In vivo and in vitro inhibition of IL-12, IL-23, and IL-10 stimulation by IFNβ.
Keywords: Type I interferons, STAT1/STAT2, IRF1/IRF7, PI3K/GSK3, cytokines
Abstract
MS is an autoimmune disease characterized by immune cell infiltration in the CNS, leading to cumulative disability. IFN-β, used clinically in RR-MS reduces lesion formation and rates of relapse. Although the molecular mechanisms are not entirely elucidated, myeloid cells appear to be a major target for the therapeutic effects of IFN-β. DCs have a critical role in experimental models of MS through their effect on encephalitogenic Th1/Th17 cell differentiation and expansion. Here we focused on the effects of IFN-β on DC expression of cytokines involved in the control of Th1/Th17 differentiation and expansion. Administration of IFN-β to mice immunized with MOG35–55 inhibited IL-12 and IL-23 expression in splenic DC and reduced in vivo differentiation of Th1/Th17 cells. IFN-β affected cytokine expression in TLR-stimulated DC in a similar manner in vitro, inhibiting IL-12 and IL-23 and stimulating IL-10 at both mRNA and protein levels, by signaling through IFNAR. We investigated the role of the signaling molecules STAT1/STAT2, IRF-1 and IRF-7, and of the PI3K→GSK3 pathway. IFN-β inhibition of the IL-12 subunits p40 and p35 was mediated through STAT1/STAT2, whereas inhibition of IL-23 was STAT1 dependent, and the stimulatory effect on IL-10 expression was mediated through STAT2. IFN-β induces IRF-7 and, to a lesser degree, IRF-1. However, neither IRF mediated the effects of IFN-β on IL-12, IL-23, or IL-10. We found that the PI3K pathway mediated IL-12 inhibition but did not interfere with the inhibition of IL-23 or stimulation of IL-10.
Introduction
IFN-β therapy is used to reduce the frequency and severity of relapses in patients with RR-MS [1, 2]. The beneficial role of IFN-β was reported in EAE models, where IFN-β treatment, including systemic stem cell-based IFN-β delivery, suppressed relapses, and IFN-β- or IFNAR-deficient mice developed exacerbated disease [3–8]. The beneficial effect of IFN-β in EAE appears to be directly associated with myeloid cells because conditional deletion of IFNAR in myeloid cells, but not in T cells or in neuroectodermal CNS cells, exacerbates EAE [5]. IFN-β engagement of IFNAR on both peripheral and CNS myeloid cells has been reported to modulate EAE by reducing MHCII expression in CNS myeloid cells, suppressing Th1/Th17 maintenance and expansion and inhibiting the reactivation of encephalitogenic T cells in the CNS [4, 5, 9–12].
DCs have been reported to have a critical role in MS/EAE neuroinflammation (reviewed in [13, 14]). Following detection of products of microbial origin or derived from damaged cells, cDCs upregulate MHCII and costimulatory molecules, activate CD4 and CD8 T cells, and secrete proinflammatory cytokines, such as IL-12, IL-1β, IL-23, which promote Th1/Th17 differentiation. cDCs are excellent IFN-β producers and respond to IFN-β following signaling through IFNAR. In EAE, cDCs participate in the initial peripheral CD4 T cell activation and differentiation into encephalitogenic Th1/Th17 cells and also in their reactivation in the CNS perivascular space. This contributes to the expansion of encephalitogenic T cells and the maintenance of their functional phenotype and enables T cells to enter the CNS parenchyma [15–17]. However, cDCs can also act as regulatory/tolerogenic cells, secreting anti-inflammatory cytokines, such as IL-10 and TGFβ, and inducing functional Tregs (reviewed in [18, 19]). Tolerogenic DC have been generated through pharmacological or biologic treatments with vitamin D3, rapamycin, IL-10, VIP, and TGF-β or through genetic engineering (IL-4, IL-10, CTLA-4, or VIP-secreting DC) (reviewed in [19]). IFN-β might also act as an endogenous or exogenous regulator (or both) of the cDC functional phenotype. Here, we report on the effects of IFN-β on TLR4-induced cDC maturation and cytokine production, with emphasis on IL-12 and IL-23 cytokines, essential for the differentiation and expansion of pathogenic Th1 and Th17 cells, and on the anti-inflammatory cytokine IL-10. We also investigated the role of signaling pathways, including the relevant STATs and IRFs, and the involvement of the PI3K→GSK3 in the effects of IFN-β on cDC IL-12, IL-23, and IL-10 expression.
MATERIALS AND METHODS
Mice
C57BL/6 mice (6–10 wk old), B6.129S2-Irf1tm1Mak (Irf1−/− mice), B6.129S2-Ifnar1tm1Agt/Mmjax (Ifnar1−/− mice), B6.129P2-Il10tm1Cgn/J (Il10−/− mice), and corresponding WT mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Stat1−/− mice (129S6/SvEv-Stat1tm1Rds) and the corresponding WT mice (129S6/SvEv) were purchased from Taconic Farms (Germantown, NY, USA). Stat2−/− mice were generously provided by Dr. Ana Gamero (Temple University, Philadelphia, PA, USA), and Irf7−/− mice were generously provided by Dr. Luis Sigal (Fox Chase Cancer Center, Philadelphia, PA, USA). C57BL/6-Tg (Tcra2D2, Tcrb2D2)1Kuch/J (2D2 TCR transgenic mice, MOG35–55 specific) were initially obtained from Jackson Laboratory, and homozygous TCR transgenic mice were generated in our mouse colony. Mice were handled and housed in accordance with the guidelines of the Temple University Animal Care and Use Committee.
Reagents
LPS (Escherichia coli O55:B5), FITC-conjugated dextran, PGN, and poly-IC were purchased from Sigma-Aldrich (St. Louis, MO, USA). Streptavidin-peroxidase was purchased from BioLegend (San Diego, CA, USA). IFN-β was purchased from PBL Assay Science (Piscataway, NJ, USA). Pam3CSK4 was purchased from InvivoGen (San Diego, CA, USA). CD11c MicroBeads were purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). Recombinant murine GM-CSF, IL-12p70, and IFN-γ were purchased from PeproTech, Inc (Rocky Hill, NJ, USA). FITC-conjugated anti-mouse CD80, CD86, CD40, and MHCII; PE-conjugated anti-mouse IL-17; APC-conjugated IFN-γ; recombinant mouse IL-10 and IL-23; capture and biotinylated anti-mouse IL-12p40, IL-12p70, IL-10, IFN-γ, and IL-17; and tetramethylbenzidine substrate reagent set were purchased from BD Biosciences (San Jose, CA, USA). Capture and biotinylated anti-mouse IL-27 antibody, recombinant mouse IL-27 and IL-17, isotype goat IgG, and APC-conjugated anti-mouse TLR4 were purchased from R&D Systems (Minneapolis, MN, USA). Anti-Akt and anti-phospho-Akt (Ser473) were purchased from Cell Signaling Technology (Beverly, MA, USA). Capture and biotinylated anti-mouse IL-23 antibody, PE-conjugated anti-mouse PD-L1, and FITC-conjugated anti-mouse PD-L2 were purchased from eBioscience (San Diego, CA, USA). PI3K inhibitor LY294002 was purchased from Cayman Chemical (Ann Arbor, MI, USA). Fast SYBR Green Cells-to-Ct kit was purchased from Life Technologies (Carlsbad, CA, USA).
Bone marrow-derived DC
Mouse primary DCs were generated from BM in the presence of 20 ng/ml rGM-CSF, as previously described [20]. On day 7, nonadherent cells were harvested and purified by immunomagnetic sorting with anti-CD11c-coated magnetic beads using the autoMACS system according to the manufacturer’s instructions (Miltenyi Biotec). The purity of the sorted cells was determined by FACS analysis (>96% for CD11c+ cells). The cells were cultured in RPMI 1640 medium, supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 50 μM β-mercaptoethanol, and 1% MEM nonessential amino acids. For generation of pDCs (CD11c+B220+), we differentiated BM precursors in medium containing Flt3L [21] (7.5% supernatant from the Flt3L cell line, a kind gift from Dr. Stefania Gallucci, Temple University School of Medicine).
Isolation of peritoneal macrophages
C57BL/6 mice were injected i.p. with 1.5 ml of 3% thioglycollate (BD Diagnostic Systems, Sparks, MD, USA) to recruit macrophages into the peritoneal cavity. Four days later, peritoneal macrophages were collected by washing the peritoneal cavity with RPMI 1640 medium. Macrophages were cultured at 2 × 106 cells/ml in 12-well plates at 37°C in the presence of 5% of CO2. After 4 h incubation, the nonadherent cells were removed by washing with PBS. The adherent cells were cultured in X-VIVO 15 medium (Lonza, Basel, Switzerland) overnight and subjected to stimulation with various TLR ligands.
FACS analysis
DCs were cultured in 12-well culture plates (1 × 106 cells/ml), and pretreated for 1 h with IFN-β, followed by LPS (1 μg/ml) for an additional 24 h. DCs were collected, washed with PBS, and incubated for 30 min at 4°C with anti-CD80 FITC, anti-CD86 FITC, anti-CD40 FITC, anti-MHCII (I-A/I-E) FITC, anti-TLR4 APC, anti-PD-L1 PE, anti-PD-L2 FITC, and appropriate isotype-matched controls. Data were collected for 10,000 cells and analyzed using Cellquest software from BD Biosciences. To detect phosphorylated Akt and total Akt, DCs were starved for 12 h. The cells were pretreated with IFN-β for 1 h, followed by LPS stimulation for 15 min. DCs were fixed, permeabilized, stained with anti-pAkt and anti-Akt according to manufacturer’s protocol (Cell Signaling Technology), and flow cytometry was used to detect intracellular pAkt and Akt.
Endocytosis
Endocytosis was measured as cellular uptake of FITC-dextran (molecular mass, 40 kDa; Sigma-Aldrich) and was quantified by flow cytometry, as previously described [20].
qRT-PCR
The expression of Ccr5, Ccr7, Il23a (p19), Il12a (p35), Il12b (p40), Il10, Irf1, Irf7, and Irf8 was detected by SYBR green-based qRT-PCR, as previously described [20]. RT-PCR was performed using StepOnePlus RT-PCR system (Applied Biosystems, Foster City, CA, USA). The following primers were used: Ccr5 sense, 5′-CATCGATTATGGTATGTCAGCACC-3′ and antisense, 5′-CAGAATGGTAGTGTGAGCAGGAA-3′; Ccr7 sense 5′-CCAGGAAAAACGTGCT GGTG-3′ and antisense 5′-GGCCAGGTTGAGCAGGTAGG-3′; Il23b (p19) sense, 5′-TGCTGGATTGCAGAGCAGTAA-3′ and antisense, 5′-ATGCAGAGATTCCGAGAGA-3′; Il12a (p35) sense, 5′-GAGGACTTGAAGATGTACAG-3′ and antisense, 5′-TTCTATCTGTGT GAGGAGGGC-3′; Il12b (p40) sense, 5′-GACCCTGCCGATTGAACTGGC-3′ and antisense, 5′-CAACGTTGCATCCTAGGATCG-3′; Il10 sense, 5′-CCTGGTAGAAGTGATGCCCC-3′ and antisense, 5′-TCCTTGATTT CTGGGCCATG-3′; Irf1 sense, 5′-CCCACAGAAGAGCATAGCAC-3′ and antisense, 5′-AGCAGTTCTTTGGGAATAGG-3′; Irf7 sense, 5′-CCAGCTCTCACCGA GCG-3′ and antisense, 5′-GTTCTTACTGCTGGGGCCAT-3′; Irf8 sense, 5′-GTTCCGTATCC CCTGGAAGC-3′ and antisense, 5′-TCAGAGCACAGCGTAACCTC-3′; ACTB (β-actin) sense, 5′-TCCACCACCACAGCTGAG AGG-3′ and antisense, 5′-CAGCTTCTCTTTGATGTCACG-3′; and Gapdh sense, 5′-GGAGCGAGACCCCACTAA-3′ and antisense, 5′-ACATACTCAGCACCGGC CTC-3′. The expression level of each gene is indicated by the number of cycles needed for the cDNA amplification to reach a threshold. The amount of DNA is calculated from the number of cycles by using standard curves, and the results are normalized to β-actin or Gapdh.
Cytokine ELISA
Cytokine production was determined by sandwich ELISA. DCs were cultured in 12-well culture plates (1 × 106 cells/ml or 2 × 106 cells/ml) and pretreated with IFN-β for 1 h, followed by LPS (1 μg/ml) stimulation for 12 (for IL-23) or 24 h (for IL-12p70, IL-10, and IL-27). Supernatants were harvested and subjected to ELISA. There were variations in IL-10 levels from experiment to experiment, although not statistically significant. The detection limits were: 15 pg/ml IL-10, 30 pg/ml for IL-23 and IL-12 p70, and 4.7 pg/ml for IL-27.
Western blot analysis
For Western blotting, cell lysates were prepared in radioimmunoprecipitation assay buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.15% SDS, 1 mM PMSF, phosphatase inhibitor, and protease inhibitor cocktail]. Proteins were separated on 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes, blotted with specific antibodies, and detected using Immobilon Western chemiluminescent HRP substrate (Millipore, Billerica, MA, USA). The following antibodies were used: anti-GSK3β (Cell Signaling) and anti-phospho-GSK3β (Ser9; Cell Signaling).
In vivo experiments
C57BL/6 mice (14 mice/group) were immunized with 100 μg MOG33–55 peptide emulsified in CFA containing 2 mg/ml of Mycobacterium tuberculosis H37 RA, s.c. on day 0 and 100 ng pertussis toxin was administered i.p. on days 0 and 2. Mice were treated with vehicle (200 μl PBS), or IFN-β 10,000 units (in 200 μl PBS) i.p. on days 3, 5, and 7 after immunization. On day 8, spleens and draining LNs (inguinal, axillary, and brachial) were harvested. Splenocytes and LN cells were pooled from 3 mice/group (PBS control and IFN-β treated) and were subjected to mRNA extraction and qRT-PCR to measure the expression of Ifng and Il17. CD11c+ DCs were purified from spleens, and LNs were pooled from 3 mice/group (PBS control and IFN-β treated), and subjected to qRT-PCR to measure cytokine expression [Il12a (p35), Il12b (p40), Il23a (p19), and Il10]. CD4+IL-17+ and CD4+IFN-γ+ splenocytes from 8 PBS control and 8 IFN-β-treated mice were analyzed individually by flow cytometry following staining for surface CD4 and intracellular IL-17 or IFN-γ.
Statistical analysis
Results are described as means ± sd. Comparisons among multiple groups were done by 1-way ANOVA, followed by Bonferroni post hoc test for comparison among treatments. Statistical significance was determined as values of P < 0.05. All statistical analyses were performed using Prism 5 (GraphPad Software, La Jolla, CA, USA).
RESULTS
IFN-β promotes cDC maturation
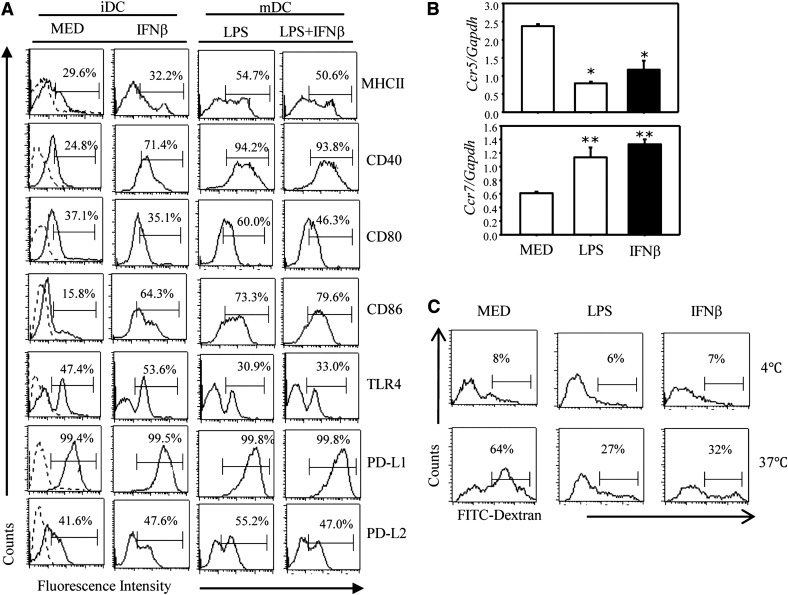
Following maturation, cDCs up-regulate expression of surface MHCII and of the costimulatory molecules CD40, CD80, and CD86, switch from CCR5 to CCR7 expression, and substantially reduce their phagocytic capacity. To determine the effects of IFN-β on DC maturation, we treated cDCs with IFN-β in the presence or absence of LPS. The LPS-induced surface MHCII and costimulatory molecule expression was not affected by IFN-β, except for a decrease in CD80 (P < 0.05). Surface TLR4, PD-L1, and PD-L2 levels were also not affected by IFN-β (Fig. 1A).
Figure 1. IFN-β promotes cDC maturation.
(A) CD11c+ DCs were treated with IFN-β (1000 U/ml), LPS (1 μg/ml), or IFN-β (for 1 h), followed by LPS for 24 h. Surface expression of MHCII, CD40, CD80, CD86, PD-L1, PD-L2, and TLR4 was measured by FACS. Results are expressed as the percentage of positive cells with 1 of 4 representative experiments shown. Comparing results from 4 independent experiments, the decrease in CD80 for mature DCs and the increase in CD40 and CD86 in immature DCs treated with IFN-β were statistically significant (P < 0.05 for CD80, P < 0.001 for CD40 and CD86). (B) CD11c+ DCs were treated with IFN-β or LPS stimulation for 12 h (Ccr7 expression) or 24 h (Ccr5 expression). Ccr5 and Ccr7 expression was determined by qRT-PCR. Data represent the means ± sd. *P < 0.05, **P < 0.01, compared with the medium. One of 3 representative experiments is shown. (C) CD11c+ DCs were treated with medium IFN-β or LPS for 24 h. The endocytic capacity of DCs was assessed by cellular uptake of FITC-dextran at 4°C (negative control) or 37°C using flow cytometry. Results are presented as the percentage of positive cells with 1 of 3 representative experiments shown.
In contrast, IFN-β significantly (P < 0.001) increased the levels of CD40 and CD86 in unstimulated immature DCs (Fig. 1A), suggesting that IFN-β could have a role in cDC maturation. The results regarding the CCR5/CCR7 switch and the reduction in FITC-dextran phagocytosis (Fig. 1B and C) support the role of IFN-β in DC maturation. In contrast to the effects on costimulatory molecules, chemokine receptors, and phagocytosis, IFN-β did not induce cytokine production (IL-12p70 and p40 or IL-10) in unstimulated DCs (results not shown).
Effects of IFN-β on DC-derived cytokine expression
In vivo experiments
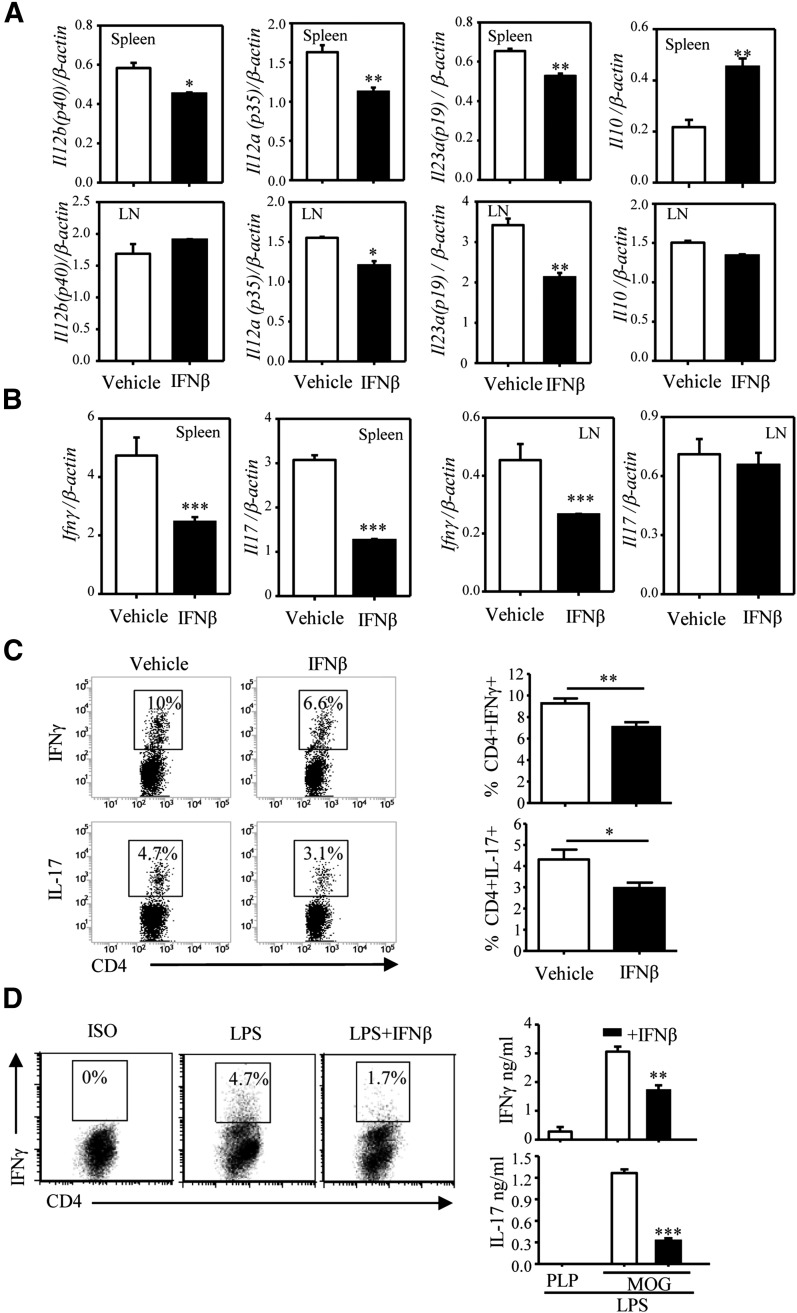
To investigate the effects of IFN-β on cytokine expression in a model relevant to MS, we used the C57BL/6 MOG35–55 EAE model. The mice were immunized with MOG/CFA and pertussis toxin, as described in the Materials and Methods section, and injected i.p. with IFN-β or vehicle on days 3, 5, and 7 postimmunization. On day 8, we assayed the expression of Il12b (IL-12/IL-23 p40), Il12a (IL-12 p35), Il23a (IL-23 p19), and Il10 (IL-10) in purified splenic and LN CD11c+ DCs, and of Ifng (IFN-γ) and Il17 (IL-17) in spleen and LN cell suspensions. Treatment with IFN-β resulted in a decrease in IL-12/IL-23 p40, IL-12 p35, and IL-23 p19, and an increase in IL-10 expression in splenic DCs (Fig. 2A). As expected from the role of IL-12 and IL-23 in Th1/Th17 differentiation and expansion, in vivo administration of IFN-β reduced splenic expression of both Th1 and Th17 signature cytokines (IFN-γ and IL-17, respectively) (Fig. 2B). We observed similar effects for Il12a, Il23a, and Ifn-γ in LN cells. In addition, lower percentages of IFN-γ+ and IL-17+ CD4 T lymphocytes were present in splenocytes from IFN-β–treated EAE mice (Fig. 2C). A similar reduction in Th1/Th17 differentiation was observed in vitro, when DCs pretreated with IFN-β and LPS and pulsed with MOG were cocultured with CD4 2D2 T cells (Fig. 2D).
Figure 2. IFN-β affects IL-12, IL-23, and IL-10 expression and Th1/Th17 differentiation in vivo and in vitro.
C57BL/6 mice (n = 14/group) were immunized with MOG33–55 emulsified in CFA as described in the Materials and Methods section. Mice were treated with the vehicle (200 μl PBS) or IFN-β 10,000 U (in 200 μl PBS) via i.p. inoculation on days 3, 5, and 7 postimmunization. On day 8, spleens and DLNs were harvested. (A) CD11c+ cells, purified from pooled spleens and LNs (3 mice/group), were subjected to RNA extraction and qRT-PCR to measure expression of Il12a (p35), Il12b (p40), Il23a (p19), and Il10. Each sample was tested in triplicate, and the results were expressed as means ± sd. (B) RNA from pooled splenocytes and LN cells (3 mice/group) was subjected to qRT-PCR to measure expression of IFN-γ and Il17. Each sample was tested in triplicate. (C) Splenocytes (from 8 mice/group) restimulated ex vivo with PMA and ionomycin for 4 h were stained for surface CD4 and intracellular IL-17 or IFN-γ and analyzed by flow cytometry. A representative experiment is shown in the left panel. Results shown in the right panel represent the combined data from all 8 individual samples as means ± sd. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the vehicle. (D) DCs were pretreated with IFN-β for 1 h, pulsed with MOG35–55, and treated with LPS (1 μg/ml) for 24 h. Following extensive washing with PBS, DCs (1 × 105 cells/ml) were cocultured with naïve 2D2 CD4+ T cells (1 × 106 cells/ml) for 3 d. Cytokine production was detected by FACS and ELISA. One of 3 independent experiments is shown. Results represent the means ± sd. **P < 0.01 and ***P < 0.001, compared with LPS alone.
In vitro experiments
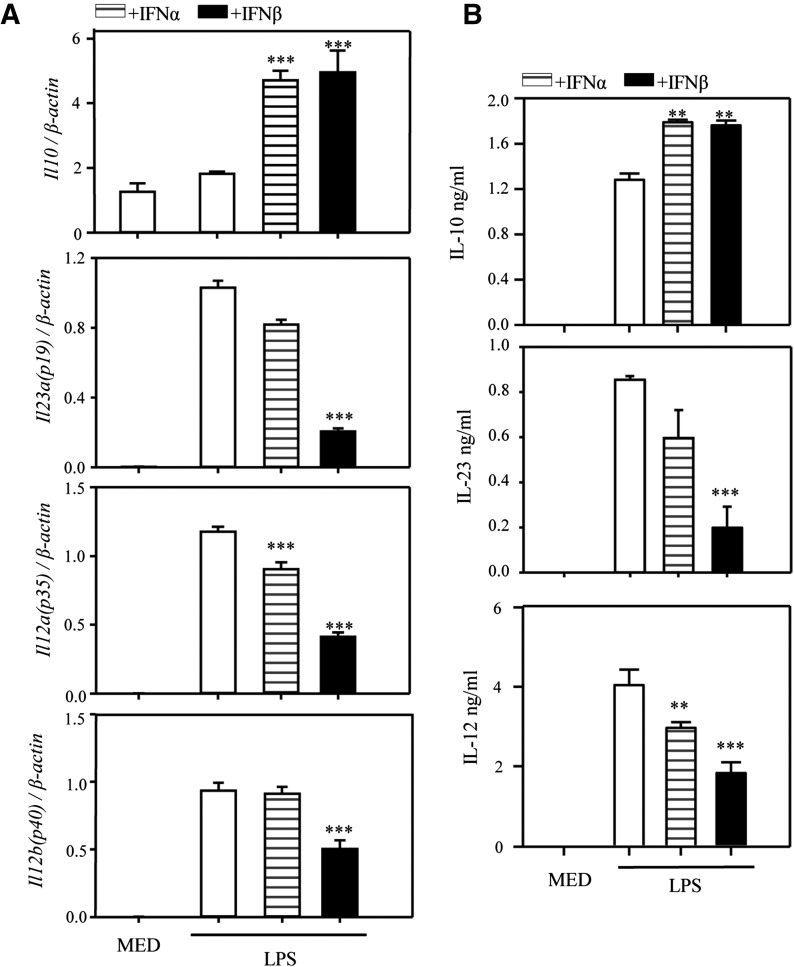
To confirm the direct effect of IFN-β on DCs, we treated BM-derived cDC with LPS in the presence of IFN-α or β and measured expression of Il23a (p19), Il12b (p40), Il12a (p35), and Il10 by qRT-PCR and the secreted IL-12 p70, IL-23, and IL-10 by ELISA. Both type-I IFNs increased IL-10 and reduced IL-12 and IL-23, with IFN-β being a much more potent inhibitor (Fig. 3A and B). In dose-dependent experiments, we treated cDCs with various doses of IFN-β, followed 1 h later, by LPS. IFN-β inhibited IL-12 and IL-23 production in the dose range of 50 to 1000 U/ml, and stimulated IL-10 production within the same range, although only 500 and 1000 U/ml were statistically significant for IL-10 (Supplemental Fig. 1A). Next, DCs were pretreated with IFN-β for 6, 3, or 1 h before LPS, or treated with both IFN-β and LPS at the same time (0 h); 1 h preincubation gave the best results in terms of IL-12 and IL-23 inhibition, as well as IL-10 stimulation (Supplemental Fig. 1B).
Figure 3. Effects of IFN-α and IFN-β on IL-12, IL-23, and IL-10 expression and production in LPS-stimulated DCs.
DCs were pretreated with IFN-α (1000 U/ml) or IFN-β (1000 U/ml) for 1 h, followed by LPS stimulation. (A) After 3 h [Il23a (p19)] or 6 h [Il10, Il12a (p35), Il12b (p40)], RNA was extracted, and the expression of Il10, Il23a, Il12a, and Il12b was measured by qRT-PCR. (B) After 12 h (IL-23) or 24 h (other cytokines), supernatants were collected and analyzed by ELISA for IL-10, IL-23, and IL-12 secretion. One of 3 independent experiments is shown. Results represent the means ± sd. **P < 0.01 and ***P < 0.001, compared with LPS alone.
Because IL-10 has been reported to inhibit IL-12 and partially reduce IL-23 production [22, 23], we used IL-10 deficient DCs to determine whether IFN-β up-regulation of IL-10 was responsible for the inhibitory effects on IL-23 or IL-12 or both. LPS induced higher levels of IL-23 and IL-12 in IL-10−/− DC, supporting the partially inhibitory role of IL-10. However, the level of IFN-β inhibition of IL-23 was similar in WT and IL-10−/− DCs, and the inhibition of IL-12 was only slightly reduced in IL-10−/− vs. WT DCs (28 vs. 35%) (Supplemental Fig. 2A). These results suggest that the IFN-β inhibition of IL-12 might be partially dependent on IL-10, whereas the inhibitory effect on IL-23 is IL-10 independent.
Recently, IL-27 was reported to mediate the inhibition of IL-23 and the up-regulation of IL-10 by IFN-β in TLR2-stimulated DCs [11]. Similar to the published report, IFN-β induced higher levels of IL-27 in our TLR4 (LPS)–stimulated DCs. However, use of neutralizing IL-27 Abs did not affect IFN-β stimulation of IL-10 or the inhibition of IL-23 and IL-12 (Supplemental Fig. 2B). Therefore, we conclude that the effects of IFN-β on IL-12, IL-23, and IL-10 production were not mediated through IL-27.
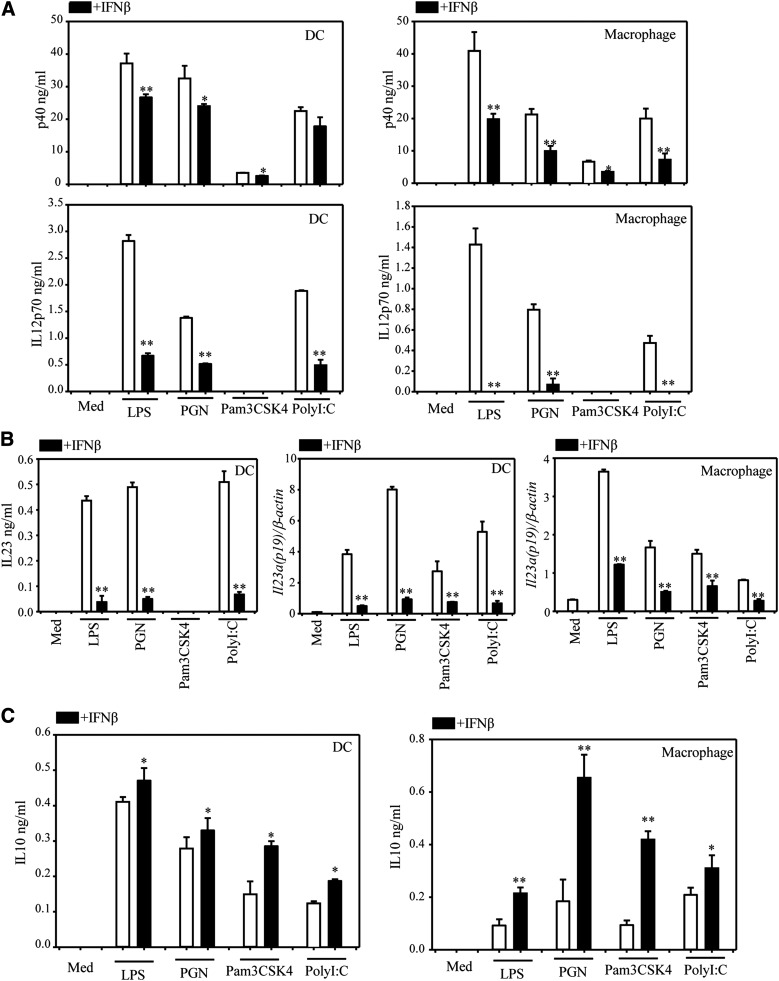
IFN-β affects cytokine production in a similar manner in DCs and macrophages stimulated with various TLR ligands
To determine whether the effects of IFN-β extend to DCs and macrophages stimulated through different TLRs, we pretreated BM-derived DC and peritoneal macrophages with IFN-β, followed by stimulation with various TLR ligands, i.e., LPS (TLR4), PGN (TLR2), Pam3CSK4 (TLR2/1) and poly-IC (TLR3). With the exception of Pam3CSK4, all other ligands induced IL-12p40 and IL-12p70 in DCs and macrophages, and IFN-β reduced or completely suppressed cytokine production (Fig. 4A). Il23a (IL23 p19) expression was induced by all ligands and substantially reduced by IFN-β in both DCs and macrophages (Fig. 4B). All 4 ligands induced IL-10 in DCs and macrophages, and IFN-β stimulated IL-10 production (Fig. 4C). Based on the similarities between DCs and macrophages stimulated with various TLR ligands, we focused on the effects of IFN-β on LPS-treated DCs for the rest of the mechanistic experiments.
Figure 4. IFN-β affects cytokine production induced by various TLR ligands.
CD11c+ DCs or peritoneal macrophages were pretreated with IFN-β (1000 U/ml) for 1 h, followed by TLR ligands: LPS (1 μg/ml), PGN (40 μg/ml), Pam3CSK4 (1 μg/ml), and poly-IC (100 μg/ml). (A) Cells were stimulated with TLR ligands for 24 h; IL-12 p40 and IL-12 p70 were detected by ELISA. (B) DCs were stimulated with TLR ligands for 12 h; IL-23 was detected by ELISA (left panel). DCs or peritoneal macrophages were also stimulated with TLR ligands for 3 h, and qRT-PCR was performed to measure Il23a (p19) (middle and right panels). (C) Cells were stimulated with TLR ligands for 24 h, and IL-10 was detected by ELISA. Results represent the means ± sd. *P < 0.05 and **P < 0.01, compared with TLR ligands alone treatment.
Involvement of IFNAR, STAT1, and STAT2 in the effects of IFN-β on cytokine expression
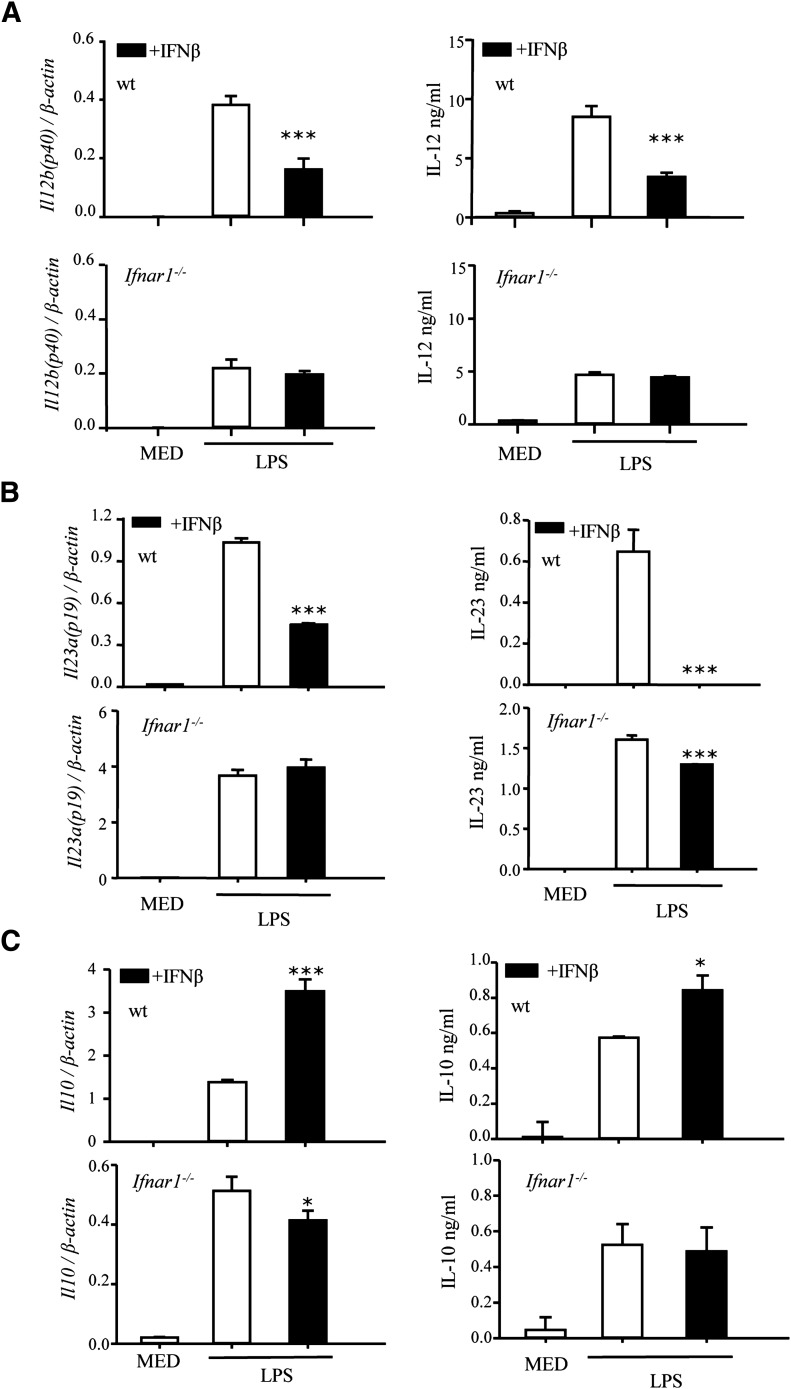
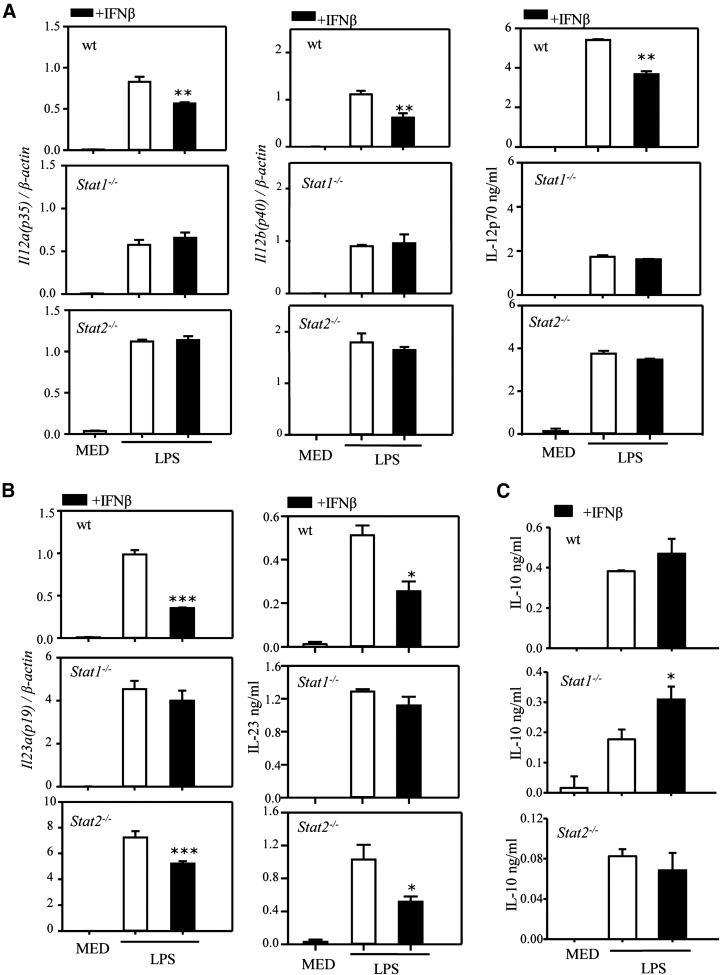
IFN-β acts through the type-I IFN receptor IFNAR. As expected, the inhibitory effect of IFN-β on IL-12 and IL-23, both at the mRNA and protein levels, as well as the stimulatory effect on IL-10, were abolished in IFNAR-deficient DCs (Fig. 5A–C). The involvement of STAT1 and STAT2 in the effects of IFN-β on IL-12, IL-23, and IL-10 expression was assessed using STAT1−/− and STAT2−/− DCs. We observed a highly statistically significant (P < 0.001) reduction in LPS-induced IL-12p70 levels in STAT1−/− DCs (Fig. 6A), which indicates that LPS induction of IL-12 is partially mediated through STAT1 signaling. The inhibitory effect of IFN-β was lost in both STAT1-deficient and STAT2-deficient DCs, suggesting that the inhibitory effect of IFN-β was dependent on both STAT1 and STAT2 (Fig. 6A). In contrast to IL-12, LPS induced statistically significant higher levels of IL-23 in both STAT1−/− (P < 0.01) and STAT2−/− (P < 0.05) DCs (Fig. 6B), suggesting a negative role for the 2 STATs in Il23a (p19) expression. In DCs treated with LPS and IFN-β, the inhibitory effect of IFN-β on Il23a was lost in STAT1−/−, but not in STAT2−/−, DCs (Fig. 6B), indicating that the inhibitory effect of IFN-β on IL-23 was mostly, if not entirely, dependent on STAT1. In terms of IL-10 production, LPS induction of IL-10 was much lower in STAT2-deficient DCs (Fig. 6C) (P < 0.001), suggesting that STAT2 activation, possibly by endogenous IFN-β, has a role in IL-10 expression. Similarly, the stimulatory effect of exogenous IFN-β appears to be mediated by STAT2 but not STAT1 (Fig. 6C).
Figure 5. The effects of IFN-β are mediated through IFNAR.
DCs were generated from WT or type-I IFN receptor–deficient (Ifnar1−/−) mice. Cells were pretreated with IFN-β (1000 U/ml) for 1 h, followed by LPS stimulation. After 3 (for Il23a) or 6 (for Il12b and Il10) h, mRNA expression was measured by qRT-PCR (A–C, left panels). After 12 (for IL-23) or 24 h (for IL-12 and IL-10), supernatants were analyzed by ELISA (right panels). A representative experiment of 3 is shown. Results represent the means ± sd. *P < 0.05, **P < 0.01, and ***P < 0.001 for LPS + IFN-β, compared with LPS alone. Statistically significant for the LPS-only treatment for WT and IFNAR KO (P < 0.01 for IL-12, P < 0.001 for IL-23; not significant for IL-10).
Figure 6. STAT1 and STAT2 involvement in the effects of IFNs on cytokine expression.
DCs were generated from WT Stat1−/− and Stat2−/− mice. Cells were pretreated with IFN-β (1000 U/ml) for 1 h, followed by LPS stimulation. After 3 or 6 h, RNA was prepared, and mRNA expression of Il12a, Il12b, and Il23a was measured by qRT-PCR (A, left and middle panels; B, left panels). After 12 or 24 h, supernatants were analyzed by ELISA for IL-12, IL-23, and IL-10 secretion (A and B, right panels, and C). One of 3 independent experiments is shown. Results represent the means ± sd. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with LPS alone. Statistically significant for the LPS-only treatment for WT and STAT1 KO (P < 0.001 for IL-12 and IL-10, P < 0.01 for IL-23) and for WT and STAT2 KO (P < 0.01 for IL-12, P < 0.05 for IL-23, and P < 0.001 for IL-10).
Role of IRFs in the effects of IFN-β on cytokine expression
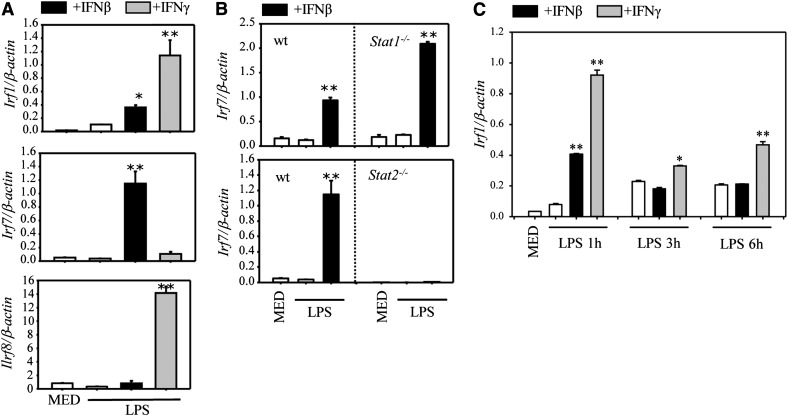
ISGF3 induced by IFN-α/β translocates to the nucleus, binds to IFN-stimulated regulatory element sequences, and regulates the expression of specific genes, including Irf7 (reviewed in [24]). The IRFs, in turn, regulate expression of various immune and nonimmune genes. To investigate the role of various IRFs, we performed first an RT2 Profiler PCR array (QIAGEN, Valencia, CA, USA) and compared levels of gene expression in DCs treated with LPS, LPS, and IFN-β, and with LPS and IFN-γ (used as control). As expected, IFN-γ induced expression of Irf1, whereas IFN-β induced Irf7 (Supplemental Table 1). We confirmed these data by qRT-PCR, adding Irf8, which was not included in the PCR array. At 1 h posttreatment, IFN-γ induced both IRF-1 and IRF-8, whereas IFN-β induced low levels of IRF-1, and high levels of IRF-7 in a STAT2-dependent manner (Fig. 7A and B). To determine the role of IRF-1 and IRF-7 in the effects of IFN-β on IL-12, IL-23, and IL-10 production, we compared WTs with the corresponding IRF-deficient DC.
Figure 7. IFN-β and IFN-γ induce differential IRF expression in LPS-treated DCs.
(A) DCs were pretreated with IFN-β (1000 U/ml) or IFN-γ (100 ng/ml) for 1 h, followed by 1 h of LPS (1 µg/ml) stimulation. The expression of Irf1, Irf7, and Irf8 was detected by qRT-PCR. (B) DCs were generated from WT Stat1−/− and Stat2−/− mice. Cells were pretreated with IFN-β (1000 U/ml) for 1 h, followed by 1 h LPS stimulation. The expression of Irf7 was determined by qRT-PCR. (C) DCs were pretreated with IFN-β or IFN-γ for 1 h, followed by LPS (1 µg/ml) for 1, 3, and 6 h. Irf1 expression was determined by qRT-PCR. One out of 2 independent experiments is shown. Results represent the means ± sd. *P < 0.05, **P < 0.01, compared with LPS alone.
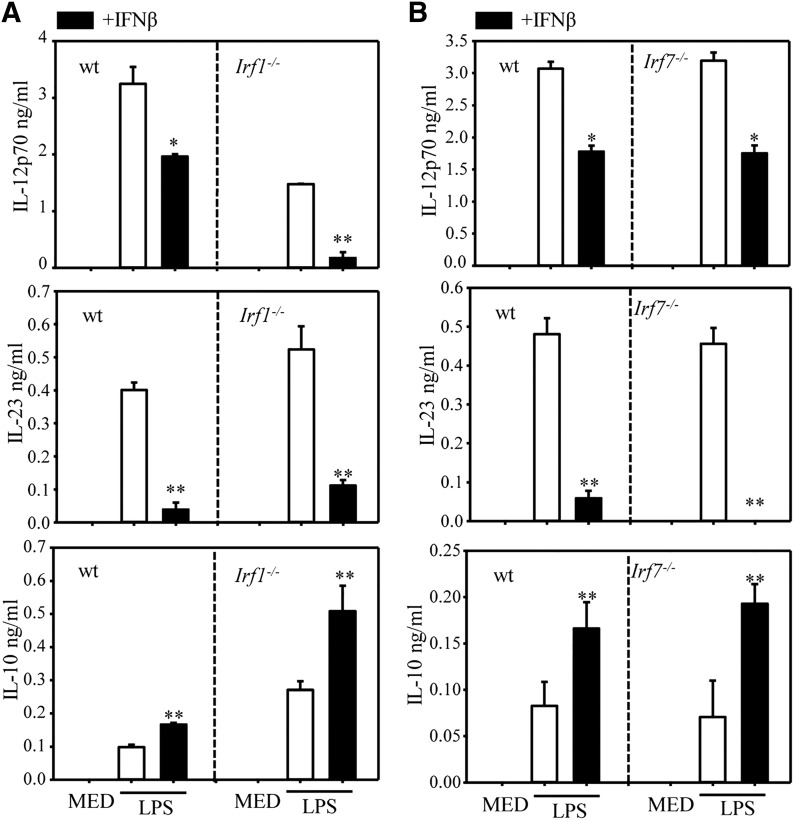
IRF-1 has been previously reported to induce p40 expression in macrophages and DCs [25, 26]. Indeed, LPS induction of IL-12p70 was significantly reduced in IRF-1–deficient mice. IFN-β induces low levels of IRF-1, which might interfere with the inhibitory effect of IFN-β on IL-12. This appears to, indeed, be the case, because the percentage of inhibition in the IRF-1 KO DC is higher than in WT (90 vs. 50%) (Fig. 8A, upper panel). In contrast to its positive role in IL-12 production, IRF-1 was reported to act as a negative regulator for Il23a (p19) expression [27]. Indeed, we observed an increase in the level of LPS induced IL-23 in IRF-1−/− DCs (Fig. 8A, middle panel). However, the inhibitory effect of IFN-β was similar to that of WT and IRF-1-deficient DCs, indicating that IRF-1 does not have a role in the inhibitory effect of IFN-β on IL-23 production. IRF-1 has been previously described as a negative regulator of Il10 expression [28]. Indeed, we observed increased LPS-induced IL-10 production in IRF-1−/− vs. WT DCs. However, the stimulatory effect of IFN-β, compared with LPS alone, was not affected by the lack of IRF-1 (Fig. 8A, lower panel).
Figure 8. IRF1 and 7 involvement in the effects of IFN-β on cytokine production.
DCs were generated from WT Irf1−/− (A) and Irf7−/− (B) mice. Cells were pretreated with IFN-β for 1 h, followed by LPS stimulation for 12 h (for IL-23) and 24 h (for IL-12 and IL-10). Supernatants were analyzed for cytokine secretion by ELISA. One of 3 independent experiments is shown. Results represent the means ± sd. *P < 0.05, **P < 0.01, compared with LPS alone.
Because IFN-β is a strong inducer of Irf7, we compared its effects in WT and Irf7 KO DCs. The levels of LPS-induced IL-12, IL-23, and IL-10 were similar in WT and IRF-7−/− DCs. In addition, IFN-β inhibited IL-12 and IL-23 and stimulated IL-10 at similar levels in WT and IRF-7−/− DCs (Fig. 8B), indicating that IRF-7 does not have a role in the regulatory effect of IFN-β on IL-12, IL-23, and IL-10 production in LPS-stimulated DCs.
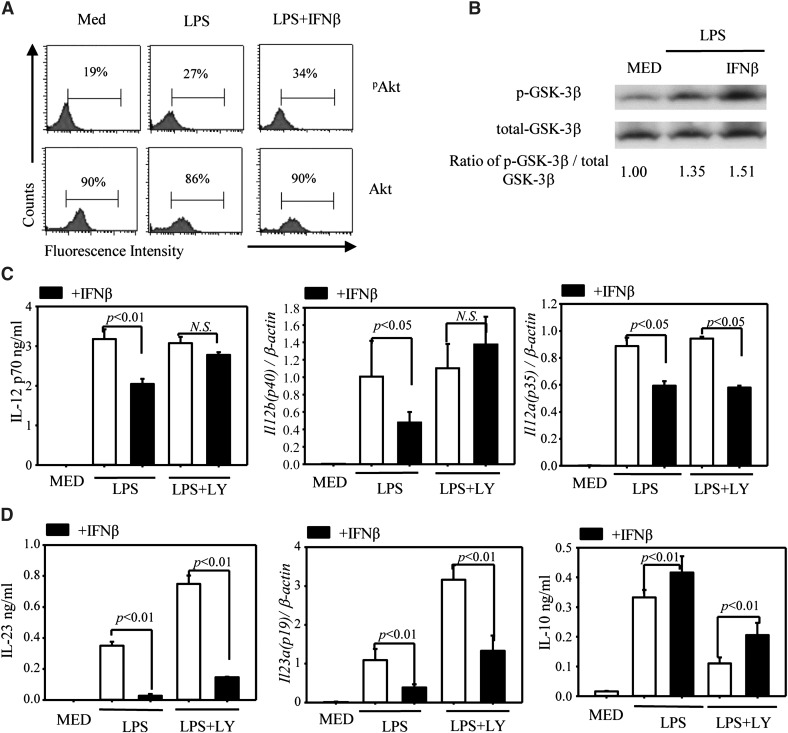
Role of the PI3K→Akt→GSK3 signaling pathway in the effects of IFN-β on cytokine expression
In addition to the JAK→STAT pathway, signaling through TLRs and IFNAR was reported to activate the PI3K→Akt pathway leading to GSK3 inactivation with subsequent up-regulation of IL-10 and inhibition of proinflammatory cytokines, such as IL-12 [29, 30]. Treatment of DC with LPS showed an increase in the percentage of pAkt-expressing cells, with no change in total Akt. IFN-β further increased (although not in a statistically significant way) the percentage of pAkt-expressing DCs (Fig. 9A). In agreement with its stimulatory effect on PI3K/Akt, IFN-β induced increased GSK3β phosphorylation (Fig. 9B). To evaluate the role of the PI3K pathway in the effects of IFN-β on cytokine expression, we treated DCs with the PI3K inhibitor LY294002. Inhibition of PI3K resulted in the reversal of the IFN-β inhibitory effect on IL-12, mediated primarily through the reversal in IL-12 p40 inhibition (Fig. 9B). In contrast to IL-12, the inhibitory effect of IFN-β on IL-23 and the stimulatory effect on IL-10 were not affected by PI3K inhibition (Fig. 9C). Based on these results, we concluded that activation of PI3K by IFN-β has a major role in its inhibitory effect on IL-12 production but not in IL-23 inhibition or IL-10 up-regulation.
Figure 9. Involvement of the PI3K signaling pathway in IFN-β inhibition of IL-12 production.
(A) DCs were serum-starved for 12 h, treated with IFN-β for 1 h, followed by LPS stimulation for 15 min. Phospho-Akt (Ser473) and total Akt were detected by flow cytometry. One of 2 independent experiments is shown. (B) DCs were serum-starved for 4 h and treated with LPS or LPS + IFN-β for 30 min. Phospho-GSK3β (Ser9) and total-GSK3β were detected by Western blot. One of 2 independent experiments is shown. (C) DCs were pretreated with the PI3K inhibitor LY294002 (10 µM) for 2 h, followed by IFN-β for 1 h and LPS for 6 h (for Il12a and Il12b expression by qRT-PCR) and 24 h for IL-12 p70 secretion by ELISA. (D) DCs were pretreated with the PI3K inhibitor LY294002 (10 µM) for 2 h, followed by IFN-β for 1 h and LPS for 3 h (for Il23a q-RT-PCR), 12 h (for IL-23 ELISA), and 24 h for IL-10 ELISA. One of 3 independent experiments is shown, and results are presented as means ± sd.
Inflammatory cDCs generated from BM cells in the presence of GM-CSF mirrored the dominant functional DC subtype in EAE. In contrast, DC generated from the BM in the presence of Flt3L resembled steady-state DCs, consisting of a substantial percentage of CD11c+B220+ pDCs. We tested the effects of IFN-β on IL-12, IL-23, and IL-10 production in Flt3L-generated DCs in the presence and absence of the PI3K inhibitor LY294002. DCs generated in the presence of Flt3L contained approximately the same number of pDCs (CD11c+B220+) and cDCs (CD11c+B220−) (Supplemental Fig. 3A). Similar to its effects on GM-CSF generated cDCs, IFN-β reduced expression of Il12a (p35), Il12b (p40), and Il23a (p19) and up-regulated Il10 (Supplemental Fig. 3B). However, in contrast to the effect of PI3K inhibitors in reversing IFN-β inhibition of IL-12 in GM-CSF cDCs, LY294002 did not affect the inhibitory effect of IFN-β on IL-12 production in Flt3-generated DC (Supplemental Fig. 3C).
DISCUSSION
MS is an autoimmune disease characterized by immune cell infiltration into the CNS and axonal damage leading to cumulative disability. In clinical trials, IFN-β showed efficacy in RR-MS, reducing lesion formation, rates of relapse, and progression of sustained disability [31, 32]. The molecular mechanisms involved in the effect of IFN-β in MS are not entirely elucidated. The fact that EAE is exacerbated following conditional deletion of IFNAR in myeloid cells, but not in T cells or in neuroectodermal CNS cells [5], suggests that the beneficial effect of IFN-β is mediated through myeloid immune cells. Indeed, signaling through IFNAR does not affect demyelination/remyelination in the nonimmune cuprizone model [33], arguing against direct effects of IFN-β on oligodendrocytes. However, determining the singular contribution of DCs, monocyte/macrophages, and microglia requires future, conditional deletions of IFNAR in each of these myeloid cell subsets. In EAE, IFN-β engagement of IFNAR on both the peripheral and CNS myeloid cells has been reported to reduce MHCII expression in CNS myeloid cells, to suppress Th1/Th17 maintenance and expansion, and to inhibit encephalitogenic T cell reactivation in the CNS [4, 5, 9–12].
Among myeloid cells, DCs were reported to have a critical role in MS/EAE neuroinflammation, primarily through their effect on the differentiation and expansion of Th1/Th17 cells (reviewed in [13, 14]. Here we focused on the effects of IFN-β on cDC expression of cytokines involved in the control of Th1/Th17 differentiation and expansion. Although IFN-β did not induce cytokine expression in the absence of TLR signaling, it affected cytokine production in TLR2-, 3-, and 4-stimulated DCs in a similar manner, inhibiting IL-12 and IL-23, and stimulating IL-10 production. The effects of IFN-β on IL-12, IL-23 and IL-10 production correlated with effects on the expression of Il12b (p40), Il12a (p35), Il23a (p19), and Il10 (IL-10), which were mediated through IFNAR. Similar effects of IFN-β on IL-12, IL-23, and IL-10 following stimulation of DCs with TLR2, 3, and 4 ligands suggests downstream effects affecting major transcriptional factors controlling cytokine gene expression. Although previous results from our laboratory indicated no direct effect of IFN-β on NF-κB activation in cDCs treated with inflammatory cytokines [34], the question of whether IFN-β affects NF-κB following TLR stimulation remains to be answered. In addition, because IFN-β was recently reported to promote recruitment of histone deacetylase 1 to the matrix metalloproteinase 9 promoter, resulting in decreased transcriptional activation [35], epigenetic regulation of cytokine gene expression preventing or promoting transcriptional activation becomes a strong possibility.
The major signaling pathway downstream of IFNAR involves ISGF3 heterodimers, composed of STAT1, STAT2, and IRF9 (reviewed in [24]). In addition to the canonical Jak/STAT pathway, IFN-β also activated the PI3K→Akt pathway resulting in GSK3 phosphorylation at Ser9 and Ser21, and subsequent GSK3 inactivation (reviewed in [36]). We analyzed the role of STAT1, STAT2, and PI3K activation in the effects of IFN-β on cytokine production by LPS-stimulated DCs. Because IFN-β-induced Irf7 expression is dependent on STAT-2 and Irf7-deficient mice develop more severe EAE [37], we also examined the possible role of IRF-7 in the effects of IFN-β on IL-12, IL-23, and IL-10 production.
IFN-β inhibition of IL-12, resulting from Il12a (p35) and Il12b (p40) down-regulation, was dependent on both STAT1 and STAT2 but not on IRF-7. Instead, the inhibitory effect of IFN-β on IL-12 appears to be mediated through activation of the PI3K signaling pathway. We confirmed that IFN-β induced Akt phosphorylation and found that the inhibitory effect of IFN-β was reversed by pharmacologic inhibition of PI3K. The inhibition of IL-12 by IFN-β is STAT1/STAT2 dependent, which is interesting because a direct connection between STAT1/2 and GSK3 has not been described. In addition to the PI3K pathway, it is possible other IFN-β signature genes controlled by STAT1/STAT2 mediate the inhibitory effect on Il12a and Il12b expression.
IL-23 functions as an essential factor for the maintenance and expansion of pathogenic Th17 cells involved in autoimmune diseases, including MS/EAE (reviewed in [38, 39]). We observed a profound inhibitory effect of IFN-β on Il23a (p19) gene expression and IL-23 production from activated DCs. Inhibition of Il23a by IFN-β was STAT1 dependent, with only a partial reversal of the inhibitory effect in STAT2-deficient DCs, suggesting that STAT1 homodimers have the primary role in the inhibitory effect. IFN-β inhibition of IL-23 was previously reported in human monocyte-derived DCs and was shown to be mediated through the induction of IL-27 [11]. In our case, however, neither IL-27 nor IL-10 mediated the inhibitory effect of IFN-β on IL-23. The therapeutic significance of IFN-β inhibition of IL-23 is supported by reports of IL-23 decrease in patients with RR-MS following IFN-β treatment [40, 41]. The molecular mechanisms involved in the inhibition of Il23a expression by IFN-β remain to be elucidated. We observed a substantial increase in IL-23 upon PI3K inhibition, indicating that the PI3K→GSK3 axis has an important role in IL-23 production. However, because IFN-β reduced IL-23 at similar levels in the presence or absence of PI3K inhibitors, we concluded that PI3K does not have a role. IRF-1 has the opposite effects on Il12b (p40) and Il23a (p19) expression, functioning as a positive regulator for Il12b and a negative regulator for Il23a [27]. Because p19 is the limiting subunit in IL-23, effects on p19 supersede those on p40, and the final effect of IRF-1 is the inhibition of IL-23 production. Indeed, we observed an increase in IL-23 in Irf1−/− DCs. However, IFN-β inhibited IL-23 at the same levels in Irf1−/− and WT DCs. The same result was obtained for Irf7−/− DCs, leading to the conclusion that neither IRF-1 nor IRF-7 was responsible for the inhibitory effect of IFN-β on IL-23.
IL-10, the classic anti-inflammatory cytokine, has an essential role in limiting the immune response against pathogens. IL-10 production by APCs, including cDCs, is triggered by TLR signaling and is modulated by IFNs. We report, here, on the stimulation of IL-10 by IFN-β, an effect mediated through IFNAR and dependent primarily on STAT2. Although a previous report identified STAT1 as a negative regulator of Il10 expression in monocytes in vivo [42], this is the first report, to our knowledge, of STAT2 involvement in the up-regulation of IL-10. Higher expression of IL-10 in splenic DCs from Irf1−/− mice [28] strongly suggests that IRF-1 is a negative regulator for Il10 gene expression. We also observed that LPS induced higher levels of IL-10 in Irf1−/− DCs and that IFN-β had a more pronounced stimulatory effect. This might be because, in WT DCs, IFN-β induces IRF-1, although less efficiently than IFN-γ does. The IFN-β–induced IRF-1 could counteract the stimulatory effect of IFN-β on IL-10 in WT DCs but not in Irf1−/− DCs. We also established that the IFN-β stimulatory activity does not involve IRF-7 as positive regulator of Il10 expression. The PI3K→GSK3 pathway regulated IL-10 production with GSK3 inhibiting Il10 expression through suppression of CREB and AP-1 binding to the Il10 promoter [29, 43]. The stimulatory effect of IFN-β on IL-10 in human monocyte-derived DCs was attributed partially to activation of PI3K and the subsequent inhibition of GSK3 [30]. We confirmed the inhibitory effect of GSK3 on IL-10 production in LPS-treated cDCs through reduction in IL-10 following PI3K inhibition and enhanced IL-10 expression and the production in the presence of the GSK3 inhibitors SB415286 and LiCl (results not shown). However, IL-10 up-regulation by IFN-β was not affected by GSK3 or PI3K inhibitors. Whether the effects of IFN-β are mediated through the ERK and p38 MAPK, previously shown to participate in the control of IL-10 expression (reviewed in [44]), remains to be ascertained.
AUTHORSHIP
J.-H.Y. and W.K. contributed equally to the experimental design and to performing most of the experiments, as well as writing and editing the manuscript, together with D.G., the corresponding author. K.M.H., F.E., and K.M.R. performed experiments related to Irf1 and Irf8 expression and involvement of the PI3K signaling pathway. P.-C.K. and B.A.S. performed experiments related to Akt and GSK3 activation.
ACKNOWLEDGMENTS
This work was supported by U.S. National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases Grants R01AI084065 and R01AI47325 (to D.G.), NIH National Institute of General Medical Sciences 5T34 GM087239 (to K.M.R.), American Heart Association Grant 12SDG8170005 and the Anna Yoder MS Fund (to J.-H.Y.). We thank Drs. Ana Gamero (Department of Biochemistry, Temple University School of Medicine, Philadelphia, PA, USA) for the Stat2−/− mice, Luis Sigal (Fox Chase Cancer Center, Philadelphia, PA, USA) for the Irf7−/− mice, and Paul Gallo (Department of Microbiology and Immunology, Temple University School of Medicine) for his help in generating Flt3L pDCs.
Glossary
- Akt
protein kinase B
- BM
bone marrow
- cDC
conventional dendritic cells
- DC
dendritic cells
- DLN
draining lymph nodes
- EAE
experimental autoimmune encephalomyelitis
- Flt3L
fms-like tyrosine kinase 3 ligand
- GSK3
glycogen synthase kinase 3
- IFNAR
interferon-α/β receptor
- IRF
IFN regulatory factor
- ISGF3
IFN-stimulated gene factor 3
- KO
knockout
- LiCl
lithium chloride
- LN
lymph node
- MOG35–55
myelin oligodendrocyte glycoprotein peptide (amino acids 35–55)
- MS
multiple sclerosis
- pAkt
phosphorylated Akt
- Pam3CSK4
synthetic bacterial lipopeptide Pam3-Cys-Ser-Lys4
- pCD
plasmacytoid DC
- PGN
peptidoglycan
- poly-IC
polyinosinic-polycytidylic acid
- qRT-PCR
quantitative real-time polymerase chain reaction
- RR-MS
remitting relapsing multiple sclerosis
- Tregs
regulatory T cells
- VIP
vasoactive intestinal peptide
- WT
wild-type
Footnotes
SEE CORRESPONDING EDITORIAL ON PAGE 683
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
DISCLOSURES
The authors declare no competing financial interests.
REFERENCES
- 1.IFNB Multiple Sclerosis Study Group (2001) Interferon beta-lb is effective in relapsing-remitting multiple sclerosis, I: clinical results of a multicenter, randomized, double-blind, placebo-controlled trial. 1993 [classical article]. [classical article] Neurology 57(12, Suppl 5)S3–S9. [PubMed] [Google Scholar]
- 2.Bendtzen K. (2010) Critical review: assessment of interferon-β immunogenicity in multiple sclerosis. J. Interferon Cytokine Res. 30, 759–766. [DOI] [PubMed] [Google Scholar]
- 3.Brod S. A., Burns D. K. (1994) Suppression of relapsing experimental autoimmune encephalomyelitis in the SJL/J mouse by oral administration of type I interferons. Neurology 44, 1144–1148. [DOI] [PubMed] [Google Scholar]
- 4.Guo B., Chang E. Y., Cheng G. (2008) The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J. Clin. Invest. 118, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prinz M., Schmidt H., Mildner A., Knobeloch K. P., Hanisch U. K., Raasch J., Merkler D., Detje C., Gutcher I., Mages J., Lang R., Martin R., Gold R., Becher B., Brück W., Kalinke U. (2008) Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity 28, 675–686. [DOI] [PubMed] [Google Scholar]
- 6.Teige I., Treschow A., Teige A., Mattsson R., Navikas V., Leanderson T., Holmdahl R., Issazadeh-Navikas S. (2003) IFN-beta gene deletion leads to augmented and chronic demyelinating experimental autoimmune encephalomyelitis. J. Immunol. 170, 4776–4784. [DOI] [PubMed] [Google Scholar]
- 7.Yu M., Nishiyama A., Trapp B. D., Tuohy V. K. (1996) Interferon-beta inhibits progression of relapsing-remitting experimental autoimmune encephalomyelitis. J. Neuroimmunol. 64, 91–100. [DOI] [PubMed] [Google Scholar]
- 8.Makar T. K., Trisler D., Bever C. T., Goolsby J. E., Sura K. T., Balasubramanian S., Sultana S., Patel N., Ford D., Singh I. S., Gupta A., Valenzuela R. M., Dhib-Jalbut S. (2008) Stem cell based delivery of IFN-beta reduces relapses in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 196, 67–81. [DOI] [PubMed] [Google Scholar]
- 9.Axtell R. C., de Jong B. A., Boniface K., van der Voort L. F., Bhat R., De Sarno P., Naves R., Han M., Zhong F., Castellanos J. G., Mair R., Christakos A., Kolkowitz I., Katz L., Killestein J., Polman C. H., de Waal Malefyt R., Steinman L., Raman C. (2010) T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat. Med. 16, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dann A., Poeck H., Croxford A. L., Gaupp S., Kierdorf K., Knust M., Pfeifer D., Maihoefer C., Endres S., Kalinke U., Meuth S. G., Wiendl H., Knobeloch K. P., Akira S., Waisman A., Hartmann G., Prinz M. (2012) Cytosolic RIG-I-like helicases act as negative regulators of sterile inflammation in the CNS. Nat. Neurosci. 15, 98–106. [DOI] [PubMed] [Google Scholar]
- 11.Sweeney C. M., Lonergan R., Basdeo S. A., Kinsella K., Dungan L. S., Higgins S. C., Kelly P. J., Costelloe L., Tubridy N., Mills K. H., Fletcher J. M. (2011) IL-27 mediates the response to IFN-β therapy in multiple sclerosis patients by inhibiting Th17 cells. Brain Behav. Immun. 25, 1170–1181. [DOI] [PubMed] [Google Scholar]
- 12.Teige I., Liu Y., Issazadeh-Navikas S. (2006) IFN-β inhibits T cell activation capacity of central nervous system APCs. J. Immunol. 177, 3542–3553. [DOI] [PubMed] [Google Scholar]
- 13.Galicia G., Gommerman J. L. (2014) Plasmacytoid dendritic cells and autoimmune inflammation. Biol. Chem. 395, 335–346. [DOI] [PubMed] [Google Scholar]
- 14.Mohammad M. G., Hassanpour M., Tsai V. W., Li H., Ruitenberg M. J., Booth D. W., Serrats J., Hart P. H., Symonds G. P., Sawchenko P. E., Breit S. N., Brown D. A. (2012) Dendritic cells and multiple sclerosis: disease, tolerance and therapy. Int. J. Mol. Sci. 14, 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goverman J. (2009) Autoimmune T cell responses in the central nervous system. Nat. Rev. Immunol. 9, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ransohoff R. M., Engelhardt B. (2012) The anatomical and cellular basis of immune surveillance in the central nervous system. Nat. Rev. Immunol. 12, 623–635. [DOI] [PubMed] [Google Scholar]
- 17.Zozulya A. L., Clarkson B. D., Ortler S., Fabry Z., Wiendl H. (2010) The role of dendritic cells in CNS autoimmunity. J. Mol. Med. (Berl). 88, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svajger U., Rozman P. (2014) Tolerogenic dendritic cells: molecular and cellular mechanisms in transplantation. J. Leukoc. Biol. 95, 53–69. [DOI] [PubMed] [Google Scholar]
- 19.Van Brussel I., Lee W. P., Rombouts M., Nuyts A. H., Heylen M., De Winter B. Y., Cools N., Schrijvers D. M. (2014) Tolerogenic dendritic cell vaccines to treat autoimmune diseases: can the unattainable dream turn into reality? Autoimmun. Rev. 13, 138–150. [DOI] [PubMed] [Google Scholar]
- 20.Kong W., Yen J. H., Vassiliou E., Adhikary S., Toscano M. G., Ganea D. (2010) Docosahexaenoic acid prevents dendritic cell maturation and in vitro and in vivo expression of the IL-12 cytokine family. Lipids Health Dis. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brasel K., De Smedt T., Smith J. L., Maliszewski C. R. (2000) Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood 96, 3029–3039. [PubMed] [Google Scholar]
- 22.Chang J., Voorhees T. J., Liu Y., Zhao Y., Chang C. H. (2010) Interleukin-23 production in dendritic cells is negatively regulated by protein phosphatase 2A. Proc. Natl. Acad. Sci. U. S. A. 107, 8340–8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Andrea A., Aste-Amezaga M., Valiante N. M., Ma X., Kubin M., Trinchieri G. (1993) Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J. Exp. Med. 178, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takaoka A., Yanai H. (2006) Interferon signalling network in innate defence. Cell. Microbiol. 8, 907–922. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Cao S., Herman L. M., Ma X. (2003) Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-gamma-primed IL-12 production by IFN regulatory factor 1. J. Exp. Med. 198, 1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura T., Ozato K. (2002) ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interferon Cytokine Res. 22, 145–152. [DOI] [PubMed] [Google Scholar]
- 27.Sheikh S. Z., Kobayashi T., Matsuoka K., Onyiah J. C., Plevy S. E. (2011) Characterization of an interferon-stimulated response element (ISRE) in the Il23a promoter. J. Biol. Chem. 286, 1174–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriele L., Fragale A., Borghi P., Sestili P., Stellacci E., Venditti M., Schiavoni G., Sanchez M., Belardelli F., Battistini A. (2006) IRF-1 deficiency skews the differentiation of dendritic cells toward plasmacytoid and tolerogenic features. J. Leukoc. Biol. 80, 1500–1511. [DOI] [PubMed] [Google Scholar]
- 29.Martin M., Rehani K., Jope R. S., Michalek S. M. (2005) Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 6, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H., Brown J., Garcia C. A., Tang Y., Benakanakere M. R., Greenway T., Alard P., Kinane D. F., Martin M. (2011) The role of glycogen synthase kinase 3 in regulating IFN-β-mediated IL-10 production. J. Immunol. 186, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drulovic J., Kostic J., Mesaros S., Dujmovic Basuroski I., Stojsavljevic N., Kisic-Tepavcevic D., Pekmezovic T. (2013) Interferon-beta and disability progression in relapsing-remitting multiple sclerosis. Clin. Neurol. Neurosurg. 115 (Suppl 1), S65–S69. [DOI] [PubMed] [Google Scholar]
- 32.Durelli L., Verdun E., Barbero P., Bergui M., Versino E., Ghezzi A., Montanari E., Zaffaroni M.; Independent Comparison of Interferon (INCOMIN) Trial Study Group (2002) Every-other-day interferon beta-1b versus once-weekly interferon beta-1a for multiple sclerosis: results of a 2-year prospective randomised multicentre study (INCOMIN). Lancet 359, 1453–1460. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt H., Raasch J., Merkler D., Klinker F., Krauss S., Brück W., Prinz M. (2009) Type I interferon receptor signalling is induced during demyelination while its function for myelin damage and repair is redundant. Exp. Neurol. 216, 306–311. [DOI] [PubMed] [Google Scholar]
- 34.Yen J. H., Ganea D. (2009) Interferon beta induces mature dendritic cell apoptosis through caspase-11/caspase-3 activation. Blood 114, 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittelstadt M. L., Patel R. C. (2012) AP-1 mediated transcriptional repression of matrix metalloproteinase-9 by recruitment of histone deacetylase 1 in response to interferon β. PLoS One 7, e42152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortés-Vieyra R., Bravo-Patiño A., Valdez-Alarcón J. J., Juárez M. C., Finlay B. B., Baizabal-Aguirre V. M. (2012) Role of glycogen synthase kinase-3 beta in the inflammatory response caused by bacterial pathogens. J. Inflamm. (Lond). 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salem M., Mony J. T., Løbner M., Khorooshi R., Owens T. (2011) Interferon regulatory factor-7 modulates experimental autoimmune encephalomyelitis in mice. J. Neuroinflammation 8, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haines C. J., Chen Y., Blumenschein W. M., Jain R., Chang C., Joyce-Shaikh B., Porth K., Boniface K., Mattson J., Basham B., Anderton S. M., McClanahan T. K., Sadekova S., Cua D. J., McGeachy M. J. (2013) Autoimmune memory T helper 17 cell function and expansion are dependent on interleukin-23. Cell Reports 3, 1378–1388. [DOI] [PubMed] [Google Scholar]
- 39.Zúñiga L. A., Jain R., Haines C., Cua D. J. (2013) Th17 cell development: from the cradle to the grave. Immunol. Rev. 252, 78–88. [DOI] [PubMed] [Google Scholar]
- 40.Krakauer M., Sorensen P., Khademi M., Olsson T., Sellebjerg F. (2008) Increased IL-10 mRNA and IL-23 mRNA expression in multiple sclerosis: interferon-beta treatment increases IL-10 mRNA expression while reducing IL-23 mRNA expression. Mult. Scler. 14, 622–630. [DOI] [PubMed] [Google Scholar]
- 41.Ulusoy C., Tüzün E., Kürtüncü M., Türkoğlu R., Akman-Demir G., Eraksoy M. (2012) Comparison of the cytokine profiles of patients with neuronal-antibody-associated central nervous system disorders. Int. J. Neurosci. 122, 284–289. [DOI] [PubMed] [Google Scholar]
- 42.VanDeusen J. B., Shah M. H., Becknell B., Blaser B. W., Ferketich A. K., Nuovo G. J., Ahmer B. M., Durbin J., Caligiuri M. A. (2006) STAT-1-mediated repression of monocyte interleukin-10 gene expression in vivo. Eur. J. Immunol. 36, 623–630. [DOI] [PubMed] [Google Scholar]
- 43.Liu J., Guan X., Tamura T., Ozato K., Ma X. (2004) Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J. Biol. Chem. 279, 55609–55617. [DOI] [PubMed] [Google Scholar]
- 44.Saraiva M., O’Garra A. (2010) The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181. [DOI] [PubMed] [Google Scholar]