Abstract
Cell type-specific transcriptional regulators play critical roles in the generation and maintenance of multicellularity. Since they are often expressed at low levels, in vivo DNA-binding studies of these regulators by standard chromatin immunoprecipitation (ChIP) assays are technically challenging. We describe here an optimized ChIP protocol termed Maximized Objects for Better Enrichment (MOBE)-ChIP, which enhances the sensitivity of ChIP assays for detecting cell type-specific signals. The protocol, which is based on the disproportional increase of target signals over background at higher scales, uses substantially larger starting materials than conventional ChIPs to achieve high signal enrichment. This technique can capture weak binding events that are ambiguous in standard ChIP assays and is useful both in gene-specific and whole-genome analysis. This protocol has been optimized for Arabidopsis, but should be applicable to other model systems with minor modifications. The full procedure can be completed within 3 days.
Keywords: Chromatin immunoprecipitation (ChIP) assay, ChIP-Seq, cell type-specific transcription factor/regulator, Arabidopsis thailiana, technical advance
INTRODUCTION
Lineage or cell type-specific transcriptional regulators control specific nuclear programs that generate the diversity of cell types in multicellular organisms. Defining how these regulators program the genome in a cell type-specific manner is crucial in understanding the molecular basis of cellular differentiation. Chromatin immunoprecipitation (ChIP) is a powerful technique to study protein-DNA interactions in vivo (Solomon et al., 1988; Orlando, 2000; Gendrel et al., 2005; Bowler et al., 2004) and in principle, a useful tool for identifying where these transcriptional regulators bind in the genome. However, due to their low expression levels and/or presence in limited number of cells, standard ChIP protocols are suboptimal for studying these regulators (Figure 1a). To overcome this technical hurdle, we recently developed an improved ChIP assay termed Maximized Objects for Better Enrichment (MOBE)-ChIP that significantly enhances ChIP sensitivity, and when combined with high-throughput sequencing, allows the genome-wide mapping of binding targets of cell type-specific transcription factors at high resolution (Lau et al., 2014; Matos et al., 2014).
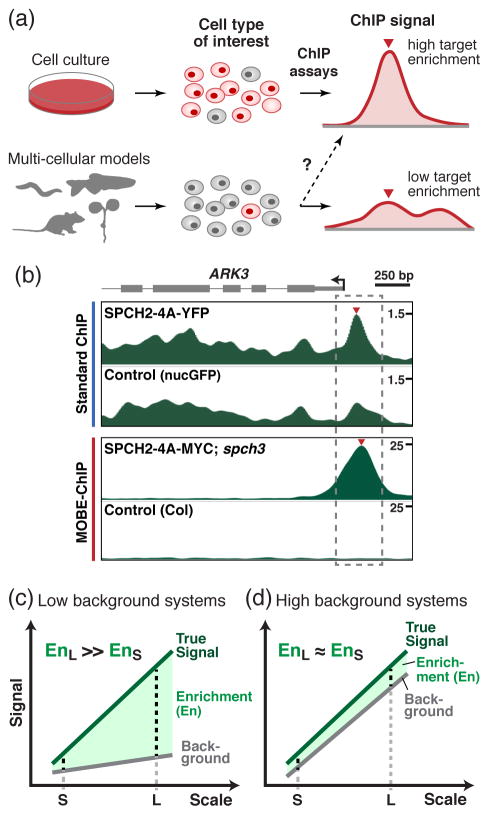
Figure 1. Enhancement of ChIP signal through Maximized Objects for Better Enrichment (MOBE)-ChIP.
(a) In contrast to the large populations of specific cell types available in cell or tissue culture, small populations of target cell types in multi-cellular model organisms typically yield low target enrichments in standard ChIP assays. (b) ChIP-Seq profiles generated from standard ChIP (blue; scale: 4×) or MOBE-ChIP (red; scale: 16×) of an Arabidopsis cell type-specific transcription factor at the ARK3 loci (Lau et al., 2014). The y-axis represents the enrichment values; note the difference in scale of the enrichment values. Grey dash box marks the binding region on the ARK3 promoter. Inverted triangles (red) mark the summit of the binding peaks. (c, d) Hypothetical model of the relationship between target signal enrichment (En) and experimental scale in ChIP systems that generate low (c) or high (d) background signals. Note the effect on enrichment at small (S) and large (L) scales in the two systems.
Overview of MOBE-ChIP
The MOBE-ChIP technique is an optimized protocol based on standard ChIP assays (Gendrel et al., 2005; Bowler et al., 2004) that improves target enrichment. The key feature of the method is to develop a ChIP system with low background noise and perform ChIP experiments at a substantially larger scale than regular ChIPs. Importantly, samples are processed in smaller-sized aliquots starting from cross-linking, through nuclei isolation and chromatin fragmentation to maintain efficiency in these steps. Only at the immunoprecipitation step are all the chromatin extracts pooled. Using a previously identified transcription factor-target interaction (typically enriched in the 2–4 fold range), we optimized the protocol to allow processing of 16-times the starting materials of standard ChIPs in Arabidopsis thaliana and obtained an improvement to 600-fold enrichment of the target. Pooling several MOBE-ChIPs provided sufficient materials for high-quality ChIP-Seq profiles (Figure 1b) (Lau et al., 2014).
Principle of MOBE-ChIP
To improve sensitivity of a ChIP assay, the goal is to increase signals from the DNA target (true signal) while minimize signals from background. In a given ChIP system, assuming the efficiency of the ChIP procedures is maintained, true signal should hypothetically display a linear relationship with the experimental scale, i.e., the more the target protein-DNA complexes are available for immunoprecipitation, the more signals from the complex could be captured by the system (Figure 1c,d). Thus, how background signal changes with experimental scale becomes a key determinant on improving target enrichment (true signal over background) through scale increase. In a system where background noise can be maintained at a low level even with increasing experimental scales, e.g. with the use of highly specific monoclonal antibody, performing ChIP at a larger scale should yield better enrichment of target signals (Figure 1c, enrichment at L vs. S). In contrast, in high background ChIP systems, where background signal increases proportionally with experimental scale in a manner similar to true signal, conducting ChIP at a larger scale might not yield the same benefit (Figure 1d). This may be the case when antibodies with low specificity are used. Thus, the success of the MOBE-ChIP technique requires not only the increase in scale, but also the development of a low background ChIP system with highly specific antibodies (see Experimental Design).
Applications
MOBE-ChIP is most useful for the in vivo DNA-binding study of cell type-specific transcriptional regulators that express transiently and at a low level, where standard ChIP assays do not have the sensitivity for their detection. As an example, we have employed this technique, in conjunction with high-throughput sequencing, to generate a high-quality genome-wide binding map of SPEECHLESS (SPCH), a transient and cell type-specific transcription factor in the Arabidopsis leaf. Functional full length SPCH was tagged with the Myc epitope and driven under its native promoter in a transcript-null spch mutant background (Figure 1b) (Lau et al., 2014).
In addition to tissue-specific factors, broadly expressed regulators play important functions in gene regulation in multicellular organisms. For these factors, it is often desirable to obtain information about their targets in specific cell types. Thus, to probe for these targets through MOBE-ChIP, a broadly expressed transcriptional regulator can likewise be epitope-tagged and targeted to express under cell type-specific promoters. We have successfully used this technique to study the binding of the essential and broadly expressed transcriptional co-regulator RBR (the Retinoblastoma (Rb) homolog in plants), in a cell type-specific manner by expressing a tagged version solely in the terminal stage of an epidermal lineage (Matos et al., 2014).
Besides cell type-specific assays, MOBE-ChIP can be used as a general strategy to enhance ChIP experiments, e.g. for validating weak binding events when conventional ChIP assays produce ambiguous results. ChIPs that are intended for genome-wide studies will especially benefit from the technique, since, to capture the genome-wide binding landscape of the regulator with high fidelity, these studies require high target enrichment and confidence that regions not enriched in the assay truly represent non-binding. Thus, our technique could be valuable to large-scale initiatives, such as ENCODE and modENCODE, in defining transcriptional network, where high-quality data is critical. Furthermore, MOBE-ChIP may also be incorporated in other ChIP-based techniques, such as ChIP-exonuclease (ChIP-exo) (Rhee and Pugh, 2011), to improve signal enrichment in these modified assays.
Although the procedure outlined here is optimized for Arabidopsis tissues, the rationale of MOBE-ChIP could be applicable to all ChIP experiments and thus can be adapted to other plants or to animals (For details, see Adaptation of MOBE-ChIP to other model systems).
Comparisons with other methods and limitations of MOBE-ChIP
Previous ChIP studies on transcriptional regulators whose expression is restricted spatially and temporally, have often required overexpression in the organism, or tissue culture of the cell types in which their activity is important (Winter et al., 2011; Brandt et al., 2012; Cao et al., 2010). However, the drawbacks of these in vitro or ectopic methods are the identification of false-positives or -negatives. This is due to differences in chromatin state, absence of native binding partners, or inappropriate interaction with protein(s) in these non-native contexts. Furthermore, in plants, cell culture systems that maintain specific lineage identities are nearly non-existent, with Zinnia xylem development being a notable exception (Fukuda and Komamine, 1980). The MOBE-ChIP technique, on the other hand, allows the study of cell type-specific transcriptional regulators in their native environments and should be applicable to all model systems amenable to ChIP procedures.
A recently developed technique, named Batch Isolation of Tissue-Specific Chromatin for Immunoprecipitation (BiTS-ChIP), also offers in vivo study of transcription factor binding and chromatin marks in a cell/tissue-specific manner (Bonn et al., 2012). It utilizes fluorescence-activated cell sorting (FACS) to isolate fixed nuclei that are labeled with cell type-specific nuclear marker for ChIP assays. Limitations for the method, however, include the need for a cell sorter and the expertise in operating it. In addition, if the cell types of interests are rare, it may not be practical to sort enough nuclei for ChIPs. In contrast to BiTS-ChIP, our technique, which involves relatively simple increase in experimental scale, does not require extra equipment and should be compatible with existing setup and expertise in most laboratories.
Limitations for MOBE-ChIP include the requirement of relatively large amounts of starting material, so studies involving unique or rare samples may not be practical with the technique. Our method also requires antibodies with high specificity (see below). In our case, we used an epitope tag for which there are commercially available monoclonal antibodies, so this limitation may be overcome in most situations. Finally, since the cell type specificity of our technique comes from the target proteins (either natively or driven by a cell type-specific promoter), MOBE-ChIP is not suitable to detect cell type-specific histone marks, as they are post-translational modifications that are present organism-wide. To study them in their native environment, isolation of the targeted cell types (e.g. FACS) or their nuclei (e.g. BiTS-ChIP or INTACT) would be needed before a ChIP assay (Bonn et al., 2012; Deal and Henikoff, 2011).
Experimental Design
Achieving a low background ChIP system
An important factor for the success of MOBE-ChIP is the establishment of a ChIP system that produces minimal background noise. Signals from background in ChIP are mainly the result of non-specific binding of proteins and/or chromatin during the immunoprecipitation step, and thus the choice of antibody and antibody-binding beads is critical in controlling background.
An ideal antibody should bind to its target protein with high affinity and specificity. For MOBE-ChIP, the specificity of an antibody is particularly important because it helps maintain background level when experimental scale increases. Thus, monoclonal antibodies, which generally have higher specificity than polyclonal ones, are better candidates for MOBE-ChIP. Nevertheless, polyclonal antibodies with validated low cross-reactivity should also be effective. As discussed below, the specificity of a candidate antibody can be assessed initially through western and immunoprecipitation analyses. A useful strategy to employ monoclonal antibodies in a ChIP experiment is through epitope-tagging of the target protein if transgenic method is possible in the model system. Epitope tags such as Myc, FLAG® and HA, which are routinely used in immunoprecipitation studies, have high-quality monoclonal antibodies against them commercially available and are good options for tagging. It is recommended, however, to confirm the function of the epitope-tagged transcriptional regulator by mutant complementation. As suggested from our study on SPCH (Lau et al., 2014), this approach may also help improve ChIP signals, presumably through eliminating competition from the endogenous TF for binding to the target sites (Kaufmann et al., 2010). Similarly, beads used in the capture of the DNA-protein immunocomplex in the immunoprecipitation step should ideally bind minimally to non-specific proteins or DNA. In our hands, we found magnetic beads, such as Dynabeads®, perform consistently and have low non-specific binding.
Detection of target protein
Before starting a ChIP experiment, it is critical to perform a western blot analysis or whole-mount immunostaining to detect the transcriptional regulator of interest. There are several reasons for this. First, they confirm the presence of the protein (particularly important for recombinant epitope-tagged targets) and provide an estimate of its expression level for determining the starting scale for MOBE-ChIP (see Scale consideration). Second, they aid in the selection of antibody for ChIP. In this regard, an additional immunoprecipitation analysis would be valuable as it also assays for the pull-down efficiency of an antibody. Although an antibody suitable for western/immunoprecipitation analyses and/or immunostaining (i.e. produces high target signal and low background) may not translate directly to success in ChIP, the results should reveal the specificity of the antibody and its potential suitability for MOBE-ChIP. Finally, expression of cell type-specific proteins is often tightly controlled by developmental time and growth conditions. Being able to detect the protein of interest allows optimization of these conditions, which would be important for determining peak expression of the protein for the ChIP experiment.
Scale consideration
For MOBE-ChIP, our hypothetical model suggests that the larger the experimental scale, the higher the target enrichment could be achieved (Figure 1c). However, for practical reasons, the final scale of the experiment would depend on the purpose of the study (gene-specific vs. genome-wide) and also the real-world constraints in carrying out the large-scale experiment, such as equipment capability and processing time of multiple aliquots. We recommend conducting a pilot ChIP assay with the standard scale as in established protocols specific to the model system, prior to working with larger starting materials. If the enrichment level is not satisfactory, the scale can be doubled (2×) in subsequent trials (i.e. 2×, 4×, 8×, and 16×) until the target enrichment level is achieved (see Anticipated results). For example, for a standard Arabidopsis ChIP using 1.5 g of tissue samples, the starting materials in the subsequent trials would be 3, 6, 12 and 24 g. If the expression level of the transcriptional regulator is especially low (as determined by western blotting), we recommend a starting scale of 4×.
An important factor when scaling up is to maintain efficacy in the crosslinking, chromatin isolation and sonication steps. This can be achieved by working with smaller aliquots as illustrated in the Experimental Procedures. Overloading these steps with extra samples may result in under-crosslinked samples, chromatin fraction with low purity, and/or insufficient sonication—all of which will negatively impact ChIP results.
Experimental controls
Experimental controls in ChIP assays are critical for distinguishing true binding events from background signals. “Input chromatin” or “no antibody control” (or beads alone) are frequently used as the negative controls. However, they do not account for non-specific binding of the antibody, which could contribute to the majority of the background noise in a ChIP assay, and thus, these controls are prone to identify false-positive binding regions. Thus, although “Input chromatin” or “no antibody control” are helpful controls to include, we recommend having either an untagged wild type as the control for an epitope-tagged transcriptional regulator, or if the tag is a larger protein, such as green fluorescence protein (GFP), a tag alone control. When an antibody against the endogenous protein is used, a mutant lacking the regulator of interest is the ideal control. These “antibody controls” should be processed in parallel with the experimental samples and would serve as the ideal negative controls for ChIP experiments.
Adaptation of MOBE-ChIP to other model systems
Since MOBE-ChIP is based on the intrinsic properties of the ChIP procedure, this technique should be readily applicable in other systems besides Arabidopsis. The initial crosslinking and chromatin isolation steps (step 1–16) are likely model system-specific because tissue composition and properties among different organisms are often different. However, once the chromatin is isolated, immunoprecipitation (step 17 onwards) can be performed as described in this protocol. Thus, when scaling up for MOBE-ChIP, researchers working on other systems are advised to optimize the crosslinking and chromatin isolation steps based on established ChIP protocols specific to these models and use our protocol as a guide in processing the extra materials. As a reminder, when scaling up the experiments, sample overload during these steps should be avoided (see Scale consideration).
MATERIALS
Reagents
Antibody (Varies with target proteins or tags; For our work with Myc-tagged SPCH or RBR, we used Cell Signaling Technology, Myc-Tag (71D10) Rabbit mAb, cat. no. 2278)
β-mercaptoethanol (Bio-Rad, cat. no. 161-0710)
DNA purification kit (Zymo, ChIP DNA Clean & Concentrator™, cat. no. D5205, or equivalent)
EDTA (0.5 M, pH 8) (Life technologies, cat no. 15575-020)
Formaldehyde, 37% (wt/vol) (Fisher Scientific, cat. no. F79-500) CAUTION Flammable and poisonous liquid and vapor. Handle with care and in a fume hood.
Glycine (Life technologies, cat. no. 15527-013)
Lithium chloride (Sigma-Aldrich, cat. no. L9650)
Magnetic beads (Life technologies, Dynabeads® Protein A, cat. no. 10001D, or equivalent)
Magnesium chloride (MgCl2; Sigma-Aldrich, cat. no. M8266)
Miracloth (pore size: 20–25 μm; EMD Millipore, cat. no. 475855-1R)
NP-40 (IGEPAL® CA-630; Sigma-Aldrich, cat. no. I8896)
Protease inhibitors (Roche, cOmplete and cOmplete Mini protease inhibitor, cat. no. 04693116001 and 04693124001, or equivalent)
Proteinase K (Qiagen, 20 mg/μl, cat. no. 19131)
Sodium bicarbonate (NaHCO3; Sigma-Aldrich, cat. no. S4772)
Sodium chloride (NaCl; EMD Millipore, cat. no. SX0420-3)
Sodium deoxycholate (Sigma-Aldrich, cat. no. D6750)
Sodium dodecyl sulfate (SDS; Bio-Rad, cat. no. 161-0302)
Sucrose (Sigma-Aldrich, cat. no. S3929)
Tris-HCl (1 M, pH 8) (Life technologies, cat no. 15568-025)
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
Optional (for sequencing library construction):
Agencourt® AMPure® XP beads (Beckman Coulter, cat. no. A63880)
ChIP-Seq library construction kit (NuGen, Ovation® Ultralow library system, cat. no. 0303, or equivalent)
Equipment
1.5-ml and 2-ml microcentrifuge tubes (Eppendorf, DNA LoBind Tube 1.5 and 2.0 ml, cat. no. 022431021 and 022431048, or equivalent; these tubes are compatible with Bioruptor® sonication)
25-ml pipette (VWR, cat. no. 89130-900, or equivalent)
50-ml tubes (VWR, cat. no. 21008-216, or equivalent)
2100 Bioanalyzer (Agilent, cat. no. G2939AA, with High Sensitivity DNA kit, cat. no. 5067-4626) or equivalent
Heat block (Eppendorf, Thermomixer® R, cat. no. 022670107, or equivalent)
Magnetic stand (EMD Millipore, Magna GrIP™ Rack, cat. no. 20–400, or equivalent)
Mortar and pestle (CoorsTek, Porcelain Mortar 275 ml Capacity and Pestle 180 mm Length, cat. no. 60319 and 60320, or equivalent)
Qubit® Fluorometer (Life Technologies, cat. no. Q32866)
Refrigerated centrifuge (Beckman Coulter, Allegra® X-22 Refrigerated Benchtop Centrifuge, cat. no. 392189, or equivalent)
Rotor for 50-ml tubes (Beckman Coulter, SX4250, cat. no. 392252)
Rotor for microcentrifuge tubes (Beckman Coulter, F2402H, cat. no. 361171)
Strainer (tea strainer with a diameter of 85 mm and a mesh size of 1 mm, or equivalent)
Sonicator (Diagenode, Bioruptor® Standard, cat. no. B01010001, or equivalent)
Tube rotator (18 rpm; VWR, cat. no. 13916-822, or equivalent)
Vacuum chamber (Bel-Art Products, cat. no. 420270000, connected to house vacuum lines or pump, or equivalent)
Reagent setup
Cross-linking buffer
This buffer is composed of 0.4 M Sucrose, 10 mM Tris-HCl (pH 8), 10 mM MgCl2 and 1% (wt/vol) formaldehyde. CRITICAL Add formaldehyde fresh before each experiment.
Extraction buffer 1
The buffer is composed of 0.4 M Sucrose, 10 mM Tris-HCl (pH 8) and 10 mM MgCl2. Add β-mercaptoethanol (final concentration: 5 mM). Dissolve one cOmplete protease inhibitor tablet per 50 ml of buffer before each experiment. For long-term storage of the base components, filter-sterilized the buffer and store at 4 °C.
Extraction buffer 2
The buffer is composed of 0.25 M Sucrose, 10 mM Tris-HCl (pH 8), 10 mM MgCl2 and 1% (vol/vol) Triton X-100. Add β-mercaptoethanol (final concentration: 5 mM). Dissolve one cOmplete mini protease inhibitor tablet per 10 ml of buffer before each experiment. For long-term storage of the base components, filter-sterilize the buffer and store at 4 °C.
Extraction buffer 3
The buffer is composed of 0.4 M Sucrose, 10 mM Tris-HCl (pH 8), 2 mM MgCl2 and 0.15% (vol/vol) Triton X-100. Add β-mercaptoethanol (final concentration: 5 mM). Dissolve one cOmplete mini protease inhibitor tablet per 10 ml of buffer before each experiment. For long-term storage of the base components, filter-sterilize the buffer and store at 4 °C.
Nuclei lysis buffer
The buffer is composed of 50 mM Tris-HCl (pH 8), 10 mM EDTA and 1% (wt/vol) SDS. Dissolve one cOmplete mini protease inhibitor tablet per 10 ml of buffer and pre-chilled to 4 °C before each experiment. For long-term storage of the base components, filter-sterilize the buffer and store at room temperature.
ChIP dilution buffer
The buffer is composed of 16.7 mM Tris-HCl (pH 8), 167 mM NaCl, 1.2 mM EDTA and 1.1% (vol/vol) Triton X-100. Dissolve one cOmplete mini protease inhibitor tablet per 10 ml of buffer before each experiment. For long-term storage of the base components, filter-sterilize the buffer and store at 4 °C.
Low salt wash buffer
The buffer is composed of 20 mM Tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, 0.1% (wt/vol) SDS and 1% (vol/vol) Triton X-100. For long-term storage, filter-sterilize the buffer and store at 4 °C.
High salt wash buffer
The buffer is composed of 20 mM Tris-HCl (pH 8), 500 mM NaCl, 2 mM EDTA, 0.1% (wt/vol) SDS and 1% (vol/vol) Triton X-100. For long-term storage, filter-sterilize the buffer and store at 4 °C.
LiCl wash buffer
The buffer is composed of 10 mM Tris-HCl (pH 8), 250 mM LiCl, 1 mM EDTA, 1% (vol/vol) NP-40 and 0.5% (wt/vol) sodium deoxycholate. For long-term storage, filter-sterilize the buffer and store at 4 °C.
TE buffer
The buffer is composed of 10 mM Tris-HCl (pH 8) and 1 mM EDTA. For long-term storage, filter-sterilize the buffer and store at 4 °C.
ChIP elution buffer
Dissolve 42 mg of NaHCO3 in 4.75 ml of ddH2O and add 250 μl of 20% (wt/vol) SDS (final concentration: 0.1 M NaHCO3 and 1% (wt/vol) SDS). Prepare fresh for each experiment.
Protease buffer
The buffer is composed of 385 mM Tris-HCl (pH 6.5) and 96 mM EDTA and 0.77 mg/_l of Proteinase K. Prepare fresh for each experiment.
EXPERIMENTAL PROCEDURES
The following procedure is based on conventional ChIP protocols in Arabidopsis (Gendrel et al., 2005; Bowler et al., 2004) but with modification to process the substantially larger starting materials (16 times) effectively. For simplicity, the processing of only one sample (a genotype) is shown. However, it is highly recommended that a control sample is included (e.g. wild-type control for an epitope-tagged sample; see Experimental controls). Thus, in most cases, twice the amount of reagents listed below will be needed and the two samples (e.g. the epitope-tagged sample and the wild-type control) should be processed in parallel.
Tissue harvesting and formaldehyde cross-linking TIMING 2–2.5 h
-
1
Grow and harvest around 24 g (fresh weight) of plant tissues (e.g. for 4-day-old seedlings, this is equivalent to around sixteen 150×15 mm plates of plant materials). Divide the tissues into 8 aliquots and add each aliquot to 37 ml cross-linking buffer in a 50-ml tube (i.e. 8 tubes in total; ~3 g per tube).
CRITICAL STEP Samples are divided into standard-sized aliquots for efficient cross-linking.
-
2
Place the tissue under vacuum for 5 min. Slowly release vacuum and re-submerge the tissue by inverting the tubes. Reapply vacuum for another 5 min.
CRITICAL STEP The infiltration time here only serves as a reference point. It may need to be optimized empirically (see Table 1 for troubleshooting).
-
3
Add 2.5 ml of 2 M glycine to each tube (final concentration of 0.125 M). Mix and apply vacuum for 5 min.
-
4
Pool all the tissues in a strainer and rinse them thoroughly with distilled water (500 ml to 1 L).
-
5
Remove as much water as possible from the tissue by blotting it with paper towels. If tissues stick to the paper towels, it may be helpful to place a piece of Miracloth (or other type of mesh) between the two for this step.
PAUSE POINT The cross-linked tissue can be frozen in liquid nitrogen and stored at −80 °C.
TABLE 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 15 | Size of DNA above 1 kb | Insufficient sonication | Increase sonication cycles |
| Inefficient sonication due to over-cross-linking | Reduce the incubation time in the cross-linking step (step 2). The concentration of formaldehyde can also be reduced | ||
| 32 | No PCR signal | Failed DNA purification due to over-cross-linking or mistakes in the purification procedure | Check the presence of DNA in the Input control by gel electrophoresis (see step 15) or other methods. If no DNA is detected, reduce the incubation time in the cross-linking step (step 2) and/or optimize the DNA purification step (28–31) with the Input control |
| Failed PCR reaction | Optimize primers and PCR conditions with the Input control or validated genomic DNA | ||
| No/low signal enrichment | High background | Use antibodies with high specificity and high quality magnetic beads. Increase the washing time in the immunoprecipitation steps (22–23), e.g. to 10 min, and/or the stringency of the wash solutions | |
| Extremely low amount of the target protein | After taking measures to control background signals (discussed above), double or quadruple the experimental scale |
Chromatin isolation and sonication TIMING 6 to 6.5 h
-
6
Grind the tissue into fine powder with a pre-cooled mortar and pestle in liquid nitrogen.
CRITICAL STEP Avoid thawing the tissue before and during grinding.
-
7
Divide the powder into 6 aliquots and add each aliquot to 40 ml of extraction buffer 1 (pre-chilled to 4 °C) in a 50-ml tube (6 tubes in total). Mix the powder and solution for 10 min on a tube rotator (~15 rpm).
CRITICAL STEP Samples are divided into standard-sized aliquots for efficient nuclei isolation.
-
8
Filter the solution in each tube with two layers of Miracloth into a new 50-ml tube on ice (6 tubes in total).
-
9
Centrifuge the filtered solution for 20 min at 3,000g at 4 °C (preferably with a swing-bucket rotor).
-
10
Remove and discard the supernatant with a 25-ml pipette without disturbing the pellet. Resuspend each pellet in 1.3 ml of extraction buffer 2 (4 °C).
-
11
Transfer the solution in each tube to a 1.5-ml microcentrifuge tube and centrifuge for 10 min at 12,000g at 4 °C (6 tubes in total).
-
12
Pipette off and discard the supernatant without disturbing the pellet. Resuspend each pellet in 400 μl of pre-chilled extraction buffer 3 (4 °C). Gently pipette up and down to avoid foaming.
-
13
Prepare 6 new 1.5-ml microcentrifuge tubes containing 400 μl of extraction buffer 3 (4 °C). Overlay each of the 6 resuspensions from step 12 over one of the newly prepared 400 μl extraction buffer 3 and centrifuge for 1 h at 16,000g at 4 °C (Total: 6 tubes with 800 μl of solutions).
-
14
Pipette off and discard the supernatant without disturbing the pellet. Resuspend each pellet in 300 μl of nuclei lysis buffer (4 °C; place on ice right before the step to prevent precipitation of SDS) by gently pipetting the solution up and down. Incubate on ice for 10 min.
-
15
Sonicate the resuspended chromatin with a Bioruptor® sonicator for 3 × 7.5 min and 1 × 2.5 min with a 30 s ON and a 30 s OFF cycle (total ON cycles: 27) at HIGH setting.
CRITICAL STEP Keep the chromatin resuspension cool (ideally at 4 °C and < 10 °C at all times) by adding crushed ice to the water bath every 7.5 min or use a water cooler. The sonication time here serves as a reference point and should be optimized empirically. Other types of sonicator, such as probe-based or Focused-ultrasonicators from Covaris®, can also be employed. The degree of DNA fragmentation (ideally < 1 kb for general downstream analysis and 200 to 600 bp for sequencing library construction) can be checked by analyzing the de-cross-linked Input control (step 31) by gel electrophoresis or with a BioAnalyzer (see Table 1 for troubleshooting).
-
16
Centrifuge the sonicated chromatin solution for 10 min at 12,000g at 4 °C. Save the 6 supernatants and combine them in a new 2-ml microcentrifuge tube (total volume: 6 × 300 μl or ~1.8 ml). Remove 10 μl of the combined solution into a new tube and store at −20 °C, which serves as the Input control for later steps.
CRITICAL STEP Avoid disturbing the pellet by careful pipetting and leaving some supernatant behind. Another round of centrifugation may be performed if the insoluble materials were still present in the saved supernatant.
PAUSE POINT The fragmented chromatin can be frozen in liquid nitrogen and stored at −80 °C.
Immunoprecipitation TIMING Overnight (or 5 h) plus 3 h
-
17
Measure the volume of the combined chromatin solution (~1.6 to 1.7 ml) and dilute it 10 times with ChIP dilution buffer (4 °C) in a 50-ml tube (Total volume: ~17 ml).
CRITICAL STEP The combined chromatin extract is used for a single immunoprecipitation.
-
18
Add 90 μg of the desired antibody to the diluted chromatin solution. Incubate overnight (or >5 h) at 4 °C with gentle agitation or rotation.
CRITICAL STEP The amount of antibody will need to be determined empirically. A 1:200 dilution or 5 μg per ml is a good starting point. Depending on the scale of the experiment, the amount of antibody is adjusted accordingly. For example, for 12 g of starting materials (8×), we use 45 μg of antibody. Likewise, the amount of beads and buffers in the subsequent steps are also scaled.
-
19
Transfer 400 μl of well-suspended Dynabeads® Protein A into a new 1.5-ml microcentrifuge tube. Place on a magnetic stand for 20–30 s on ice and discard the supernatant with a pipette. Completely resuspend the beads in 1 ml of ChIP dilution buffer (4 °C) with a pipette, place on a magnetic stand for 20–30 s on ice and discard the supernatant with a pipette. Repeat the ChIP dilution buffer wash one more time and resuspend the beads in 400 μl ChIP dilution buffer (4 °C).
-
20
Add 400 μl of the pre-washed Dynabeads® Protein A (step 19) to the chromatin-antibody mixture in step 18 and incubate for 1 h at 4 °C on a rotator.
-
21
Capture the magnetic beads with a magnetic stand on ice and discard the supernatant with a pipette. Depending on the size of the magnetic stand available, the mixture may need to be transferred successively to a smaller-sized tube (e.g. a 15-ml tube) for the capture.
-
22
Resuspend the beads in 8 ml of low salt wash buffer in a 15-ml tube and incubate for 5 min at 4 °C with gentle rotation. Alternatively, the solution can be processed as 4 × 2 ml samples in 2-ml microcentrifuge tubes. Centrifuge briefly (<3,800g and ~ 10 s) after the wash. Place the tube on a magnetic stand for 20–30 s and discard the supernatant with a pipette.
-
23
Repeat step 22 with successive washes in cold (i) High salt wash buffer, (ii) LiCl wash buffer and (iii) TE buffer for a total of 4 washes (one wash per buffer). Discard the supernatant with a pipette in each wash.
-
24
Elute the immune complexes by adding 1.6 ml of freshly prepared ChIP elution buffer. Vortex to mix and divide the sample into 2 × 0.8 ml sample in 1.5-ml microcentrifuge tubes for easy handling. Incubate for 40 min at 65 °C on a heat block with mixing every 5 min. Alternatively, the incubation can be done on a ThermoMixer® (at 65 °C) with mixing frequency at 1,200 rpm.
-
25
Centrifuge the tubes briefly (<3,800g and ~ 10 s) and place the tubes on a magnetic stand for 20–30 s. Save and transfer the eluates into new 1.5-ml microcentrifuge tubes.
Reverse cross-linking and purification of DNA TIMING Overnight (or 6 h) plus 3 h
-
26
Add 32 μl of 5 M NaCl to each of the two 800 μl eluates and mix. Thaw the Input sample from step 16, add 190 μl of ChIP elution buffer and 8 μl of 5 M NaCl to this sample and mix. Incubate all the tubes for at least 6 h to overnight at 65 °C.
-
27
Add 83.2 and 20.8 μl of protease buffer to the eluates and the Input sample, respectively. Incubate for 1 h at 45 °C.
-
28
Purify DNA from the samples with the ChIP DNA Clean & Concentrator™ kit or an equivalent product. Add 5 volumes of ChIP DNA Binding Buffer to 1 volume of eluate or the Input sample. Mix and apply <750 μl of the mixture to a Zymo-Spin™ column. Centrifuge for 30 s at >10,000g and discard the flow-through. Repeat the loading and centrifugation steps until all volume of a sample has passed through the column.
CRITICAL STEP Only one column is used per sample, i.e. for the 8 ml of mixture derived from the 1.6 ml eluate, the sample is applied to the same column 12 times.
-
29
Add 250 μl of wash buffer (from the kit in step 28) to the column. Centrifuge for 30 s at >10,000g and discard the flow-through. Repeat once.
-
30
Centrifuge the empty column for 1 min at >10,000g.
-
31
To elute DNA, add 15 μl of elution buffer (from the kit in step 28) to the column matrix. Transfer the column to a new 1.5-ml microcentrifuge tube, incubate for 1 min at room temperature and centrifuge for 1 min at >10,000g. Repeat once with another 15 μl-elution to the same tube. If not use immediately, store purified DNA at −80 °C.
CRITICAL STEP The choice of elution buffer and the elution volume can be adjusted according to downstream needs.
-
32
Analyze the success of the ChIP assay by quantitative PCR for the enrichment of known genomic regions with specific primers (see Table 1 for troubleshooting).
-
33
For subsequent sequencing library construction, it is advisable to quantify the yield of the ChIP sample using fluorescence-based method, e.g. Qubit® quantitation assay. Since background signals are minimal in a successful MOBE-ChIP, DNA yield may be quite low (< 1 ng). Thus, several MOBE-ChIPs may need to be performed to obtain enough ChIPed DNA (1–10 ng) for library construction. ChIPed DNAs can be pooled and eluted in suitable volume by Agencourt® AMPure® XP DNA binding beads or column-based methods. We have had success combining 6–8 independent MOBE-ChIPs for sequencing (Lau et al., 2014), using library preparation kits designed for low input (see Reagents).
TIMING
Day 1
Step 1–5, tissue harvesting and formaldehyde cross-linking: 2–2.5 h (or Day 0)
Step 6–16, chromatin isolation and sonication: 6 to 6.5 h
Step 17–18, immunoprecipitation: overnight (or 5 h)
Day 2
Step 19–25, immunoprecipitation (continued): 3 h
Step 26, reverse cross-linking and purification of DNA, overnight (or 6 h)
Day 3
Step 27–32, reverse cross-linking and purification of DNA (continued): 3 h
ANTICIPATED RESULTS
Improvement of target enrichment (as judged by qPCR in step 32) over standard ChIP protocols indicates the success of MOBE-ChIP. As a reference, we observed doubling the experiential scale leads to three- to ten-fold increase in enrichment of targets (Lau et al., 2014). In the experiment, MOBE-ChIP of a cell type-specific transcription factor in Arabidopsis performed at scales of 4×, 8× and 16× resulted in fold enrichment of its target (compared to WT) at 19, 183 and 613, respectively. Furthermore, the ChIP-Seq profile derived from multiple MOBE-ChIPs at 16× has exceptionally high signal-to-noise ratio (Figure 1b). However, for some very challenging transcriptional regulators, whose target enrichment is not detectable at standard scale, initial scale increase may not lead to noticeable improvement until the scale becomes substantially large. For example, the binding of the lineage-specific expressed RBR to its targets was only detectable at 16× (Matos et al., 2014). Therefore, the scale of MOBE-ChIP will vary depending on the nature of the transcriptional regulator of interest and the purpose of the ChIP assays (gene-specific vs. whole-genome studies).
Acknowledgments
We thank members of our laboratory and the Arabidopsis community for comments on the manuscript. Funding for this work was provided by NIH 1R01GM086632. OSL was a Croucher Fellow. DCB is a Gordon and Betty Moore Foundation Investigator of the Howard Hughes Medical Institute. The authors declare no competing financial interests.
References
- Bonn S, Zinzen RP, Perez-Gonzalez A, Riddell A, Gavin AC, Furlong EEM. Cell type-specific chromatin immunoprecipitation from multicellular complex samples using BiTS-ChIP. Nature Protocols. 2012;7:978–994. doi: 10.1038/nprot.2012.049. [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. Chromatin techniques for plant cells. The Plant Journal. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72:31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nature Protocols. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H, Komamine A. Establishment of an Experimental System for the Study of Tracheary Element Differentiation from Single Cells Isolated from the Mesophyll of Zinnia elegans. Plant Physiol. 1980;65:57–60. doi: 10.1104/pp.65.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP) Nature Protocols. 2010;5:457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science. 2014;345:1605–1609. doi: 10.1126/science.1256888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos JL, Lau OS, Hachez C, Cruz-Ramírez A, Scheres B, Bergmann DC. Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. Elife. 2014:3. doi: 10.7554/eLife.03271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, et al. LEAFY Target Genes Reveal Floral Regulatory Logic, cis Motifs, and a Link to Biotic Stimulus Response. Dev Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]