ABSTRACT
It is widely accepted that bacterial endophytes actively colonize plants, interact with their host, and frequently show beneficial effects on plant growth and health. However, the mechanisms of plant-endophyte communication and bacterial adaption to the plant environment are still poorly understood. Here, whole-transcriptome sequencing of B. phytofirmans PsJN colonizing potato (Solanum tuberosum L.) plants was used to analyze in planta gene activity and the response of strain PsJN to plant stress. The transcriptome of PsJN colonizing in vitro potato plants showed a broad array of functionalities encoded in the genome of strain PsJN. Transcripts upregulated in response to plant drought stress were mainly involved in transcriptional regulation, cellular homeostasis, and the detoxification of reactive oxygen species, indicating an oxidative stress response in PsJN. Genes with modulated expression included genes for extracytoplasmatic function (ECF) group IV sigma factors. These cell surface signaling elements allow bacteria to sense changing environmental conditions and to adjust their metabolism accordingly. TaqMan quantitative PCR (TaqMan-qPCR) was performed to identify ECF sigma factors in PsJN that were activated in response to plant stress. Six ECF sigma factor genes were expressed in PsJN colonizing potato plants. The expression of one ECF sigma factor was upregulated whereas that of another one was downregulated in a plant genotype-specific manner when the plants were stressed. Collectively, our study results indicate that endophytic B. phytofirmans PsJN cells are active inside plants. Moreover, the activity of strain PsJN is affected by plant drought stress; it senses plant stress signals and adjusts its gene expression accordingly.
IMPORTANCE
In recent years, plant growth-promoting endophytes have received steadily growing interest as an inexpensive alternative to resource-consuming agrochemicals in sustainable agriculture. Even though promising effects are recurrently observed under controlled conditions, these are rarely reproducible in the field or show undesirably strong variations. Obviously, a better understanding of endophyte activities in plants and the influence of plant physiology on these activities is needed to develop more-successful application strategies. So far, research has focused mainly on analyzing the plant response to bacterial inoculants. This prompted us to study the gene expression of the endophyte Burkholderia phytofirmans PsJN in potato plants. We found that endophytic PsJN cells express a wide array of genes and pathways, pointing to high metabolic activity inside plants. Moreover, the strain senses changes in the plant physiology due to plant stress and adjusts its gene expression pattern to cope with and adapt to the altered conditions.
INTRODUCTION
Burkholderia phytofirmans PsJN (1) is a naturally occurring endophyte isolated from onion roots (2) that is able to establish nonpathogenic relationships with a wide range of plant species, both monocotyledons and dicotyledons (3, 4). Numerous studies have reported beneficial effects of strain PsJN on host plants such as increased plant growth (for a review, see reference 5) and enhanced biotic and abiotic stress tolerance (4, 5).
There are increasing efforts to understand the plant physiological and genetic response to inoculation with B. phytofirmans PsJN. Bordiec and colleagues (6) compared local defense reactions in grapevine cell cultures inoculated with either strain PsJN or the nonhost pathogen Pseudomonas syringae pv. pisi. The authors found that strain PsJN does not induce the defense events commonly found after pathogen attack in plants. Infection with the pathogen led to a two-phase oxidative burst and a hypersensitive response (HR)-like reaction, whereas no oxidative burst or cell death was observed in cells treated with strain PsJN (6). Theocharis et al. (7) demonstrated that inoculation of Chardonnay grapevine plantlets with B. phytofirmans PsJN speeds up the plant response to chilling and plant adaption to cold temperatures. Numbers of cold stress-related gene transcripts and metabolites increased earlier and faster and reached higher levels in bacterized plantlets than in control plants. Fernandez et al. (8) demonstrated that the higher tolerance of chilling of PsJN-colonized grapevine plantlets could be related to changes in plant photosynthesis and sugar metabolism. More recently, Poupin and colleagues (9) studied the response of Arabidopsis thaliana to inoculation with B. phytofirmans PsJN. The bacterium affected the whole life cycle of Arabidopsis plants, increasing plant growth, especially at the early stage of ontogeny, and speeding up maturity. These physiological changes correlated with altered expression of plant growth regulator genes; i.e., genes involved in auxin and gibberellin pathways were induced in PsJN-inoculated plants, and flowering-control genes were induced earlier in PsJN-inoculated plants than in control plants.
Whereas the plant response to beneficial bacteria has been described in several studies, very little is known about bacterial physiology, response, and adaptation processes in planta. Efforts to characterize the effects of the plant environment on endophytic bacteria have been rare (10–12), and information on in planta gene expression of B. phytofirmans PsJN is missing. For example, how does B. phytofirmans PsJN recognize the plant environment? Does the bacterium respond to changing physiological conditions, e.g., due to plant stress, in plants?
Therefore, the aim of this study was to investigate gene expression patterns of B. phytofirmans PsJN cells colonizing potato (Solanum tuberosum L.) plants and, furthermore, to assess the effect of plant drought stress on the transcriptome of strain PsJN. In vitro-grown potato plants of two varieties (Russet Burbank and Bionta) were inoculated with B. phytofirmans PsJN, and drought stress was induced by application of polyethylene glycol (PEG). Bacterial transcriptomes of cells colonizing potato plants (cv. Bionta) were analyzed under nonstressed conditions (control) and at three different time points after drought stress induction by direct short-read deep sequencing (Illumina RNA-seq). Differentially expressed genes included genes for extracytoplasmatic function (ECF) group IV sigma factors. TaqMan quantitative PCR (TaqMan-qPCR) assays were performed to quantitatively assess ECF sigma factor activation in B. phytofirmans PsJN colonizing potato plants of two varieties (Russet Burbank and Bionta) in response to plant stress.
RESULTS
Detection of B. phytofirmans PsJN in plants.
Six weeks after inoculation with B. phytofirmans PsJN, potato plants of two varieties (Bionta and Russet Burbank) showed increased shoot and root length in response to colonization by PsJN (see Fig. S1 in the supplemental material). Application of PEG caused visible symptoms of water deficiency in potato plants (see Fig. S2). PCR amplification with primers targeting bacterial 16S rRNA genes and plant 18S rRNA genes resulted in two bands in all inoculated plants, the mitochondrial band and a band of about 720 bp representing the bacterial 16S rRNA gene. No amplification of the bacterial fragment was found in control plants (see Fig. S3). Isolation and sequencing of the bacterial bands confirmed the presence of B. phytofirmans PsJN in all inoculated potato plants.
Transcriptome sequencing.
Sequencing of cDNA samples yielded 35.7 to 42.2 million reads of cDNA, corresponding to over 2 billion nucleotides of cDNA per sample (Table 1). Around 10% of the reads were removed by initial quality filters to trim poor-quality data. After poly(A) tail removal and length trimming, more than 37% of the reads were removed, mainly because the bacterial mRNA molecules were poly(A) tailed during cDNA library preparation. Around 61% to 63% of total reads were considered for further analysis. By removal of rRNA sequences, the data set was further reduced by 5%. Of the remaining sequences, 0.3% to 1.95% of the reads mapped to the genome of PsJN (Table 1). Normalization of transcript levels in control and stressed plants was done by RPKM (reads per kilobase per million mapped) normalization using NOISeq. For verification of this procedure, we used the expression data of selected housekeeping genes which were shown by qPCR to be stably expressed (see Fig. S4 in the supplemental material).
TABLE 1 .
Statistics of cDNA sequencing reads and their alignments on the B. phytofirmans PsJN genome
| Sample | Total no. of nucleotides | Total no. of RNA-seq reads | Total no. of trimmed readsa | Total no. of rRNA reads | Total no. of readsb | Total no. (%) of mapped Ps readsc |
|---|---|---|---|---|---|---|
| Control | 1,865,634,927 | 36,581,077 | 20,759,363 | 1,141,647 | 19,617,716 | 93,404 (0.48) |
| Stress 1 | 2,156,419,995 | 42,282,745 | 24,171,466 | 1,358,716 | 22,812,750 | 382,477 (1.70) |
| Stress 6 | 1,825,100,280 | 35,786,280 | 20,023,318 | 1,036,065 | 18,987,253 | 56,604 (0.30) |
| Stress 12 | 1,926,749,451 | 37,779,401 | 20,048,099 | 1,266,245 | 18,781,854 | 64,047 (0.34) |
Data indicate the total numbers of reads after quality assessment, trimming, and poly(A) removal.
Data indicate the total numbers of reads after removal of the rRNA sequences which proceed to the mapping step.
Data indicate the total numbers (percentages) of reads mapped on the B. phytofirmans PsJN (Ps) genome.
Transcriptomic profile of B. phytofirmans PsJN in potato plants.
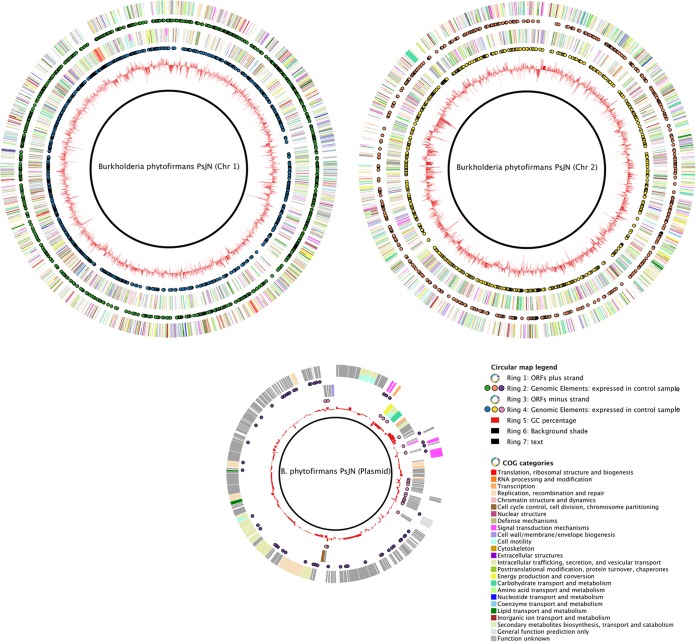
A total number of 4,591 different transcripts of B. phytofirmans PsJN were detected in PsJN-colonized potato plants. The expressed genes were evenly distributed on both chromosomes, and 76 of 167 genes carried on the plasmid were active in this experiment (Fig. 1). Among the latter, we found genes for type II B. phytofirmans 7353 (Bphyt_7353) and type IV (Bphyt_7341, Bphyt_7342, Bphyt_7347, Bphyt_7350, and Bphyt_7351) secretion system proteins.
FIG 1 .
Circular maps representing the two chromosomes and the plasmid of Burkholderia phytofirmans PsJN. The following rings are included in each map: open reading frames (ORFs) on the plus (rings 1) and minus (rings 3) strands of the genome of strain PsJN (color by COG categories). Transcripts expressed on the plus (rings 2) and minus (rings 4) strands of the genome of strain PsJN during colonization of unstressed in vitro potato plants are indicated. The images were generated with a microbial genome context viewer (MgcV; http://mgcv.cmbi.ru.nl/) (53).
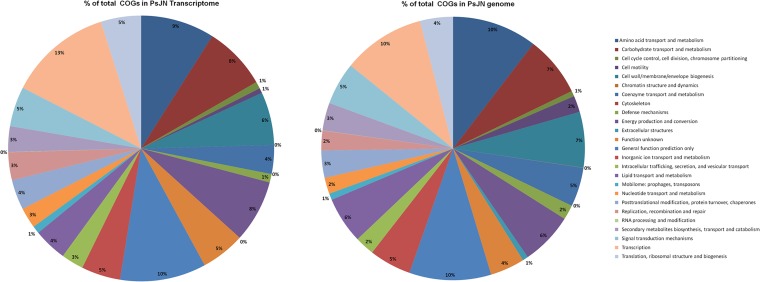
Figure 2 shows a comparison of the genome of B. phytofirmans PsJN and the transcriptome of PsJN colonizing in vitro potato plants on the basis of the relative distributions of clusters of orthologous group (COG) categories. Overall, the functional categories of the in planta transcriptome and the genome of strain PsJN were highly similar. Few differences were found in the following COG categories: cell motility, defense mechanisms, and extracellular structures. The relative abundance of cell motility- and defense mechanism-related genes was reduced from 2% in the genome to 1% in the transcriptome. Genes encoding proteins in the COG category of “extracellular structures” covered 1% of the genes in the genome of PsJN but were not found in the transcriptome.
FIG 2 .
Relative distributions of functional COG categories in the in planta transcriptome and genome of B. phytofirmans PsJN.
Expressed genes with an RPKM value of ≥55 were grouped in 74 clusters representing 354 functional classes by gene ontology (GO) analysis (see Table S1 in the supplemental material). The majority of functions were related to transcription regulation, general metabolism (sugars, amino acids, lipids, and nucleotides), energy production, and cellular homeostasis. Furthermore, we found a high number of transcripts for signal transduction mechanisms such as two-component systems and extracytoplasmatic (ECF) sigma factor genes (Bphyt_1327, Bphyt_1666, Bphyt_1784, Bphyt_2906, Bphyt_3189, Bphyt_4397, Bphyt_4574, Bphyt_4980, Bphyt_5021, Bphyt_5131, and Bphyt_5142).
GO functions that were found in the PsJN transcriptome included functions generally considered important for endophytic plant colonization and plant growth promotion such as cell motility and chemotaxis, cellular iron homeostasis, and photosynthesis (see Table S1 in the supplemental material). Cellular iron homeostasis was represented mainly by ferritin (Bphyt_0714, Bphyt_2657, and Bphyt_5727) and bacterioferritin (Bphyt_1412 and Bphyt_2740) genes. By analyzing genes represented by the GO function “photosynthesis,” we found NADH dehydrogenase subunit A, B, C, and D (Bphyt_1343 to Bphyt_1346) and polyprenyl synthetase (Bphyt_3450) genes, which do not clearly indicate putative photosynthetic activity. Furthermore, we found expression of an N-acyl homoserine lactone (AHL) synthase gene (bpI.2; Bpyht_4275), a quinolinate phosphoribosyl transferase gene (Bphyt_3152), indole-3-acetic acid (IAA) synthesis genes such as those encoding nitrile hydrolase (Bphyt_7182 and Bphyt_7181), and a gene encoding an IAM hydrolase of the indole-3-acetamide (IAM) pathway as well as aldehyde dehydrogenase (Bphyt_5803) of the tryptophan side-chain oxidase pathway bypassing IPyA. Transcripts of IAA degradation (aromatic ring hydroxylating dioxygenase; Bphyt_2156) and 1-aminocyclopropane-1-carboxylate (ACC) deaminase (Bphyt_5397) genes were not detected.
Transcriptional response of B. phytofirmans PsJN to plant stress. (i) One hour after stress induction.
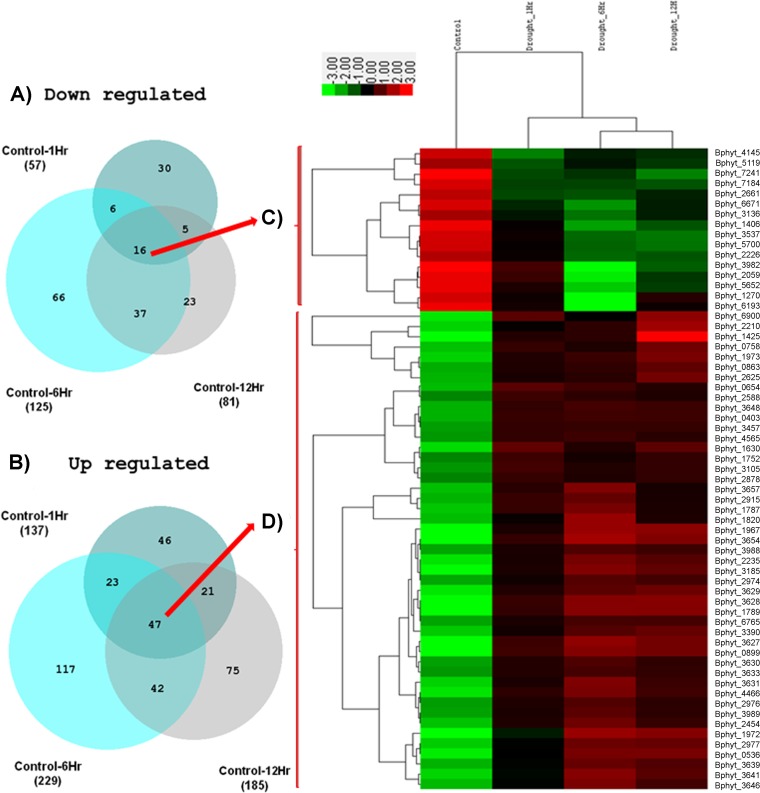
Analysis of differentially expressed genes in B. phytofirmans PsJN colonizing nonstressed and drought-stressed potato plants identified 194 genes with modified expression 1 h after stress induction (see Table S2 in the supplemental material). Among these, 137 genes were upregulated and 57 genes were downregulated (Fig. 3).
FIG 3 .
(Left) Venn diagrams illustrating numbers of downregulated (A) and upregulated (B) differentially expressed (DE) genes either shared or unique at all time points after drought stress induction from pairwise comparisons of control samples at each of the drought stress time points (control at 1 h [1Hr], control at 6 h [6Hr], and control at 12 h [12Hr]). The total numbers of up- or downregulated DE genes in each time point are indicated in parentheses. (Right) Hierarchical clustering heat map of expression changes for downregulated (C) and upregulated (D) differentially expressed genes that are common across all time points (16 genes in downregulated genes and 47 genes in upregulated genes). The heat map was constructed based on RPKM expression values. Rows correspond to differentially expressed genes, and columns represent control and drought-stressed samples at the indicated time points (in hours) of drought stress. Green and red boxes represent differentially expressed genes that decreased and increased their expression levels at the indicated time points of drought stress, respectively. The identifiers (ID) and descriptions of differentially expressed genes are listed at the right in the table.
For a better understanding of the genetic traits involved in the response of B. phytofirmans PsJN to drought stress of its host plant, the differentially expressed genes were affiliated to biological processes using gene ontology (GO) analysis (see Table S3 in the supplemental material). The differentially expressed genes represented eight different GO biological processes at 1 h after inducing drought stress. Genes that were upregulated in the transcriptome of B. phytofirmans PsJN belonged mostly to the following functional categories: cellular homeostasis, homeostatic process, and cell redox homeostasis. Among them were genes such as those encoding bacterioferritins, glutaredoxin, redoxin domain protein, thioredoxin, and RNA polymerase factor sigma 70 (ECF sigma factor; Bphyt_1327) (see Table S3). The downregulated genes represented regulation of transcription and DNA-dependent functions (see Table S3) such as those encoding various types of transcriptional regulators.
(ii) Six hours after stress induction.
A total number of 354 genes were differentially expressed compared to the control at this time point, with 229 genes being upregulated and 125 genes being downregulated (Fig. 3). The complete list of differentially expressed genes is available (see Table S2 in the supplemental material). GO analysis grouped these genes into 79 different biological processes (see Table S3). The induced functions with the highest enrichment score were oxidative phosphorylation, hydrogen transport, proton transport, ATP synthesis-coupled proton transport, energy-coupled proton transport, ion transmembrane transport, ATP biosynthetic and metabolic processes, purine nucleoside triphosphate biosynthetic processes, and ribonucleoside triphosphate biosynthetic and metabolic processes. The most enriched genes were those encoding ATP synthase C and gamma chain, ATP synthase subunit a/b/alpha/beta, sulfate adenylyltransferase, cytochrome o ubiquinol oxidase subunit III, succinate dehydrogenase, and acetolactate synthase (see Table S2).
(iii) Twelve hours after stress induction.
At this time point, potato plants treated with PEG showed severe wilting and 266 genes were differentially expressed in B. phytofirmans PsJN (Fig. 3). The complete list of differentially expressed genes is available (see Table S2 in the supplemental material). Twelve hours after stress induction, 185 genes were upregulated and 81 genes were downregulated. These genes were grouped by GO analysis into 52 different biological processes (see Table S3). The GO functions showing highest enrichment at this time point were those corresponding to positive regulation of the cellular biosynthetic process, transcription, the macromolecule biosynthetic process, gene expression, the nitrogen compound metabolic process, and the macromolecule metabolic process. Peptidylprolyl isomerase FK506-binding protein (FKBP), UTP-glucose-1-phosphate uridylyltransferase, and glucose-6-phosphate dehydrogenase genes were upregulated at this time point under conditions of drought stress. Genes encoding histone family protein, transcriptional regulator GntR, transposase mutator type, peroxidases, and catalase/peroxidase (HPI) were downregulated at this time point (see Table S3).
(iv) Transcriptional response of B. phytofirmans PsJN to plant stress at all three time points.
As shown by comparisons of the genes that are expressed in control plants and stressed plants, 47 genes were upregulated and 16 genes were downregulated in B. phytofirmans PsJN in response to plant stress at all three time points (Fig. 3). These genes were subjected to further hierarchical clustered analysis and were classified into four groups by Cluster 3.0 (13). The functions of these genes correspond to the functions with the highest enrichment score at all time points obtained using David (Database for Annotation, Visualization, and Integrated Discovery) software.
The first group consisted of two oxidoreductase activity-related genes (glutaredoxin and alkyl hydroperoxide reductase subunit) that are involved in cellular homeostasis and cell redox homeostasis. The second group represented genes involved in regulation of transcription activity and consisted of, among others, those encoding a GntR family transcriptional regulator and a cold-shock DNA-binding protein. The third group consisted of UTP-glucose-1-phosphate uridylyltransferase and glyceraldehyde-3-phosphate dehydrogenase, which showed similar (upregulated) expression patterns at all time points. The proteins encoded by those genes represent glucose metabolic processes. In the fourth group, we found an elongation factor, Tu, and ribosomal protein S7 (Fig. 3C and D).
The roles of differentially expressed genes in cellular metabolic pathways were analyzed using the KEGG database. Seven pathways were found to be differentially expressed under conditions of plant stress. Genes corresponding to four pathways, namely, those corresponding to oxidative phosphorylation, sulfur metabolism, pentose and glucuronate interconversions, and aminosugar and nucleotide sugar metabolism, were upregulated. The genes corresponding to the KEGG pathway for glutathione metabolism, a two-component system, and the pentose phosphate pathway were downregulated.
Furthermore, KEGG analysis revealed that the oxidative phosphorylation pathway was the only metabolic pathway that was activated under conditions of drought stress at all three different time points (see Fig. S5 in the supplemental material). This pathway was activated at 1 h after drought stress induction with upregulation of ATP synthase subunit delta. After the organism had been maintained under conditions of drought stress for 6 h, the number of expressed genes reached 16 and included genes for cytochrome o ubiquinol oxidase subunit III, succinate dehydrogenase hydrophobic membrane anchor protein (SdhC), NADH-quinone oxidoreductase, and several ATP synthase subunits and NADH dehydrogenase subunits. This pathway was still active after 12 h, and the genes expressed included those encoding protoheme IX farnesyltransferase, succinate dehydrogenase, cytochrome b556 subunit (SdhD), NADH-quinone oxidoreductase, and several ATP synthase subunits and NADH dehydrogenase subunits.
Expression of ECF sigma factor genes in B. phytofirmans PsJN colonizing potato plants.
Expression of extracytoplasmatic function (ECF) sigma factor genes in B. phytofirmans PsJN colonizing potato plants (cv. Bionat and Russet Burbank) with and without drought stress was tested by real-time qPCR with cDNA of four biological replicates per treatment. Transcripts of six ECF sigma factor genes (ECF_164, ECF_886, ECF_429, ECF_718, and ECF_474) were detected in PsJN-colonized plants (see Fig. S6 in the supplemental material). The number of expressed ECF sigma factor genes in stressed plants was different from the number in control plants, and the numbers varied over time in stressed plants. ECF_164, ECF_429, and ECF_886 were expressed in control plants of both cultivars and were also active in stressed plants at all time points. The number of expressed ECF sigma factor genes increased under conditions of drought stress to a maximum of five genes after 1 h in Bionta and after 6 h in Russet Burbank (see Fig. S6). Transcripts of ECF_718 were detected in both cultivars, whereas ECF_474 was found in Russet Burbank and ECF_230 in Bionta only.
Differential expression of ECF sigma factor genes in B. phytofirmans PsJN colonizing potato plants.
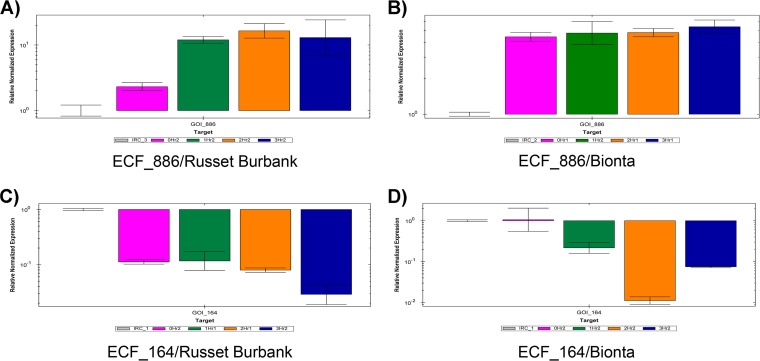
The expression levels of ECF sigma factor genes were normalized to those of the most stably expressed control gene, Bphyt_2615 (glutamine synthetase). Relative expression levels of ECF sigma factor genes under conditions of drought stress are shown in Fig. 4. ECF_886 was significantly upregulated in B. phytofirmans PsJN colonizing cv. Russet Burbank potato plants under conditions of drought stress. The expression of ECF_886 reached a maximum (3×) after 6 h of drought stress. In cultivar Bionta, ECF_886 expression did not change significantly after stress induction compared to control results. Expression of ECF_164 was relatively constant in Russet Burbank but was clearly downregulated in Bionta under conditions of plant drought stress. The transcript levels of the other expressed ECF sigma factor genes (ECF_429, ECF_718, ECF_230, and ECF_474) remained relatively constant throughout the different time points of drought stress in both cultivars.
FIG 4 .
Relative expression levels of ECF sigma factors with modulated expression in B. phytofirmans PsJN in response to plant drought stress. (A) ECF_886 (Bphyt_1327) in Russet Burbank. (B) ECF_886 (Bphyt_1327) in Bionta. (C) ECF_164 (Bphyt_4063) in Russet Burbank. (D) ECF_164 (Bphyt_4063) in Bionta.
DISCUSSION
The genome of B. phytofirmans PsJN encodes 7,405 genes, 4,591 (62%) of which were expressed in PsJN colonizing cv. Bionta potato plants in vitro. The active genes were evenly distributed across both chromosomes and the plasmid. Interestingly, about 46% of the coding sequences (CDS) located on the plasmid were expressed by PsJN inside potato plants. The genome of B. phytofirmans PsJN shows a high degree of similarity to that of B. xenovorans LB400, but the pBPHYT01 plasmid is different from the megaplasmid of B. xenovorans. In B. xenovorans LB400, the small chromosome is the “lifestyle-determining” replicon, whereas strain-specific functions are encoded on the megaplasmid (14). In B. phytofirmans PsJN, only 29% of the CDS on pBPHYT01 could be functionally described (15); consequently, the majority of plasmid-carried genes that are active in PsJN colonizing cv. Bionta potato plants are of unknown function. Several reports of studies have proposed that plasmids are genetic determinants of functional diversification and niche adaptation (16, 17). We can only speculate on the role of pBPHYT01 in the endophytic lifestyle of B. phytofirmans PsJN. Experiments designed to test plasmid-free PsJN for the ability to colonize plants and to establish endophytic population could give further insights.
The transcriptome profile of B. phytofirmans PsJN colonizing potato plants indicates that the bacterium is metabolically active in plants. The majority of expressed traits were related to transcription regulation, general metabolism (sugars, amino acids, lipids, and nucleotides), energy production, and cellular homeostasis. The overall patterns of functions encoded on the genome of B. phytofirmans PsJN and expressed in in vitro potato plants were highly similar. We conclude from this that the plant interior as a habitat for bacteria does not require very selected and specialized functionalities. Only a few differences in the COG patterns were found, with cell motility and defense mechanisms being less represented in the transcriptome than in the genome of B. phytofirmans PsJN. Cell motility might be important in plant invasion and in the spreading of endophytic microorganisms throughout plant organs and tissues (18). Recently, Balsanelli and colleagues showed that motility-related functions such as chemotaxis and type VI pilus functions play an important role in the initial contact with plants and the epiphytic colonization of maize roots by Herbaspirillum seropedicae (19). Our data indicate that active movement is less important once a bacterial population is successfully established inside plants. Interestingly, defense-related traits seem to play a minor role in the endophytic life of B. phytofirmans PsJN also. A possible explanation is that the plant endosphere is a protected niche allowing endophytes to escape the high competitive pressure in the rhizosphere and soil.
B. phytofirmans PsJN stimulates plant growth in many of its host plants. Metabolic properties suggested to be involved in this activity include the production and degradation of auxin phytohormone indole-3-acetic acid (IAA) (20), ACC deaminase activity (21), quinolinate phosphoribosyl transferase (QPRTase) or nicotinate-nucleotide pyrophosphorylase activity (22), and siderophore production (1). In our experiment, in vitro potato plants colonized by B. phytofirmans PsJN showed increased growth compared to an untreated control. By showing the expression of quinolinate phosphoribosyl transferase and indole-3-acetic acid (IAA) synthesis genes of strain PsJN colonizing potato plantlets, the results of the present study support previous reports of the importance of these functions for the beneficial interaction between PsJN and plants.
Plants colonized by B. phytofirmans PsJN often show increased tolerance of abiotic stress such as chilling (23) and drought (4). One of the main ambitions of this study was to elucidate whether and how endophytic B. phytofirmans PsJN is affected by host plant drought stress. Analysis of differentially expressed genes in B. phytofirmans PsJN colonizing nonstressed and drought-stressed potato plants showed that 137, 229, and 185 genes were upregulated and 57, 125, and 81 genes were downregulated in response to host plant drought stress at 1, 6, and 12 h after PEG application. This indicates that B. phytofirmans PsJN adjusts gene expression to physiological conditions that have been altered in host plants due to plant stress responses. Genes that were significantly upregulated in B. phytofirmans PsJN in response to host plant stress are mostly involved in transcription regulation, cellular homeostasis, and cell redox homeostasis, indicating a stress response in B. phytofirmans PsJN. Drought stress provokes an oxidative burst in plants as a primary immune response. This increase in the production of reactive oxygen species (ROS) serves on the one hand as an alarm signal that triggers acclimation and defense reactions and is kept in tight balance by the antioxidant system in plants (24). If, on the other hand, the drought stress continues for a certain period of time, the oxidative burst may lead to extensive cellular damage and finally to cell death (24). Upregulation of genes involved in detoxification of ROS in strain PsJN colonizing potato plants suffering from drought stress led us to the assumption that endophytic B. phytofirmans PsJN senses and is affected by plant-produced ROS. We propose the following scenario. Water limitation leads to ROS burst in plants. Endophytic PsJN cells respond with the expression of genes involved in the defense against oxidative stress in order to prevent cell damage. ROS scavenging by endophytes is also very important during the early stage of plant colonization, as previously shown for Gluconacetobacter diazotrophicus PAL5 (11). Whether bacterial antioxidant activity may help to maintain the redox balance in plants and thus reduce the effects of drought stress on plants remains elusive and merits further investigation.
Oxidative phosphorylation was found to be activated in B. phytofirmans PsJN over time during plant drought stress. In the process of cellular respiration, aerobic bacteria pass electrons from oxidizable substances to molecular oxygen via the so-called electron transport chain. The released energy is used to produce energy-rich ATP from ADP by phosphorylation. Oxidative phosphorylation generates the energy needed for almost all vital processes (25). Apart from this, pentose and glucuronate interconversions and amino sugar and nucleotide sugar metabolism were also activated. Upregulation of genes involved in these processes in B. phytofirmans PsJN colonizing drought-stressed potato plants could indicate activation of bacterial growth. At least for Epichloe endophytes, it is well documented that the mutualistic interaction of fungi and host plant is tightly regulated. Perturbations of this balance result in a switch from restricted to proliferative growth of the endophyte inside the plant and a breakdown from mutualistic to pathogenic behavior (26). Furthermore, anarchic proliferation of otherwise asymptomatic bacterial endophytes is a common phenomenon in in vitro plant propagation when cultures are under stress (27). We therefore quantified PsJN in the control and stressed potato plants by performing qPCR with the selected housekeeping genes used for data normalization but did not find a significant increase in copy numbers over time under conditions of drought stress (data not shown). It is possible that the time span (12 h) was too short to observe a significant increase in cell numbers, but it is also likely that the increase in metabolic activity in B. phytofirmans PsJN under conditions of host plant drought stress was not coupled with proliferated growth.
One way that bacteria sense and react to the extracellular environment is by the so-called cell surface signaling-employing extracytoplasmatic function (ECF) sigma factors (28). This signal transduction system consists of an outer membrane receptor, an inner membrane-bound sigma factor regulator (anti-sigma factor), and, bound to that, an ECF sigma factor. In the absence of a signal, the anti-sigma factor tightly binds the ECF sigma factor, thereby keeping it in its inactive state. The anti-sigma factor is proteolytically degraded in the presence of a stimulus. As a result, the sigma factor is released and activates expression of its target genes (28). The ECF subfamily is the largest group in the sigma 70 family, and its members are involved in a wide range of environmental responses, such as metal homeostasis, starvation, and resistance to antimicrobial peptides, and are also required for pathogenesis in some cases (28). ECF sigma factors may also play a role in the establishment of plant-microbe interactions. Gourion and colleagues (29) showed that an extracytoplasmatic sigma factor is involved in symbiotic efficiency in the plant symbiont Bradyrhizobium japonicum USDA110. The genome of B. phytofirmans PsJN harbors eighteen different CDS putatively coding for extracytoplasmatic function (ECF) sigma factors. Analysis of the in planta transcriptome of B. phytofirmans PsJN revealed the expression of eleven extracytoplasmatic sigma factor genes, and the expression of one of these genes was upregulated upon plant stress induction. TaqMan-quantitative PCR experiments were performed to quantitatively assess ECF sigma factor activation in B. phytofirmans PsJN in response to plant stress. Six ECF sigma factor genes were expressed in B. phytofirmans PsJN colonizing drought-stressed potato plants. One of these genes (Bphyt_1327; ECF_886) was significantly upregulated in response to plant stress. This gene has orthologs in 36 other sequenced Burkholderia strains (Burkholderia Genome Database [30]) with similar genetic neighborhoods. Little is known of the function of this ECF sigma factor in Burkholderiaceae; only the ortholog in Burkholderia cenocepacia, EcfD, was found to be involved in the response to chlorhexidine (31). The biological role of Bphyt_1327 in B. phytofirmans PsJN remains unclear and requires further investigation.
Another ECF sigma factor (Bphyt_4063; ECF_164) was downregulated in B. phytofirmans PsJN. It is orthologous to EcfI in Burkholderia cenocepacia and can be found in 31 other Burkholderia genomes in similar genetic neighborhoods. In B. cenocepacia, EcfI is involved in the synthesis of ornibactin siderophores and, thus, iron transport (32). Downregulation of this ECF sigma factor in B. phytofirmans PsJN colonizing drought-stressed potato plants could be directly linked to oxidative stress response. In bacteria, the regulation of iron homeostasis is coordinated with defense against oxidative stress (33). In many bacteria, Fur-like transcription regulators act as kind of master regulator in this regulatory network. Fur-like proteins control iron supply in dependence on the redox status of cells, either directly by repressing iron acquisition genes or indirectly by repression of other regulators such as ECF sigma factors (32, 33). Consequently, it seems likely that Bphyt_4063 in B. phytofirmans PsJN is involved in the regulation of iron uptake.
Interestingly, we found differences in the intensities of ECF sigma factor activation in PsJN in the two potato cultivars (Bionta and Russet Burbank). In agreement, it is well known that the plant growth-promoting effect of PsJN in potato is cultivar dependent (34, 35). In previous studies, the intensity of the effects correlated with the PsJN titer in the plants, which was found to be much higher in cultivars showing a greater increase in growth (35). We therefore quantified PsJN in the potato plants used in this study by performing qPCR with the selected housekeeping genes used for data normalization but did not find significant differences in copy numbers in cv. Bionta and Russet Burbank (data not shown). Moreover, the differences in response may have been due to differences in DNA methylation in plants, which was found to be enhanced in poorly responsive potato cultivars (34). Together with the previous observations, our findings indicate that the plant genotype-dependent plant growth-promoting effect of B. phytofirmans PsJN is accompanied by differences in the responsiveness of the strain to plant physiology.
Summarizing our data, one of the main outcomes of this study is that endophyte-plant interactions are not a one-way relationship in which the plant responds to the endophyte but represent a complex interplay in which each partner is affected by the other. This may hold true, and may become even more complicated under natural conditions, when plants are colonized by a rich microbial community consisting of bacteria, fungi, and viruses. The term “hologenome” has been introduced to describe the sum of the genetic information corresponding to an organism and its microbiota, which function in consortium (36). Our findings very much support this theory, and it is obvious that we need to develop a better understanding of the plant phenotype as an outcome of the interplay between inoculants and the host plants and their endogenous microbiota to be able to fully explain beneficial plant-microbe interactions.
Our data provide novel insights into the response of the plant growth-promoting endophyte B. phytofirmans PsJN to the plant host but also raise many issues such as those concerning the role of the plasmid in the endophytic lifestyle of strain PsJN and whether endophytes are involved in maintaining redox and energy balance in plants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
In this study, the plant growth-promoting rhizosphere bacterium and endophyte Burkholderia phytofirmans PsJN (= LMG 22146 T = CCUG 49060 T) was used (1). The bacterial strain was grown by loop inoculation of a single colony in LB broth. The bacterial culture was incubated at 28 ± 2°C for 2 days at 180 rpm in a shaking incubator.
Plant experiment.
Two potato varieties (Solanum tuberosum cv. Russet Burbank and Bionta) were grown in vitro in a growth chamber at 20°C with a 16-h-light/8-h-dark-photoperiod cycle. Four-week-old elongated apical shoots (10 cm in length) were used directly as explants. They were inoculated with B. phytofirmans PsJN by dipping for some seconds in a bacterial culture (1.3 × 108 CFU/ml). Inoculated plants were grown in 10 ml solid Murashige-Skoog (MS) medium containing 8% Duchefa Daishin agar (pH 5.8) and 20% saccharose for 4 weeks in glass tubes (2.5-cm diameter). Rooted potato plantlets were transferred into custom-tailored glass tubes with a narrow neck allowing hydroponic plant culturing. The plantlets were grown in 15 ml liquid MS medium containing 20% saccharose for two more weeks. Drought stress was induced by adding polyethylene glycol (PEG) (molecular weight [MW], 6,000) to reach a final concentration of 45%. Control plants were not treated. Shoots of stressed and control plantlets of each variety (6 replicates per treatment) were harvested at 1, 6, and 12 h after PEG application, immediately frozen in liquid nitrogen, and stored at −80°C for further analysis. To evaluate the presence of B. phytofirmans PsJN in inoculated potato plantlets, 16S rRNA gene PCR was performed using universal primers 799F [5′-AAC(AC)GGATTAGATACCC(GT)-3′] (37) and 1520R (5′-AAGGAGGTGATCCAGCCGCA-3′) (38). Amplification with this primer pair allows exclusion of chloroplast 16S rRNA gene-based amplicons but results in coamplification of plant mitochondrial small-subunit rRNA gene fragments and the bacterial 16S rRNA gene (see Fig. S1 in the supplemental material). Each PCR (50 µl) contained 30 to 50 ng/µl bacterial or potato DNA as the template, 1× PCR buffer, 2.5 mM MgCl2, 200 µM deoxynucleoside triphosphate (dNTP) mix (Thermo Scientific), a 150 nM concentration of each forward and reverse primers, 2.5 U of Firepol DNA polymerase (Solis Biodyne, Estonia), and PCR-grade water. Cycling conditions were as follows: initial denaturation for 5 min at 95°C; 34 cycles of 30 s at 95°C, 1 min at 60°C, 2 min at 72°C; and final elongation for 4 min at 72°C. Amplified PCR products (5 µl) were separated by electrophoresis (80 V) on 1% (wt/vol) agarose gels. Agarose gels were stained with ethidium bromide. Bacterial PCR amplicons were sequenced, making use of the sequencing service at LGC Genomics (Germany). Retrieved sequences were visualized and aligned with ClustalW as implemented in BioEdit v7.1.3 (39). For identification, sequences were subjected to BLAST analysis with the NCBI database.
Total RNA isolation and cDNA synthesis from plant tissue and bacterial cells.
Frozen plant material (100 mg) was prechilled with liquid nitrogen in 2-ml Safe-Lock tubes (Greiner Bio-One, Germany) and homogenized by the use of a ball mill MM301 mixer (Retsch GmbH & Co., Germany) at 30 Hz for 2 min using a single steel ball (5-mm diameter). Afterward, the material was immediately subjected to RNA isolation as described by Chang et al. (40). Extraction of total RNA from pure bacterial cultures was done using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions, and the RNA was used as a control in qPCR experiments. RNA samples were treated with DNase (Turbo DNA-free kit; Ambion, USA) according to the manufacturer’s protocol for purification from DNA contamination. RNA samples were tested for contaminating DNA by 16S rRNA gene PCR. RNA was analyzed at a 260-nm/280-nm ratio using a NanoDrop 1000 spectrophotometer (Thermo Scientific, USA), and the integrity was checked by electrophoresis in a 1% agarose gel. Purified RNA samples were reverse transcribed to cDNA with an iScript cDNA synthesis kit (BioRad Inc., USA) using random hexamers according to the manufacturer’s instructions.
Transcriptome sequencing.
In order to get a comprehensive image of the total transcriptome of B. phytofirmans PsJN colonizing potato plants (cv. Bionta), the total RNAs of four biological replicates for each treatment and control samples were pooled at equal concentrations to obtain approximately 48 µg RNA per treatment. rRNA depletion, cDNA library preparation, and sequencing on an Illumina HiSeq 2000 system with a 50-bp single-end read length were outsourced to Vertis Biotechnologie AG (Germany). In brief, plant rRNA molecules were depleted from the total RNA using a Ribo-Zero rRNA removal kit (Epicentre, USA). Plant mRNA molecules were removed by the use of oligo(dT) magnetic beads. The bacterial RNA samples were poly(A) tailed using poly(A) polymerase, and the RNA species which carried a 5′ monophosphate were degraded with Terminator exonuclease (Epicentre, USA). First-strand cDNA synthesis was performed using an oligo(dT)-adapter primer and Moloney murine leukemia virus (MMLV) reverse transcriptase. The resulting cDNA was PCR amplified to about 20 to 30 ng/µl using high-fidelity DNA polymerase. The cDNA was then purified using an Agencourt AMPure XP kit (Beckman Coulter Genomics, USA) and analyzed by capillary electrophoresis. The raw data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3524.
Preprocessing of sequencing data and mapping of reads.
RNA-seq reads were subjected to quality filtering using Prinseq (41) on the basis of the criterion of a minimum read length of 40, and good-quality reads were obtained with Q20 (sequencing error rate lower than 1%) for all reads. The poly(A) tails with a minimum length of 10 bp were removed by the use of Prinseq . Reads longer than 30 bp were considered for further analysis. Before read alignment, rRNA fragments were filtered from the transcriptomics data by the use of SortMeRNA software and the default rRNA database included in the software package (42). Reads were aligned to the genome of B. phytofirmans PsJN, and gene annotations were obtained from the NCBI database of bacterial genomes (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/Burkholderia_phytofirmans_PsJN_uid58729/).
Transcriptome analysis.
RNA-seq reads were aligned to the genome of B. phytofirmans PsJN using the Burrows-Wheeler Alignment Tool (BWA v0.6.2) (43). Transcript abundance was calculated using in-house Python scripts. Differential gene expression levels were analyzed and visualized with NOIseq (44). Gene expression levels were normalized using the number of reads per kilobase of coding sequence per million mapped reads (RPKM) in B. phytofirmans PsJN. As there was no replicate available for our data set, NOISeq-sim was used with the highest threshold (q = 0.9) to compute the probability of differential expression of genes under different conditions. To determine the variations of the differentially expressed genes which were expressed across all the time points from control compared to drought-stressed samples in B. phytofirmans PsJN, hierarchical cluster analysis was performed using Cluster 3.0 (13). The clustering results were visualized using TreeView (http://jtreeview.sourceforge.net/). Functional annotation of target genes based on Gene Ontology terms was performed using David v6.7 software (45) and all PsJN genome data available at NCBI RefSeq database (46) as the background. Venn diagrams were drawn using BioVenn software (47). Functional COG (clusters of orthologous groups) categories of the transcripts expressed in the control were listed using the COG database (48) and compared to the COG categories of the B. phytofirmans PsJN genome obtained from Integrated Microbial Genomes (IMG) systems (http://img.jgi.doe.gov).
Design of primers and probes for amplification of ECF sigma factor genes.
Oligonucleotides and probes for amplification of extracytoplasmatic function (ECF) sigma factor genes (see Table S4 in the supplemental material) were designed on the basis of the genome sequence of B. phytofirmans PsJN (GenBank project accession no. CP001052, CP001053, and CP001054) by making use of the ARB software package with its subfunction “Probe design,” version ARBuntu 2.0 (49). Probes were labeled with the reporter dye 6-carboxyfluorescein (FAM) at the 5′ end and with black hole quencher 1 (BHQ-1) fluorophore at the 3′ end.
Test for specificity of primers and probes for ECF sigma factor genes.
Primer specificity was checked by PCR amplification using genomic DNA of B. phytofirmans PsJN. DNA was extracted from bacterial cell pellets using a FastDNA Spin kit for soil (MP Biomedicals, LLC). DNA concentrations were measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE). PCR amplification was performed in a T-gradient PCR thermocycler (Biometra, Germany). PCR and PCR-amplicon sequencing as well as sequence analysis and identification were done as described above. Three primer sets (Bphyt_5131, Bphyt_7017, and Bphyt_1666) were not specific for their target ECF sigma factor genes and were removed from further analysis.
Selection of housekeeping genes for normalization of expression data.
Primers and probes were designed for 12 (see Table S4 in the supplemental material) gene candidates using the ARB software package (49), and probes were labeled with the 6-FAM reporter dye at the 5′ end and with black hole quencher 1 (BHQ-1) at the 3′ end. The primers were designed to be specific for B. phytofirmans PsJN and to avoid amplification of host plant material. The specificity of primers was tested by PCR, using genomic DNA of B. phytofirmans PsJN and potato plants as the template in separate reactions. The analysis of PCR amplification, sequencing, and sequences was performed as described above. Expression of all housekeeping genes was checked by TaqMan-qPCR using cDNA synthesized from total RNA isolated from stressed and control plants. The expression stability of selected housekeeping genes under conditions of drought stress and the variation in quantitative PCR efficiency in B. phytofirmans PsJN were calculated using qBasePLUS software with the geNormPLUS algorithm implemented (50, 51). Glutamine synthetase (Bphyt_2615) was the candidate housekeeping gene product that showed the most stable expression at different time points of drought stress and in both potato varieties (see Fig. S4).
TaqMan real-time PCR assays.
Quantitative PCR was carried out using the TaqMan-qPCR assay and a Bio-Rad CFX-96 real-time detection system (Bio-Rad, Hercules, CA). Triplicate qPCR reactions were performed with 1 µl of cDNA as the template, 1× BioRad SsoFast probe mix (BioRad Inc., Hercules, CA), 10 µM of forward and reverse primers, and 5 µM probe in a final volume of 10 µl. Cycling conditions were as follows: a hot start at 95°C for 2 min, 69 cycles of denaturation at 95°C for 5 s, and 20 s of elongation at 60°C. In each run, 2 negative controls were used, one as a no-template control performed with PCR-grade water instead of cDNA and another that included cDNA from non-PsJN-inoculated control plant samples. Four biological replicates of both potato varieties were tested. Data were analyzed with Bio-Rad CFX Manager software (version 3.0). Based on the Pfaffl equation (52) implemented in this software, normalized relative quantity (NRQ) values of ECF sigma factor genes in comparison to the most stably expressed reference gene (Bphyt_2615) were determined.
SUPPLEMENTAL MATERIAL
In vitro potato plants before stress application. Six weeks after inoculation with B. phytofirmans PsJN potato plants of both cultivars (Russet Burbank and Bionta) showed increased shoot and root length in response to colonization by strain PsJN. +, plants inoculated with B. phytofirmans PsJN; −, control plants treated with sterile growth media. Download
Application of polyethylene glycol (PEG) caused visible symptoms of water deficiency in in vitro potato plants at 1, 6, and 12 h after stress induction. Download
Detection of B. phytofirmans PsJN in inoculated potato plants. Lanes 1, molecular marker (GeneRuler; 100 bp); lanes 2, 3, 4, 11, 12, and 13, PCR products from PsJN-inoculated plant samples (Russet Burbank [RB] and Bionta [bi] varieties); lanes 5 to 10, PCR products from noninoculated Russet Burbank [RB] and Bionta [bi] plant samples; lanes 14 to 16, PCR products of genomic DNA of B. phytofirmans PsJN; lanes 17, PCR-negative control. Download
Evaluation of the gene expression stability of housekeeping genes (geNorm M value) from the most unstable genes (high M value on the left side) to the most stable ones (low M value on the right side) in Russet Burbank (A) and Bionta (B). In both potato cultivars, glutamine synthetase (Bphyt_2615) was the candidate gene with the most stable expression at the different time points tested, with an average geNorm M value of <0.2 (lowest M value). Download
Activation of oxidative phosphorylation in B. phytofirmans PsJN colonizing drought-stressed potato plants over time. Genes that are upregulated at a certain time point are labeled with a red asterisk. Download
Range of mean quantification cycle (Cq) values for expressed ECF sigma factor genes in B. phytofirmans PsJN colonizing potato plants under conditions of drought stress at different time points in cultivar Russet Burbank (panels A and C and panels E and G) and in potato cultivar Bionta (panels B and D and panels F and H). Each box indicates the 25% and 75% percentiles of mean Cq values. Whiskers represent the maximum and minimum Cq values. The median is depicted as a line across the box. Download
Examples of biological functions expressed in B. phytofirmans PsJN colonizing in vitro potato plants (cv. Bionta).
Complete list of genes that are differentially expressed in B. phytofirmans PsJN after 1 h (A), 6 h (B), and 12 h (C) of host plant drought stress.
Examples of biological functions expressed in B. phytofirmans PsJN under conditions of drought stress at three different time points.
Oligonucleotide primer and probe sequences (5′ → 3′) for ECF sigma factor genes (A) and housekeeping genes (B) of B. phytofirmans PsJN (FWD, forward primer; RWD, reverse primer; FAM, reporter dye 6-carboxyfluorescein; BHQ-1, black hole quencher 1 fluorophore).
ACKNOWLEDGMENT
This work was supported by a grant provided by the Austrian Science Fund (FWF): P22867-B16.
Footnotes
Citation Sheibani-Tezerji R, Rattei T, Sessitsch A, Trognitz F, Mitter B. 2015. Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 6(5):e00621-15. doi:10.1128/mBio.00621-15.
Contributor Information
Anton Hartmann, Helmholtz Zentrum Muenchen, German Research Center for Environmental Health.
Janet K. Jansson, Pacific Northwest National Laboratory.
REFERENCES
- 1.Sessitsch A, Coenye T, Sturz AV, Vandamme P, Barka EA, Salles JF, van Elsas JD, Faure D, Reiter B, Glick BR, Wang-Pruski G, Nowak J. 2005. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant beneficial properties. Int J Syst Evol Bacteriol 55:1187–1192. doi: 10.1099/ijs.0.63149-0. [DOI] [PubMed] [Google Scholar]
- 2.Frommel MI, Nowak J, Lazarovits G. 1991. Growth enhancement and developmental modifications of in vitro grown potato (Solanum tuberosum ssp. tuberosum) as affected by a nonfluorescent Pseudomonas sp. Plant Physiol 96:928–936. doi: 10.1104/pp.96.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compant S, Nowak J, Coenye T, Clément C, Ait Barka E. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev 32:607–626. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 4.Naveed M, Mitter B, Reichenauer TG, Wieczorek K, Sessitsch A. 2014. Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD17. Environ Exp Bot 97:30–39. doi: 10.1016/j.envexpbot.2013.09.014. [DOI] [Google Scholar]
- 5.Mitter B, Weilharter A, Chain PSG, Trognitz F, Nowak J, Compant S, Sessitsch A. 2013. Genome analysis, ecology, and plant growth promotion of the endophyte Burkholderia phytofirmans strain PsJN, p 865–874. In de Bruijn F (ed), Molecular microbial ecology of the rhizosphere. Wiley-Blackwell, , NJ. [Google Scholar]
- 6.Bordiec S, Paquis S, Lacroix H, Dhondt S, Ait Barka E, Kauffmann S, Jeandet P, Mazeyrat-Gourbeyre F, Clément C, Baillieul F, Dorey S. 2011. Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J Exp Bot 62:595–602. doi: 10.1093/jxb/erq291. [DOI] [PubMed] [Google Scholar]
- 7.Theocharis A, Bordiec S, Fernandez O, Paquis S, Dhondt-Cordelier S, Baillieul F, Clément C, Barka EA. 2012. Burkholderia phytofirmans PsJN primes Vitis vinifera L. and confers a better tolerance to low nonfreezing temperatures. Mol. Plant Microb Interact 25:241–249. doi: 10.1094/MPMI-05-11-0124. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez O, Theocharis A, Bordiec S, Feil R, Jacquens L, Clément C, Fontaine F, Barka EA. 2012. Burkholderia phytofirmans PsJN acclimates grapevine to cold by modulating carbohydrate metabolism. Mol Plant Microb Interact 25:496–504. doi: 10.1094/MPMI-09-11-0245. [DOI] [PubMed] [Google Scholar]
- 9.Poupin MJ, Timmermann T, Vega A, Zuñiga A, González B. 2013. Effects of the plant growth-promoting bacterium Burkholderia phytofirmans PsJN throughout the life cycle of Arabidopsis thaliana. PLoS One 8:e69435. doi: 10.1371/journal.pone.0069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shidore T, Dinse T, Öhrlein J, Becker A, Reinhold-Hurek B. 2012. Transcriptomic analysis of response to exudates reveals genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ Microbiol 14:2775–2787. doi: 10.1111/j.1462-2920.2012.02777.x. [DOI] [PubMed] [Google Scholar]
- 11.Alquéres S, Meneses C, Rouws L, Rothballer M, Baldani I, Schmid M, Hartmann A. 2013. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol Plant Microbe Interact 26:937–945. 10.1094/MPMI-12-12-0286-R. [DOI] [PubMed] [Google Scholar]
- 12.Drogue B, Sanguin H, Borland S, Prigent-Combaret C, Wisniewski-Dyé F. 2014. Genome wide profiling of Azospirillum lipoferum 4B gene expression during interaction with rice roots. FEMS Microbiol Ecol 87:543–555. doi: 10.1111/1574-6941.12244. [DOI] [PubMed] [Google Scholar]
- 13.De Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 14.Chain PS, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, Hauser L, Córdova M, Gómez L, González M, Land M, Lao V, Larimer F, LiPuma JJ, Mahenthiralingam E, Malfatti SA, Marx ZJ, Parnell JJ, Ramette A, Richardson P, Seeger M, Smith D, Spilker T, Sul WJ, Tsoi TV, Ulrich LE, Zhulin IB, Tiedje AM. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.79-Mbp genome shaped for versatility. Proc Natl Acad Sci U S A 103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitter B, Petric A, Shin MW, Chain PSG, Hauberg-Lotte L, Reinhold-Hurek B, Nowak J, Sessitsch A. 2013. Bacterial genomes reveal a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci 4:120. doi: 10.3389/fpls.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Maayer P, Chan WY, Rubagotti E, Venter SN, Toth IK, Birch PR, Coutinho TA. 2014. Analysis of the Pantoea ananatis pan-genome reveals factors underlying its ability to colonize and interact with plant, insect and vertebrate hosts. BMC Genomics 15:404. doi: 10.1186/1471-2164-15-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuer H, Smalla K. 2012. Plasmids foster diversification and adaption of bacterial populations in soil. FEMS Microbiol Rev 36:1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 18.Compant S, Clément C, Sessitsch A. 2010. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 19.Balsanelli E, Tadra-Sfeir MZ, Faoro H, Pankievicz VC, de Baura VA, Pedrosa FO, de Souza EM, Dixon R, Monteiro RA. 29 April 2015. Molecular adaptations of Herbaspirillum seropedicae during colonization of the maize rhizosphere. Environ Microbiol. doi: 10.1111/1462-2920.12887. [DOI] [PubMed] [Google Scholar]
- 20.Zúñiga A, Poupin MJ, Donoso R, Ledger T, Guiliani N, Gutiérrez RA, González B. 2013. Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol Plant Microb Interact 26:546–553. doi: 10.1094/MPMI-10-12-0241-R. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Cheng Z, Glick BR. 2009. The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol Lett 296:131–136. doi: 10.1111/j.1574-6968.2009.01625.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Conn K, Lazarovits G. 2006. Involvement of quinolinate phosphoribosyl transferase in promotion of potato growth by a Burkholderia strain. Appl Environ Microbiol 72:760–768. doi: 10.1128/AEM.72.1.760-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ait Barka E, Nowak J, Clément C. 2006. Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans PsJN. Appl Environ Microbiol 72:7246–7252. doi: 10.1128/AEM.01047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F. 2000. Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stryer L. 1988. Biochemistry, 3rd ed. W. H. Freeman and Company, New York, NY. [Google Scholar]
- 26.Eaton CJ, Cox MP, Scott B. 2011. What triggers grass endophytes to switch from mutualism to pathogenism? Plant Sci 180:190–195. doi: 10.1016/j.plantsci.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Fki L, Nasir O, Chkir M, Maalej M, Shumacher HM, Drira N. Risk evaluation of endophytic bacteria's anarchic proliferation within in vitro tissue cultures. COST Action FA 1103: Endophytes in Biotechnology and Agriculture, 3-7 November 2014, Izmir, Turkey http://endophytes.eu/wp/wp-content/uploads/2015/01/ABSTRACT-BOOK-Izmir.pdf. [Google Scholar]
- 28.Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110. doi: 10.1016/S0065-2911(02)46002-X. [DOI] [PubMed] [Google Scholar]
- 29.Gourion B, Sulser S, Frunzke J, Francez-Charlot A, Stiefel P, Pessi G, Vorholt JA, Fischer HM. 2009. The PhyR-sigma(EcfG) signalling cascade is involved in stress response and symbiotic efficiency in Bradyrhizobium japonicum. Mol Microbiol 73:291–305. doi: 10.1111/j.1365-2958.2009.06769.x. [DOI] [PubMed] [Google Scholar]
- 30.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coenye T, Van Acker H, Peeters E, Sass A, Buroni S, Riccardi G, Mahenthiralingam E. 2011. Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother 55:1912–1919. doi: 10.1128/AAC.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agnoli K, Lowe CA, Farmer KL, Husnain SI, Thomas MS. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function sigma factor which is a part of the Fur regulon. J Bacteriol 188:3631–3644. doi: 10.1128/JB.188.10.3631-3644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 34.Da K, Nowak J, Flinn B. 2012. Potato cytosine methylation and gene expression changes induced by a beneficial bacterial endophyte, Burkholderia phytofirmans strain PsJN. Plant Physiol Biochem 50:24–34. doi: 10.1016/j.plaphy.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Nowak J, Veilleux RE, Nowak J, Turgeon S. 2007. Priming for transplant stress in in vitro propagation via plantlet bacterization. Acta Hort 748:65–75. [Google Scholar]
- 36.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 37.Chelius MK, Triplett EW. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol 41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 38.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. 1989. Isolation and direct complete nucleotide determination of entire genes—characterisation of a gene coding for 16S-ribosomal RNA. Nucleic Acids Res 17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 40.Chang S, Puryear J, Cairney J. 1993. Simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 41.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. BioInformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows–Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-seq: a matter of depth. Genome Res 21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Tatusova T, Ciufo S, Federhen S, Fedorov B, McVeigh R, O'Neill K, Tolstoy I, Zastavsky L. 2015. Update on RefSeq microbial genomes resources. Nucleic Acids Res 43:D599–D605. doi: 10.1093/nar/gku1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn—a Web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar A, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K.-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Overmars L, Kerkhoven R, Franke C. 2013. MGcV: the microbial genomic context viewer for comparative genome analysis. BMC Genomics 14:209. doi: 10.1186/1471-2164-14-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro potato plants before stress application. Six weeks after inoculation with B. phytofirmans PsJN potato plants of both cultivars (Russet Burbank and Bionta) showed increased shoot and root length in response to colonization by strain PsJN. +, plants inoculated with B. phytofirmans PsJN; −, control plants treated with sterile growth media. Download
Application of polyethylene glycol (PEG) caused visible symptoms of water deficiency in in vitro potato plants at 1, 6, and 12 h after stress induction. Download
Detection of B. phytofirmans PsJN in inoculated potato plants. Lanes 1, molecular marker (GeneRuler; 100 bp); lanes 2, 3, 4, 11, 12, and 13, PCR products from PsJN-inoculated plant samples (Russet Burbank [RB] and Bionta [bi] varieties); lanes 5 to 10, PCR products from noninoculated Russet Burbank [RB] and Bionta [bi] plant samples; lanes 14 to 16, PCR products of genomic DNA of B. phytofirmans PsJN; lanes 17, PCR-negative control. Download
Evaluation of the gene expression stability of housekeeping genes (geNorm M value) from the most unstable genes (high M value on the left side) to the most stable ones (low M value on the right side) in Russet Burbank (A) and Bionta (B). In both potato cultivars, glutamine synthetase (Bphyt_2615) was the candidate gene with the most stable expression at the different time points tested, with an average geNorm M value of <0.2 (lowest M value). Download
Activation of oxidative phosphorylation in B. phytofirmans PsJN colonizing drought-stressed potato plants over time. Genes that are upregulated at a certain time point are labeled with a red asterisk. Download
Range of mean quantification cycle (Cq) values for expressed ECF sigma factor genes in B. phytofirmans PsJN colonizing potato plants under conditions of drought stress at different time points in cultivar Russet Burbank (panels A and C and panels E and G) and in potato cultivar Bionta (panels B and D and panels F and H). Each box indicates the 25% and 75% percentiles of mean Cq values. Whiskers represent the maximum and minimum Cq values. The median is depicted as a line across the box. Download
Examples of biological functions expressed in B. phytofirmans PsJN colonizing in vitro potato plants (cv. Bionta).
Complete list of genes that are differentially expressed in B. phytofirmans PsJN after 1 h (A), 6 h (B), and 12 h (C) of host plant drought stress.
Examples of biological functions expressed in B. phytofirmans PsJN under conditions of drought stress at three different time points.
Oligonucleotide primer and probe sequences (5′ → 3′) for ECF sigma factor genes (A) and housekeeping genes (B) of B. phytofirmans PsJN (FWD, forward primer; RWD, reverse primer; FAM, reporter dye 6-carboxyfluorescein; BHQ-1, black hole quencher 1 fluorophore).