Key Points
AAV- and ZFN-mediated targeting of the albumin locus corrects disease phenotype in mouse models of hemophilia A and B.
Robust expression from the albumin locus provides a versatile platform for liver-directed protein replacement therapy.
Abstract
Site-specific genome editing provides a promising approach for achieving long-term, stable therapeutic gene expression. Genome editing has been successfully applied in a variety of preclinical models, generally focused on targeting the diseased locus itself; however, limited targeting efficiency or insufficient expression from the endogenous promoter may impede the translation of these approaches, particularly if the desired editing event does not confer a selective growth advantage. Here we report a general strategy for liver-directed protein replacement therapies that addresses these issues: zinc finger nuclease (ZFN) –mediated site-specific integration of therapeutic transgenes within the albumin gene. By using adeno-associated viral (AAV) vector delivery in vivo, we achieved long-term expression of human factors VIII and IX (hFVIII and hFIX) in mouse models of hemophilia A and B at therapeutic levels. By using the same targeting reagents in wild-type mice, lysosomal enzymes were expressed that are deficient in Fabry and Gaucher diseases and in Hurler and Hunter syndromes. The establishment of a universal nuclease-based platform for secreted protein production would represent a critical advance in the development of safe, permanent, and functional cures for diverse genetic and nongenetic diseases.
Introduction
Adeno-associated viral (AAV) vectors are showing great promise in clinical trials for delivering therapeutic genes to treat monogenic disorders.1,2 For gene transfer to the liver, the standard approach is to deliver an expression cassette that persists primarily in the form of extrachromosomal episomes. Episomal expression faces 2 major limitations: (1) the dilution of expression in proliferating cells, and (2) the restricted packaging capacity of the AAV vector. Site-specific integration of a corrective donor cassette into the genome allows therapeutic gene expression to persist through cell divisions and increases the effective carrying capacity of the vector by obviating the need for enhancer and/or promoter elements within the exogenous donor.
Genome editing has been successfully applied in a variety of preclinical models, both ex vivo and in vivo.3-7 Historically, the efficiency of gene-specific editing in mammalian cells has been very low, which limits its therapeutic potential. The targeting process is known to be greatly enhanced (100- to 1000-fold) by the induction of a DNA double-strand break at the target site.8,9 The development of customized DNA-cleaving enzymes, such as zinc finger nucleases (ZFNs), has made it possible to achieve far greater genome editing efficiencies. ZFNs function as dimers by coupling DNA binding motifs from transcription factors with the FokI endonuclease domain to generate double-stand breaks at their target site.
A typical genome editing approach is to target the disease locus itself; however, the proportion of alleles successfully edited may not express sufficient levels of protein to alleviate the disease phenotype. Alternatively, integration into a locus with high transcriptional activity (safe harbor) would address this limitation and provide a versatile platform for expressing various proteins, substituting the donor for each respective therapeutic transgene.
For our studies, serum albumin was chosen as the genomic safe harbor because of its very high expression level and the tractability of liver for gene delivery and in vivo editing relative to other tissues. In addition, the albumin gene structure is well suited for transgene targeting into intronic sequences because its first exon encodes a secretory peptide that is cleaved from the final protein product. By analogy to our previous work on human factor 9 (hF9),7,10 we reasoned that integration of a promoterless cassette bearing a splice acceptor and therapeutic transgene would support expression and secretion of many different proteins, because signal peptides are often functionally interchangeable.11,12
Materials and methods
Animal experiments
AAV vector was diluted to 200 µL with phosphate-buffered saline plus 0.001% Pluronic F68 before being injected into the tail vein. Plasma for human factor IX (hFIX) enzyme-linked immunosorbent assay (ELISA) was obtained by retro-orbital bleeding into heparinized capillary tubes. Plasma for activated partial thromboplastin time (aPTT) was obtained by tail bleeding into 3.8% sodium citrate at a 9:1 ratio. Tissue for nucleic acid analysis was immediately frozen on dry ice after necropsy. Wild-type C57BL/6 mice were purchased from The Jackson Laboratory. Hemophilia B and hemophilia A/CD4 null mice have been previously described.10,13 Minimum sample sizes were determined by estimating mean values and standard deviations based on previous in vivo ZFN studies that used an α of .05 and a power of .80. Randomization and blinding were not conducted.
ZFN reagents and targeting vectors
Heterodimeric ZFNs targeting the mouse albumin (mAlb) locus contain the ELD:KKR mutations14 in the FokI domain (Figures 1-3; sequences provided in supplemental Figure 6, available at the Blood Web site). The experiment described in Figure 4 uses a modified pair of ZFNs that retain the same target sequence within mAlb and similar cleavage efficiency (48641/31523 ZFNs; supplemental Figure 6). The promoterless hF9 donor vector containing a complementary DNA (cDNA) cassette with exons 2 to 8 of the hF9 gene has been previously described.10 All the hF8 donors carry a splice acceptor sequence derived from the hF9 gene followed by a B-domain-deleted F8 cDNA (devoid of the first 57 nucleotides encoding the signal peptide). hF8 donor 1 contains the SQ amino acid sequence15 and was codon optimized by DNA2.0. hF8 donor 2 was codon optimized and contains a putative glycosylation sequence and a shorter polyA sequence as described16 (summarized in supplemental Figure 4). hF9 and hF8 donors do not contain arms of homology to the target site. The donors used in Figure 3 carried an hF9 splice acceptor sequence followed by the cDNA encoding for α-galactosidase A, acid β-glucosidase (GBA), α-l-iduronidase, and iduronate-2-sulfatase. These donors contain arms of homology of approximately 600 bp to the mouse albumin target site.
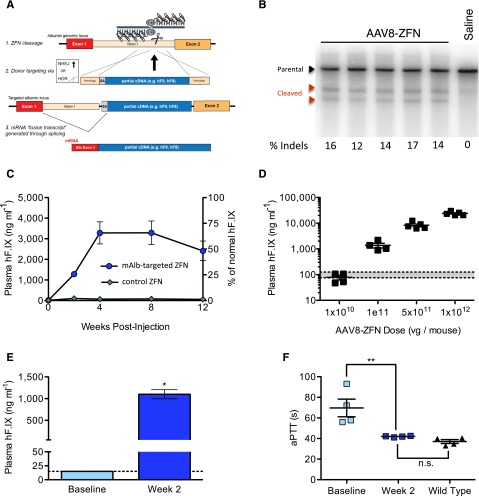
Figure 1.
Hepatic gene targeting of the mouse albumin locus results in phenotypic correction of hemophilia B. (A) Schematic illustrating albumin targeting strategy. (B) Cel I nuclease assay from liver DNA measuring ZFN-induced indels within albumin intron 1. Lanes represent individual mice at day 7 after AAV8-ZFN treatment. (C) hFIX in mouse plasma after treatment with AAV8-hF9-donor and either AAV8-ZFN (blue circles) or AAV8-hF9-ZFN (green diamonds) with a target sequence not present in the mouse genome; n = 3 mice per group. (D) hFIX levels at week 2 after treatment are proportional to AAV dose (1:5 ZFN to donor). Gray bar: normal levels. Points represent individual mice. (E) hFIX levels in hemophilia B mice 2 weeks after treatment with AAV8-mAlb-ZFN and AAV8-hF9-donor (n = 4 mice per group). *P = .029, Fisher's exact test. (F) Clot formation in mice depicted in panel E, measured by aPTT prior to and 2 weeks after treatment. The aPTTs of wild-type mice are shown for comparison. **P < .01, 2-tailed Mann-Whitney test. HDR, homology directed repair; n.s., nonsignificant; SA, splice acceptor.
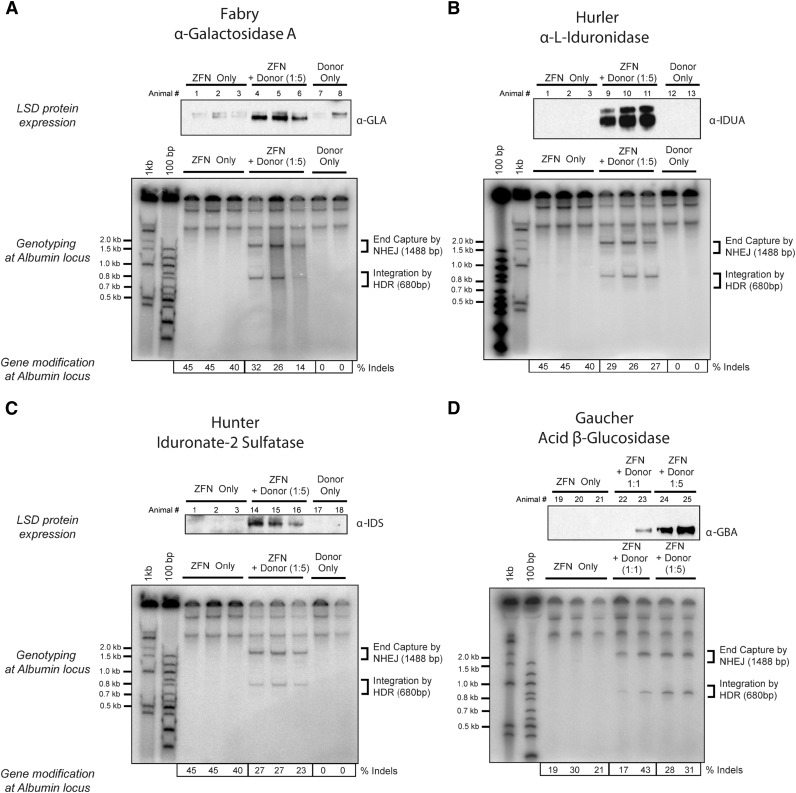
Figure 3.
Expression of lysosomal enzymes deficient in Fabry and Gaucher diseases and Hurler and Hunter syndromes. Top panels of (A-D) Western blot detection of (A) α-galactosidase A, (B) α-l-iduronidase, (C) iduronate-2 sulfatase, and (D) acid β-glucosidase in liver lysates of mice 30 days after treatment with 3 × 1011 vg of AAV8-ZFN and AAV8 of the appropriate donor at the indicated ratio of 1:1 or 1:5 (see “Methods” section for details). Middle panels (A-D) PCR detection of bands consistent with homology directed (HDR) and homology independent (NHEJ) integration of donor at the albumin locus. Lower panels (A-D) Indel formation as measured by MiSeq sequencing (n = 3 mice per group). Each lane represents an individual mouse. LSD, lysosomal storage disease.
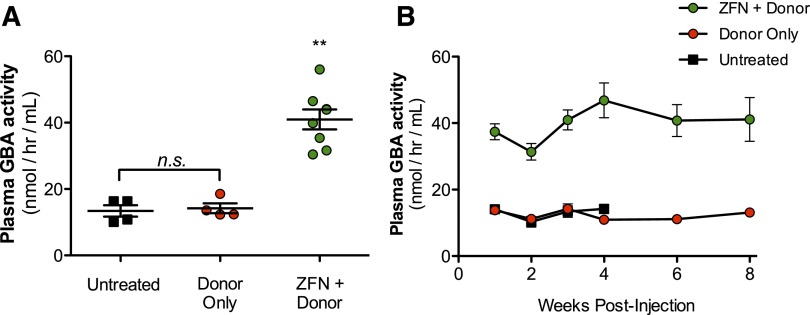
Figure 4.
Targeting of albumin locus promotes stable supraphysiological activity of GBA in mouse plasma. (A) GBA activity as determined by enzymatic activity assay in wild-type mice 3 weeks after treatment with either 1.2 × 1012 vg AAV8-GBA donor alone (red circle) or in combination with 1.5 × 1011 vg of each individual AAV8-ZFN (green circle). Untreated wild-type mice (▪) shown as controls. (B) Time course of GBA activity in the mice treated in (A). **P < .01, 2-tailed Mann-Whitney test comparing ZFN + donor group to untreated or donor only groups.
AAV vectors used in Figures 1 and 2 and supplemental Figures 2B-D and 3 were produced and titered as previously described.17,18 Experiments in Figures 3 and 4, and in supplemental Figures 1, 2A, and 5 were conducted by using AAV vectors prepared by triple-transfection of 293 cells in 10-layer CellSTACK chambers (Corning). After 3 days, cells were lysed and recombinant AAV (rAAV) was purified by a single cesium chloride gradient followed by dialysis. The titer of these vectors was measured by quantitative polymerase chain reaction (qPCR) (forward primer 5′-GTTGCCAGCCATCTGTTGTTT, reverse primer 5′-GACAGTGGGAGTGGCACCTT, and probe 5′-CTCCCCCGTGCCTTCCTTGACC). It appeared that 1 × 1011 vector genomes (vg) as determined by silver staining is equivalent to 3 × 1011 vg as determined by qPCR.
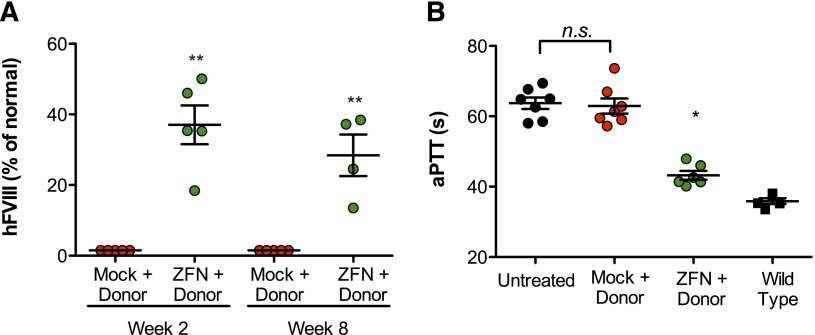
Figure 2.
Targeting albumin supports production of therapeutic levels of FVIII and functional correction of hemophilia A phenotype. (A) FVIII activity as determined by chromogenic assay in hemophilia A/CD4-deficient mice 2 and 8 weeks after treatment with 1 × 1011 vg of AAV8-mock (red circles), 5 × 1010 vg of each individual AAV8-ZFN (green circles), and 1 × 1011 vg of donor 2 (see “Methods” section for details). **P = .008, Fisher's exact test. (B) Measurement of clot formation by aPTT prior to and 11 weeks after AAV administration. The aPTT of wild-type (▪) and untreated (●) mice are shown for comparison. **P < .01, 2-tailed Mann-Whitney test.
FIX antigen and activity
An hFIX ELISA kit (Affinity Biologicals) was used to quantify plasma hFIX. All readings below the last value of the standard curve (15 ng/mL) were arbitrarily given the value of 15 ng/mL. hFIX activity levels in citrated mouse plasma were determined by using a chromogenic assay (A221802; Aniara).
hFVIII activity and aPTT assay
Coagulant activity of human factor VIII (hFVIII) in citrated mouse plasma was determined by Coatest SP4 FVIII (Chromogenix). The aPTT assay was performed by mixing sample plasma 1:1:1 with pooled hemophilia B (Figure 1F) or hemophilia A (Figures 2 and 4B) human plasma (George King Biomedical, Inc.) and aPTT reagent (Trinity Biotech) followed by a 180-second incubation period at 37°C. Coagulation was initiated after the addition of 25 mM calcium chloride. Time to clot formation was measured by using a STart 4 coagulation instrument (Diagnostica Stago).
Western blot analysis of liver homogenates
Western blot detection from liver homogenates was carried out 30 days after treatment. Highly abundant proteins (albumin and immunoglobulin G) were depleted from liver homogenates in radioimmunoprecipitation assay buffer. Primary antibodies were α-l-iduronidase (MAB4119; 1:1000; R&D Systems), iduronate-2-sulfatase (MAB2449; 1:500; R&D Systems), α-galactosidase A (12078-R001; 1:1000; Sino Biological), and acid β-glucosidase (sc-100544; 1:500; Santa Cruz Biotechnology). Secondary antibodies were from Santa Cruz Biotechnology (mouse, sc-2005; rabbit, sc-2004).
Surveyor nuclease (Cel I) assay
Genomic DNA from mouse liver was isolated by using the MasterPure complete DNA purification kit (Epicentre Biotechnologies), and the assay was performed as described previously.7,14 Loci were amplified for 30 cycles (60°C annealing and 30-inch elongation at 68°C). The following primers were used to detect DNA cleavage at the albumin locus: 5′-CCTGCTCGACCATGCTATACT-3′ and 5′-CAGGCCTTTGAAATGTTGTT-3′.
RT-PCR and qPCR
Reverse-transcription PCR (RT-PCR) followed by qPCR was performed by using Ambion’s High Capacity RNA-to-cDNA and Fast SYBR Green Master Mix (Applied Biosystems) according to manufacturer’s instructions. The following primers were used: mAlb Fw1 (5′-tgggtaacctttctcctcctc-3′) with mAlb Rv (5′-gggaaaaggcaatcaggact-3′) and mAlb Fw2 (5′-gtctccggctctgctttttc-3′) with hF9 Rv (5′-caggattttgttggcgtttt-3′). Cycling conditions were 95°C for 2 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. To quantify the relative levels of transcript expression, the threshold cycle (Ct) was determined by the 2-ΔΔCt method described by Livak.19
In vitro AAV transduction of primary human hepatocytes
Cell culture dishes (48-well; CM1048; Lifetech) were purchased precoated or plates (3548; VWR) were coated with a mixture of 250 µL BD Matrigel (BD Biosciences) in 10 mL hepatocyte basal medium (CC-3199; Lonza) at 150 µL per well. Plates were incubated for 1 hour at 37°. Thawing/plating media was prepared by combining 18 mL InVitroGRO CP medium (BioreclamationIVT) and 400 µL Torpedo antibiotic mix (Celsis In Vitro Technologies). Once the plates were prepared, the female plateable human hepatocytes (lot# AKB; cat# F00995-P) were transferred from the liquid nitrogen vapor phase directly into the 37° water bath. The vial was stirred gently until the cells were completely thawed. The cells were transferred directly into a 50-mL conical tube containing 5 mL of prewarmed thawing/plating medium. To transfer cells completely, the vial was washed with 1 mL of thawing/plating medium. The cells were resuspended by gently swirling the tube. A small aliquot (20 µL) was removed to perform a cell count and to determine cell viability by using trypan blue solution 1:5 (25-900-C1; Cellgro). The cells were then centrifuged at 75g for 5 minutes. The supernatant was decanted completely and the cells were resuspended at 1 × 106 cells/mL. The matrigel mixture was aspirated from the wells, and cells were seeded at 2 × 105 cells per well in a 48-well dish. Cells were then incubated in a 5% CO2 incubator at 37°C. At the time of transduction, cells were switched to hepatocyte culture medium (HCM) for maintenance (hepatocyte basal medium, CC-3199, Lonza; HCM, CC-4182, SingleQuots). AAV6 particles were diluted in HCM and added to cells at indicated multiplicities of infection. Transfection of mAlb ZFN messenger RNA (mRNA) was carried out with Lipofectamine RNAiMAX (Lifetech). After 24 hours, the medium was replaced by fresh HCM, which was done daily to ensure maximal health of the primary hepatocyte cultures. For experiments in which hFIX detection by ELISA was required, sometimes the medium was not exchanged for several days to allow hFIX to accumulate in the supernatants.
GBA enzymatic activity
Enzymatic activity of GBA in citrated mouse plasma was determined by mixing 50 μL of plasma diluted 5× or 10× in assay buffer (0.1 M citrate/0.2 M phosphate buffer [pH 5.4] containing 0.25% sodium taurocholate and 0.25% Triton X-100) with 50 μL of 10 mM synthetic substrate (4-methylumbelliferyl-β-d-glucopyranoside; Sigma-Aldrich) also dissolved in assay buffer and incubated for 2 hours at 37°C. Reactions were then terminated by the addition of 50 μL of 1 M glycine buffer (pH 12.5), and the amount of cleaved 4-methylumbelliferone (4-MU) produced was determined by measuring fluorescence using a SpectraMax Gemini XS fluorescent reader (Ex365/Em450; Molecular Devices) and interpolating on a 4-MU standard curve. Enzymatic activity is expressed as nmol of 4-MU produced per hour of assay incubation time per mL of plasma. Statistical analysis was performed by using a 2-tailed Mann-Whitney test. GBA activity increase in ZFN + donor group is significant for days 7 to 21 compared with untreated and donor only groups. For week 4 and later, only 2 mice remained in the untreated and donor only groups because of scheduled mouse euthanasia, leaving insufficient mice for statistical analysis.
Indel detection using next-generation sequencing
Loci were PCR amplified from genomic DNA, and the levels of modification were determined by paired-end deep sequencing on an Illumina MiSeq sequencing system. Paired sequences were merged via SeqPrep (jstjohn; https://github.com/jstjohn/SeqPrep). For the analysis of off-target activity, genomic DNA was isolated from mouse liver 30 days after transduction with mAlb ZFNs and GBA donor (animal 22; Figure 3D) or from a control mouse injected with formulation buffer. ZFN activity was determined by deep sequencing at either on-target (mouse albumin) or off-target (rank 1-40 off-target sites as predicted by the systematic evolution of ligands by exponential enrichment (SELEX) profile of mAlb ZFNs) sites for both the phosphate-buffered saline control and the ZFN + donor–treated animal. The statistical test described in Pattanayak et al20 was applied to quantify insertions and deletions (indels) by adjusting for MiSeq/PCR-induced indels (P < .05 was considered significant). Complete filter criteria are described in supplemental Data. Primers used to amplify genomic DNA for indel quantification (Table 1) are provided in supplemental Table 1.
Table 1.
mALB ZFN SELEX-based off-target analysis of in vivo mouse study on day 30
| Rank | Gene/chromosome | % Indels |
|---|---|---|
| On target | 31.4 | |
| 1 | Nfia | 1.92 |
| 2 | Stk40 | 0.85 |
| 3 | Rab9 | 0.70 |
| 4 | Ccdc101 | 0.62 |
| 5 | Vstm4 | 0.53 |
| 6 | Intergenic region, chr1 | 0.27 |
| 7 | Il17rd | 0.26 |
| 8 | Intergenic region, chr14 | 0.25 |
| 9 | Ppp1r12b | 0.24 |
| 10 | Tirap | 0.23 |
| 11 | Intergenic region, chr1 | 0.12 |
| 12 | Tiam1 | N.S. |
| 13 | Lrp2 | N.S. |
| 14 | Spata16 | N.S. |
| 15 | Intergenic region, chr8 | N.S. |
| 16 | Intergenic region, chr3 | N.S. |
| 17 | Nfib | N.S. |
| 18 | Intergenic region, chr1 | N.S. |
| 19 | Hs3st3b1 | N.S. |
| 20 | Intergenic region, chr7 | N.S. |
| 21 | Gabrb2 | N.S. |
| 22 | B4galt1 | N.S. |
| 23 | Arhgef16 | N.S. |
| 24 | Intergenic region, chr9 | N.S. |
| 25 | Parva | N.S. |
| 26 | Pigu | N.S. |
| 27 | Intergenic region, chr8 | N.S. |
| 28 | 1810013L24Rik | N.S. |
| 29 | Intergenic region, chr2 | N.S. |
| 30 | Arl8b | N.S. |
| 31 | Rptor | N.S. |
| 32 | Cd96 | N.S. |
| 33 | Barx2 | N.S. |
| 34 | Kcnj6 | N.S. |
| 35 | Dpp10 | N.S. |
| 36 | Fam49b | N.S. |
| 37 | Rbms3 | N.S. |
| 38 | Intergenic region, chr9 | N.S. |
| 39 | Sin3a | N.S. |
| 40 | Exoc4 | N.S. |
N.S., not significant.
Statistics
GraphPad Prism was used to perform all statistical tests. For comparisons with groups in which values were unmeasurable (below limit of detection), a two-sided Fisher’s exact test was used. If data passed the D’Agostino and Pearson normality test, a 2-sided Student t test was used. Otherwise, the nonparametric Mann-Whitney test (2-tailed) was used. In all tests, P < .05 was considered significant.
Results
As an initial test of the feasibility of the strategy outlined in Figure 1A, we transduced human primary hepatocytes with an AAV6 vector that contained the hF9 donor sequence together with transfection of mRNA that encoded a ZFN pair targeting a site within the first intron of human albumin. Hepatocytes treated with donor and ZFNs exhibited measurable hFIX in the culture supernatant (supplemental Figure 1).
We next sought to demonstrate this approach in vivo in the mouse. To accomplish this, we first engineered a ZFN pair targeting an analogous site in mAlb intron 1 and confirmed its activity in vitro in murine hepatoma cells (supplemental Figure 2A). Next, we assessed activity in vivo via tail vein injection of 8-week-old C57BL/6 mice with 1 × 1011 vg of an AAV8 vector encoding the ZFN pair (AAV8-ZFN) followed by Cel I assay of the target albumin locus from liver genomic DNA 7 days after vector administration. We observed cleavage at frequencies ranging from 12% to 17% (Figure 1B), indicative of small indels characteristic of break repair by nonhomologous end joining (NHEJ). This result demonstrated that these ZFNs can efficiently cleave their endogenous target in the livers of adult mice and established their suitability for studies of the albumin locus for therapeutic transgene expression.
Hemophilia B represents an ideal disease for a liver-directed genome editing strategy because modest levels of hFIX activity (>1% of normal) can greatly ameliorate the disease phenotype. To determine whether ZFN-mediated insertion of an hF9 therapeutic donor could yield stable hFIX expression, we treated wild-type mice with 1 × 1011 vg of AAV8-ZFN and 5 × 1011 vg of AAV8-hF9-donor in which the donor construct encoded a promoterless hF9 cassette containing exons 2 through 8 of the hF9 gene flanked by a splicing acceptor signal and a poly A sequence.7,10 Consistent with ZFN-driven targeted integration, mice receiving the hF9 donor and the mAlb-targeted ZFNs exhibited high circulating human hFIX levels (>3000 ng/mL; Figure 1C). Our protocol was well tolerated, and follow-up studies revealed stable hFIX expression levels (beyond 1 year; supplemental Figure 2B) as well as no significant alterations in levels of serum alanine aminotransferase (supplemental Figure 2C) or plasma albumin (data not shown). Although substantial levels of hFIX were obtained, the hybrid mAlb-hF9 mRNA represented a small fraction (0.5%) of total wild-type mAlb transcript, as determined by qRT-PCR analysis on RNA samples extracted from liver. This indicates that only a small fraction of hepatocytes need to be modified to achieve high levels of hFIX in the blood (supplemental Figure 2D). We also demonstrated that hFIX levels could be adjusted by varying the dose of AAV. At a fixed 1:5 ratio of ZFN to donor, transgene expression was proportional to the AAV dose within a range of more than 2 orders of magnitude, yielding ∼100 to 15 000 ng/mL of hFIX 2 weeks after treatment (Figure 1D).
Our strategy relies on splicing between albumin exon 1 and the integrated donor and is predicted to create a hybrid mRNA, which results in the substitution of a novel tripeptide for the 2 amino terminal residues of the hFIX propeptide. However, once processed, the resulting mature polypeptide should be identical to wild-type hFIX (supplemental Figure 3A). To test whether these substitutions affected enzyme function, we assessed clotting activity in a mouse model of hemophilia B. We observed a correction of the hemophilic phenotype in mice treated with AAV8-ZFN and AAV8-hF9-donor as measured by aPTT (Figure 1E-F). By using a chromogenic activity assay, we then confirmed that the FIX enzymatic activity in plasma correlated with the antigen levels in a 1:1 ratio (supplemental Figure 3B), indicating that the activity of the mature protein is not compromised. Collectively, these data show that long-term corrective levels of hFIX can be achieved after a single treatment with ZFN and therapeutic donor vectors.
One of the main advantages of our targeting strategy is that it allows production of any secretable protein without the need to change the ZFN reagent for each specific disease. To test the generalizability of the approach, we pursued a similar strategy for therapeutic expression of hFVIII in a mouse model of hemophilia A (HA/CD4null). The use of hemophilia A mice in a CD4null background allowed us to measure circulating human hFVIII without interference from endogenous mouse FVIII or the development of inhibitors against the human protein. Because the length of the coding sequence for this gene (7 kb) substantially exceeds the packaging capacity of AAV (∼4.7 kb), an important aspect of these studies involved reducing the donor size to a length approaching this threshold. Accordingly, our donor encoded a truncated hFVIII variant that has also been engineered for reduced size and more efficient expression.16 The resulting donor is summarized in supplemental Figure 4 (hF8 donor 2). To further increase integration activity, we also delivered ZFNs individually by using separate vectors (rather than a single vector encoding a dual-expression cassette) because, in a preliminary study, this yielded a greater than threefold increase in ZFN potency in vivo at equivalent vector doses (supplemental Figure 5). Combining these improvements to ZFN delivery and donor design, injection of 5 × 1010 vg of each individual AAV8-ZFN and 1 × 1011 vg of AAV8-hF8-donor 2 resulted in hFVIII activity levels that were 37% ± 5.5% of normal (Figure 2A). Of note, the wild-type FVIII protein does not contain a propeptide, whereas the predicted hybrid mAlb-FVIII fusion will contain the murine albumin propeptide at the N terminus. To demonstrate full functionality of the mature hFVIII protein in vivo, we performed an aPTT assay in mice treated with AAV8-hF8-donor 2 and either AAV8-ZFN or AAV8-mock vectors. Importantly, the observed hFVIII levels were able to correct the aPTT in treated animals with hemophilia A (Figure 2B), demonstrating that in vivo genome editing targeting the albumin locus is able to restore hemostasis in mouse models of both hemophilia A and B.
Liver-directed gene transfer is attractive for the treatment of lysosomal storage diseases because of the liver’s ability to secrete large amounts of protein into the blood and the ability of many lysosomal enzymes to be taken up by cells in the periphery (cross correction21). It is anticipated that treating these progressive diseases as early as possible will provide the greatest therapeutic benefit. However, long-term expression after conventional, predominantly nonintegrating AAV administration in young patients may be compromised because of episomal dilution as hepatocytes divide.22 Integration of a donor transgene into the albumin locus could potentially address this limitation. We treated adult wild-type mice with AAV8-ZFN and 4 donors that encoded human α-galactosidase A, acid β-glucosidase, iduronate-2 sulfatase, or α-l-iduronidase (ie, the genes that are deficient in patients with Fabry and Gaucher diseases and Hunter or Hurler’s syndromes, respectively). Wild-type mice were treated with 3 × 1011 vg of AAV8-ZFN and 3 × 1011 or 1.5 × 1012 vg of AAV8-donor for each transgene as indicated (Figure 3A-D). Four weeks after administration, all 4 lysosomal enzymes were detectable by western blot in liver lysates of treated mice. To assess the level of enzyme secretion more quantitatively, we treated wild-type mice with either 1.2 × 1012 vg AAV8-GBA donor alone or in combination with 1.5 × 1011 vg of each individual AAV8-ZFN (48641/31523 pair; supplemental Figure 6). We determined the enzymatic activity of GBA in plasma from treated mice as well as from untreated controls (Figure 4A). Plasma GBA activity in the ZFN + donor–treated group was threefold greater than in untreated and/or donor only groups. These experiments indicate that GBA can be integrated and expressed from the albumin locus in vivo (Figure 3D) and that the resulting enzyme is secreted in an active form and can be readily detected even in the context of normal GBA activity levels in wild-type mice. These supraphysiological levels of GBA remained consistent throughout the course of the experiment (Figure 4B), demonstrating the stability of both albumin modification and GBA expression and secretion. Together, these data provide a proof of principle that demonstrates the versatility of albumin as a targeting platform for various transgenes.
To assess the in vivo specificity of our mAlb-targeted ZFNs, we performed deep sequencing analysis on genomic DNA from animals treated with vehicle only or AAV8-ZFN and the GBA donor (animal 22; Figure 3D). We quantified indels at 40 genomic targets that composed the most likely sites of off-target cleavage for homodimers and heterodimers as gauged by SELEX analysis of the mAlb-ZFNs. Encouragingly, despite the unoptimized nature of the ZFNs that were used in this study (compared with clinical leads23), this analysis revealed highly efficient in vivo modification of the intended target (mAlb; 31.4% indels) with much lower indel levels observed at a minority of queried off-target loci (<2% at 11 loci; Table 1).
Discussion
Recent clinical trials that used AAV-mediated gene transfer have highlighted the tremendous potential of gene therapy.2,24 However, an unanswered question is whether episome-derived liver expression will be sustained in a setting of substantial liver proliferation, as in pediatric patients (the liver quadruples in size during the first 4-5 years of development25) or those with liver disease (eg, hepatitis and/or cirrhosis). For these patients, site-specific integration of the transgene to avoid AAV dilution and loss of expression could be especially beneficial. We have previously shown that in vivo genome editing can be applied successfully with therapeutic benefit in an engineered Rosa26 locus.7,10 Here, we demonstrate the therapeutic potential of targeting the endogenous albumin locus by insertion of a variety of transgenes in wild-type C57BL/6 mice.
In the specific context of in vivo gene targeting, in which it may not be possible to positively select corrected cells, targeting a limited number of cells may not result in enough secreted protein to correct a disease phenotype. In addition, targeted integration of a therapeutic transgene may not be a viable solution for some diseased loci because of mutations in the regulatory elements that control gene expression (eg, hemophilia B Brandenburg). The results presented here support the notion that a ZFN pair targeting a highly expressed locus such as mouse albumin may be used to overcome these limitations, representing an attractive platform for expression of multiple therapeutic genes.
It has recently been reported that AAV-mediated targeting of the albumin locus with no nuclease may be sufficient to correct disease.26 We have not observed measurable hFIX protein in mice that did not receive a nuclease; however, these differences may be attributable to differences in our systems. For example, our donors do not contain the full hFIX open reading frame but rather exons 2 through 8 preceded by a splice acceptor. It is extremely unlikely that a mature protein would be produced as a result of the basal promoter activity of the AAV inverted terminal repeats or any cryptic promoter sequences in the donor construct. In addition, the length of homology arms and precise region of albumin targeted in the 2 studies were different. Nonetheless, on the basis of our results as well as the genome editing literature, we predict that adding a targeted nuclease to this strategy would substantially improve the efficiency of successful genome editing. This is an important consideration because it is well known that transduction with AAV in mouse liver is particularly efficient compared with that in large animals (∼50 to 100 times greater at equivalent vg/kg doses27). An unanswered question is whether genome editing in the absence of the nuclease would be efficient enough to achieve robust levels of protein production in larger animals. We believe it is reasonable to expect that adding a nuclease would allow the use of considerably lower donor-AAV doses to achieve therapeutically relevant levels of transgene expression compared with those required by the approach of Barzel and colleagues.26 With regard to safety, the critical question will be whether a high dose of donor vector alone, which presumably relies on spontaneous DNA damage to initiate targeting, is preferred over a potentially lower overall vector dose of donor and nuclease. In addition, strategies based on donor integration via nonhomologous end joining following nuclease cleavage are a promising approach for diseases such a hemophilia A, in which the transgene, even without flanking arms of homology, pushes the limits of AAV packaging.
Cleavage specificity of designer nucleases remains an area of active inquiry. It is clear that patterns of unintended cleavage depend on the specific enzyme used, target tissue (and species of genome), and magnitude and/or duration of nuclease expression. An optimal delivery vector would permit short-lived nuclease expression in a large majority of target cells, mediating donor integration through DNA break-repair mechanisms while minimizing risk of off-target effects. In the proof-of-concept study presented here, we used AAV vectors for nuclease and donor delivery to achieve efficient gene targeting and stable expression of therapeutic transgenes from the mAlb locus in vivo. The translation of this approach will require the use of highly optimized specific nucleases along with a comprehensive analysis of potential off-target modification in the human genome. Characterization of a single universal reagent as opposed to multiple enzymes represents a critical advance for maximizing the safety profile of gene editing technologies.
In summary, our results demonstrate phenotypic correction of mouse models of hemophilia A and B after the administration of AAV vectors that encode a donor construct and a ZFN pair targeting the albumin locus. We also show that this strategy can be extended to indications beyond hemophilia, demonstrating the potential for therapeutic use in diverse protein replacement therapies.
Acknowledgments
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute (HL64190 and HL078810 [K.A.H.] and T32-HL007971 [R.S.]), the Howard Hughes Medical Institute, and the Center for Cellular and Molecular Therapeutics at the Children’s Hospital of Philadelphia. Shire supported this research through a grant under the terms of a collaboration agreement between Shire and Sangamo BioSciences, Inc.
Footnotes
Presented as an oral abstract at the 54th American Society of Hematology (ASH) Annual Meeting and Exposition, Atlanta, GA, December 8-11, 2012, and 55th ASH Annual Meeting and Exposition, New Orleans, LA, December 7-10, 2013.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.S., X.M.A., Y.D., T.W., D.E.P., P.D.G., E.J.R., M.C.H., and K.A.H. designed the experiments; R.S., X.M.A., Y.D., T.W., R.C.D., S.S., D.E.P., J.C.M., E.J.R., R.J.D., D.S., S.Z., J.R., P.D.G., and M.C.H. generated reagents and performed the experiments; and R.S., X.M.A., Y.D., E.J.R., M.C.H., P.D.G., and K.A.H. analyzed the data and wrote the manuscript.
Conflict-of-interest disclosure: K.A.H. has consulted for companies that develop adeno-associated viral–based gene therapeutics and is an inventor on issued and pending patents on zinc finger nuclease and adeno-associated viral gene transfer technologies. Y.D., T.W., R.C.D., S.S., D.E.P, J.C.M., E.J.R., D.S., P.D.G. and M.C.H. are employees of Sangamo BioSciences. The remaining authors declare no competing financial interests.
Correspondence: Katherine A. High, 3737 Market St, Suite 1300, Philadelphia, PA 19104; e-mail: kathy.high@sparktx.com.
References
- 1.Nathwani AC, Reiss UM, Tuddenham EGD, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371(21):1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett J, Ashtari M, Wellman J, et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med. 2012;4(120):120ra15. doi: 10.1126/scitranslmed.3002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 4.Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genovese P, Schiroli G, Escobar G, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510(7504):235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porteus MH, Cathomen T, Weitzman MD, Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol. 2003;23(10):3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14(12):8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anguela XM, Sharma R, Doyon Y, et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood. 2013;122(19):3283–3287. doi: 10.1182/blood-2013-04-497354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan NS, Ho B, Ding JL. Engineering a novel secretion signal for cross-host recombinant protein expression. Protein Eng. 2002;15(4):337–345. doi: 10.1093/protein/15.4.337. [DOI] [PubMed] [Google Scholar]
- 12.Gierasch LM. Signal sequences. Biochemistry. 1989;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- 13.Siner JI, Iacobelli NP, Sabatino DE, et al. Minimal modification in the factor VIII B-domain sequence ameliorates the murine hemophilia A phenotype. Blood. 2013;121(21):4396–4403. doi: 10.1182/blood-2012-10-464164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyon Y, Vo TD, Mendel MC, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 15.Lind P, Larsson K, Spira J, et al. Novel forms of B-domain-deleted recombinant factor VIII molecules. Construction and biochemical characterization. Eur J Biochem. 1995;232(1):19–27. doi: 10.1111/j.1432-1033.1995.tb20776.x. [DOI] [PubMed] [Google Scholar]
- 16.McIntosh J, Lenting PJ, Rosales C, et al. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood. 2013;121(17):3335–3344. doi: 10.1182/blood-2012-10-462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayuso E, Mingozzi F, Montane J, et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2010;17(4):503–510. doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- 18.Wright JF, Zelenaia O. Vector Characterization Methods for Quality Control Testing of Recombinant Adeno-Associated Viruses. In: Merten O-W and Al-Rubeai M, eds. Methods in Molecular Biology. New York, NY: Humana Press; 2011:247-278. [DOI] [PubMed]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8(9):765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fratantoni JC, Hall CW, Neufeld EF. Hurler and Hunter syndromes: mutual correction of the defect in cultured fibroblasts. Science. 1968;162(3853):570–572. doi: 10.1126/science.162.3853.570. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Wang H, Bell P, McMenamin D, Wilson JM. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum Gene Ther. 2012;23(5):533–539. doi: 10.1089/hum.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathwani AC, Tuddenham EGD, Rangarajan S, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365(25):2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stocker JT, Dehner LP, Husain AN. Philadelphia, PA: Lippincott Williams & Wilkins; 2012. Stocker and Dehner’s Pediatric Pathology. [Google Scholar]
- 26.Barzel A, Paulk NK, Shi Y, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517(7534):360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mingozzi F, Anguela XM, Pavani G, et al. Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci Transl Med. 2013;5(194):194ra92. doi: 10.1126/scitranslmed.3005795. [DOI] [PMC free article] [PubMed] [Google Scholar]