Abstract
Although Fourier transform (FT) and tryptophan-scanning mutagenesis (TrpScanM) have been extremely useful for predicting secondary structures of membrane proteins, they are deemed to be low-resolution techniques. Herein, we describe the combined use of FT and TrpScanM (FT-TrpScanM) as a more reliable approach for the prediction of secondary structure. Five TrpScanM studies of the acetylcholine receptor lipid-exposed transmembrane domains (LETMDs) were revisited and analyzed by FT-TrpScanM. FT analysis of the raw data from the aforementioned TrpScanM studies supports and validates the conclusions derived from their tryptophan-periodicity profiles. Furthermore, by FT-TrpScanM, we were able to determine the minimum number of consecutive tryptophan substitutions necessary for more robust prediction of α-helical secondary structures and evaluate the quality of structure predictions by α-helical character curves. Finally, this study encourages future utilization of FT-TrpScanM to more reliably predict secondary structures of the membrane protein LETMDs.
Keywords: Tryptophan-scanning mutagenesis, Fourier transform, Ion channel, Lipid-exposed transmembrane domain, Secondary structure, Two-electrode voltage clamp
1. Introduction
Unlike globular proteins, high-resolution three-dimensional structures of membrane proteins as determined by x-ray crystallography are seldom found in protein structure databases, reflecting the difficulties associated with their crystallization. As preferred alternatives to x-ray crystallography for structural assessment, NMR and cryo-electron microscopy have been typically used; however, they are limited by protein size and reachable resolution, respectively. In this scenario, the tryptophan-scanning mutagenesis (TrpScanM) has emerged as another option for predicting the secondary structure and packing arrangement of the lipid-exposed transmembrane domains (LETMDs), which can then be used to model their overall spatial orientation [1]. Although TrpScanM has been successfully applied to several ion-channel proteins such as nicotinic AChR channels [1-5], inward rectifier potassium channels [6-8], voltage-activated potassium channels [9-15], glutamate receptor channels [16], γ-aminobutyric acid type A receptor channels [17, 18], voltage-gated sodium channels [19], N-methyl-D-aspartate receptor channels [20], P2X4 receptor channels [21] mechanosensitive channels MscL [22, 23], human ether-a-go-go-related gene (HERG) K+ channels [24] and epithelial Na+ channels [25], the TrpScanM is, nevertheless, deemed a low-resolution method for predicting secondary structure and packing arrangement because it does not provide direct structural information. The rationale for choosing TrpScanM is that tryptophan systematic substitutions in the transmembrane domain of the ion-channel protein should result in a loss of function and/or functional expression when facing the interior of the protein. Conversely, when the tryptophan substitution faces the lipid environment the mutation could be tolerated, thus, the tryptophan systematic substitutions give rise to periodic patterns of perturbations in ion-channel function and functional expression. Herein, we coupled Fourier transform (FT) to TrpScanM (FTTrpScanM) to predict reliable secondary structures of the acetylcholine receptor (AChR) LETMDs. The focus of the present study is not to determine the secondary structure of the AChR LETMDs because these have already been determined. Indeed, the main objective of this study is to convey the use of FT-TrpScanM as a valuable alternate tool to make reliable predictions of secondary structures of the membrane protein LETMDs for which high-resolution structures are not yet available. It is noteworthy that we took advantage of the recently reported structure of the Torpedo AChR, which was determined at 4 Å resolution using cryo-electron microscopy in order to corroborate our predictions [26]. Interestingly, the TrpScanM predicted α-helical structures for the LETMDs [1-5] even when a lower-resolution cryo-electron microscopy structure mistakenly predicted the LETMDs to be arranged as β-sheets [27]. Thus, the AChR serves as an excellent model to test the validity of the FT-TrpScanM approach and provides reassurance on its capacity to generate trustworthy predictions of the secondary structures of membrane protein LETMDs in general. Furthermore, FT-TrpScanM supports and validates the conclusion from previous TrpScanM [1-5] and biochemical [28-30] studies, which predicted that the LETMDs are arranged as α-helical structures. In addition, using this approach, we were able to provide additional structural information, estimate mean periodicities, determine the minimum number of consecutive tryptophan substitutions required to reliably predict α-helical structures, and assess the quality of structure predictions.
2. Materials and methods
2.1. General Experimental Procedures
Xenopus laevis oocytes were microinjected with complementary RNAs from mouse muscle adult-type or Torpedo californica AChR (see also refs. [1-5]). Mutations were engineered with the QuikChange® Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA) and were confirmed by automated DNA sequencing. All mutagenic primers were designed with the tryptophan codon (TGG) instead of the wild type (WT) codon at the desired position. Muscle-type or Torpedo AChR cRNA transcripts were synthesized with the mMESSAGE mMACHINE® kit (Ambion, Inc., Austin, TX, USA). Oocytes were incubated for 3-5 days with fresh liquid medium at 19 °C.
2.2. Voltage Clamp
Macroscopic ACh-induced currents were recorded at room temperature with a whole-cell two-electrode voltage clamp configuration using a Gene Clamp 500B amplifier (Axon Instruments, Inc., Union City, CA, USA). Electrodes were filled with 3 M KCl and had resistances of < 2 mega ohms. Impaled oocytes were automatically perfused with a MOR-2 buffer [82.5 mM NaCl, 2.5 mM KCl, 5 mM MgCl2, 1 mM Na2HPO4, 0.2 mM CaCl2, 5 mM HEPES, and 0.5 mM EGTA (pH 7.4)] at a rate of 0.43 mL/s and a solution exchange of < 3 s using a Perfusion Valve Controller VC-8 (Warner Instruments, Inc., Hamden, CT, USA). Membrane potential was held at −70 mV. Membrane currents were filtered at 100 Hz and digitized at 1 kHz using Gene Clamp 500B amplifier and DigiData 1322A interface (Axon Instruments, Inc., Union City, CA, USA), respectively. Data acquisition was conducted through the Clampex 9.2 program (Axon Instruments, Inc., Union City, CA, USA). Dose-response curves were generated from macroscopic peak currents (I) obtained from seven different ACh concentrations (1, 3, 10, 30, 55, 100, and 300 μM ACh). Dose-response curves were fitted through a sigmoidal dose-response equation with variable slope using GraphPad Prism 4 program (GraphPad Software, Inc., San Diego, CA, USA),
| (1) |
where I is the macroscopic peak ionic current at a given ACh concentration, Imin and Imax are the smallest and the largest currents observed, respectively, EC50 is the concentration of acetylcholine that provokes a response halfway between Imin and Imax, and the Hill Slope is the steepness of the dose-response curve [31].
2.3. Periodicity Profiles
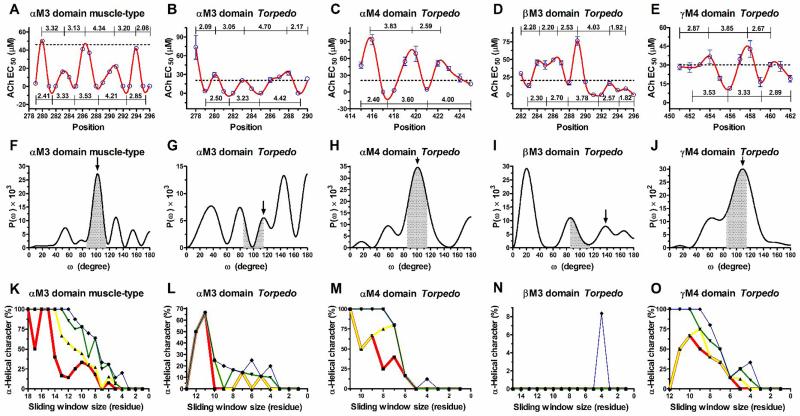
The oscillation patterns of the TrpPPs were generated from the reported ACh EC50 values from five LETMDs (αM4, βM3, γM4, and two αM3 domains). TrpPPs were plotted with ACh EC50 values as a function of the tryptophan substitution positions. TrpPP curves were fitted through a cubic spline method with 3,000 segments to interpolate the points using GraphPad Prism 4 program. The number of residues per helical turn of the periodicity profiles was estimated as the number of amino acids between adjacent maximums and minimums peaks (see Fig. 1 A-E).
Figure 1. Tryptophan-periodicity profiles, Fourier transform power spectra, and α-helical character curves generated from tryptophan-scanning mutagenesis data of the AChR lipid-exposed transmembrane domains.
(A-E) are TrpPPs. Values along the lines indicate the number of residues per helical turn between adjacent maximums and minimums peaks. The black dashed line indicates the ACh EC50 value of the wild-type AChR. (F-J) are FT power spectra from entire sequences of the ACh EC50 values shown in the TrpPPs (A-E). The black headed arrows indicate the peaks that correspond to mean periodicities of the TrpPPs (A-E). Grey shadings demarcate the region 85° ≤ ω ≤ 115° that is used to calculate peak ratio values. (K-O) are α-helical character curves of different periodicity intervals as a function of the number of residues in the sliding window; red line (3.5 – 3.7) residues/turn; yellow line (3.4 – 3.8) residues/turn; green line (3.3 – 3.9) residues/turn; or blue line (3.2 – 4.0) residues/turn.
2.4. Fourier Transform
The FT power spectra from ACh EC50 data were generated using a least-squares discrete FT equation [13, 21, 32-36]. The FT power spectrum P(ω) were plotted as a function of angular frequency ω (rotation angle between residue around a helical axis) using,
| (2) |
where Vj is the ACh EC50 values at a given position j, <Vj> is the mean value of Vj in the sliding window, and N is the number of ACh EC50 values in the sliding window (see Fig. 1 F-J). The sliding windows of the ACh EC50 values must follow the order presented in the original sequence. For instance, the FT analysis of a sequence of values from an ideal α-helical pattern (3.6 residues per turn) should give a prominent peak at ~100° in the FT power spectrum, given that the ω parameter is related to the number of residues per turn (d) by the expression ω = 360°/d. The mean periodicities of the sequences of ACh EC50 values were estimated from the peak localized in the vicinity of the rotation angle as indicated by the mean periodicities of their TrpPPs. The expected rotation angles (ω) of the TrpPPs’ mean periodicities were calculated by the abovementioned equation, ω = 360°/d. The reliability of the sequences of ACh EC50 values for predicting α-helical periodicities was evaluated by calculating the ratio of P(ω) in the α-helical range (85° ≤ ω ≤ 115°) relative to P(ω) over the whole spectrum using,
| (3) |
where P(ω) is the FT power spectrum. Peak ratio values greater than 2, which represent at least 33% of the whole power spectrum, have been considered as a very good indicator of an α-helical secondary structure [21, 32-34, 36]. The algorithm templates developed to plot the FT power spectra and calculate the peak ratio values were generated using the MATLAB® 7.4 program (The MathWorks, Inc., Natick, MA, USA) with a precision of 1×10−4. The algorithm templates are available in the appendix 1.
2.5. α-Helical character curves
To generate the α-helical character curves, different sequences of ACh EC50 values were partitioned as a squared triangle to form the sliding windows of different sizes. The number of sliding windows was determined using a triangular number equation; number of sliding windows = n(n+1)/2, where n is the number of values in the sequence. The partitions of sequences, as triangular numbers, satisfy the condition that the sub-sequences generated should be unique and also follow the consecutive order of the original sequences. The FT power spectra generated from different sliding windows were evaluated to determine the percent of FT power spectra per sliding window size that fulfilled two characteristic parameters of α-helical secondary structures: (i) peak ratio > 2 and (ii) prominent peak inside several frequency intervals such as 102.9° ≤ prominent peak ≤ 97.3° for 3.5-3.7 periodicities; 105.9° ≤ prominent peak ≤ 94.7° for 3.4-3.8 periodicities; 109.1° ≤ prominent peak ≤ 92.3° for 3.3-3.9 periodicities; or 112.5° ≤ prominent peak ≤ 90° for 3.2-4.0 periodicities. The percent of the FT power spectra that fulfilled the aforementioned parameters were plotted as a function of sliding window size to generate α-helical character curves of the different frequency intervals (see Fig. 1 K-O). For instance, a value of 100% α-helical character for a sliding window size of 12 residues implies that each one of the seven FT power spectra (see Fig. 1 K; blue curve), which were generated from seven partitions composed of 12 consecutive and different values, has a peak ratio value > 2 and a prominent peak maximum inside the range 90°-112.5°. In this example, the sliding window size of 12 values was moved seven times along a sequence of 18 values in total to generate the seven different partitions that were used to generate the seven FT power spectra (see Appendix 2; Table A2).
3. Results and discussion
In this study, we revisited the reported TrpScanM from five LETMDs (αM4, βM3, γM4, and two αM3 domains) of two different AChR species (Table 1). The raw data of these studies, which predicted helical structures with diverse periodicities and packing arrangements for the LETMDs [1-5], were used to generate the FT power spectra and the α-helical character curves. By coupling FT to TrpScanM, we achieve a more reliable prediction for secondary structures of the LETMDs and provide initial support of this valuable alternative tool to the future scanning mutagenesis studies.
Table 1.
Amino acid sequences of the studied AChR LETMDs.
3.1. Tryptophan-periodicity profiles of the αM4, βM3, γM4, and two αM3 domains
We used TrpScanM to predict the secondary structures and packing arrangements of the LETMDs. The periodic patterns of the functional consequences due to the tryptophan substitutions were displayed in the TrpPPs (Fig. 1 A-E). The TrpPPs were plotted with ACh EC50 values derived from dose-response curves of the mutations generated in the AChR LETMDs [1-5]. The TrpPPs, which have been previously reported, display different oscillation patterns and different frequencies, indicating that LETMDs form helical structures with different periodicities and different helical packings. These findings have been supported by a refined structure of the Torpedo marmorata nicotinic acetylcholine receptor at 4 Å resolution determined by cryo-electron microscopy method [26].
3.2. Fourier transform power spectra of the αM4, βM3, γM4, and two αM3 domains
Generally, the discrete Fourier transform analysis is used for converting a sequence of values into a frequency spectrum and thus, it may be used to detect periodic variations in the sequence. Therefore, we coupled FT to TrpScanM to estimate mean periodicities and peak ratios of the sequences of ACh EC50 values of the LETMDs that are displayed in the TrpPPs (Fig. 1 A-E). Using FT equation (Eq. 2), the sequences of ACh EC50 values were decomposed into multiple peaks with different frequencies and abundances at the FT power spectra, suggesting that TrpPPs may be divided as well into several periodicities with diverse oscillation patterns (Fig. 1 F-J). The similarities in the periodicity values from the mean periodicities determined by rotation angle of the peaks and the mean periodicities of the TrpPPs reveal that FT supports and validates TrpScanM data (see Table 2). Interestingly, the prominent peaks of the Torpedo αM3 and βM3 AChRs were not found in the expected vicinity of the rotation angle indicated by the mean periodicities of their TrpPPs. The unexpected location of these prominent peaks could be due to the contribution of anomalous oscillations in their TrpPPs. For instance, the anomalous amplitudes displayed by the values in the 278 and 284-287 positions in the TrpPPs (Fig. 1 B and D, respectively), have considerable weight on the fitting of the sequences of values, causing a bias on the abundance of the peaks. Therefore, these anomalous oscillations populate peaks at frequencies which are not characteristic of the periodicity of an α-helical secondary structure. A notorious example of this is the abovementioned value in the 278 position. The value in the 278 position contributes extensively to delocalizing the prominent peak from the α-helical frequency intervals (see Appendix 2; Fig. A1 A, B, and D). However, this contribution is not reflected in the FT power spectra generated from sub-series (partitions) that do not bear the value of the 278 position (see Appendix 2; Fig. A1 C, E, and F).
Table 2.
Secondary structure parameters of AChR LETMDs determined by TrpPPs and FT power spectra.
| TrpPPs | FT power spectra | |||||
|---|---|---|---|---|---|---|
| AChR type | No residues | Periodicity | Expected rotation angle | Rotation angle | Peak ratio | Mean periodicity |
| residues/turn | Degree | degree | residues/turn | |||
| αM3 Muscle-type | 18 | 3.24 ± 0.70 | 111.11 | 102.25 | 2.94 | 3.52 |
| αM3 Torpedo | 13 | 3.17 ± 1.04 | 113.56 | 115.00 | 0.53 | 3.13 |
| αM4 Torpedo | 11 | 3.28 ± 0.74 | 109.76 | 100.76 | 3.15 | 3.57 |
| βM3 Torpedo | 15 | 2.61 ± 0.74 | 137.93 | 138.06 | 0.75 | 2.61 |
| γM4 Torpedo | 12 | 3.19 ± 0.45 | 112.85 | 108.88 | 2.67 | 3.31 |
Given values correspond to analysis performed on the entire sequence of ACh EC50 values from the AChR LETMDs. Periodicities of the TrpPPs are given as mean ± SD. Expected rotation angles of the TrpPPs and the mean periodicities of the peaks that correspond to mean periodicities of the TrpPPs were calculated using periodicity (residues/turn) = 360°/rotation angle.
3.3. α-helical character curves of the αM4, βM3, γM4, and two αM3 domains
We used α-helical character curves as complementary tool to the prediction of secondary structure by TrpScanM. The peak ratio from FT power spectra has been used as a parameter of reliability for predicting α-helical structures, however, it is not enough for a clear prediction because the maximum of the prominent peak could be located outside the α-helical frequency interval (85° ≤ ω ≤ 115°) and yet have a peak ratio > 2. In order to avoid this problem and to make more reliable predictions of secondary structures, we have generated the α-helical character curves (Fig. 1 K-O). The α-helical character curves provide us a tracking quality map of structure predictions. The α-helical character curves of the muscle-type αM3 domain, and the Torpedo αM4 and γM4 domains show that the α-helical structures predicted by FT-TrpScanM method were more reliable than those of the Torpedo αM3 and βM3 domains because they maintain a level of 100% α-helical characters even while reducing the size of the sliding windows at different periodicity intervals. A level of 100% α-helical character indicates that all the FT power spectra, generated from TrpScanM data, for a given sliding window size and periodicity interval fulfill both parameters that characterize an α-helix structure (see “2. Material and methods”). The Torpedo αM3 and βM3 domains displayed low percents in the α-helical character curves, presumably due to two factors: (i) insufficient amount of tryptophan substitutions to predict an α-helix structure and/or (ii) the sequence of values predicted a mixture of α- and 310-helix structures or a 310-helix structure. The size of the smallest sliding window displaying a level of 100% α-helical character can be considered indicative of the minimum number of consecutive tryptophan substitutions necessary to predict reliable α-helix structures at a given periodicity interval. For instance, the amount of 12 residues is suitable to predict a reliable α-helical structure that would have periodicities between 3.2 and 4.0 residues per turn (Fig. 1 K; blue curve), whereas in the Fig. 1 L more residues are required to predict a reliable α-helix structure using the studied periodicity intervals (Appendix 2; Table A1 and A2). Also, from the α-helical character graphs, it is evident that the prediction of extremely reliable α-helical structures requires shorter periodicity intervals and larger sliding windows. In addition, the predictions of the LETMDs could be ranked according to the following order: αM3 muscle-type > αM4 Torpedo ≈ γM4 Torpedo > αM3 Torpedo >> βM3 Torpedo through the use of the α-helical character curves. In essence, the quality of structure predictions by TrpScanM could be examined by α-helical character curves at different periodicity intervals.
4. Conclusions
FT-TrpScanM may be a useful tool for predicting reliable secondary structures of membrane protein LETMDs, shedding light into their overall spatial orientation. The α-helical character curves may be used to fingerprint the quality of structure predictions and determine the minimum of substitutions required for reliable structure predictions. Finally, this work corroborates the predictions from previous TrpScanM studies, demonstrates that similar results could have been reached with even fewer substitutions and encourages future utilization of FTTrpScanM as a means of predicting the secondary structure of LETMDs.
Acknowledgments
We especially thank Dr. Kenton J. Swartz and Dr. Shai D. Silberberg for kindly providing the program used to generate the Fourier transform power spectra. This work was supported by the NIH grants 2RO1GM56371-10, GM08102-27, and the University of Puerto Rico Institutional Funds for Research grant (to J.A.L.-D.), and the NIH-NIGMS-MBRS-Research Initiative for Scientific Enhancement fellowship 2R25GM61151 (to J.D.O.-C. and C.A.B.-P.).
Abbreviations
- AChR
acetylcholine (ACh) receptor
- FT
Fourier transform
- FT-TrpScanM
Fourier transform coupled to tryptophan-scanning mutagenesis
- LETMDs
lipid-exposed transmembrane domains
- TrpPPs
tryptophan-periodicity profiles
- TrpScanM
tryptophan-scanning mutagenesis
APPENDIX 1
The script 1 of the algorithm template to plot the FT power spectra, calculate the peak ratios values and determine the maximum of the prominent peaks using the MATLAB® program with a precision of 1×10−4is located below. This script is designed to use with the Microsoft Office Excel program.
%------------------------------------------------------------------------------------------------------------------
function[]=FOURIERTRANSFORMRESULTS(DATAFILENAME,DATARANGE,STORAGE FILE)
clc;
dirtemp=cd;FILENAME=strcat(cd,'\',STORAGEFILE);
data=xlsread(strcat(cd,'\',DATAFILENAME),DATARANGE);
n=length(data);N=n*(n+1)/2;K=cell(n-1,1);
[omega p]=fftspectra(data');T=[omega'p'];MM=length(omega);
for j=1:n-1
[T1]=windowareafft(data',j);K(j)={T1};
end
DK=[{'Window Size'};{'Average'};{'Peak Ratio'};{'Max omega'};{'P(Max omega)'}];
HK=cell2mat(K);[a b]=size(HK);H=cell2mat(K);
for jj=2:n
C1(jj-1)=length(find(H(:,4)<=360/3.5&H(:,4)>=360/3.7&
H(:,3)>=2&H(:,1)==jj))/length(find(H(:,1)==jj));
C2(jj-1)=length(find(H(:,4)<=360/3.4&H(:,4)>=360/3.8&
H(:,3)>=2&H(:,1)==jj))/length(find(H(:,1)==jj));
C3(jj-1)=length(find(H(:,4)<=360/3.3&H(:,4)>=360/3.9&
H(:,3)>=2&H(:,1)==jj))/length(find(H(:,1)==jj));
C4(jj-1)=length(find(H(:,4)<=360/3.2&H(:,4)>=360/4.0&
H(:,3)>=2&H(:,1)==jj))/length(find(H(:,1)==jj));
end
Tabla=[[2:n]' C1' C2' C3' C4'];[c d]=size(Tabla);
DKTabla=[{'Window Size'};{'3.5 - 3.7'};{'3.4 - 3.8'};{'3.3 - 3.9' };{'3.2 - 4.0'}];
xlswrite(FILENAME,T,strcat('A2:B',num2str(MM+1)));
xlswrite(FILENAME,[{'omega'};{'P(omega)'}]','A1:B1');
xlswrite(FILENAME,DK','D1:H1');
xlswrite(FILENAME,HK,strcat('D2:H',num2str(a+1)));
xlswrite(FILENAME,DKTabla','J1:N1');
xlswrite(FILENAME,Tabla,strcat('J2:N',num2str(c+1)));disp('Finish');
end
function[omega,P]=fftspectra(data)
omega=0:.0001:180;n=length(data);dbar=sum(data)/n;
h=data-dbar;P=(h*cosd([1:n]'*omega)) ^2+(h*sind([1:n]'*omega)) ^2;
end
function[M]=windowareafft(v,k)
n=length(v);m=n-k+1;M=zeros(n-m+1,5);P=partition(n,m);
for i=1:(n-m+1)
data=v(P(i,:));
if ~(sum(data~=0)==0)
[omega p]=fftspectra(data);
Area=6*trapz(omega(find(omega>=85&omega<=115)),
p(find(omega>=85&omega<=115)))/trapz(omega,p);
Average=mean(data);M(i,:)=[m Average Area omega(find(p==max(p))) max(p)]
else
M(i,:)=[m 0 0 0 0];end;end;
function[H]=partition(n,m)
Residue=1:n;t=1;
while t<=(n-m)+1
tmp=(1:m)+t-1;H(t,:)=Residue(tmp);t=t+1;
end;end;end
%------------------------------------------------------------------------------------------------------------------
The script 2 for the directory of the files is located below. All files should be inside a folder. Replace “Put you data file name” and “Put you storage file name” with the desired data file name and storage file name, respectively. The data should be placed and saved in a Microsoft Office Excel file. The data should be along the column B beginning with the cell B2. The number signs “##”should be replaced with the number of the cell from the last data.
%------------------------------------------------------------------------------------------------------------------
DATAFILENAME='Put you data file name.xls';
DATARANGE='B2:B##';
STORAGEFILE='Put you storage file name.xls';
FTRESULTS(DATAFILENAME,DATARANGE,STORAGEFILE)
%------------------------------------------------------------------------------------------------------------------
APPENDIX 2
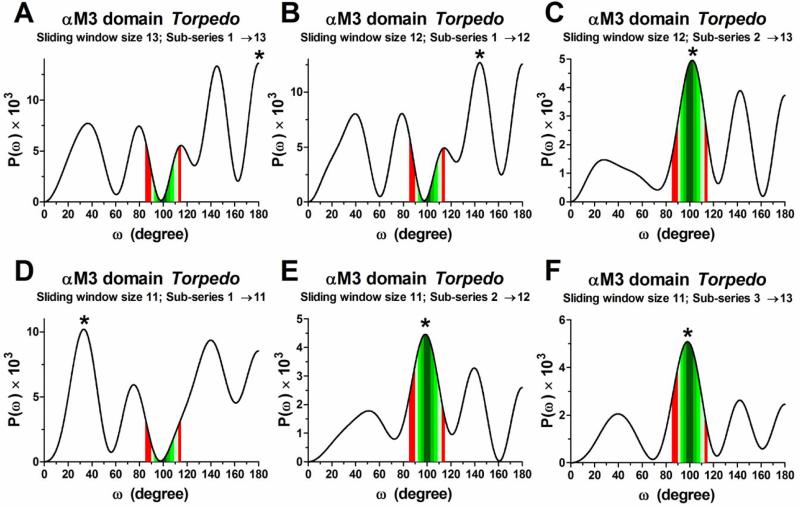
In this appendix, we illustrate extracts of the Fourier transform power spectra data from αM3 Torpedo AChR and the α-helical character data from αM3 Torpedo and αM3 Muscle-type AChRs for a clear comprehension of the FT-TrpScanM method. In Fig. 1A, we show the FT power spectra of the sliding window sizes 11 through 13. When the value of the 278 position is included on the partition to generate a FT power spectrum (i.e. the sub-series 1→13, 1→12, and 1→11), the prominent peak is not localized inside α-helical frequency intervals and neither into peak ratio interval (see Fig. A1 A, B, and D). In Table A1, we show the contribution of the value in the 278 position of the αM3 Torpedo to the α-helical character curves. The contribution of the value in the 278 position to the α-helical character curves at the sliding window sizes of 11 to 13 residues is shown by the sub-series 1→13, 1→12, and 1→11 (see red shading boxes; Table A1). In these sub-series the value of the 278 position is included to generate their respective FT power spectrum. From these FT power spectra, it is determined the peak ratio value and rotation angle of the prominent peak resulting on FT power spectra not characteristics of an α-helical secondary structure (see No=0; Table A1). Hence, these sub-series do not contribute positively to α-helical character from their sliding window size. Overall, the anomalous oscillations in the TrpPPs take more notoriety in sliding window sizes that are composed of few sub-series (e.g. the sub-series 1→13, 1→12, and 1→11 from αM3 Torpedo; see Table A1) because these sub-series individually represent a portion larger than in sliding windows of many sub-series (i.e. the increase in the number of sub-series decrease the proportion that is contributed by each sub-series in the sliding window). In the Table A2, we show the minimum number of substitutions that is required to predict a reliable α-helical secondary structure at a level of 100% α-helical character. The αM3 Muscle-type data show that to predict an α-helical secondary structure of mean periodicities between 3.2 and 4.0 residues per turn at a level of 100% α-helical character at least 12 substitutions are necessary, while for mean periodicities between 3.3 and 3.9, 3.4 and 3.8, or 3.5 and 3.7 at least 13, 14, or 15 substitutions, respectively, are necessary (see red shading boxes; Table A2).
Note: Sub-series are partitions from the sequence of values. Out or In represents wheather the localization of the maximum of the prominent peak is outside or inside of the frequency interval. 0=No or 1=Yes represent if the FT power spectrum generated from the sub-series fulfill the two parameters characteristic of an α-helical secondary structure: i) Peak ratio value > 2 and ii) Localization of the maximum of the prominent peak is inside frequency interval. 0=No represents that one of the two parameters is not fulfilled. 1=Yes represents that both parameters are fulfilled (see gray shading boxes; Table A1 and A2). % is the percent of α-helical character per sliding window. % represents the percent of sub-series per sliding window that fulfill the characteristic parameters of an α-helical structure.
Figure 1A. Extract of the Fourier transform power spectra data from αM3 Torpedo AChR.
(A-F) are FT power spectra generated from sub-series 1→13, 1→12, 2→13, 1→11, 2→12, and 3→13 of the αM3 Torpedo data. Asterisks indicate the localization of the prominent peaks. Different intensities of green shadings demarcate the different frequency intervals; dark green (97.3°-102.9°); green; (94.7°-105.9°); bright green (92.3°-109.1°); light green (90.0°-112.5°). Red shadings demarcate the region 85° ≤ ω ≤ 115° that is used to calculate peak ratio values.
Table A1.
Extract of the α-helical character data from αM3 Torpedo AChR.
| Sliding window | Fourier transform power spectra | α-Helical character curves | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Partition | Peak ratio | Prominent Peak | Periodicity interval (3.5-3.7) | Periodicity interval (3.4-3.8) | Periodicity interval (3.3-3.9) | Periodicity interval (3.2-4.0) | |||||||||
| Sub series | Rotation angle | Frequency interval (97.3°-102.9°) | Frequency interval (94.7°-105.9°) | Frequency interval (92.3°-109.1°) | Frequency interval (90.0°-112.5°) | ||||||||||
| Residue | Position | Degree |
Out
or In |
0=No
1=Yes |
% |
Out
or In |
0=No
1=Yes |
% |
Out
or In |
0=No
1=Yes |
% |
Out
or In |
0=No
1=Yes |
% | |
| 13 | 1→13 | 0.53 | 180 | Out | 0 | 0 | Out | 0 | 0 | Out | 0 | 0 | Out | 0 | 0 |
| 12 | 1→12 | 0.50 | 144.22 | Out | 0 | 50 | Out | 0 | 50 | Out | 0 | 50 | Out | 0 | 50 |
| 12 | 2→13 | 2.12 | 102.17 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 11 | 1→11 | 0.28 | 33.18 | Out | 0 | 66 | Out | 0 | 66 | Out | 0 | 66 | Out | 0 | 66 |
| 11 | 2→12 | 2.06 | 98.64 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 11 | 3→13 | 2.36 | 98.17 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 10 | 1→10 | 0.35 | 130.15 | Out | 0 | 0 | Out | 0 | 25 | Out | 0 | 25 | Out | 0 | 25 |
| 10 | 2→11 | 1.60 | 99.36 | In | 0 | In | 0 | In | 0 | In | 0 | ||||
| 10 | 3→12 | 2.28 | 95.09 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 10 | 4→13 | 1.88 | 94.40 | Out | 0 | Out | 0 | In | 0 | In | 0 | ||||
| 9 | 1→9 | 0.29 | 134.08 | Out | 0 | 0 | Out | 0 | 0 | Out | 0 | 0 | Out | 0 | 20 |
| 9 | 2→10 | 2.57 | 110.58 | Out | 0 | Out | 0 | Out | 0 | In | 1 | ||||
| 9 | 3→11 | 1.83 | 93.54 | Out | 0 | Out | 0 | In | 0 | In | 0 | ||||
| 9 | 4→12 | 1.71 | 88.69 | Out | 0 | Out | 0 | Out | 0 | Out | 0 | ||||
| 9 | 5→13 | 1.69 | 154.98 | Out | 0 | Out | 0 | Out | 0 | Out | 0 | ||||
Table A2.
Extract of the α-helical character from αM3 Muscle-type AChR.
| Sliding window | Fourier transform power spectra | α-Helical character curves | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Partition | Peak ratio | Prominent Peak | Periodicity interval (3.5-3.7) | Periodicity interval (3.4-3.8) | Periodicity interval (3.3-3.9) | Periodicity interval (3.2-4.0) | |||||||||
| Sub series | Rotation angle | Frequency interval (97.3°-102.9°) | Frequency interval (94.7°-105.9°) | Frequency interval (92.3°-109.1°) | Frequency interval (90.0°-112.5°) | ||||||||||
| Residue | Position | Degree |
Out
or In |
0=No
1=Yes |
% |
Out
or In |
0=No
1=Yes |
% |
Out
or In |
0=No
1=Yes |
% |
Out
or In |
0=No
1=Yes |
% | |
| 18 | 1→18 | 2.94 | 102.25 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 |
| 17 | 1→17 | 2.75 | 103.24 | Out | 0 | 50 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 |
| 17 | 2→18 | 3.08 | 101.26 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 16 | 1→16 | 2.75 | 101.82 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 |
| 16 | 2→17 | 2.90 | 102.11 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 16 | 3→18 | 3.37 | 98.02 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 15 | 1→15 | 2.51 | 102.31 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 |
| 15 | 2→16 | 2.93 | 100.41 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 15 | 3→17 | 3.11 | 98.69 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 15 | 4→18 | 3.23 | 98.94 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 14 | 1→14 | 2.62 | 105.38 | Out | 0 | 40 | In | 1 | 100 | In | 1 | 100 | In | 1 | 100 |
| 14 | 2→15 | 2.70 | 100.60 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 14 | 3→16 | 3.18 | 96.66 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 14 | 4→17 | 2.91 | 100.14 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 14 | 5→18 | 3.32 | 95.99 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 13 | 1→13 | 2.31 | 107.10 | Out | 0 | 16 | Out | 0 | 66 | In | 1 | 100 | In | 1 | 100 |
| 13 | 2→14 | 2.85 | 103.59 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 13 | 3→15 | 2.80 | 94.40 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 13 | 4→16 | 2.95 | 97.49 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 13 | 5→17 | 2.97 | 96.43 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 13 | 6→18 | 3.23 | 95.80 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 12 | 1→12 | 2.30 | 109.32 | Out | 0 | 14 | Out | 0 | 57 | Out | 0 | 86 | In | 1 | 100 |
| 12 | 2→13 | 2.54 | 104.32 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 12 | 3→14 | 2.97 | 98.12 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 12 | 4→15 | 2.52 | 94.42 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 12 | 5→16 | 3.02 | 92.65 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 12 | 6→17 | 2.91 | 96.29 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 12 | 7→18 | 3.23 | 96.43 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 11 | 1→11 | 2.26 | 109.41 | Out | 0 | 25 | Out | 0 | 50 | Out | 0 | 75 | In | 1 | 87 |
| 11 | 2→12 | 2.53 | 106.47 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 11 | 3→13 | 2.59 | 96.69 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 11 | 4→14 | 2.66 | 100.66 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 11 | 5→15 | 2.59 | 88.16 | Out | 0 | Out | 0 | Out | 0 | Out | 0 | ||||
| 11 | 6→16 | 3.01 | 92.61 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 11 | 7→17 | 2.90 | 97.37 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 11 | 8→18 | 2.87 | 95.09 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 10 | 1→10 | 2.09 | 112.6 | Out | 0 | 33 | Out | 0 | 44 | Out | 0 | 77 | Out | 0 | 77 |
| 10 | 2→11 | 2.52 | 106.48 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 10 | 3→12 | 2.59 | 101.2 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 10 | 4→13 | 2.16 | 100.56 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 10 | 5→14 | 2.80 | 92.71 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 10 | 6→15 | 2.44 | 87.23 | Out | 0 | Out | 0 | Out | 0 | Out | 0 | ||||
| 10 | 7→16 | 3.01 | 93.74 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 10 | 8→17 | 2.42 | 95.86 | Out | 0 | In | 1 | In | 1 | In | 1 | ||||
| 10 | 9→18 | 3.35 | 101.19 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 9 | 1→9 | 1.79 | 112.61 | Out | 0 | 30 | Out | 0 | 30 | Out | 0 | 40 | Out | 0 | 70 |
| 9 | 2→10 | 2.37 | 109.8 | Out | 0 | Out | 0 | Out | 0 | In | 1 | ||||
| 9 | 3→11 | 2.39 | 100.72 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 9 | 4→12 | 2.16 | 107.3 | Out | 0 | Out | 0 | In | 1 | In | 1 | ||||
| 9 | 5→13 | 2.22 | 82.29 | Out | 0 | Out | 0 | Out | 0 | Out | 0 | ||||
| 9 | 6→14 | 2.71 | 92.2 | Out | 0 | Out | 0 | Out | 0 | In | 1 | ||||
| 9 | 7→15 | 2.44 | 88.03 | Out | 0 | Out | 0 | Out | 0 | Out | 0 | ||||
| 9 | 8→16 | 2.46 | 90.79 | Out | 0 | Out | 0 | Out | 0 | In | 1 | ||||
| 9 | 9→17 | 2.86 | 102.83 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
| 9 | 10→18 | 2.80 | 98.15 | In | 1 | In | 1 | In | 1 | In | 1 | ||||
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Otero-Cruz JD, Báez-Pagán CA, Caraballo-González IM, Lasalde-Dominicci JA. Tryptophan-scanning mutagenesis in the αM3 transmembrane domain of the muscle-type acetylcholine receptor. A spring model revealed. J Biol Chem. 2007;282:9162–9171. doi: 10.1074/jbc.M607492200. [DOI] [PubMed] [Google Scholar]
- 2.Tamamizu S, Guzmán GR, Santiago J, Rojas LV, McNamee MG, Lasalde-Dominicci JA. Functional effects of periodic tryptophan substitutions in the αM4 transmembrane domain of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry. 2000;39:4666–4673. doi: 10.1021/bi992835w. [DOI] [PubMed] [Google Scholar]
- 3.Guzmán GR, Santiago J, Ricardo A, Martí-Arbona R, Rojas LV, Lasalde-Dominicci JA. Tryptophan scanning mutagenesis in the αM3 transmembrane domain of the Torpedo californica acetylcholine receptor: Functional and structural implications. Biochemistry. 2003;42:12243–12250. doi: 10.1021/bi034764d. [DOI] [PubMed] [Google Scholar]
- 4.Santiago J, Guzmán GR, Torruellas K, Rojas LV, Lasalde-Dominicci JA. Tryptophan scanning mutagenesis in the TM3 domain of the Torpedo californica acetylcholine receptor beta subunit reveals an α-helical structure. Biochemistry. 2004;43:10064–10070. doi: 10.1021/bi0362368. [DOI] [PubMed] [Google Scholar]
- 5.Ortiz-Acevedo A, Meléndez M, Asseo AM, Biaggi N, Rojas LV, Lasalde-Dominicci JA. Tryptophan scanning mutagenesis of the αM4 transmembrane domain of the acetylcholine receptor from Torpedo californica. J Biol Chem. 2004;279:42250–42257. doi: 10.1074/jbc.M405132200. [DOI] [PubMed] [Google Scholar]
- 6.Choe S, Stevens CF, Sullivan JM. Three distinct structural environments of a transmembrane domain in the inwardly rectifying potassium channel ROMK1 defined by perturbation. Proc Natl Acad Sci U S A. 1995;92:12046–12049. doi: 10.1073/pnas.92.26.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins A, Chuang H, Jan YN, Jan LY. Scanning mutagenesis of the putative transmembrane segments of Kir2.1, an inward rectifier potassium channel. Proc Natl Acad Sci U S A. 1997;94:5456–5460. doi: 10.1073/pnas.94.10.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cukras CA, Jeliazkova I, Nichols CG. Structural and functional determinants of conserved lipid interaction domains of inward rectifying Kir6.2 channels. J Gen Physiol. 2002;119:581–591. doi: 10.1085/jgp.20028562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monks SA, Needleman DJ, Miller C. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J Gen Physiol. 1999;113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong KH, Miller C. The lipid-protein interface of a Shaker K(+) channel. J Gen Physiol. 2000;115:51–58. doi: 10.1085/jgp.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li-Smerin Y, Hackos DH, Swartz KJ. A localized interaction surface for voltage-sensing domains on the pore domain of a K+ channel. Neuron. 2000;25:411–423. doi: 10.1016/s0896-6273(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 12.Li-Smerin Y, Hackos DH, Swartz KJ. alpha-helical structural elements within the voltage-sensing domains of a K(+) channel. J Gen Physiol. 2000;115:33–50. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li-Smerin Y, Swartz KJ. Helical structure of the COOH terminus of S3 and its contribution to the gating modifier toxin receptor in voltage-gated ion channels. J Gen Physiol. 2001;117:205–218. doi: 10.1085/jgp.117.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hackos DH, Chang TH, Swartz KJ. Scanning the intracellular S6 activation gate in the shaker K+ channel. J Gen Physiol. 2002;119:521–532. doi: 10.1085/jgp.20028569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry SN, Kutluay E, Drews G, Hart SJ, Heginbotham L. Opening the KcsA K+ channel: tryptophan scanning and complementation analysis lead to mutants with altered gating. Biochemistry. 2002;41:13653–13662. doi: 10.1021/bi026393r. [DOI] [PubMed] [Google Scholar]
- 16.Panchenko VA, Glasser CR, Mayer ML. Structural similarities between glutamate receptor channels and K(+) channels examined by scanning mutagenesis. J Gen Physiol. 2001;117:345–360. doi: 10.1085/jgp.117.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueno S, Lin A, Nikolaeva N, Trudell JR, Mihic SJ, Harris RA, Harrison NL. Tryptophan scanning mutagenesis in TM2 of the GABA(A) receptor alpha subunit: effects on channel gating and regulation by ethanol. Br J Pharmacol. 2000;131:296–302. doi: 10.1038/sj.bjp.0703504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins A, Andreasen A, Trudell JR, Harrison NL. Tryptophan scanning mutagenesis in TM4 of the GABA(A) receptor alpha1 subunit: implications for modulation by inhaled anesthetics and ion channel structure. Neuropharmacology. 2002;43:669–678. doi: 10.1016/s0028-3908(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 19.Wang SY, Bonner K, Russell C, Wang GK. Tryptophan scanning of D1S6 and D4S6 C-termini in voltage-gated sodium channels. Biophys J. 2003;85:911–920. doi: 10.1016/S0006-3495(03)74530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honse Y, Ren H, Lipsky RH, Peoples RW. Sites in the fourth membrane-associated domain regulate alcohol sensitivity of the NMDA receptor. Neuropharmacology. 2004;46:647–654. doi: 10.1016/j.neuropharm.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Silberberg SD, Chang TH, Swartz KJ. Secondary structure and gating rearrangements of transmembrane segments in rat P2X4 receptor channels. J Gen Physiol. 2005;125:347–359. doi: 10.1085/jgp.200409221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powl AM, Wright JN, East JM, Lee AG. Identification of the hydrophobic thickness of a membrane protein using fluorescence spectroscopy: studies with the mechanosensitive channel MscL. Biochemistry. 2005;44:5713–5721. doi: 10.1021/bi047338g. [DOI] [PubMed] [Google Scholar]
- 23.Powl AM, East JM, Lee AG. Heterogeneity in the binding of lipid molecules to the surface of a membrane protein: Hot spots for anionic lipids on the mechanosensitive channel of large conductance MscL and effects on conformation. Biochemistry. 2005;44:5873–5883. doi: 10.1021/bi047439e. [DOI] [PubMed] [Google Scholar]
- 24.Subbiah RN, Kondo M, Campbell TJ, Vandenberg JI. Tryptophan scanning mutagenesis of the HERG K+ channel: the S4 domain is loosely packed and likely to be lipid exposed. J Physiol. 2005;569:367–379. doi: 10.1113/jphysiol.2005.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashlan OB, Maarouf AB, Kussius C, Denshaw RM, Blumenthal KM, Kleyman TR. Distinct structural elements in the first membrane-spanning segment of the epithelial sodium channel. J Biol Chem. 2006;281:30455–30462. doi: 10.1074/jbc.M604615200. [DOI] [PubMed] [Google Scholar]
- 26.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Unwin N. Nicotinic acetylcholine receptor at 9 A resolution. J Mol Biol. 1993;229:1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- 28.Blanton MP, Cohen JB. Mapping the lipid-exposed regions in the Torpedo californica nicotinic acetylcholine receptor. Biochemistry. 1992;31:3738–3750. doi: 10.1021/bi00130a003. [DOI] [PubMed] [Google Scholar]
- 29.Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: Secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- 30.Blanton MP, Dangott LJ, Raja SK, Lala AK, Cohen JB. Probing the structure of the nicotinic acetylcholine receptor ion channel with the uncharged photoactivable compound [[r]3[/r]H]diazofluorene. J Biol Chem. 1998;273:8659–8668. doi: 10.1074/jbc.273.15.8659. [DOI] [PubMed] [Google Scholar]
- 31.Motulsky HJ, Christopoulos A. Fitting models to biological data using linear and nonlinear regression: a practical guide to curve fitting. Oxford University Press; Oxford ; New York: 2004. [Google Scholar]
- 32.Cornette JL, Cease KB, Margalit H, Spouge JL, Berzofsky JA, DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987;195:659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- 33.Komiya H, Yeates TO, Rees DC, Allen JP, Feher G. Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: Symmetry relations and sequence comparisons between different species. Proc Natl Acad Sci U S A. 1988;85:9012–9016. doi: 10.1073/pnas.85.23.9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li-Smerin Y, Hackos DH, Swartz KJ. α-Helical structural elements within the voltage-sensing domains of a K+ channel. J Gen Physiol. 2000;115:33–50. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rees DC, DeAntonio L, Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989;245:510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 36.Rees DC, Komiya H, Yeates TO, Allen JP, Feher G. The bacterial photosynthetic reaction center as a model for membrane proteins. Annu Rev Biochem. 1989;58:607–633. doi: 10.1146/annurev.bi.58.070189.003135. [DOI] [PubMed] [Google Scholar]