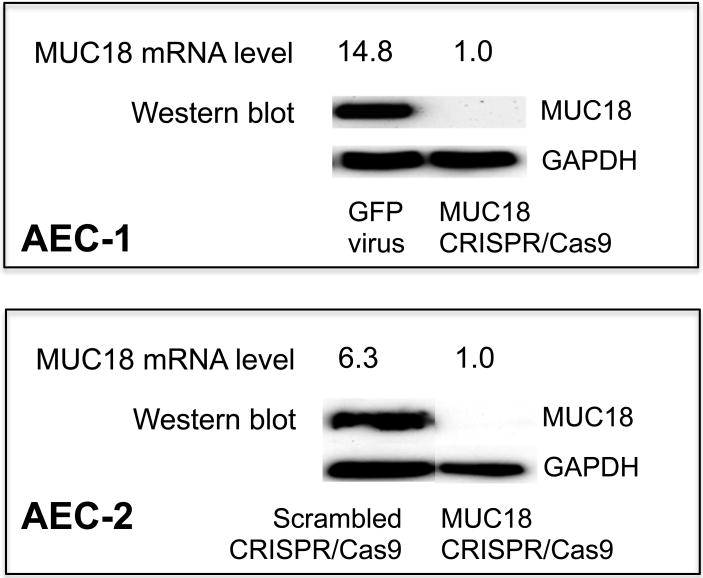
Examining MUC18 expression in both donors by qPCR, we found 93% and 84%lower mRNA levels for the AEC-1 and AEC-2 donors, respectively, when comparing MUC18 CRISPR-treated vs control virus-treated cells (Fig. 4). These results indicate the genomic edits interfere with either expression or stability of the edited transcripts. MUC18 protein expression was determined by western blot and revealed no detectable MUC18 protein in CRISPR-Cas9 treated and selected cells from either donor, in contrast to control virus infected and selected cells, which had robust MUC18 expression (Fig. 4). These results show the MUC18 CRISPR-Cas9 treated and selected primary cell populations are completely deficient in MUC18 protein, results consistent with our sequence analysis of these cell populations.
Figure 4. MUC18 mRNA and Protein Expression in MUC18 CRISPR-Cas9 treated AECs.
The two MUC18 knockout cell populations (AEC-1 and AEC-2) were cultured under submerged conditions without any TLR agonist stimulation. Cells were harvested for detection of MUC18 mRNA by real-time PCR and MUC18 protein by Western blot.
Effects of MUC18 knockout on pro-inflammatory cytokine production in Toll-like Receptor agonist-stimulated AECs
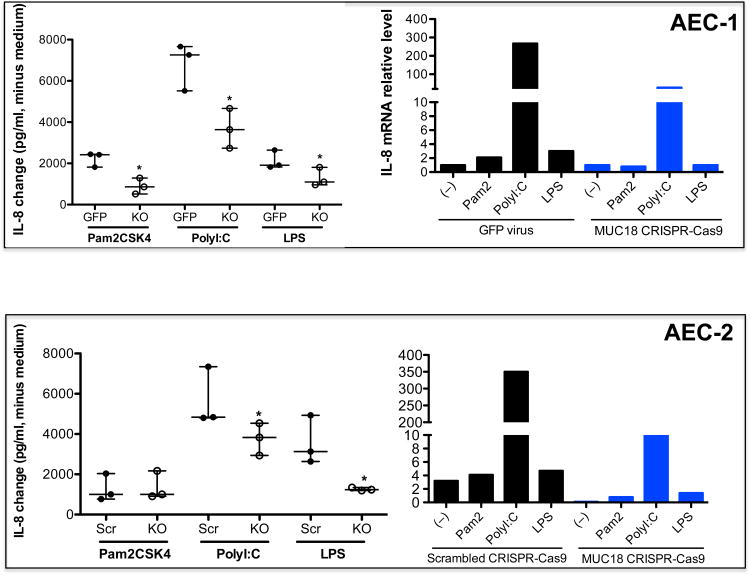
The function of MUC18 in airway epithelial cells remains unclear. In our previous studies in mouse lung macrophages, we found that MUC18 exerts a pro-inflammatory function by promoting TNF-α production.16 IL-8 is one of the major pro-inflammatory cytokines in the airways. Therefore, we determined if MUC18 knockout affects airway epithelial cell IL-8 responses to stimulation by the agonists of three major toll-like receptors (TLR). These agonists mimic microbial components of bacteria and viruses. Importantly, we observed that production of IL-8 protein by agonists of TLR2 (Pam2CSK4), TLR3 (polyI:C) and TLR4 (LPS) was significantly dampened in MUC18 knockout cells as compared to the GFP control cells, in donor AEC-1 (Fig. 5). These results were consistent with IL-8 mRNA expression (Fig. 5). MUC18 knockout cells from the second donor (AEC-2) had a similar decrement in IL-8 protein response for polyI:C and LPS stimulation, as compared to the cells expressing the scrambled MUC18 CRISPR-Cas9 (Fig. 5). IL-8 mRNA expression response to all agonists was reduced in the MUC18 AEC-2 knockout cells (Fig. 5).
Figure 5. Pro-inflammatory response of MUC18 knockout AECs to TLR agonist stimulation.
The two MUC18 knockout cell populations (AEC-1 and AEC-2) were incubated in triplicate with PBS (medium control) or agonists of TLR2 (Pam2CSK4), TLR3 (polyI:C) and TLR4 (LPS) for 24 hours. Cell supernatants were harvested for detection of IL-8 protein by ELISA (n = 3 replicates for each condition) and IL-8 mRNA by real-time PCR (n=1). IL-8 protein data (median/interquartile range) are expressed as changes of IL-8 in TLR agonist stimulated cells minus IL-8 in medium control cells. *, p< 0.05 comparing KO supernatants to GFP virus or scrambled MUC18 CRISPR-Cas9. Src = scrambled MUC18 CRISPR-Cas9. Pam 2 = Pam2CSK4. KO = MUC18 CRISPR-Cas9.
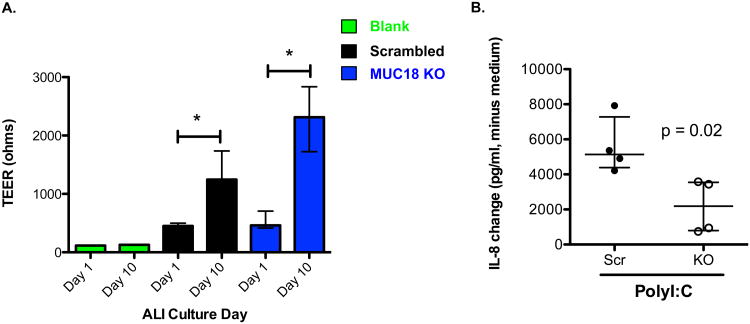
We were able to form polarized air-liquid interface (ALI) cultures from these MUC18 knockout cells. We confirmed cell polarization by Transepithelial electrical resistance (TEER) measurements (Fig. 6A). These ALI cultures were used to test the TLR3 agonist response. As shown in Fig. 6B when MUC18 knockout cells (AEC-2) were stimulated with polyI:C they produced significantly lower levels of IL-8 than the control cells (scrambled CRISPR), consistent with submerged culture results. These results strongly suggest a pro-inflammatory function of MUC18 in airway epithelial cells in the context of both bacterial and viral lung infections.
Figure 6. Air-liquid interface polarized MUC18 knockout AECs havedecreased IL-8 production in response to TLR3 agonist polyI:C.
A. MUC18 knockout and scrambled MUC18 CRISPR-Cas9 treated AECs were cultured at air-liquid interface (ALI) for 10 days. Transepithelial electrical resistance(TEER) measurements are shown before and after polarization. Bars represent median and interquartile range for 8 ALI inserts at two different time points. A single empty insert was used to produce the blank measurement at both time points. *, p<0.001. B. Polarized ALI day 10 cells were stimulated in triplicate with polyI:C for 24 hrs. IL-8 protein data (median/interquartile range) are expressed as changes of IL-8 in polyI:C stimulated cells minus IL-8 in medium control cells. Src = scrambled MUC18 CRISPR-Cas9 treated cells. KO = MUC18 knockout cells.
Discussion
The ability to specifically edit the genome of human cells would permit gene/sequence variant functional analyses and allow determination of their contribution to human disease pathology. The emergence of CRISPR-Cas9 technology with its efficiency of editing, lower cost, and open-source availability of reagents has seemingly put the goal of generating cell lines harboring gene knockouts within reach of most investigators. Lentiviral-delivered CRISPR-Cas9 technology has recently been used to successfully perform genome-wide gene knockout screens and correct genetic sequence defects.4, 5, 19 However, most of these studies were performed in cells with an enhanced capacity to proliferate, namely iPS cells or transformed cell lines. These studies are limited, in the case of transformed cells, by the degree to which the transformed cells represent the primary cells from which they were derived, and for iPS cells by the requirement to differentiate these cells into the specific cell type important to the disease being studied. Therefore, the application of editing methods to the various primary cell types currently being used to study cellular mechanisms of disease is highly desirable. Traditional limitations to primary cell editing include low transfection efficiency, limited proliferative capacity, and sensitivity to selection methods.
To overcome these limitations in primary human nasal airway epithelial cells, we applied recently developed culture methods to conditionally reprogram epithelial cells.17, 18 As previously reported, this reprogramming allows a large number of cell passages and extensive proliferation of primary epithelial cells, with retention of specific primary cell character. Importantly, this process does not involve cellular transformation or cancerous phenotypic changes (such as loss of contact inhibition and mutations).18 The application of these culture conditions resulted in low cell toxicity while achieving near complete lentiviral transduction, as has recently been reported in AECs.20 Additionally, these methods allowed us to sub-culture cells at low density in selective media for multiple passages, aiding in the generation of edited cells. Importantly, we demonstrated that gene knockout in primary cells can be maintained not only in un-differentiated cells, but can be achieved in polarized cells. Considering this conditional reprogramming method has been successful in multiple other epithelial cell types 17, 21, we believe the methods we outline here may be broadly applicable to various other primary cells. Supporting this CRISPR-Cas9 gene targeting with homology directed repair was recently used to correct the CFTR del508 mutation in intestinal stem cells, which were then grown into organoids9.
Our editing efficiency compares favorably to recent editing studies in other primary mammalian cells both in vitro and in vivo. Specifically, our second attempt at editing by this method resulted in 95.6% of cells edited after just 8 days of selection. In fact, this editing rate and selection timeframe is comparable to the 92% indel rate in 11 days of selection achieved in transformed HEK293T cells.4 Bone marrow derived dendritic cells have been edited in culture using lentiviral delivery of CRISPR machinery achieving 67-78% indel rate, 7 days posttransduction.6 Non-integrating adeno-associated viral (AAV) vectors have also recently seen great success in both an in vitro and in vivo settings in primary mouse neurons (65% idel rate in 7 days) and brain tissue in live mice (68% indel rate of transduced cells in 2 weeks).22 In vivo adenoviral delivery of CRISPRs to the mouse liver and subsequent indel generation was used successfully as well, achieving indel rates of 90% after 7 days.23 Additionally, an AAV9 virus was used to deliver sgRNA's to the in vivo lung for gene knockout and introduction of a mutation.6 A non-viral delivery method, hydrodynamic injections have similarly been used in mice for in vivo delivery of CRISPR and homologous plasmids to hepatocytes, resulting in the correction of a modeled human genetic disease.8 Our results combined with these indicate the CRISPR-Cas9 machinery is extremely efficient in primary cell genome targeting, and amenable to various viral and non-viral delivery methods.
We acknowledge several limitations to this approach. First, the batch selection and random nature of non-homologous double strand break repair and indel generation creates a mixed population of edited cells, some of which may express functional or altered functional protein. Therefore, multiple selected populations should be generated with verification of protein loss and agreement in functional consequences between selected populations. Secondly, the integration and continued expression of the CRISPR-Cas9 machinery may result in phenotypic changes unrelated to the gene knockout. Again, these problems are partially addressed by generation of multiple knockout cell populations. Alternative strategies to develop knockouts include “knockin-knockout's,”that is gene interruption by homologous insertion of a selection cassette, which would directly address this issue. The efficiency of CRISPR machinery delivery and cutting we achieved here, increase the likelihood this more complex strategy will be fruitful. Despite these limitations we believe our study represents a substantial initial step towards the realization of edited primary human cells.
Although the major focus of this report was on the successful application of CRISPR-Cas9 technology to knockout a gene in human primary airway epithelial cells, our results have provided information on MUC18 function. We selected the MUC18 gene for study as we recently found that MUC18 protein was increased in alveolar macrophages from both asthma and COPD patients.16 Moreover, study of MUC18 knockout mice indicated a pro-inflammatory function of MUC18, as KO mice displayed reduced inflammation and neutrophil recruitment in response to lung bacterial infections.16 To date, the functional relevance of airway epithelial MUC18 expression to inflammation in the lung remains unclear. Our MUC18 knockout of human AECs reveal this molecule has pro-inflammatory function under stimulation with various TLR agonists, mimicking bacterial and viral infections, and have brought these findings into the context of human cells. These results suggest the need for future studies using live respiratory bacteria or viruses to clearly demonstrate MUC18 function under these disease conditions. Given the importance of bacterial and viral infections in triggering asthma/COPD exacerbations and lung inflammation, MUC18 may play a significant role in these processes.
We also realize that further dissection of the molecular mechanisms whereby MUC18 exerts a broad spectrum of pro-inflammatory functions will be important. Nonetheless, in a recent paper by our group24, we determined the mechanism by which MUC18 amplifies the pro-inflammatory response to the TLR3 agonist polyI:C. Our previous data suggest that MUC18 serine phosphorylation particularly at the cytoplasmic tail is critical to MUC18 pro-inflammatory function in response to various TLR agonists. In response to polyI:C, increased phosphorylation of MUC18 serines was observed. Reduction of MUC18 serine phosphorylation by inhibiting ERK activity was associated with less production of IL-8 following polyI:C stimulation.
In summary, our results indicate epithelial-derived MUC18 contributes to airway inflammation in response to important microbial innate immunity triggers. Additionally, we suggest the methods outlined herein to achieve CRISPR-Cas9 mediated gene knockout can be applied to the study of other genes in primary human airway epithelial cells and potentially other primary cell types.
Material and Methods
Nasal brushing, cell recovery and expansion
Human nasal airway epithelial cells were collected from 2 healthy subjects, by brushing the region posterior to the inferior turbinate with a cell cytology brush using a nasal illuminator, as previously described.25 The epithelial cells from the brush were dispersed in PBS, washed twice, and cultured using a modified Schlegel method. 17, 18 Briefly, cells were cultured on a monolayer of irradiated NIH 3T3 fibroblasts in F-Media 18 plus 10 μM ROCK inhibitor (Y-27632). Double trypsinization was used to separate epithelial cells from fibroblasts at passage. Specifically, trypsin is added to the cultures and the fibroblasts typically release quickly, ∼30 seconds to a 1 minute. Complete release of fibroblasts is monitored by microscope. The epithelial colonies are then washed and trypsinized a second time to remove epithelial colonies. This trypsinization is longer; it takes 5-7 minutes depending on the size of the colonies. The study was approved by the institutional review board at National Jewish Health. The two study subjects provided written informed consent.
Design of MUC18 gRNA and lentiviral CRISPR-Cas9 vector
We used the lentiviral expressing CRISPR-Cas9 vector generated by the Zhang lab, plentiCRISPRv1.4 This one vector system expresses the gRNA, Cas9 protein, and puromycin resistance gene from one virus. The MUC18 gRNA was designed using the Zhang lab software available at http://crispr.mit.edu. The gRNA sequence used was 5′CTGCTGCTGTCCTCGCGTCG3′. According to Zhang lab protocols (http://www.genome-engineering.org/crispr/?page_id=23), DNA oligonucleotides for the gRNA and reverse complement sequence plus adapters needed for ligation were synthesized and cloned into the plentiCRISPRv1 vector (Forward oligo, 5′CACCGCTGCTGCTGTCCTCGCGTCG3′; Reverse oligo, 5′AAACCGACGCGAGGACAGCAGCAGC3′). We also scrambled the MUC18 gRNA sequence and cloned this ligated oligo (Forward oligo, 5′CACCGCGTGCTCCGTTCGCGCTTC3′; 5′AAACGAAGCGCGAACGGAGCACGC3′) into the same vector to serve as a control for characterization experiments. Correct cloning and sequence was confirmed by capillary resequencing of the plentiCRISPRv1 constructs.
Generation of MUC18 CRISPR-Cas9 and GFP lentiviral particles
Lentivirus particles were generated in the Lenti-X 293T packaging cell line by transfecting the psPAX2 packaging plasmid (Addgene Plasmid #12260), the pCMV-VSV-G pseudotyping plasmid (Addgene Plasmid #8454), and either the plenty CRISPRv1-MUC18, plentiCRISPRv1-MUC18_Scr or pLJM1-EGFP (Addgene Plasmid #19319)viral vector plasmids, to produce MUC18 CRISPR-Cas9, MUC18_Scr CRISPR-Cas9, and GFP expressing viruses, respectively. Transfections were performed with liptofectamine 2000 in 100mm dishes using 7μg of the viral vector, 9μg of the viral packaging plasmid, and 0.9 μg of the pseudotyping vector. Virus containing media was collected 72 hours later and centrifuged. Adequate titer was determined by positive Lenti-X GoStix test, which indicates a titer >5× 105 IFU/ml.The pLJM1-EGFP viral vector also expresses a puromycin resistance gene.
Viral transduction and selection of AECs
Nasal AECs were passaged twice using the modified Schlegel culture method17, 18. Prior to transduction, the nasal AECs were sub-cultured on collagen-coated 100mm dishes in BEGM growth media (CC-4175, Lonza) plus 10 μ M ROCK inhibitor for one to two days. Cells were then infected with either GFP, MUC18 CRISPR-Cas9, MUC18_Scr CRISPR-Cas9 lentiviral media (viral media diluted in BEGM media (total volume 8ml) plus 10 μ M ROCK inhibitor, 16 mM HEPES buffer, and 12 μg/ml polybrene) by spin infection (920 g at 25 degrees C) for 1 hour. At 24 hours post-infection puromycin (1 μg/ml final concentration) was added to the culture media. In parallel we transduced 3T3 fibroblasts with the empty plentyCRISPR vector and selected these cells (1 μg/ml puromycin) to generate feeder cells resistant to puromycin. The viral transduced nasal AECs were grown to 80-90% confluence and passaged using the modified Schlegel culture method and the puromycin resistant 3T3 cells. These modified Schlegel culture conditions were maintained throughout the selection period to generate transduced GFP expressing, MUC18 scrambled gRNA expressing, and MUC18 KO cells, as described in Figure 3 and Supplementary Figure 1.
High resolution melt (HRM) analysis of MUC18 CRISPR-Cas9 target site and potential off target site amplicons
PCR primers were designed to specifically amplify the chromosome 11 genome region 119187608-119187798 that spans the MUC18 CRISPR-Cas9 cut site (primer sequences listed below). The same CRISPR gRNA design sequence provides a list of the top potential off-target sites for the gRNA that it designs. We designed primers to amplify the regions containing the overall top three off-target sites and top two genic off-target sites (4 in total) for cutting by HRM assay. The off-target locations and primers to amplify these regions are listed in supplementary table 1 and 2. These primers were used to perform a PCR reaction and subsequent HRM analysis of the amplicons using the MeltDoctor HRM Master Mix (Life Tech, Part #4415440) and standard protocol. PCR reaction and HRM analysis were performed on a Life Technologies QuantStudio6 quantitative PCR instrument.
Sequence analysis of MUC18 CRISPR cut site
Library generation and sequencing
We generated Ion Torrent PCR amplicon libraries using a fusion method (Ion Torrent User Guide 468326 Rev. B). Namely, the primers used to perform HRM analysis were used to amplify the MUC18 CRISPR-Cas9 cut site. The forward primer contains a 5′ universal adapter sequence followed by the MUC18 priming sequence 5′CTGCTGT-ACGCAGCGTGGGGATCGGGGACCCAGGGAGGAGGC3′. The reverse primer contains the 5′ trP1 sequencing adapter followed by the MUC18 priming sequence CCTCTCTATGGGCAGTCGGTGATGGCTGGTCTGCGCCTTCTTGCTCG. This primary PCR reaction was performed with Pyromark PCR reagents (Qiagen, Cat#978703) according to standard protocols. These PCR products were used to template a secondary PCR reaction, which introduces the A-sequencing adapter and barcodes specific to each sample through the forward primer, 5′CCATCTCATCCCTGCGTGTCTCCGACTCAGX-XXXXXXXXXCTGCTGTACGCAGCGT3′ (X= Ion Torrent barcode sequences), and maintains the trP1 sequencing adapter with a trP1 reverse primer, 5′CCTCTCTA-TGGGCAGTCGGTGAT3′. Secondary PCR reactions were completed with Phusion PCR reagents using HF buffer (New England Biolabs, Cat#M0530). Secondary PCR products representing the sequencing libraries were gel-purified and combined in equi-molar amounts to form the sequencing library pool. Standard Ion Torrent templating of Ion Spheres was completed followed by sequencing on a 314 chip with the Ion Torrent Personal Genome Machine (PGM).
Bioinformatic analysis of sequencing reads
Sequencing reads were mapped to the PCR amplicon using the default settings of the Ion Torrent mapper, TMAP. We interrogated the cut site (3-4 bp upstream of the gRNA PAM sequence) within sequencing reads for insertions and deletions (indels). The boundaries up- and down-stream of the cut site defining the area within which indels were counted, were determined by the last amplicon position which exhibited an indel frequency of at least 0.5% in the CRISPR-Cas9 treated sample. Additionally, only indels of at least 2 bp in length were considered for this purpose, in order to eliminate possible sequencing error artifacts. The total indel rate for each sample was estimated as the number of reads harboring any >=2 bp long indel within the defined cut site divided by the total number of reads spanning the entire cut site. All the described calculations were performed using custom written Perl scripts using mapped sam files as input, and confirmed by visual inspection using the Integrative Genomic Viewer.26 Using the above described criteria, in both samples we observed a significant number of reads (∼8%) which were not scored as containing an indel. Investigating these reads we discovered that the majority harbored one of two 1bp insertions at exactly the same positions in close proximity to the expected cut site. Frequency of these particular 1 bp insertions in the control samples were negligible; therefore we believe this event to be a direct effect of the CRISPR treatment. Therefore indel frequencies listed in Figure 2 reflect this insertion.
Gene and protein expression analysis of MUC18 CRISPR-Cas9 treated AECs
MUC18 and IL-8 mRNA in epithelial cells was determined by real-time quantitative PCR (qPCR) and analyzed using the comparative threshold cycle method by normalizing to the housekeeping gene GAPDH. Western blots were used to examine epithelial MUC18 protein (Epitomics, Burlingame, CA, Catalog #2505-1). Densitometry was performed to quantify the protein in relation to GAPDH. IL-8 protein levels in culture supernatants were determined by using the IL-8 enzyme-linked immunosorbent assay (ELISA) DuoSet development kit (R&D Systems, Minneapolis, MN, Cat# D8000C).
TLR agonist treatment of submerged Nasal AECs
MUC18 knockout, GFP control, and MUC18 scrambled CRISPR-Cas9 nasal AECs were seeded in collagen-coated 12-well plates at 2×105 cells/well in standard BEGM media on collagen. After the cells reached 80 to 90% confluence, they were treated with agonists of TLR2 (Pam2CSK4, 100 ng/ml, InvivoGen), TLR3 (polyI:C, 2.5 μg/ml, InvivoGen) and LPS (10 μg/ml, Sigma). After 24 hrs of TLR agonist stimulation or cell culture medium alone (control), supernatants were harvested for measurement of the pro-inflammatory cytokine, IL-8. Cells were collected for Western bot of MUC18 protein, and for real-time qPCR to measure MUC18 as well as IL8 mRNA levels.
Air-liquid interface growth and TLR agonist treatment of Nasal AECs
MUC18 knockout and MUC18 scrambled CRISPR/Cas-9 passage 7 nasal AECs were grown on air liquid interface as described in Karp et al.27 Specifically, 6.5mm inserts (Costar 3470) were pre-coated with 2.7 mg/ml collagen (Advanced BiomatrixPurecol Bovine Collagen #5005-B). Knockout and MUC18 scrambled cells were plated in airway medium 127 on coated inserts at 1.2×105 cells per insert. At 24 hours post-plating airway media 1 was replaced with airway medium 227 (containing 2% Ultroser G). Air-lift (removal of apical media) was performed 72 hours post-plating. Cells were maintained air-lifted for 10 days. Transepithelial electrical resistance (TEER) was determined using the Millicell ERS-2 Epithelial Volt-Ohm Meter (Millipore). Day 10 ALI cultures were stimulated at both apical and basolateral sides with polyI:C, (2.5 μg/ml) for 24 hrs. The combined supernatants from both apical and basolateral sides were collected for measuring IL-8 protein.
Statistical Analysis
Comparison of KO and control cells was performed by Wilcoxon signed rank test for IL-8 protein data and Mann-Whitney test for TEER measurements. Data are presented as medians ± interquartile ranges. Sample sizes were not chosen based on a pre-specified effect, as this was an exploratory analysis. A P Value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by R01 AI106287 and R01 HL122321 to HWC and NJH startup funds to MAS.
Footnotes
Supplementary information is available at Gene Therapy's website.
Conflicts of Interest: The authors have no conflicts of interest to disclose.
References
- 1.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428(6981):431–7. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 2.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. Rna. 2006;12(7):1179–87. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–4. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–3. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature medicine. 2013;19(7):939–45. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 10.Jean D, Gershenwald JE, Huang S, Luca M, Hudson MJ, Tainsky MA, et al. Loss of AP-2 results in up-regulation of MCAM/MUC18 and an increase in tumor growth and metastasis of human melanoma cells. J Biol Chem. 1998;273(26):16501–8. doi: 10.1074/jbc.273.26.16501. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann JM, Riethmuller G, Johnson JP. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(24):9891–5. doi: 10.1073/pnas.86.24.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie S, Luca M, Huang S, Gutman M, Reich R, Johnson JP, et al. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 1997;57(11):2295–303. [PubMed] [Google Scholar]
- 13.Sers C, Kirsch K, Rothbacher U, Riethmuller G, Johnson JP. Genomic organization of the melanoma-associated glycoprotein MUC18: implications for the evolution of the immunoglobulin domains. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(18):8514–8. doi: 10.1073/pnas.90.18.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson JP, Bar-Eli M, Jansen B, Markhof E. Melanoma progression-associated glycoprotein MUC18/MCAM mediates homotypic cell adhesion through interaction with a heterophilic ligand. Int J Cancer. 1997;73(5):769–74. doi: 10.1002/(sici)1097-0215(19971127)73:5<769::aid-ijc26>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Simon GC, Martin RJ, Smith S, Thaikoottathil J, Bowler RP, Barenkamp SJ, et al. Up-regulation of MUC18 in airway epithelial cells by IL-13: implications in bacterial adherence. American journal of respiratory cell and molecular biology. 2011;44(5):606–13. doi: 10.1165/rcmb.2010-0384OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Case SR, Minor MN, Jiang D, Martin RJ, Bowler RP, et al. A novel function of MUC18: amplification of lung inflammation during bacterial infection. The American journal of pathology. 2013;182(3):819–27. doi: 10.1016/j.ajpath.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. The American journal of pathology. 2012;180(2):599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suprynowicz FA, Upadhyay G, Krawczyk E, Kramer SC, Hebert JD, Liu X, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(49):20035–40. doi: 10.1073/pnas.1213241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, et al. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome research. 2014;24(9):1526–33. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horani A, Nath A, Wasserman MG, Huang T, Brody SL. Rho-associated protein kinase inhibition enhances airway epithelial Basal-cell proliferation and lentivirus transduction. American journal of respiratory cell and molecular biology. 2013;49(3):341–7. doi: 10.1165/rcmb.2013-0046TE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. The Journal of clinical investigation. 2010;120(7):2619–26. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–6. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng R, Peng J, Yan Y, Cao P, Wang J, Qiu C, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588(21):3954–8. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Berman R, Huang C, Jiang D, Finigan JH, Wu Q, Chu HW. MUC18 Differentially Regulates Pro-Inflammatory and Anti-Viral Responses in Human Airway Epithelial Cells. Journal of clinical & cellular immunology. 2014;5(5) doi: 10.4172/2155-9899.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O'Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. The Journal of allergy and clinical immunology. 2014;133(3):670–8 e12. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14(2):178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, et al. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol. 2002;188:115–37. doi: 10.1385/1-59259-185-X:115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.