Abstract
The CpxAR two-component system is present in many Proteobacteria. It controls expression of genes required to maintain envelope integrity in response to environmental injury. Consequently, this two-component system was shown to be required for virulence of several zoo-pathogens but it has never been investigated in phyto-pathogens. In this paper, we investigate the role of the CpxAR two-component system in vitro and in vivo in Dickeya dadantii, an enterobacterial phytopathogen that causes soft-rot disease in a large variety of plant species. cpxA null mutant displays a constitutively phosphorylated CpxR phenotype as shown by direct analysis of phosphorylation of CpxR by a Phos-Tag retardation gel approach. Virulence in plants is completely abolished in cpxA or cpxR mutants of D. dadantii. In planta, CpxAR is only activated at an early stage of the infection process as shown by Phos-Tag and gene fusion analyses. To our knowledge, this is the first time that the timing of CpxAR phosphorelay activation has been investigated during the infection process by direct monitoring of response regulator phosphorylation.
Keywords: CpxAR, Virulence, stress, plant pathogen, D. dadantii
Introduction
For bacteria, for which rapid division is a key component of fitness, the adaptation to environmental variations, but also to various hosts for pathogens, is essential. These adaptations required selective expression of a set of genes depending on the environmental injury. Two-component systems are the main systems sensing these variations directly in the border between the cell and the environment. They are also responsible for the plasticity of gene expression in response to these environmental variations.
Typical two component systems are characterized by a transmembrane sensor histidine kinase/phosphatase and by a cytoplasmic cognate response regulator. Under stimuli, often unknown, the sensor histidine kinase autophosphorylates and transfers its phosphate group to its cognate response regulator, which in turn regulates the expression of different target genes (Hoch, 2000). Dephosphorylation is thought to occur via the reverse way in absence of stimulus. Several two component systems, such as CpxAR (Suntharalingam et al. 2003; Ronnebaumer et al. 2009; Labandeira-Rey et al. 2010), are implicated in virulence and in stress response of many pathogens.
The CpxAR system regulates expression of genes required for the envelope stress response (Raivio and Sylhavy, 1997) and for motility, is required for resistance against antimicrobial peptides (Audrain et al. 2013) and is consequently required for virulence (Raivio 2005, Ravio 2014). The Cpx system includes the transmembrane sensor histidine kinase CpxA and its cytoplasmic response regulator CpxR. In E. coli, this system was shown to be involved in folding and quality control of periplasmic proteins, particularly via the regulation of the expression of spy or degP (Price and Raivio 2009). The spy gene encodes a periplasmic ATP-independent chaperone that prevents protein aggregation and aids in the folding of proteins. DegP, a periplasmic protease, degrades all the misfolded proteins in the periplasm. The CpxAR system is also involved in the regulation of the porin OmpF (Batchelor et al., 2005). Surprisingly, despite its important role in envelope integrity maintenance and in virulence, this two-component system was often studied in vitro in E. coli or in zoopathogenic bacterial species, a few times in vivo in zoopathogenic bacterial species (Humphreys et al., 2004, Spinola et al. 2010) but never in phytopathogenic bacterial species.
Our model, Dickeya dadantii EC3937 (formerly Erwinia chrysanthemi) is a necrotrophic phytopathogenic Enterobacteria and causative agent of soft-rot diseases affecting a wide range of plant species, especially crops. Dickeya spp. are directly responsible for from 5 to 25% of the losses of potato crops in Europe and Israel (Toth et al., 2011). The pathogen is list as a A2 quarantine organism by the European and Mediterranean Plant Protection Organization (OEPP/EPPO, 1982, 1988, 1990).
The infection process occurs as follows. D. dadantii colonize the surface of the plant via the motility. Bacteria penetrate at a wounded site of the plant. Once in the apoplast, bacteria fight against plant defenses mainly acidic stress, oxidative stress, and antimicrobial peptides (Plessis et al., 2011; Reverchon and Nasser, 2013). Bacteria synthesize a set of plant cell wall degrading enzymes (PCWDEs), particularly pectinases and cellulases (Barras et al., 1994; Collmer and Keen, 1986), allowing growth of bacteria on degraded plant polymers and spread of the disease throughout the whole plant leading to maceration, the visible symptom of the disease.
In this study, we characterized the CpxAR system in D. dadantii. Data indicated that this two component system is essential for virulence because cpxA or cpxR mutant strains are completely non virulent, and that activation of the CpxAR two-component system is required during the early steps of the virulence process.
Results
Putative structural features and phylogeny of CpxAR
The putative cpxR and cpxA genes of D. dadantii are annotated on its genome and are classically organized in an operon of two genes cpxRA (Glasner et al, 2011). We analyzed the family and domain of CpxA and CpxR proteins by using Pfam databases (Finn et al., 2014). The CpxA (Dda3937_03052) is the putative sensor protein of 457 amino acids with 2 transmembrane segments, from amino acid 6 to amino acid 27 and from amino acid 165 to amino acid 184 separated by a periplasmic domain of 138 amino acids. The first transmembrane segment is preceded by 5 cytoplasmic amino acids. The second transmembrane segment is followed by a cytoplasmic amino acids sequence (amino acids 185 to 457) carrying three domains. A so called HAMP domain overlaps the second transmembrane segment and the beginning of the cytoplasmic amino acid sequence (amino acids 164 to 234). A histidine kinase is located between amino acids 238 and 300 and its ATPase domain is located between amino acids 345 and 454. The CpxR (Dda3937_03053) encodes the putative 232 amino acid cognate cytoplasmic regulator protein containing the response regulator domain (amino acid 4 to 112) containing the phosphorylatable D51 residue and the transregulator domain required for fixation to DNA regulatory sequences (amino acid 151 to 228). A phylogenic tree of the CpxR protein obtained by the maximum-parsimony method was constructed with all the bacterial species in which the Cpx system has been studied (in red) and with a selection of the most important pectinolytic phytopathogenic bacterial species (in green) (Fig. 1A). A response regulator from Gloeobacter violaceus was used as outgroup (in black) and indicate the possible location of the root. No separated group could be observed between the CpxR sequences from zoopathogens and the CpxR sequences from phytopathogen. A similar tree was observed for the CpxA homologues (Fig. 1B). No synteny was observed among species. This in silico analysis strongly suggests that this operon encodes the CpxAR two-component system of D. dadantii.
Figure 1. Rooted phylogenetic tree of CpxR (A) and CpxA (B) based on the mamixum likelihood.
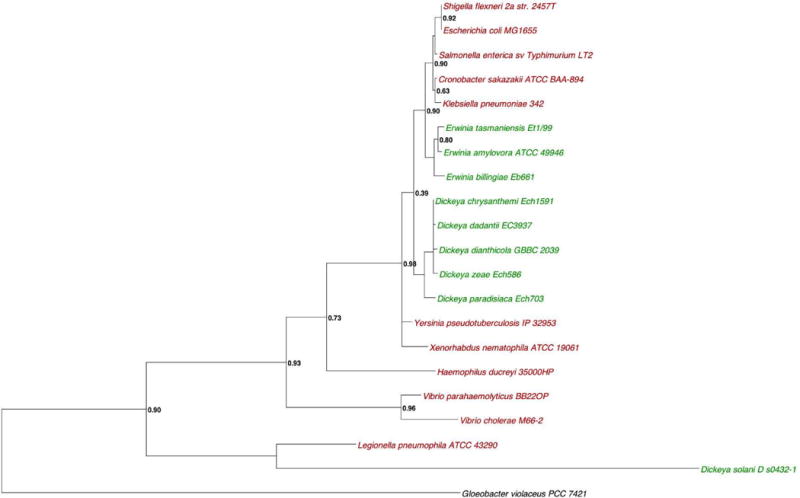
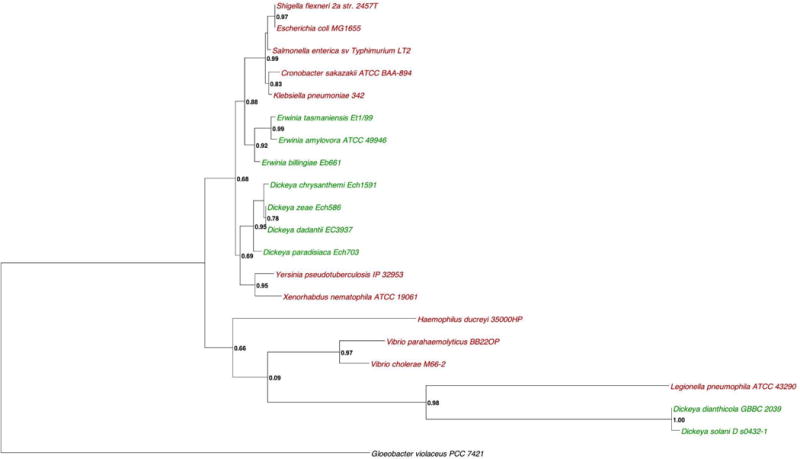
Numbers on knot are bootstrap confidence levels. Zoopathogen are in red. Phytopathogen are in green. In black, Gloeobacter violaceus used as outgroup. Both trees were constructed with the entire protein sequence from Uniprot.
The Cpx two component system controls expression of the degP, ompF and spy genes in D. dadantii
To confirm that the genes found in silico encode the CpxRA two component system of D. dadantii, expression of degP, spy and ompF were analyzed. These three genes are known to be regulated by the CpxRA two-component system in other related bacterial species (see introduction). The cpxA and the cpxR genes were inactivated by reverse genetics and we constructed and introduced an ectopic copy of the degP::uidA, ompF::uidA, and spy::uidA transcriptional fusions into these cpxA and cpxR null mutant strains (see materials and methods). The activity of the 3 transcriptional fusions was measured (Fig. 2 A–C). As compared to the wild-type strain, the cpxA mutant strain displays a 3-fold increase in the expression of the 3 genes while the cpxR mutant strain displays a 1.5-fold increase in degP expression, and a 0.8 and 4-fold decrease in expression of the spy and the ompF expressions, respectively. To confirm that the change in the regulation of the degP, ompF and spy genes was the result of the inactivation of the sensor histidine kinase cpxA or the response regulator cpxR, the plasmids pNFW512 (harboring the cpxA wild-type gene, cpxA+) and pNFW460 (harboring the cpxR wild-type gene, cpxR+), were constructed and complementation was performed. Plasmid pCpxA (harboring the wild-type cpxA gene) was introduced in the cpxA mutant strains harboring the degP, spy and ompF fusions and plasmid pCpxR (harboring the wild-type cpxR gene) was introduced in the cpxR mutant strains harboring the same fusions (respectively pCpxA/cpxA and pCpxR/cpxR strains). Complementation occurred, because expression was restored at a level similar to the one observed in the wild-type strain for all the fusions. Taken together, these results strongly suggest that the inactivated genes are the cpxA and cpxR genes of D. dadantii. The CpxA and CpxR are known to activate expression of the genes of its regulon. The hypothesis generally given to explain the increased expression of the genes of the cpx regulon in cpxA null mutant strain is the hyperphosphorylation of the CpxR regulator protein (Klein et al., 2007, Wolfe et al., 2008 Labandeira-Rey et al., 2010, Wolfe, 2010).
Figure 2. Effect of the cpxA or cpxR inactivation on the degP (A), spy (B) and ompF (C) gene expression and on the phosphorylation level of the CpxR regulator (D-F).
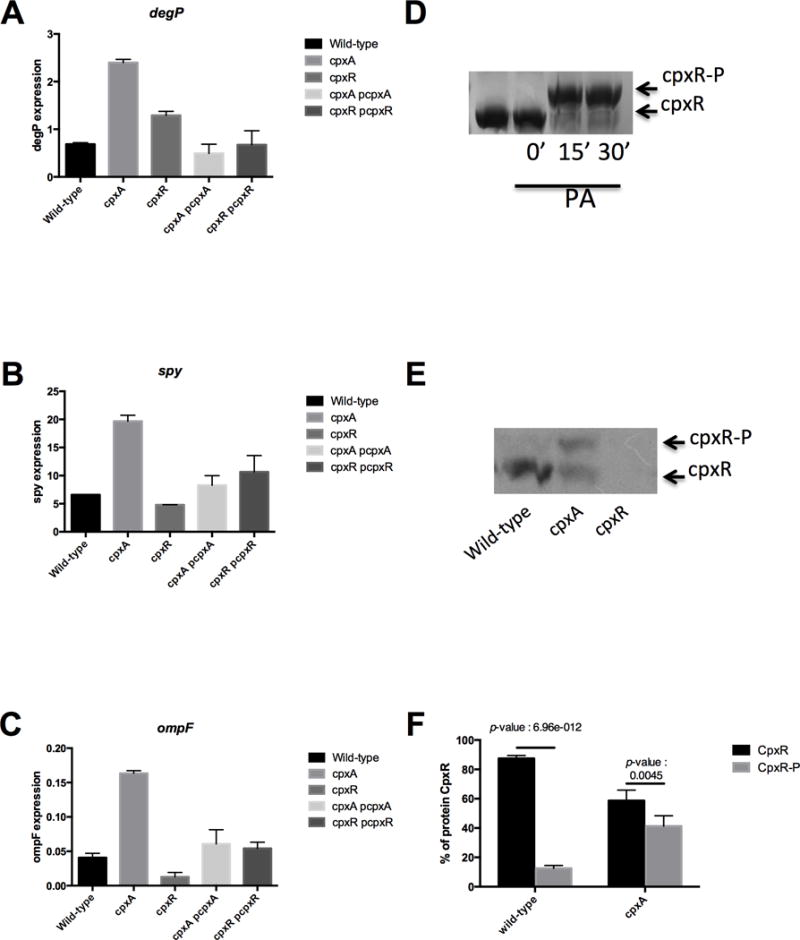
(A–C) For expression of the degP::uidA, spy::uidA and ompF::uidA gene fusions, bacteria were grown to the mid-log phase and lysed by sonication. B-glucuronidase activity was measured with PNPU as a substrate. Specific activity was expressed as the change in OD405 per minute and per milligram of protein. Results are the average of four independent experiments. (D) Six micrograms of purified CpxR regulator was incubated in the presence of 15 mM phosphoramidate (PA) for 0 min, 15 min and 30 min or without PA for 30 min before loading onto the Phos-tag acrylamide gel (125 μM). After migration, the gel was stained with Coomassie Blue Brilliant to revealed the CpxR and CpxR-P. (E) Separation of CpxR and CpxR-P by Phos-Tag™ gel after extraction from bacteria in vivo. Cell lysate of wild-type, cpxA null mutant, and cpxR null mutant of D. dadantii were loaded into Phos-Tag acrylamide gel (35 μM). Both forms of CpxR were revealed by Western-blot. This results presented are from one of the three independent experiments performed. (F) Quantification of CpxR and CpxR-P extracted from the wild-type and the cpxA null mutant strains. Results are the average of four independent experiments.
The CpxR regulator protein is constitutively phosphorylated in a cpxA null mutant
To examine directly the hyperphosphorylation hypothesis of CpxR in cpxA null mutants, we carry out Phos-Tag™ analysis. Phos-Tag™ analysis is based on a dinuclear metal complex linked to acrylamide molecules, which binds phosphate groups. In a Phos-Tag™ acrylamide gel, the Phos-tag™ linked to acrylamide molecules interacts with the phosphorylated form of the protein, which migrates more slower than the non-phosphorylated protein, allowing the separation of both forms of the protein. This method has to be adapted to each regulator (Barbieri and Stock, 2008). Purified CpxR was phosphorylated in vitro by the phosphor-donor phosphoramidate (PA). 5 ug of CpxR was incubated different times with or without PA, then the samples were loaded onto a Phos-Tag™ acrylamide gel and stained with coomassie blue (Fig. 2 D). Without PA, a single band corresponding to the non-phosphorylated CpxR form (called CpxR) was observed (Fig. 2 D). In contrast, a retardation of migration was observed when PA was added during 15 min and 30 min (Fig. 2 D). This shifted band corresponds to the phosphorylated CpxR form (called CpxR-P). The in vivo phosphorylation level was performed using the same approach (Fig. 2 E). In the wild-type strain, almost all the CpxR (89%) was not phosphorylated (Fig. 2 E). As expected, no band could be observed in cpxR null mutant strain (Fig. 2 E). In the cpxA null mutant strain, almost half (41%) of the CpxR was phosphorylated (Fig. 2 E, and Fig. 2 F). Thus, Phos-Tag data agrees with fusion data. There are yet no studies reporting direct observation of CpxR phosphorylation levels in various genetic backgrounds.
The CpxAR two-component system is required for virulence in D. dadantii
The impact of the inactivation of the Cpx system on the virulence of D. dadantii was evaluated in chicory leaves, carrots and potatoes. Each plant host was inoculated with the D. dadantii wild-type, cpxA and cpxR mutant strains (EC3937, NFB7515 and NFB7532, respectively) (Fig. 3). After 72h of incubation, no maceration could be observed with mutant strains in any plant host (Fig. 3). To confirm that the inactivation of the sensor histidine kinase cpxA or the response regulator cpxR provokes a loss of virulence in the three plant hosts, the cpxA/pCpxA or cpxR/pCpxR complemented strains were inoculated in to various plants. In each kind of plant, the virulence was restored to a level similar to that seen when using the wild-type strain (Fig. 3). To understand the total loss of virulence, the impact of the inactivation of the Cpx system on two of the major virulence factors (the motility and the PCWDE production) was observed.
Figure 3.
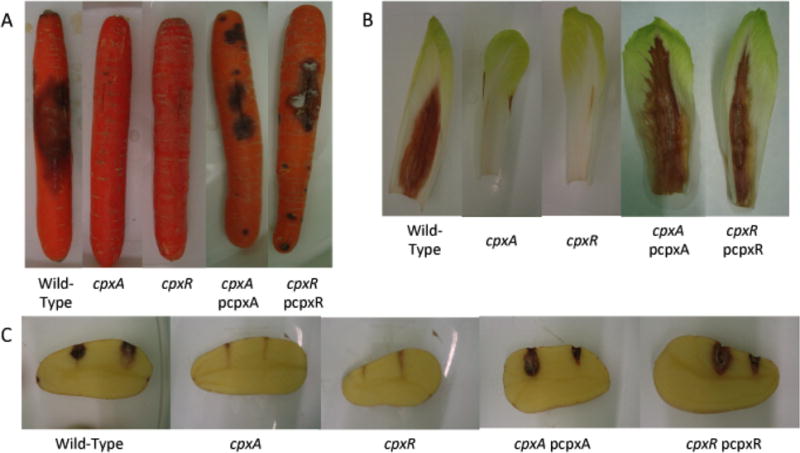
Pathogenicity of the wild-type, cpxA null mutant, cpxR null mutant and complemented strains on carrots (A), chicory leaves (B) and potato tubers (C) after 3 days.
The motility is affected by the inactivation of the CpxAR two-component system
Motility is required for full virulence, especially for the spread of the disease throughout the plant. The impact of the inactivation of the CxpAR two-component system on motility was measured on soft agar plates (Fig 4 A). The cpxA and cpxR null mutant strains showed 2-fold lower swim diameters than the wild-type strain (Fig. 4 A). The motility was restored in the pCpxA/cpxA and pCpxR/cpxR complemented strains since similar swim diameters as the wild-type strain were observed (Fig. 4 A).
Figure 4. Motility (A), pectate-lyase (B) and cellulase activities (C) of the wild-type, cpxA null mutant, cpxR null mutant and complemented strains.
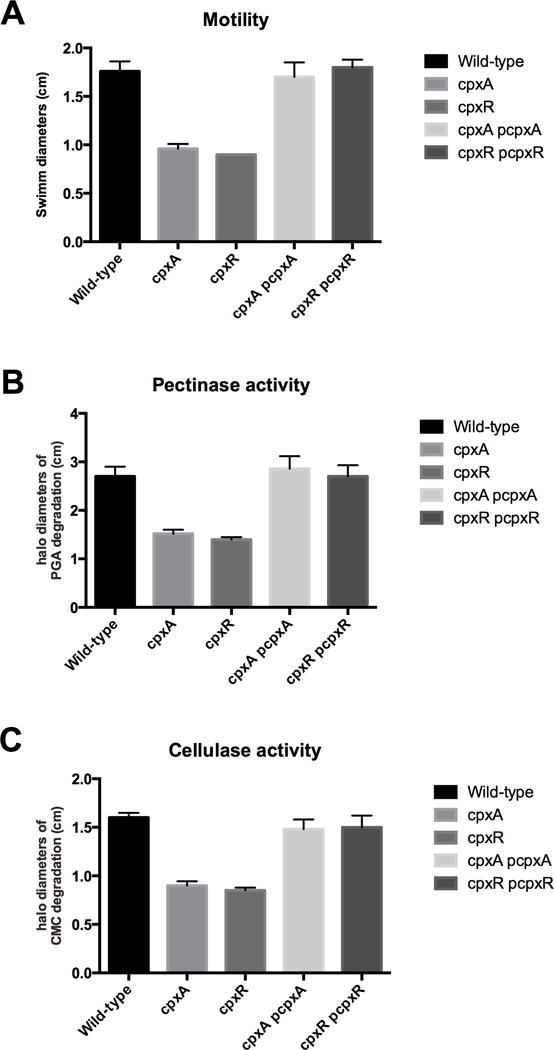
(A) Motility was measured in LB semisolid plates. Swim diameters were measured after 30h of incubation at 30°C. (B-C) Exoenzyme activities were estimated on plates by measurement of the halo diameters, expressed in cm of substrate degradation.
The production of the PCWDEs is reduced by the inactivation of the CpxAR two-component system
The PCWDEs are essential factors of virulence and are directly responsible for the damage (maceration) caused by D. dadantii. Global pectinase and cellulase activities were estimated on plates (Fig. 4 B–C).
In the pectinase plate test, haloes derived from polygalacturonate degradation by the cpxA and cpxR null mutant strains were severely reduced as compared to the wild-type strain (Fig. 4 B). The complementation with the pCpxA/cpxA and pCpxR/cpxR strains allowed restoration of halos similar to those seen in the wild-type strain.
In the cellulase plate test, haloes derived from carboxylmethylcellulase degradation by the cpxA and cpxR null mutant strains were severely reduced as compared to the wild-type strain (Fig. 4 C). The complementation with the pCpxA/cpxA and pCpxR/cpxR strains allowed restoration of halos similar to those seen in the wild-type strain.
Thus, two major virulence factors of D. dadantii, i. e. motility and PCWDEs production, are severely affected by loss of the CpxAR two component system.
The CpxAR system is required in the resistance of stresses delivered by plant cells against D. dadantii.
As a pathogen, D. dadantii has to defend against the plants defense. During its infectious cycle, D. dadantii has to protect against acidic stress, oxidative stress and antimicrobial peptide (Plessis et al., 2011). To investigate the role of the CpxAR system in the response to these stresses, the resistance of the cpxA and cpxR null mutant strains was compared to the wild type strain for each stress.
No difference in resistance to acidic stress, encountered by bacteria during maceration, was observed (data not shown) assayed by comparing the growth rates of the mutant strains in M63 medium at pH 4.5 and 7. For each pH, no significant growth rate difference was observed between the two different mutant strains with growth rates values of 0.63 and 0.67 at pH 4.5 and 7 respectively.
Resistance to the oxidative stress was also assayed. Stationary-phase culture of the wild-type, the cpxA and cpxR null mutant strains were exposed to 6 mM H2O2 and the number of viable cells was counted after plating serial dilution on LB plates. The survival of the cpxA and cpxR null mutant strains was significantly lower than the wild-type strain (Fig. 5A). The resistance to H2O2 was similar to the wild-type strain for the two complemented strains pCpxA/cpxA and pCpxR/cpxR (Fig. 5A).
Figure 5. Sensitivity tests of the wild-type, cpxA null mutant, cpxR null mutant and complemented strains to H2O2 or polymyxin B.
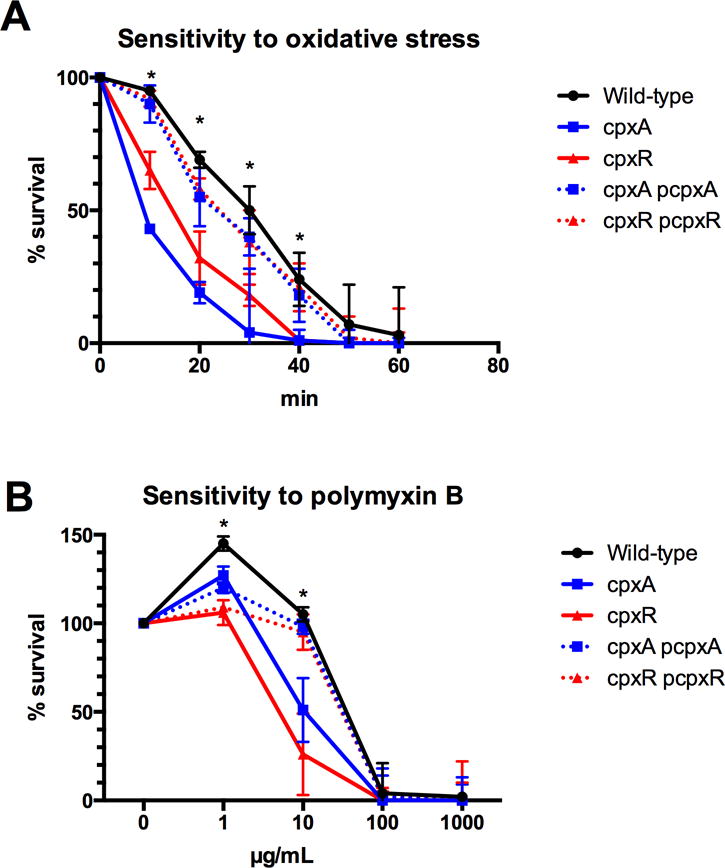
(A) For sensitivity to H2O2, bacteria were grown until stationary-phase in M63 glycerol medium. The culture was incubated with 3 mM of H2O2 and survivals were determined by numeration. Survival rate was expressed as the number of CFU counted at each time/the number of CFU before addition of H2O2. Results are the average of three independent experiments (B) For sensitivity to polymyxin B, bacteria were grown until stationary phase in M63 glycerol medium. The culture was incubated with the indicated concentrations and survivals were determined by numeration. Survival rate was expressed as the number of CFU counted at each time/the number of CFU before addition of polymyxin B. Results are the average of three independent experiments. Asterisks indicate that a significant difference exists only between the wild-type and the cpxA or cpxR null mutants.
Resistance to the antimicrobial peptide was evaluated with polymixin B. The cpxA and cpxR null mutant strains were significantly more sensitive than the wild-type strain (Fig. 5 B). At 10 μg/ml, only half of the initial population was still alive for the cpxA and cpxR null mutants while all the population of the wild-type strain was still alive (Fig. 6B). The wild-type resistance level resumed for the pCpxA/cpxA and pCpxR/cpxR complemented strains.
Figure 6. Expression of degP (A), spy (B) and ompF (C) gene and phosphorylation level of CpxR (D-E) during infection in chicory leaves.
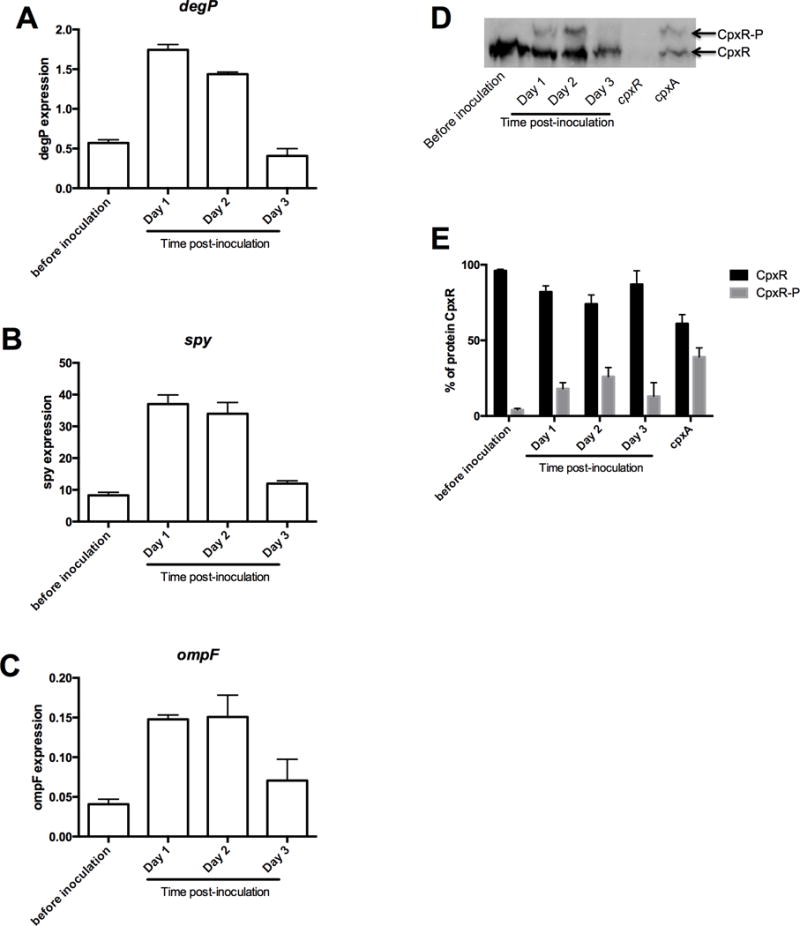
(A–C) B-glucuronidase activity was measured with PNPU as a substrate. Specific activity was expressed as the change in OD405 per minute and per milligram of protein. Results are the average of three independent experiments. (D) Separation of CpxR and CpxR-P by Phos-Tag™ gel after extraction from bacteria during the infectious cycle. Cell lysate of wild-type strain before inoculation and after 1, 2 or 3 days after the inoculation, cpxR null mutant, cpxA null mutant from in vitro stationary growth phase culture of D. dadantii were loaded into Phos-Tag™ acrylamide gel (35 μM). Both forms of CpxR were revealed by Western-blot. (E) Quantification of CpxR and CpxR-P extracted from wild-type before inoculation and after 1, 2 or 3 days after the inoculation, cpxR null mutant, cpxA null mutant from in vitro culture of D. dadantii. Results are the average of three independent experiments.
These data indicate that the Cpx system is involved in the response to the plant defense and suggest that the Cpx system is activated at an early stage of the infectious process.
Activation of the CpxAR two-component system is required in the early stage of the infectious process
To investigate when D. dadantii needs to activate the CpxAR two component system during its infectious process, we measured the expression level of the degP, spy and ompF gene before inoculation and after one, two, and three days of incubation in chicory leaves (Fig. 6). The expression level increased 4-fold for degP and spy (Fig. 6AB) and 3-fold for ompF (Fig. 6C) one day after inoculations and stayed at the same level during the second day after inoculation as compared to the levels observed before inoculation. The expression level of these 3 genes decrease in the third day of inoculation and display a level similar to the one observed before inoculation.
To strengthen these results, a Phos-tag approach was performed on bacteria extracted from chicory leaves after each of these three days of infection. Because chicory leaves are routinely inoculated with bacteria grown until their stationary phase, CpxR phosphorylation before inoculation (Fig. 6 D) was monitored in these physiological conditions unlike in Fig. 2E where CpxR phosphorylation was monitored in bacteria grown until mid-log phase. The CpxR phosphorylation level of the cpxA mutant strain was added as a control of CpxR (61%) and phosphorylated CpxR (39%). Phosphorylated CpxR (4%) was detected in bacteria extracted before inoculation (Fig. 6 D). CpxR phosphorylation increased after one day of infection (18% of phosphorylated CpxR), and after two days of infection (26% of phosphorylated CpxR) but decreased after the third day of infection (13% of phosphorylation) (Fig. 6D). Thus, the Cpx two-component system is activated at an early stage of the infectious process in response to the plants immune response of the plant, but activation decreases as the majority part of the leaf is macerated, logically suggesting a decrease in plant defense. This is the first time that the variation of the phosphorylation level of the response regulator of a two-component system has been observed in a bacterial pathogen species during its infection cycle.
Discussion
During the infectious cycle, D. dadantii goes from the surface of the leaf through the apoplast. To achieve a successful infection, bacteria needs to sense its new environment (i.e. the plant) and to adapt. Bacteria have one and two-component systems to sense and respond to environmental variations. One-component system are more likely involved in the adaptation of the metabolism to fit better to the new environment (Ulrich et al., 2005). Two-component systems are the major system able to sense stresses from the environment directly in the border between the cell and the environment. Among the 32 two-component systems of D. dadantii, only GacAS and PhoPQ systems were shown to be essential for the virulence (Llama-Palacios et al., 2003, Llama-Palacios et al., 2005, Yang et al., 2008, Lebeau et al., 2008). For both, the inactivation of the system displays a total loss or severe decreased of virulence in planta. Based on our study, the CpxAR system may be added to the list of essential two-component systems required for virulence.
Inactivation of cpxA or cpxR genes provokes a change in the expression of degP, spy and ompF. OmpF is a porin while DegP and Spy proteins are involved in folding and quality control of periplasmic proteins. These results suggest that the CpxAR system plays a classic role in the fitness of the envelope. This result can be correlated with the phylogenic tree showing the conservation of the CpxAR system among the zoopathogenic and the phytopathogenic bacterial species. In E. coli, the OmpF porin expression is regulated, to a large extent, by the EnvZ/OmpR two-component system. In D. dadantii, the EnvZ/OmpR two-omponent system is not involved in the regulation of ompF expression (Condemine and Ghazi, 2007). Our study showed that in D. dadantii, the regulation of ompF expression depends at least in part on the CpxAR two component system.
The loss of the CpxAR system results in a total loss of virulence regardless the kind of plant used. This loss of virulence is the result of at least three factors. First of all, the CpxAR system is involved in the regulation – although indirect – of the PCWDEs and motility. Second, the CpxAR system is involved in the protection of D. dadantii against plant defenses (oxidative stress, peptide antimicrobial). Our data suggest that these plant defenses are mainly effective during the early phase of the infectious cycle. Our result show both by gene expression analysis and Phos-Tag analysis, that the CpxAR system is also activated during the early phase of the infectious cycle. Taken together, these data show clearly that the CpxAR system is activated during the early phase of the infectious cycle to counteract the damage provoked by plant defenses.
A global opposite effect on the expression of the cpx regulon was often observed in other bacterial species when either CpxA or CpxR were inactivated. The hypothesis generally given to explain the increased expression of the genes of the cpx regulon in cpxA null mutant strains is the constitutive phosphorylation of the CpxR regulator protein (Klein et al., 2007, Wolfe et al., 2008 Labandeira-Rey et al., 2010, Wolfe, 2010). We demonstrate this hypothesis by a Phos-Tag gel analysis since the loss of cpxA displays an excess of the phosphorylated form of the CpxR regulator as compared to the CpxR phosphorylation level of the wild-type strain. In silico analysis indicates that CpxA possesses both kinase and phosphatase activities. The fine-tuning between both activities is required to obtain the adapted response allowing appropriate expression of genes of the cpx regulon in the different environments encountered by D. dadantii. Activation of the CpxAR two-component system is essential to fight against plant defenses. Thus, the non-virulent phenotype of the cpxA null mutant was surprising since this mutant displays a constitutive activation of CpxR. Excess of CpxR-P was previously described to affect the virulence in Haemophilys ducreyi, Salmonella typhimurium or Yersinia pseudotuberculosis (Humphreys et al., 2004; Spinola et al., 2010, Liu et al., 2012). In Salmonella typhimurium, Humphreys and collaborators proposed that excess of CpxR-P down-regulated genes involved in the virulence (Humphreys et al., 2004).
The reason of the same phenotype in cpxA and cpxR null mutant in oxidative stress or polymixin B sensitivity is not clear. Same guess can be proposed for the oxidative stress or polymixin B sensitivity. An excess of CpxR-P harm the defense against oxidative stress or polymixin B because the CpxAR system affects directly the traffic into the membrane.
Moreover a similar phenotype is observed in Salmonella typhimurium for the PhoPQ two-component system. The inactivation of the response regulator PhoP as well as a constitutive activation of PhoPQ two-component system attenuates the virulence of the bacteria (Miller, 1991).
This indicates that the fine-tuning observed in the wild-type strain is also of importance in planta for survival of D. dadantii and that phosphorylation of the CpxR regulator must be turned off as soon as possible because excess of CpxR phosphorylation becomes deleterious when not required for D. dadantii in this kind of environment. Taken together, these results strongly suggest that without stress, the phosphatase activity is the main activity of the CpxA sensor needed to maintain the CpxR regulator at a very low level of phosphorylation.
Experimental Procedures
Bacterial strains, media and growth conditions
Bacterial strains are described in Table 1. Bacteria were grown at 30°C in lysogeny broth (LB) (Bertani, 2004), or in minimal medium M63 supplemented with a carbon source at a concentration of 2 g.l−1 (Miller, 1992). Solid media were obtained by adding agar at 15 g.l−1.
Table 1.
Strain, Plasmid and Primer
| Strain | Genotype a, b | Source or Reference |
|---|---|---|
| Dickeya dadantii | ||
| EC 3937 | Wild-type | Laboratory collection |
| A 4229 | ompF::uidA-Kan | Condemine and Ghazi, 2007 |
| NFB 7012 | mTn5degP::uidA-Kan | This study |
| NFB 7039 | mTn5spy::uidA-Kan | This study |
| NFB 7515 | cpxA::gm | This study |
| NFB 7532 | cpxR::gm | This study |
| NFB 7539 | cpxA::gm ompF::uidA-Kan | This study |
| NFB 7543 | cpxR::gm ompF::uidA-Kan | This study |
| NFB 7548 | cpxR::gm mini-Tn5degP::uidA-Kan | This study |
| NFB 7549 | cpxA::gm mini-Tn5degP::uidA-Kan | This study |
| NFB 7550 | cpxR::gm mini-Tn5spy::uidA-Kan | This study |
| NFB 7551 | cpxA::gm mini-Tn5spy::uidA-Kan | This study |
| Escherichia coli | ||
| BL21(DE3) | ompT, hsdSB, gal, dcm | Invitrogen |
| Top 10 F’ | F′(lacIq, Tn10) mcrA, Δ(mrr-hsdRMS-mcrBC), Φ80lacZΔM15, ΔlacX74, recA1, araD139, Δ(ara- leu)7697, galU, galK, rpsLendA1 nupG | Invitrogen |
| S17-λpir | recA1, thi, pro, hsdR-M+, RP4:2-Tc::Mu-Kan::Tn7, λpir | De Lorenzo et al., 1994 |
| Plasmids | ||
| pUC18Not-uidA | ‘uidA, Amp | Bontemps-Gallo et al., 2014 |
| pJET 1.2 | Amp | Fermentas |
| pNFW460 | pJET2.1 cpxR | This study |
| pNFW466 | pJET2.1 cpxR::gm | This study |
| pNFW512 | pJET2.1 cpxA | This study |
| pNFW513 | pJET2.1 cpxA::gm | This study |
| pUTmini-Tn5-Kan | mini-Tn5Spe, oriR6K, Kan, Amp | De Lorenzo et al., 1990 |
| Primer sequencesc | ||
| cpxAFor | CGCGAGCTGACATCCCTATT | This study |
| cpxARev | TCGAAAAAGCTCTCCAGCGT | This study |
| cpxRFor | CTATCATCCAGCCCCTGAC | This study |
| cpxRRev | CGGATGCTGTTTCAGGTTAC | This study |
| K7gentabsrFor | TCTCTGTACACCCCCATCCCCCTGTTGAC | This study |
| K7gentabsrRev | TCTCTGTACACGCAAGCTAGCTTGGCTGC | This study |
| K7gentahpaFor | GTGTGTTAACCCCCATCCCCCTGTTGAC | This study |
| K7gentahpaRev | GTGTGTTAACCGCAAGCTAGCTTGGCTGC | This study |
| spyForKpnI | AAGGTACCTGATGGCTCCTGCCGCCGGCGA | This study |
| spyRevXbaI | GTTCTAGAAGCCGACGCTACCCAGCGCCAG | This study |
| degPForKpnI | TTGGTACCACAAACTCTCCAGCAAGCATTG | This study |
| degPRevXbaI | TGCTCTAGACTCAACGCCAACGCACTCAGC | This study |
| cpxRhisFor | CACCATGAACAAAATCCTGTTGGTTGATGACG | This study |
| cpxRhisRev | CGGTAACCCGAATCATGCGGTGG | This study |
The degP-uidA, cpxP-uidA and spy-uidA gene fusions are carried by a mini-Tn5 Kan
Amp, ampicillin resistance, Kan, kanamycin resistance, Gm, gentamicin resistance
Restriction sites are underlined
Motility tests were made on LB plates containing agar at 0.3 g.l−1. 107 bacteria in 5μL were spotted onto the plate, incubated at 30°C and swim diameters were measured after 30 hours of incubation.
The solid media used to test the pectinase and cellulases activities have been described previously (Page et al., 2001).
Antibiotics in media were used at following concentrations: kanamycin, 25 μg.ml−1; chloramphenicol, 25 μg.ml−1 and gentamycin, 2 μg.ml−1.
Cloning of the cpxA and cpxR genes
The cpxA and cpxR DNA fragments were amplified by PCR (cpxAFor and cpxARev, cpxRFor and cpxRRev primers respectively), and cloned in blunt end into pJET1.2 following the manufacturer’s recommendations (Fermentas) to give pNFW512 and pNFW460 respectively. The cloned DNA fragments were sequenced.
Construction of the cpxA and cpxR mutations
To inactivate cpxA, a gentamycin DNA cassette was amplified by PCR (K7gentabsrFor and K7gentabsrRev primers respectively) digested by BsrGI and inserted into pNFW521 digested by the same enzyme (pNFW513). To inactivate cpxR, a gentamycin DNA cassette was amplified by PCR (K7gentahpaFor and K7gentahpaRev primers respectively),digested by HpaI and inserted into pNFW460 digested by AfeI (pNFW466). After electroporation (2.5kV) of these two last plasmids, the mutations were integrated into the D. dadantii chromosome by marker exchange recombination in the presence of gentamycin after successive cultures in low phosphate medium (Roeder et al., 1985).
Construction of the spy and degP gene fusions
The spy’ and degP’ DNA fragments were amplified by PCR (spyForKpnI and spyPRevXbaI, degPForKpnI and degPRevXbaI primers respectively). The spy’ and degP’ DNA fragments were digested by KpnI and XbaI and cloned into pUC18Not-uidA digested by the same enzymes.
The NotI fragments of these plasmids encompassing spy::uidA and degP::uidA respectively, were cloned into pUTmini-Tn5 Kan.
Transduction, conjugation and transformation
Transformation of E. coli cells was carried out by the rubidium chloride technique (Miller, 1992). Construction of strains was performed by transferring genes from one strain of D. dadantii to another by generalized transduction with phage ΦEC2 as described previously (Resibois et al., 1984). Plasmids were introduced in D. dadantii by conjugation or electroporation.
Transposon mutagenesis
To allow integration of a single ectopic copy of the mini-Tn5 spy::uidA-Kan and mini-Tn5 degP::uidA-Kan genes fusions, transposon mutagenesis was performed as described previously (Bouchart et al., 2010). Briefly, after conjugation between an E. coli strain harboring the pUTmini-Tn5 Kan plasmid carrying the appropriate fusion and a D. dadantii strain, Kanr mutants were selected on M63 plates containing sucrose as a unique carbon source and kanamycin.
Determination of enzyme activities
β-glucuronidase assays were performed on crude extracts obtained from bacteria disrupted by sonication 2 × 20 s (Sonifier cell disruptor B-30, Branson, 70% duty cycle, 7 microtip limit, Hold time, continuous, appropriate probe) after growth in vitro (LB medium) or in planta and extracted from chicory leaves as described elsewhere (Bontemps-Gallo et al., 2013). β-glucuronidase activity was determined by spectrometric monitoring of the hydrolysis of PNPU (4-nitrophenyl- β-D-glucuronide) at 405 nm.
The protein concentration was determined by the Bradford assay with bovine serum albumin as a standard (Bradford, 1976).
Susceptibility to acidic stress, oxidative stress and antimicrobial peptides
The suceptibility to acidic stress was assayed by measuring and comparing the growth rates of bacteria in M63 medium at pH 4.5 and at pH7.
The susceptibility to oxidative stress (H2O2) and to antimicrobial peptides (polymixin B) was previously described (Bontemps-Gallo et al., 2014). H2O2 at 3 mM or polymyxin B at indicated concentrations was added to cells grown until stationary-phase in M63 glycerol medium. After different times of incubation, aliquot of bacteria were taken up, and the survival rates were determined by numeration of CFU after plating serial dilutions on LB plates.
Pathogenicity test
Potato tubers, chicory leaves or carrots were inoculated as previously described (Page et al., 2001). Briefly, bacteria from an overnight culture in LB medium were recovered by centrifugation and diluted in physiological water. After wounding, plants were inoculated with 107 bacteria and incubated in a dew chamber at 28°C until 48h.
Preparation of polyclonal antibodies against CpxR
A DNA fragment encoding the cpxR gene of D. dadantii was amplified by PCR using the primers cpxRhisFor and cpxRhisRev. The PCR product was cloned into a His6 tag expression vector, pET100/D-Topo® (Invitrogen Life Technologies). The resulting His-tagged CpxR was expressed in E. coli BL21(DE3) and the protein was purified by affinity chromatography according to the manufacturer’s procedure [Ni-nitrilotriacetic acid (NTA) agarose; Qiagen]. The purified CpxR was used to immunize rabbits (Eurogentec).
Analysis of CpxR phosphorylation in vitro
Phosphorylation reactions of purified His-tagged CpxR were performed with 5 µg of protein in 50 mM Tris-HCl, pH7.5, 100 mM NaCl, 10 mM MgCl2 and 2 mM β-mercaptoethanol. PA was added to a final concentration of 15 mM to initiate the reaction. After 0, 15, or 30 minutes of incubation at room temperature, the reactions were stopped by addition of SDS-PAGE loading buffer (final concentration: 50 mM Tris-HCl pH6.8, 2% (w/v) SDS, 10% (w/v) glycerol, 20 mM DTT, 0.02% bromophenol blue). The mixtures were resolved using phosphoprotein affinity gel electrophoresis as described in Barbieri and Stock, 2008, with minor modifications. Briefly, Phos-tag™ acrylamide gels were composed of a 10% resolving solution [10% (w/v) 37.5:1 acrylamide/N,N’methylenebisacrylamide, 375 mM Tris (pH8.8) and 0.1% (w/v) SDS, 125μM Phos-tag™ acrylamide and 250 μM MnCl2] and a 4% stacking solution [4% (w/v) 37.5:1 acrylamide/N,N’methylenebisacrylamide, 125 mM Tris (pH6.8) and 0.1% (w/v) SDS]. The gels were run at 4 °C under constant voltage (150 V) with standard running buffer (0.1% (w/v) SDS, 25 mM Tris and 192 mM glycine) and stained with Coomassie Blue.
Phos-Tag analysis of the CpxR phosphorylation in vivo
Gel retardation of the phosphorylated form of CpxR was performed as described previously (Madec et al, 2014). Briefly, 1.5×108 cells of D. dadantii cells were harvested by centrifugation and the pellet was lysed with 12.7 μl of 1M formic acid, solubilized by 5 μl of 4X SDS-PAGE loading buffer and neutralized by 2.8 μl 5 N NaOH. Sample was loaded and run onto gel containing 35 μM Phos-tag™ acrylamide and 70 μM MnCl2. Then, 10 min wash with transfer buffer (25 mM Tris and 192 mM glycine) supplied with 1 mM EDTA, followed by 10 min wash with transfer buffer without EDTA were performed before transferring gel to nitrocellulose membranes using a Trans-Blot® TurboTM Blotting system (Bio-Rad) with a pre-programmed protocol (2.5 A, up to 25 V, 7 min). Western blotting against CpxR was performed using standard protocols with the rabbit anti-CpxR polyclonal antibodies at a dilution of 1:1000 and anti-rabbit secondary antibody coupled to horseradish peroxidase at a dilution of 1:10 000. Blots were imaged by chemiluminescent detection (ECL kit, GE healthcare Pharmacia Biotech).
Quantification of the phosphorylated CpxR protein amount
Phosphorylated CpxR and unphosphorylated CpxR were quantified by determination of the area intensity of each band with the software Quantity One (Bio Rad) after detection by Western blot. Quantification of phosphorylated CpxR was expressed as the ratio of the phosphorylated CpxR amount divided by the sum of the phosphorylated CpxR and the unphosphorylated CpxR amounts as described elsewhere (Barbieri and Stock, 2008).
Statistical Analysis
For statistical analyses, Graph-prism6 software was used. Data were analysed by paired t-test; a value of p<0.05 was considered significant.
Phylogeny tree
The phylogenic tree was built using the Phylogeny.fr website (Dereeper et al., 2008). The protein sequences of the 21 strains were aligned with Muscle 3.6 with default parameters. The trees were generated using the maximum likelihood based on a bootstrapping procedure of 100 bootstraps. The protein sequence of the strain Gloeobacter violaceus was used as outgroup. Phylogenetic trees were displayed with Dendroscope (version 3.2.10).
Acknowledgments
The authors thank Lucas Briche for technical assistance. We are grateful to Martha Thayer for the English corrections. The ‘A strains’ were kindly provided by Dr N. Cotte-Pattat and Dr G. Condemine (CNRS UMR 5240, University of Lyon 1). This work was supported by Grants from the Centre National de la Recherche Scientifique (CNRS), the Université de Lille 1 and the Ministère de l’Enseignement Supérieur et de la Recherche. SBG was funded by post-doctoral fellowship of the Lille 1 University. The funders were not involved in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions
Conceived, designed and performed the experiments: SBG EM JML.
Analyzed the data: SBG EM JML.
Wrote the manuscript: SBG JML.
References
- Audrain B, Ferrières L, Zairi A, Soubigou G, Dobson C, Coppée JY, Beloin C, Ghigo JM. Induction of the Cpx envelope stress pathway contributes to Escherichia coli tolerance to antimicrobial peptides. Appl Environ Microbiol. 2013;79:7770–7779. doi: 10.1128/AEM.02593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tagTM -based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F, van Gijsegem F, Chatterjee AK. Extracellular Enzymes and Pathogenesis of Soft-Rot Erwinia. Annu rev phytopathol. 1994;32:201–234. [Google Scholar]
- Batchelor E, Walthers D, Kenney LJ, Goulian M. The Escherichia coli CpxA–CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol. 2005;187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Lysogeny at Mid-Twentieth Century: P1, P2, and Other Experimental Systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S, Madec E, Dondeyne J, Delrue B, Robbe-Masselot C, Vidal O, Prouvost AF, Boussemart G, Bohin JP, Lacroix JM. Concentration of osmoregulated periplasmic glucans (OPGs) modulates the activation level of the RcsCD RcsB phosphorelay in the phytopathogen bacteria Dickeya dadantii. Environ Microbiol. 2013;15:881–894. doi: 10.1111/1462-2920.12054. [DOI] [PubMed] [Google Scholar]
- Bontemps-Gallo S, Madec E, Lacroix JM. Inactivation of pecS restores the virulence of mutants devoid of osmoregulated periplasmic glucans in the phytopathogenic bacterium Dickeya dadantii. Microbiology. 2014;160:766–777. doi: 10.1099/mic.0.074484-0. [DOI] [PubMed] [Google Scholar]
- Bouchart F, Boussemart G, Prouvost AF, Cogez V, Madec E, Vidal O, Delrue B, Bohin JP, Lacroix JM. The virulence of a Dickeya dadantii 3937 mutant devoid of osmoregulated periplasmic glucans is restored by inactivation of the RcsCD-RcsB phosphorelay. J Bacteriol. 2010;192:3484–3490. doi: 10.1128/JB.00143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Collmer A, Keen NT. The Role of Pectic Enzymes in Plant Pathogenesis. Annu rev phytopathol. 1986;24:383–409. [Google Scholar]
- Condemine G, Ghazi A. Differential regulation of two oligogalacturonate outer membrane channels, KdgN and KdgM, of Dickeya dadantii (Erwinia chrysanthemi) J Bacteriol. 2007;189:5955–5962. doi: 10.1128/JB.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo V, Timmis KN. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res. 2008;36:465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPPO. Data sheets on quarantine organisms, Erwinia chrysanthemi Burkholder. Bulletin OEPP/EPPO Bulletin. 1982;12:21–5. [Google Scholar]
- EPPO. Specific Quarantine Requirements. 92 Paris, France: EPPO/EPPO Publications Series B; 1988. A1 and A2 Lists of Quarantine Pests. [Google Scholar]
- EPPO. Specific Quarantine Requirements. 1008 Paris, France: EPPO/EPPO Technical Documents; 1990. [Google Scholar]
- Fin RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: the protein families database. Nucl Acids Res. 2013;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasner JD, Yang CH, Reverchon S, Hugouvieux-Cotte-Pattat N, Condemine G, Bohin JP, Van Gijsegem F, Yang S, Franza T, Expert D, Plunkett G, San Francisco MJ, Charkowski AO, Py B, Bell K, Rauscher L, Rodriguez-Palenzuela P, Toussaint A, Holeva MC, He SY, Douet V, Boccara M, Blanco C, Toth I, Anderson BD, Biehl BS, Mau B, Flynn SM, Barras F, Lindeberg M, Birch PR, Tsuyumu S, Shi X, Hibbing M, Yap MN, Carpentier M, Dassa E, Umehara M, Kim JF, Rusch M, Soni P, Mayhew GF, Fouts DE, Gill SR, Blattner FR, Keen NT, Perna NT. Genome sequence of the plant-pathogenic bacterium Dickeya dadantii 3937. J Bacteriol. 2011;193:2076–2077. doi: 10.1128/JB.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA. Two-component and phosphorelay signal transduction. Curr Microbiol. 2000;3:165–70. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Humphreys S, Rowley G, Stevenson A, Anjum MF, Woodward MJ, Gilbert S, Kormanec J, Roberts M. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect Immun. 2004;72:4654–4661. doi: 10.1128/IAI.72.8.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate inEscherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M, Brautigam CA, Hansen E. Characterization of the CpxRA regulon in Haemophilus ducreyi. J Infect Immun. 2010;78:4779–4791. doi: 10.1128/IAI.00678-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau A, Reverchon S, Gaubert S, Kraepiel Y, Simond-Côte E, Nasser W, Van Gijsegem F. The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Env Microbiol. 2008;10:545–559. doi: 10.1111/j.1462-2920.2007.01473.x. [DOI] [PubMed] [Google Scholar]
- Llama-Palacios A, López-Solanilla E, Poza-Carrión C, García-Olmedo F, Rodríguez-Palenzuela P. The Erwinia chrysanthemi phoP-phoQ operon plays an important role in growth at low pH, virulence and bacterial survival in plant tissue. Mol, Microbiol. 2003;49:347–357. doi: 10.1046/j.1365-2958.2003.03583.x. [DOI] [PubMed] [Google Scholar]
- Llama-Palacios A, López-Solanilla E, Rodríguez-Palenzuela P. Role of the PhoP-PhoQ System in the Virulence of Erwinia chrysanthemi Strain 3937: Involvement in Sensitivity to Plant Antimicrobial Peptides, Survival at Acid pH, and Regulation of Pectolytic Enzymes. J Bacteriol. 2005;187:2157–2162. doi: 10.1128/JB.187.6.2157-2162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Thanikhal EJ, Obi IR, Francis MS. Elevated CpxR-P levels repress the Ysc-Yop type III secretion system of Yersinia pseudotuberculosis. Res in Microbiology. 2012;163:518–530. doi: 10.1016/j.resmic.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Madec E, Bontemps-Gallo S, Lacroix JM. Increased phosphorylation of the RcsB regulator of the RcsCDB phosphorelay in strains of Dickeya dadantii devoid of osmoregulated periplasmic glucans revealed by Phos-tag gel analysis. Microbiology. 2014;160:2763–2770. doi: 10.1099/mic.0.081273-0. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; New York: 1992. [Google Scholar]
- Miller SI. PhoP/PhoQ: macrophage-specific modulators of Salmonella virulence? Mol Microbiol. 1991;5(2073):2078. doi: 10.1111/j.1365-2958.1991.tb02135.x. [DOI] [PubMed] [Google Scholar]
- Page F, Altabe S, Hugouvieux-Cotte-Pattat N, Lacroix JM, Robert-Baudouy J, Bohin JP. Osmoregulated periplasmic glucan synthesis is required for Erwinia chrysanthemi pathogenicity. J Bacteriol. 2001;183:3134–3141. doi: 10.1128/JB.183.10.3134-3141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A, Cournol R, Effroy D, Silva Pérez V, Botran L, Kraepiel Y, Sotta B, Cornic G, Leung J, Giraudat J, Marion-Poll A, North HM. New ABA-Hypersensitive Arabidopsis Mutants Are Affected in Loci Mediating Responses to Water Deficit and Dickeya dadantii Infection. PLoS ONE. 2011;6(5):e20243. doi: 10.1371/journal.pone.0020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- Reverchon S, Nasser W. Dickeya ecology, environment sensing and regulation of virulence programme. Env Microbiol Reports. 2013;5:622–636. doi: 10.1111/1758-2229.12073. [DOI] [PubMed] [Google Scholar]
- Raivio TL. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta. 2014;1843:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Resibois A, Collet M, Faelen M, Schoonejans T, Toussaint A. Phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology. 1984;137:102–112. doi: 10.1016/0042-6822(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Rönnebäumer K, Sander G, Shutinoski B, Schmidt MA, Heusipp G. Controlled activation of the Cpx system is essential for growth of Yersinia enterocolitica. FEMS Microbiol Lett. 2009;296:274–281. doi: 10.1111/j.1574-6968.2009.01649.x. [DOI] [PubMed] [Google Scholar]
- Spinola SM, Fortney KR, Baker B, Janowicz DM, Zwickl B, Katz BP, Blick RJ, Munson RS., Jr Activation of the CpxRA system by deletion of cpxA impairs the ability of Haemophilus ducreyi to infect humans. Infect Immun. 2010;78:3898–3904. doi: 10.1128/IAI.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam P, Spencer H, Gallant CV, Martin NL. Salmonella enterica serovar typhimurium rdoA is growth phase regulated and involved in relaying Cpx-induced signals. J Bacteriol. 2003;185:432–443. doi: 10.1128/JB.185.2.432-443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth IK, van der Wolf JM, Saddler G, Lojkowska E, Hélias V, Pirhonen M, Tsror (Lahkim) L, Elphinstone JG. Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 2011;60:385–399. [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends in Microbiology. 2005;13(2):52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Parikh N, Lima BP, Zemaitaitis B. Signal integration by the two-component signal transduction response regulator CpxR. J Bacteriol. 2008;190:2314–2322. doi: 10.1128/JB.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol. 2010;13:204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Peng Q, Zhang Q, Yi X, Choi CJ, Reedy RM, Charkowski AO, Yang CH. Dynamic Regulation of GacA in Type III Secretion, Pectinase Gene Expression, Pellicle Formation, and Pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937) Mol Plant-Microbe Interact. 2008;21:133–142. doi: 10.1094/MPMI-21-1-0133. [DOI] [PubMed] [Google Scholar]


