ABSTRACT
An estimated one-third of the world’s population is currently latently infected with Mycobacterium tuberculosis. Latent M. tuberculosis infection (LTBI) progresses into active tuberculosis (TB) disease in ~5 to 10% of infected individuals. Diagnostic and prognostic biomarkers to monitor disease progression are urgently needed to ensure better care for TB patients and to decrease the spread of TB. Biomarker development is primarily based on transcriptomics. Our understanding of biology combined with evolving technical advances in high-throughput techniques led us to investigate the possibility of additional platforms (epigenetics and proteomics) in the quest to (i) understand the biology of the TB host response and (ii) search for multiplatform biosignatures in TB. We engaged in a pilot study to interrogate the DNA methylome, transcriptome, and proteome in selected monocytes and granulocytes from TB patients and healthy LTBI participants. Our study provides first insights into the levels and sources of diversity in the epigenome and proteome among TB patients and LTBI controls, despite limitations due to small sample size. Functionally the differences between the infection phenotypes (LTBI versus active TB) observed in the different platforms were congruent, thereby suggesting regulation of function not only at the transcriptional level but also by DNA methylation and microRNA. Thus, our data argue for the development of a large-scale study of the DNA methylome, with particular attention to study design in accounting for variation based on gender, age, and cell type.
IMPORTANCE
DNA methylation modifies the transcriptional program of cells. We have focused on two major populations of leukocytes involved in immune response to infectious diseases, granulocytes and monocytes, both of which are professional phagocytes that engulf and kill bacteria. We have interrogated how DNA methylation, gene expression, and protein translation differ in these two cell populations between healthy individuals and patients suffering from TB. To better understand the underlying biologic mechanisms, we harnessed a statistical enrichment analysis, taking advantage of predefined and well-characterized gene sets. Not only were there clear differences on various levels between the two populations, but there were also differences between TB patients and healthy controls in the transcriptome, proteome, and, for the first time, DNA methylome in these cells. Our pilot study emphasizes the value of a large-scale study of the DNA methylome taking into account our findings.
INTRODUCTION
Tuberculosis (TB) is a threatening disease, with currently 9 million new cases and 1.5 million deaths per year (1). One-third of the global population is latently infected (latent TB infection [LTBI]) with Mycobacterium tuberculosis, thus facing the risk of developing active TB during their lifetime. Effective drug treatment regimens exist, albeit they have been challenged by increasing multidrug-resistant, extensively drug-resistant, and totally drug-resistant forms of TB (2). In addition, TB diagnosis and control are still hampered by the unavailability of vaccines capable of preventing TB. The fact that only an estimated 5 to 10% of people with LTBI develop active TB disease combined with the high occurrence of infection in household contacts emphasizes that the underlying biological mechanisms remain poorly understood. A better understanding of the biological processes involved in progression from LTBI to active TB will contribute toward better intervention measures. To this end, sufficiently validated biomarkers to support development of TB vaccines, diagnostics, and drugs are needed (3, 4). To add further complexity to this scenario, the biology of and interactions between the host, microbe, and environment are highly complex and variable (5). Some factors are known to affect the host response to M. tuberculosis infection, including inherent host genetics, variability among hosts, status of the immune system, and external factors, such as nutrition, pollution, coinfection, frequency of previous infections and coinfections, stress levels, and adaptations by bacterial strains in specific host populations (6–8). External factors are also known to affect host epigenetics, prompting us to include the epigenome in future TB biomarker research. In particular, stable marks such as those derived from DNA methylation patterns are currently absent from studies of this kind, while more dynamic epigenetic marks (histone modifications) have uncovered matters relating to “trained immunity,” even in the case of recipients of the only current TB vaccine, M. tuberculosis BCG (bacillus Calmette-Guérin) (9, 10).
To date, transcriptomic profiles from peripheral blood cells have been the main focus in the exploration for biomarkers (11). In addition to easy access, blood also represents a site of dynamic exchange of chemokines, cytokines, and cells trafficking between foci of active disease and the lymphoid system (12). Within the immune system, cells have their own discrete functions, but the system as a whole exerts a concerted function with remarkable plasticity. Hence careful consideration of different cell types is required when describing either predispositions for disease development or the resultant functions following infection. Understanding how each specific cell contributes to maintenance of LTBI instead of progressing to active TB is essential. Professional phagocytes play a central role in these processes (13). Phagocytes comprise dendritic cells, monocytes (differentiating into macrophages), and granulocytes, predominantly neutrophils. Monocytes and granulocytes have epigenomes distinct from other cell types (14, 15). Due to its inherent stability, disruption of the normal DNA methylome can produce stable cell populations with prolonged aberrant phenotypes and thereby contribute to disease, which suggests a useful source of biomarkers for risk stratification and disease diagnosis (16). In primary human leukocyte subsets, single nucleotide polymorphisms (SNPs), which are associated with immune-mediated disease, preferentially map to cell-specific regulatory hypomethylated regions (HMRs) (14). Such loci, in combination with DNA methylation sites, offer hypotheses toward depicting cellular subsets in which specific epigenetic changes may drive disease.
This pilot study aims to investigate the level of differentiation between TB patients and genetically unrelated LTBI household contacts in a cell-specific manner. Isolated monocytes and granulocytes from peripheral blood were investigated to determine the extent and biological functions of differences between the DNA methylome, transcriptome (mRNA and microRNA [miR]), and proteome at a global level using high-throughput techniques.
Toward this end, we studied each platform in a descriptive manner, followed by differential analyses between LTBI and TB at each platform in monocytes and granulocytes alone and in combination. Next, we determined if these differences point toward specific functions. Finally, we analyzed these data to determine which, if any, correlations of differences between LTBI and TB exist in functions from one platform to another. Specifically, we wanted to ascertain in this very-limited-sample-size study whether a portion of the differences between LTBI and TB in global DNA methylation and miR platforms reveal gene regulation that will be reflected by the mRNA data set and in turn is translated into the proteome.
At the levels of DNA methylation, miR, mRNA, and proteins, the data allowed differentiation between LTBI and TB. Moreover, unique functions were congruent from one platform to another, suggesting a global regulation of function (or loss thereof) during TB.
RESULTS AND DISCUSSION
Global DNA methylation in LTBI and TB.
The DNA methylation status of over 485,000 CpGs was interrogated in isolated monocytes and granulocytes from LTBI and TB participants. For both cell types, a clear bimodal data distribution was evident, with no major variation of these distributions between LTBI and TB (see Fig. S1A and B in the supplemental material) either together or per cell type. Frequency distributions illustrate similar levels of hypomethylated (0 to 30%) and hypermethylated (70 to 100%) CpGs in both study groups. This observation differs from the overall spread of methylated CpGs in precursor cells, such as hematopoietic stem cells (15).
We tested whether methylation of CpGs in professional phagocytes was related to known functions in these cell types. The list of genes with fully hyper- or hypomethylated CpGs associated with promoter regions was tested for enrichment in blood transcription modules (BTMs) using the hypergeometric test, in which genes with fully hyper- or hypomethylated CpGs associated with promoter regions constituted the foreground set, while genes lacking such sites constituted the background (17, 18). Genes with promoter-associated hypermethylated sites (i.e., putatively silenced in the analyzed cell subsets) were not significantly enriched in BTMs. However, significant enrichment in BTMs for genes with hypomethylated CpGs associated with promoter regions (i.e., potentially activated genes) was observed with statistically significant enrichment in modules involved in cell cycling and transcription, as well as those involved in immune activation (see Table S1 in the supplemental material).
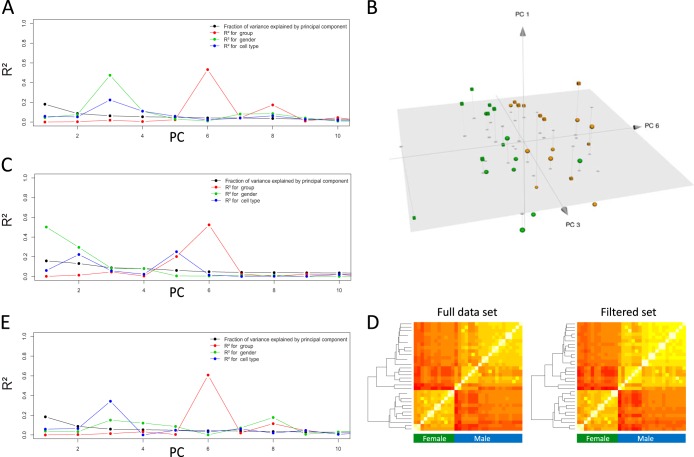
To determine which known sources of variation (that is, disease phenotype, gender, and cell type) explain the total variance of the data set, we applied principal component analysis (PCA) regression of principal components (PCs) over the independent variables. Both gender and cell type explained a substantial portion of the variance of the first two PCs, while disease phenotype explained a smaller fraction of the variance (Fig. 1A). Disease phenotype explained the majority of the variance of PC6 (Fig. 1A and B). When the global DNA methylation status of these and other cell types in peripheral blood was compared in another study of similar design, PCA indicated that DNA methylation patterns differ more profoundly between cell types than between individuals (19). These results, in combination with our findings, indicate that M. tuberculosis infection affects DNA methylation in monocytes and granulocytes to a lesser extent than differences observed in cell type at the level of the global DNA methylome.
FIG 1 .
Methylation in professional phagocytes in latent Mycobacterium tuberculosis infection (LTBI) and tuberculosis (TB) illustrating sources of variance in the methylation data following principal component analysis (PCA). (A) Variance in the full data set (black line) is broken down into known sources of variance within each component of PCA, illustrating the majority of variance being explained. (B) Mainly along the axis of principal component 6 (PC6), a distinction can be made between LTBI (green) and TB (yellow): spheres, monocytes; cubes, granulocytes. (C) Variance and sources thereof following PCA analysis of the filtered set. (D) Heat maps showing correlation coefficients between the samples, from low (red) to high (yellow). Samples clustered by hierarchical clustering indicate a primary distinction between genders. (E) Variance explained in the filtered set from which all X- or Y-chromosome-associated loci were removed.
Given the clear binomial distribution both in cell types and in disease phenotypes, a nonspecific filtered data set was defined to exclude uninformative data. This set excluded (i) CpGs for which no link to functional genes had been established thus far and (ii) CpGs that were either hypermethylated or hypomethylated in every sample. The resulting filtered data set contained 80,198 CpG sites and showed an even more concise discrimination between genders (Fig. 1C and D). Unsupervised hierarchical clustering of the methylation profiles from 36 samples confirms gender differences in these two cell types (Fig. 1E). Thus, gender accounted for higher variation between individuals than TB disease phenotype in this data set. However, when the filtered set of CpGs was evaluated for differences between LTBI and TB, a clear distinction between groups was observed in PC5 and PC6 of the PCA (Fig. 1D), suggesting that some of the epigenetic changes in monocytes and granulocytes were either the result or the cause of TB.
Second, differential analyses revealed <1% of the CpGs in the filtered set to be statistically differentially methylated between TB and LTBI (Wilcoxon q < 0.05, adjusted for multiple testing using the Benjamini-Hochberg procedure) (see Table S2 in the supplemental material). Of these, the CpGs showing most differentially methylated targets occurred in CpG islands and shores (see Fig. S1C in the supplemental material). We tested whether the effect of the disease phenotype was random by considering the predictive power of the methylated sites using a machine learning (ML) approach. The resulting models were significantly better than random at distinguishing TB from LTBI (area under the curve [AUC], 0.74; 95% confidence interval [CI], 0.57 to 0.92), with an overall error rate below 30%. Classification between cell types yielded no errors (AUC, 1.00).
As can be expected following PCA, the relative abundances of differential methylation between monocytes and granulocytes within the same individual were much larger (~20% [Fig. S1D]). This confirms a previous study reporting 22% of CpGs to be differentially methylated between these two cell types (19).
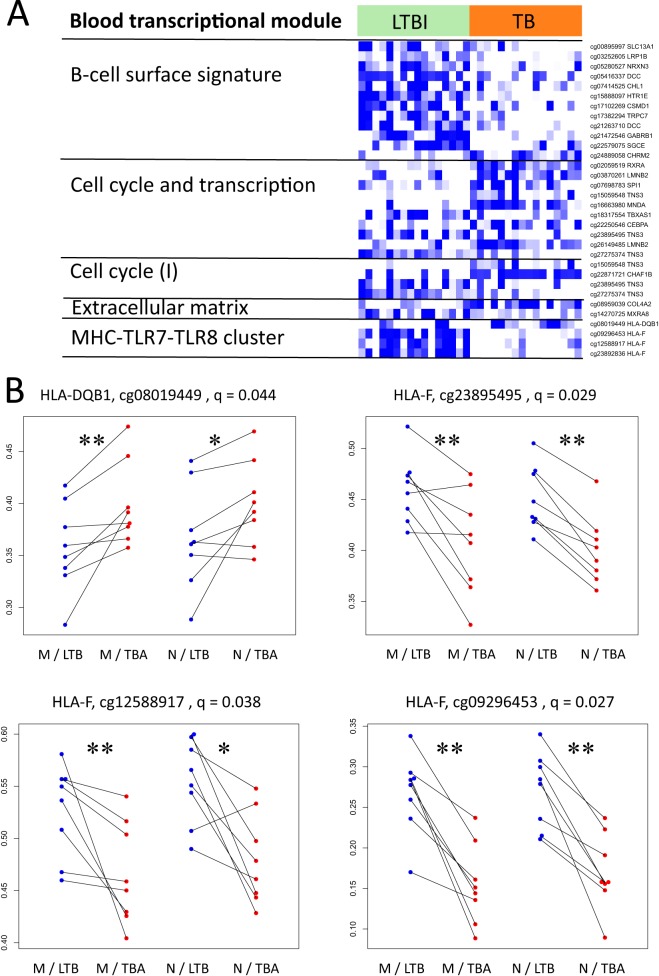
To gain insight into the functional role of CpGs with methylation differences between LTBI and TB, we tested enrichment in functional categories of the genes associated with differentially methylated CpGs. BTMs for which the differences between LTBI and TB were significantly enriched resemble a tentative signature/fingerprint with CpGs either hyper- or hypomethylated in LTBI compared to TB within five known functional modules (Fig. 2A). Of particular interest are four CpGs found to be differentially methylated in the “MHC-TLR7-TLR8 cluster,” which all occur in CpG islands (Fig. 2B). While the CpGs associated with HLA-DQB1 (coding for one HLA class II chain) did not reside in a promoter-associated area of the gene, the other three CpGs associated with HLA-F and coding for HLA class I were all located within the promoter. Moreover, these two HLA-related genes to which these CpGs have bearing had opposite effects when LTBI and TB were compared: CpGs for HLA-DQB1 were significantly hypermethylated in TB patients compared to their paired LTBI samples, while CpGs at three different loci in the promoter of HLA-F were hypomethylated (Fig. 2).
FIG 2 .
Functional association of statistically significant differentially methylated CpGs. (A) Heat map showing differentially methylated CpGs associated with genes from five blood transcriptional modules. Colors are relative as data are scaled row-wise: white indicates lowest methylation for a given CpG, while blue indicates highest methylation for a given CpG. Columns correspond to samples. Multiple CpGs can correspond to a single gene. (B) Dot plots showing changes between LTBI and TB for one CpG associated with the HLA-DQB1 gene and three CpGs associated with the HLA-F gene. Lines connect samples from a single pair of individuals. Blue indicates measurements for samples in LTBI, and red indicates measurements for samples in TB. The q values represent the P values after correction for multiple testing (Benjamini-Hochberg); *, P < 0.05; **, P < 0.01 (Wilcoxon paired test).
Hypermethylated HLA-DQB1 in TB and reduced major histocompatibility complex class II (MHC-II) expression, regardless of whether it is a trigger or a result of activation, could benefit M. tuberculosis. This resonates with reports (20, 21) indicating that M. tuberculosis interferes with antigen processing and presentation. Interestingly, an immunogenic peptide from M. tuberculosis is presented by HLA-DQB1, and some HLA-DQB1 alleles associated with TB sensitivity present such peptides resulting in suboptimal antigen-specific interferon gamma (IFN-γ) secretion by CD4 T cells (22). This underlines the relevance of methylation of disease-related MHC-II alleles and warrants further investigation. On the other hand, HLA-F, which has been associated with tumor invasiveness (23) and immune suppression (24), was hypomethylated in TB compared to LTBI-matched individuals. This, along with hypermethylated HLA-DQB1, may result in immune modulation to favor the pathogen.
Global expression of transcripts and miRs.
Transcriptomic data from the four experimental groups can be clearly distinguished according to gene expression profiles, with differences between cell types dominating over differences between study groups (see Fig. S2A in the supplemental material). PCA revealed that 33% of the overall variance in the data corresponded to PC1, which was correlated with the two cell types. A further 14% of the variance is explained by PC2 and PC5, which correlate with TB. We have trained ML models to distinguish TB from LTBI and, independently, the two cell types. TB could be distinguished from LTBI with few errors (AUC, 0.99; 95% CI, 0.96 to 1.00), while there were no errors in classification of the cell types.
Second, both cell types showed a number of significantly differentially expressed genes between LTBI and TB (see Table S3 in the supplemental material). Pairwise comparisons between monocytes and granulocytes as well as between the LTBI and TB groups demonstrated a substantial response to TB in both cell types, as well as marked differences between these cell types in both LTBI and TB samples (see Fig. S3B in the supplemental material), as can be seen on the PCA plot (see Fig. S3C). PC1 corresponded to differences between monocytes and granulocytes and was enriched in “mitochondrial and translation-related” genes, and PC2 and PC5 corresponded to differences between LTBI and TB samples.
In agreement with previous studies, TB patients showed a significant increase in expression of several genes identified as relevant in previous studies (25–27). These include significant upregulation of CD64 (Fc-γ receptor IA), and guanylate-binding proteins (GBPs) in TB. In granulocytes, we found a higher expression in TB for several MHC-II-related genes, including the CD74, HLA-DRA, and HLA-DMB genes. While the expression of these genes was significantly lower in granulocytes than in monocytes, no differences between LTBI and TB were apparent in monocytes.
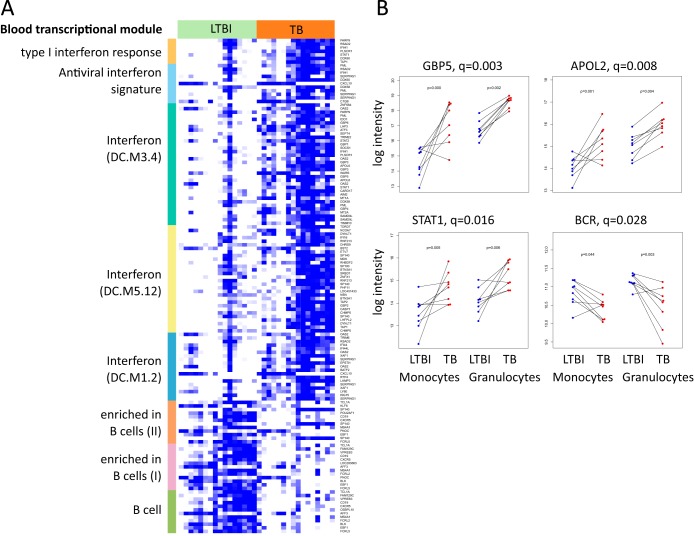
Differential transcription analysis between LTBI and TB followed by functional analysis revealed several BTMs that were statistically significantly enriched for among differentially expressed genes (q < 0.05) (Table 1; Fig. 3A). Notably, the BTM designated “B cell surface signature” was significantly enriched on both the methylation and transcription levels. Likewise, transcriptional differences between monocytes and granulocytes were coherent with the corresponding module enrichments in the methylation data set (see Table S4 in the supplemental material). The responses to TB in monocytes and granulocytes were largely similar in the mRNA data set. The overall Pearson correlation coefficient between the log2-fold changes in monocytes and granulocytes was 0.42 (q ~ 0). In line with this, there were no genes with a statistically significant interaction between cell type and disease state—that is, genes with a different response to TB in monocytes compared to granulocytes.
TABLE 1 .
Blood transcriptional module enrichment analysis of genes differentially expressed between latent M. tuberculosis infection and TBa
| Module ID | Module title | No. of genes in a module |
AUC | q value |
|---|---|---|---|---|
| DC.M3.4 | IFN | 51 | 0.88 | 2.61E−18 |
| DC.M5.12 | IFN | 57 | 0.77 | 2.48E−10 |
| DC.M1.2 | IFN | 24 | 0.87 | 4.57E−08 |
| LI.M47.0 | Enriched in B cells (I) | 47 | 0.73 | 1.94E−06 |
| LI.M75 | Antiviral IFN signature | 22 | 0.83 | 4.40E−06 |
| LI.M67 | Activated dendritic cells | 11 | 0.95 | 8.61E−06 |
| LI.M47.1 | Enriched in B cells (II) | 34 | 0.74 | 5.45E−05 |
| LI.M127 | Type I IFN response | 12 | 0.90 | 5.69E−05 |
| DC.M4.10 | B cell | 31 | 0.75 | 5.69E−05 |
| LI.M37.1 | Enriched in neutrophils (I) | 49 | 0.30 | 5.69E−05 |
| LI.M111.1 | Viral sensing and immunity; IRF2 targets network (II) | 11 | 0.89 | 2.06E−04 |
| LI.M150 | Innate antiviral response | 12 | 0.85 | 5.53E−04 |
| DC.M3.2 | Inflammation | 118 | 0.39 | 5.58E−04 |
| LI.M226 | Proteasome | 12 | 0.83 | 1.72E−03 |
| DC.M3.5 | Cell cycle | 143 | 0.41 | 4.24E−03 |
| LI.M68 | RIG-1-like receptor signaling | 10 | 0.83 | 4.27E−03 |
| DC.M6.2 | Mitochondrial respiration | 144 | 0.41 | 4.75E−03 |
| LI.M5.0 | Regulation of antigen presentation and immune response | 79 | 0.62 | 5.20E−03 |
| DC.M4.13 | Inflammation | 77 | 0.39 | 1.18E−02 |
| DC.M2.3 | Erythrocytes | 66 | 0.61 | 1.50E−02 |
| LI.M32.8 | Cytoskeletal remodeling | 10 | 0.79 | 1.69E−02 |
| LI.M69 | Enriched in B cells (VI) | 20 | 0.70 | 2.03E−02 |
| DC.M6.12 | Mitochondrial stress | 66 | 0.39 | 2.35E−02 |
| DC.M4.15 | T cells | 41 | 0.64 | 2.67E−02 |
| LI.M156.0 | Plasma cells and B cells; immunoglobulins | 24 | 0.67 | 2.88E−02 |
| LI.M111.0 | Viral sensing and immunity; IRF2 targets network (I) | 17 | 0.71 | 2.88E−02 |
| LI.M7.1 | T cell activation (I) | 48 | 0.62 | 3.09E−02 |
| LI.M14 | T cell differentiation | 12 | 0.74 | 3.09E−02 |
| LI.M112.0 | Complement activation (I) | 17 | 0.70 | 3.91E−02 |
| LI.M209 | Lysosome | 8 | 0.79 | 3.93E−02 |
| DC.M4.1 | T cell | 53 | 0.61 | 4.06E−02 |
| LI.S2 | B cell surface signature | 168 | 0.56 | 4.06E−02 |
The module title is the title of the blood transcription module according to references 17 and 18. In the module ID, the prefix “DC” refers to BTMs according to Chaussabel et al. (18) and the prefix “LI” refers to BTMs according to Li et al. (17). Only modules with a functional annotation are shown. The AUC is the area under the curve, and the q value is the adjusted P value after correction for multiple testing (Benjamini-Hochberg). E, exponential notation (e.g., E–03 represents ×10−3); IFN, interferon; IRF2, interferon regulatory factor 2.
FIG 3 .
Gene expression patterns of LTBI and TB samples in professional phagocytes. (A) Heat map showing gene expression for differentially expressed (DE) genes in eight blood transcriptional modules. Colors are relative, as data are scaled row-wise: white denotes the lowest expression for a given gene, while blue denotes the highest expression for the given gene. Colors on the left-hand bar denote modules: gold, LI.M127 (type I IFN response); light blue, LI.M75 (antiviral IFN signature); dark green, DC.M3.4 (IFN); yellow, DC.M5.12 (IFN); gray-blue, DC.M1.2 (IFN); orange, LI.M47.1 (enriched in B cells [II]); pink, LI.M47.0 (enriched in B cells [I]); green, DC.M4.10 (B cells). Blood transcription modules are from Li et al. (17), unless “DC” is noted in parentheses. “DC” refers to Chaussabel et al. (18). (B) Dot plots showing changes in expression measured by microarray as reflected in panel A. Lines connect samples from a single pair of individuals. Blue, measurements for samples in LTBI; red, measurements for samples in TB. The q values represent the P values after correction for multiple testing over both monocytes and granulocytes; the p values represent the uncorrected P values, calculated separately for monocytes (left) or granulocytes (right) for each target reported.
However, based on visual inspection and uncorrected P values, we marked several candidate genes with an apparent difference between LTBI and TB for 1 cell type only. Employing quantitative reverse transcription-PCR (qRT-PCR), we were able to confirm differential expression in both cell types for GBP5 and signal transducer and activator of transcription 1 (STAT1), while for STAT1 in separate analyses for monocytes and granulocytes, only the monocytes revealed a statistically significant difference (n = 10 per group; P < 0.05, Wilcoxon paired test) (Fig. 3B). Even though statistical analyses fail to point out singular gene products to differ between LTBI and TB after correction for multiple testing, these data were confirmed by a second technique. Moreover, this supports the importance of interrogating cellular subsets independently when investigating the transcriptome for differential markers in TB.
In the miR data set, the largest portion of variation can be explained by differences between cell type and disease phenotype (see Fig. S3 in the supplemental material). Gender played a far less significant role than DNA methylation. The separation between LTBI and TB was less clear in the PCA for mRNA. The random forest ML models showed high performance both for separation of TB from LTBI samples (1 error; AUC, 1.00) and monocytes from granulocytes (no errors; AUC, 1.00).
Several miRs were significantly differently expressed between LTBI and TB in both cell types as well as in one cell type (see Table S5 in the supplemental material). One such example is miR-146a-5p (see Fig. S4A in the supplemental material), for which the largest relative upregulation was in monocytes from LTBI compared to TB. This miR has recently been reported to play a central role in the immune response (28) and to be upregulated in peripheral blood mononuclear cells of control subjects versus TB patients (29). miR-146a is upregulated in response to microbial stimuli and proinflammatory cytokines and has also been negatively correlated with interferon (IFN) type I signaling (30). We have validated this result using a TaqMan miR real-time quantitative PCR (qPCR) assay (see Fig. S4B).
Of note is the relatively large fraction of variance explained by gender in the full data set of DNA methylation as opposed to those for mRNA and miR. Transcripts such as mRNA and miR have a very short half-life. Therefore, changes in transcription rate rapidly affect the number of transcripts, which in turn causes a relatively high baseline variation due to rapid change in regulatory signals. Relative to this high baseline variation in transcripts, the contribution of gender to the total variation will be small. On the other hand, DNA methylation is a biochemically stable modification that results in markedly less baseline variation than that in mRNA and miR transcripts.
DNA methylation at CpGs had been documented repeatedly to be influenced by gender (31–34), not only on X and Y chromosomes but also CpGs on autosomal genes (32, 34). Moreover, DNA methylation patterns are thought to be established largely during early embryonic development and then stably propagated via mitosis.
Since the samples for this study were collected from adult participants, we can expect to see a relatively small contribution to variation by gender compared to the large baseline variation in transcript data sets but a relatively high contribution of variation by gender against the low baseline variation in the DNA methylation set.
Phagocyte proteomics during active TB.
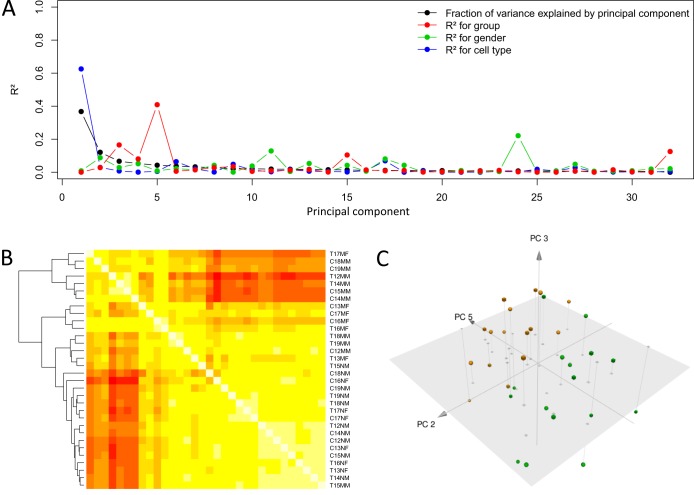
To shed light on the relationship between active disease and protein synthesis in professional phagocytes, we measured the relative abundances for 3,047 unique proteins and an additional 429 isoforms. Variance in these data was largely accounted for by differences observed in cell types (Fig. 4A). Yet, in PC3 and PC5, the majority of variance is explained by differences between LTBI and TB, revealing that at the proteomic level in professional phagocytes, LTBI and TB can be discriminated (Fig. 4C). Accordingly, ML models could correctly classify both TB versus LTBI samples (AUC, 0.93; 95% CI, 0.84 to 1.00) and monocytes/granulocytes (no errors; AUC, 1.00).
FIG 4 .
Overall analysis of proteomic data. (A) Variance decomposition of the PCA of the full data set. Variance (black line) in the protein data set is broken down into the known sources of variance within each PC of PCA, illustrating the majority of variance being explained in the first two PCs, of which gender and cell type explain most of the PCs. (B) Heat map showing correlation coefficients between the samples from low (red) to high (yellow). Samples were clustered by hierarchical clustering. Sample code: for the first digit, T represents TB and C represents LTBI, for the second-to-last digit, N represents granulocyte and M represents monocyte, and for the last digit, M represents male and F represents female. (C) PCA of protein data sets illustrating distinctions between LTBI and TB along PC5. Green, LTBI; yellow, TB active (TBA); spheres, monocytes; cubes, granulocytes.
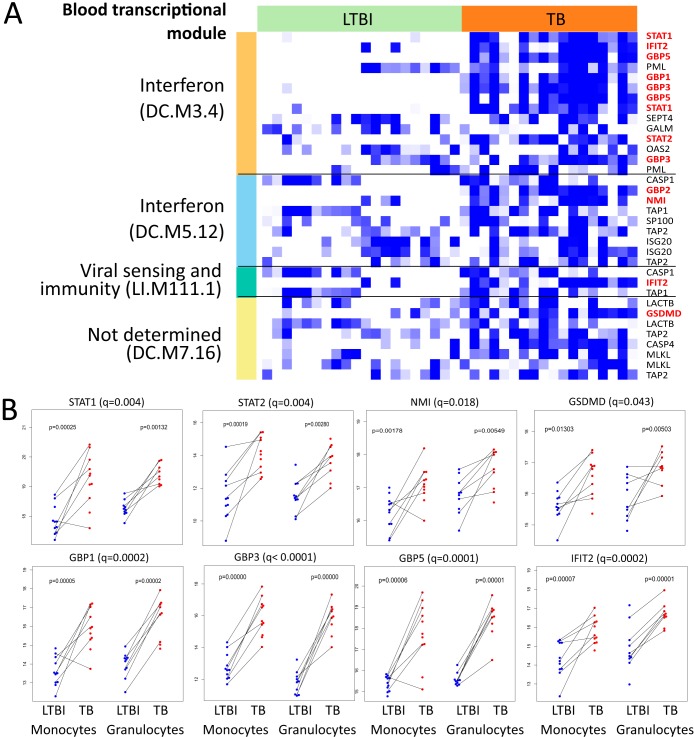
In both cell types, we found significant differences between LTBI and TB (see Table S6 in the supplemental material). Several of these proteins are functionally related, as has been revealed by interrogation for enriched BTMs. Notably, we identified enrichment of the IFN signaling modules (Fig. 5), including GBP1, GBP3, GBP5, STAT1, STAT2, and IFN-induced proteins with tetratricopeptide repeats (IFITs).
FIG 5 .
Protein levels of LTBI and TB samples in professional phagocytes. (A) Heat map showing gene expression for differentially expressed genes in four BTMs, which was enriched for, among differentially translated proteins. From these, panel B indicates only the peptides that were differentially translated between LTBI and TB in these modules. Colors are relative, as data are scaled row-wise; white denotes lowest expression for a given gene, while blue denotes highest expression for this gene. Colors on the left-hand bar denote modules. In parentheses, “LI” indicates modules from Li et al. (17) and “DC” indicates modules from Chaussabel et al. (18). Dot plots illustrate the relative pairwise abundance of peptides in monocytes and granulocytes found to be differentially translated between LTBI and TB (q < 0.05, limma) overall (B).
Effect of DNA methylation on transcription.
We next interrogated the extent to which differences in DNA methylation may influence transcription in TB. To test whether, in general, distinct methylation sites were correlated with expression of corresponding genes, we calculated row-wise correlation coefficients between CpG methylation sites and the corresponding genes (83,562 CpG gene pairs in total). The distribution of these correlation coefficients was significantly different from those of randomly paired genes and methylation sites (P ~ 0 in a two-sample Kolmogorov-Smirnov test).
We then focused on genes for which expression was strongly correlated with methylation of related CpG sites by investigating which BTMs contained such genes. In other words, for each functional group (BTM), we interrogated whether the expression of genes included in that functional group was, on average, correlated with the methylation of sites linked to that gene.
Toward this end, we tested for BTMs in which gene expression was strongly correlated with methylation of the linked CpG sites. First, we calculated correlation coefficients between values of methylation or expression for each pair of a CpG and a corresponding gene. Then, for each BTM, we performed a randomization test to determine whether the average correlation coefficient per BTM was significantly different from a random set of correlation coefficients. The existence of a correlation between the absolute methylation values and gene expression does not necessarily mean that a regulation of gene expression by differential methylation is relevant for the TB disease process. Therefore, we further calculated the correlation coefficients of paired differences in methylation/expression between LTBI and TB in monocytes and granulocytes, as well as between monocytes and granulocytes in LTBI and TB. We found that for 39 BTMs, the average correlation coefficients for the genes in that module were significantly different from 0 (at q < 0.05) in a randomization test. Several of these modules were related to the immune response, including “regulation of antigen presentation and immune response,” “enriched in neutrophils,” and “immune regulation—monocytes, T and B cells.” Interestingly, 24 annotated modules, including modules related to antigen processing and presentation, showed a significant average correlation coefficient (at q < 0.05) of differences between LTBI and TB (Table 2), indicating that disease-specific expression of genes in these modules is connected to differential methylation.
TABLE 2 .
Blood transcriptional modules with significant average correlation coefficients between the differences in DNA methylation and gene expression in comparison of LTBI and TBa
| Module ID | Module title | r | q value |
|---|---|---|---|
| LI.M200 | Antigen processing and presentation | 0.25 | 3.86E−16 |
| LI.M95.0 | Enriched in antigen presentation (II) | 0.22 | 3.43E−14 |
| DC.M8.83 | Immune responses | 0.22 | 1.68E−10 |
| LI.M71 | Enriched in antigen presentation (I) | 0.15 | 9.94E−08 |
| LI.M37.0 | Immune activation—generic cluster | −0.07 | 5.50E−06 |
| LI.M17.3 | Hox cluster IV | −0.25 | 9.40E−06 |
| LI.S2 | B cell surface signature | 0.07 | 1.06E−05 |
| LI.M146 | MHC-TLR7-TLR8 cluster | 0.09 | 0.0002 |
| LI.M168 | Enriched in dendritic cells | −0.28 | 0.001 |
| LI.M5.0 | Regulation of antigen presentation and immune response | 0.09 | 0.002 |
| LI.M17.1 | Hox cluster II | −0.03 | 0.003 |
| LI.M17.0 | Hox cluster I | −0.19 | 0.003 |
| LI.M96 | Hox cluster V | −0.03 | 0.004 |
| DC.M5.15 | Neutrophils | −0.11 | 0.005 |
| LI.M24 | Cell activation (IL-15, IL-23, TNF) | −0.18 | 0.009 |
| LI.M74 | Transcriptional targets of glucocorticoid receptor | −0.12 | 0.02 |
| DC.M4.13 | Inflammation | −0.18 | 0.02 |
| LI.M160 | Leukocyte differentiation | −0.09 | 0.02 |
| LI.M37.1 | Enriched in neutrophils (I) | −0.26 | 0.03 |
| LI.M75 | Antiviral IFN signature | −0.14 | 0.03 |
| DC.M4.2 | Inflammation | −0.22 | 0.03 |
| LI.M112.0 | Complement activation (I) | −0.11 | 0.04 |
| DC.M3.2 | Inflammation | −0.18 | 0.04 |
| LI.M57 | Immunoregulation—monocytes, T and B cells | −0.21 | 0.04 |
“Module ID” refers to the original publication, where the prefix “LI” refers to BTMs according to Li et al. (17), and the prefix “DC” refers to BTMs according to Chaussabel et al. (18). Only modules with a functional annotation are shown. r is the average correlation coefficient in the module, and the q value is the P value in a randomization test corrected for multiple testing. IFN, interferon; IL, interleukin; TNF, tumor necrosis factor.
Furthermore, we applied an alternative approach to elucidate whether a functional link between methylation and the transcriptome exists. We calculated the correlation between methylation and gene expression for each pair consisting of a CpG and a matched gene. Genes were ordered by their highest correlation coefficient with any matched CpG, and enrichment in BTMs was calculated. Thirty modules were significantly enriched at q < 0.05, including “enriched in monocytes,” “enriched in neutrophils,” and “immune activation—generic cluster.”
Using this approach, we further directly investigated whether differences in methylation between LTBI and TB within a matched-pair design had an effect on changes in gene expression. For each gene, the highest correlation of differences was calculated between matched samples in LTBI and TB in methylation and gene expression and tested for enrichment in BTMs accordingly. We found enrichment in 18 modules at q < 0.05, including “enriched in neutrophils,” “inflammation,” and “enriched in monocytes.” These results confirm our findings and demonstrate that differences in methylation between LTBI and TB are functionally linked to differences in gene expression relevant to the immune response.
Effect of DNA methylation on protein synthesis.
The effect of changes in gene expression on the abundance of the final protein product is moderated by various factors (35), and any effect of DNA methylation on protein abundance will be exerted via transcriptional changes. Therefore, we interrogated whether observed changes in DNA methylation could be linked to changes in protein abundance.
Applying the same approaches described above, we first calculated the per-BTM average correlation coefficients and identified modules showing a significant average correlation. Second, we determined module enrichment in genes ordered by their correlation with methylation sites.
The first approach (average correlations in a BTM) revealed that for the general correlation, 22 modules had significant correlation coefficients, including “cell cycle and transcription,” “enriched in monocytes,” and “interferon.” Moreover, several modules showed significant average correlations of changes in methylation and protein abundance between LTBI and TB. These modules included “cell cycle and translation,” “enriched in monocytes,” “T cell activation,” and “inflammation.”
In the second approach, we tested the genes ordered by their respective correlation coefficients for enrichment in BTMs. Here, possibly due to low statistical power, we identified “immune activation—generic cluster” for both general correlations and correlations of differences between LTBI and TB and “enriched in monocytes” for general correlation (Table 3). In any case, these results confirm that disease-specific differences in methylation are correlated with protein expression.
TABLE 3 .
Blood transcriptional modules with significant average correlation coefficients between the differences in DNA methylation and protein abundance in comparison of latent M. tuberculosis infection and TBa
| Module ID | Module title | r | q value |
|---|---|---|---|
| LI.M37.0 | Immune activation—generic cluster | −0.27 | 1.08E−06 |
| LI.M7.4 | T cell activation (III) | 0.07 | 0.01 |
| LI.M4.0 | Cell cycle and transcription | −0.15 | 0.03 |
| LI.M11.0 | Enriched in monocytes (II) | −0.14 | 0.03 |
| DC.M4.2 | Inflammation | −0.28 | 0.03 |
| DC.M4.14 | Monocytes | −0.25 | 0.04 |
“Module ID” refers to the original publication, where the prefix “LI” refers to BTMs according to Li et al. (17), and the prefix “DC” refers to BTMs according to Chaussabel et al. (18). Only modules with a functional annotation are shown. r is the average correlation coefficient in the module, and the q value is the P value in a randomization test corrected for multiple testing.
Conclusions.
To date, defining the diagnostic biosignatures of TB has been largely based on gene expression analysis. However, the complexity of gene expression regulation is greatly simplified in approaches where only the transcriptome of a mixture of cells is analyzed. We dissected a fragment of regulation of transcriptomic changes in professional phagocytes of TB patients, elucidating the underlying biological mechanisms and paths of gene regulation. For the first time, parallel analysis of the DNA methylome, transcriptome (mRNA and miR), and proteome revealed disease-specific changes permeating these levels of regulation, commencing with methylation marks on the DNA. We demonstrate that methylation events can distinguish TB disease from healthy, infected LTBI in this sample set. These events are functionally related to unique immune-relevant classes and are manifested on both transcriptomic and proteomic levels. Although the low number of samples constrained our investigation to general effects rather than specific regulatory mechanisms, this study paves the way for further detailed investigations that interrogate the DNA methylome and proteome in addition to the transcriptome of TB patients. These future investigations would necessarily entail also the analysis of professional phagocytes in healthy subjects neither latently infected nor with active TB, allowing a comparison and biomarker identification between the two healthy groups. Whether the observed differences from each of the platforms as well as the correlated platforms will be appreciated in a large sample set representative of a population remains to be shown. We cannot ascertain whether the observed changes in methylation causally contribute to risk of TB and whether the altered methylation patterns are a cause or effect of disease. Given that the differences observed in the DNA methylome are related to functional differences observed in both transcriptome and proteome, we suggest that studying the epigenome can bring us closer to defining biomarkers of predisposition to disease, as well as uniquely contribute to our understanding of TB pathogenesis.
MATERIALS AND METHODS
Ethics statement.
Blood samples (20 ml) were collected from participants following written consent (ethical approval from Stellenbosch University N10/08/274).
Clinical procedures and sample isolation.
Samples from patients diagnosed with active TB (n = 8) and LTBI participants (n = 8) were obtained from an area of high TB endemicity in Cape Town, South Africa (Ravensmead and Uitsig), conforming to International Conference on Harmonisation good clinical practice (ICH-GCP) procedures. TB participants were included following a chest X ray suggestive of active disease in combination with symptoms of active TB and a confirmed positive M. tuberculosis culture result. LTBI participants were recruited to match TB patients’ age, gender, and ethnicity and were confirmed as controls based on a negative chest X-ray, the presence of no signs of active TB, and a negative M. tuberculosis culture. Both TB and LTBI participants were HIV negative. LTBI participants for this study were not followed up longitudinally.
Blood was collected from patients presenting with TB symptoms (n = 8), who were recruited on the day of diagnosis. Subsequently, blood was collected from LTBI participants who were recruited to match the patients’ age and gender. Granulocytes and monocytes were sequentially separated from peripheral blood with magnetic beads by magnetically activated cell sorting (MACS) (Miltenyi Biotec GmbH) (CD15+ and CD14+, respectively) according to the manufacturer’s instructions. Total RNA (tRNA), genomic DNA (gDNA), and protein were isolated using TRIzol reagent (Life Technologies Corporation) according to the manufacturer’s instructions. The quality and quantity of nucleic acids were determined by electrophoresis (Agilent 2100, BioAnalyser; Agilent Technologies) and spectrophotometry (NanoDrop 2000c; Thermo Scientific).
DNA methylation.
To investigate DNA methylation marks on CpGs, genomic DNA (500 ng) was bisulfate converted using the EZ-96 DNA methylation kit (Zymo Research Corporation) and whole-genome amplified, fragmented enzymatically, and then applied to the arrays. After extension, arrays were fluorescently stained and scanned, and the intensities of the nonmethylated and methylated bead types were measured using the Infinium human methylation450K BeadChip (Illumina). DNA methylation values (described as β values and expressed as fractions of the total number of available oligomers on the bead to anneal to) were recorded for each locus in each sample and analyzed using the software GenomeStudio (Illumina, Genomestudio 2011.1, methylation module 1.9.0). The raw data of the microarrays were uploaded to Gene Expression Omnibus.
Transcription.
mRNA, long intergenic noncoding RNA (lincRNA), and miR abundances were measured using microarrays from Agilent Technologies (human 8-by-60,000 custom layout design 041580 containing the whole human genome), novel content for lincRNAs (from Agilent-028004), and Broad Institute human lincRNA together with Broad Institute TUCP transcripts (from Agilent-039494) and 8-by-60,000 (release 16) unrestricted human miR microarrays (Agilent-031181). Sample labeling and microarray processing were done according to the manufacturer’s instructions, and features were extracted with Agilent Feature Extraction 11.5.1.1 using the recommended protocols and settings. Data were background corrected and normalized using the R package limma version 3.20. To detect differentially expressed genes, we used the linear models in limma and the moderated t statistic (36). The differences included pairwise comparisons between LTBI and TB samples for monocytes and granulocytes separately and pairwise comparisons between monocytes and granulocytes for LTBI and TB separately, as well as testing the significance of the interaction between cell type and disease status. The raw data of the microarrays were uploaded to the Gene Expression Omnibus.
Proteomics.
Protein pellets were resuspended in lysis buffer containing 8 M urea, RapiGest (Waters), and ammonium bicarbonate. Proteins were reduced and alkylated, followed by a tryptic digest. The peptide solution was desalted by C18 reverse-phase chromatography, vacuum dried, and resolubilized to a final concentration of 1 mg/ml. Each peptide sample was analyzed on a Thermo Easy-nLC 1000 high-performance liquid chromatography (HPLC) system connected to an Orbitrap Elite mass spectrometer, which was equipped with a nanoelectrospray ion source (Thermo Scientific). Peptides were separated on a 15-cm Acclaim PepMap rapid separation liquid chromatography (RSLC) column (75-µm inner diameter, 2-µm particle size; Thermo, Fisher Scientific) at a flow rate of 300 nl·min−1. Mass spectrometry (MS) spectra were acquired in the Orbitrap with a resolution of 120,000, and tandem MS (MS/MS) spectra were acquired in the linear ion trap at normal scan speed following collision-induced dissociation of the 10 most abundant precursors per cycle (normalized collision energy, 35%). We performed label-free quantification (LFQ) using Progenesis 4.0 (Nonlinear Dynamics) by automatic alignment of total ion chromatograms of raw files, using imported pep.xml files from Sequest searches against the human UniProtKB/Swiss-Prot protein database. The search identifications were statistically scored using PeptideProphet (37) within the TPP (38), and all peptides with an iProbability score above 0.9 were considered resulting in a protein false discovery rate (FDR) of 1%. After manually improving the alignment, quantified peaks were filtered for identification by sequence search, and overall protein abundances were calculated thereof. The mass spectrometry discovery peptidomics data have been deposited into the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.
Statistical analysis and systems biological approaches.
Unless otherwise stated, statistical analyses were performed as follows: data were tested in each platform’s quality control pipeline. These were then tested for normality, described in general, followed by differential analyses employing Wilcoxon tests (DNA methylation) and limma (gene expression, miR, and proteomics), with correction for multiple testing according to Benjamini-Hochberg (39). For each platform, we performed a PCA and estimated the contribution of controlled variables in explaining the variance of the PCA components by applying PCA regression. For machine learning, random forests were applied as implemented in the R package randomForest version 4.10 (40). Unless otherwise stated, for statistical tests, we used the significance threshold of q < 0.05, where q is the P value corrected for the family-wise error rate using the Benjamini-Hochberg method (39).
To validate the predictive power of various platforms to differentiate between TB and LTBI, we have applied the random forest ML method (R package randomForest version 4.6 [40]), cross-validated with a modified leave-one-out (LOO) scheme. Here, at each iteration of the LOO, we have removed from the training a set of matching samples (monocytes and granulocytes) from one TB patient and the matching LTBI control. We have used the remaining samples as the training set and applied the trained model to the four test samples. The variable set used to train the model was the full set, except for the methylation platform, in which we used the filtered variable set. The results are reported as AUC and 95% CIs.
To functionally annotate results of statistical tests, we used the R software package tmod version 0.19 (available from CRAN; http://cran.r-project.org/web/packages/tmod/index.html), with BTMs as described by Li et al. (17) and Chaussabel et al. (41). Depending on context, we used either a hypergeometric test for enrichment of modules in a set of differentially regulated genes compared to the genetic background or U summed rank statistics for enrichment in modules in an ordered list of genes. All procedures and R scripts required for replication of results are available upon request.
Microarray data accession number.
The raw data from the microarrays have been uploaded to the Gene Expression Omnibus (GEO) under SuperSeries accession no. GSE70478. The mass spectrometry discovery peptidomics data have been deposited into the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001960.
SUPPLEMENTAL MATERIAL
Density distribution of methylation data in monocytes (A) and granulocytes (B). Horizontal axis, methylation, where 0 indicates no methylation at the locus and 1 indicates full methylation. Dark gray bars depict data from TB, white bars depict data from LTBI, and light gray reflects overlap of both phenotypic states. (C) Stratification of differentially methylated CpGs in the filtered set between LTBI and TB (green slice of pie); (D) stratification of CpGs differentially methylated between monocytes and granulocytes (green slice of pie). Square box, stratification of differentially methylated CpGs; red, CpG islands; green, north (N) shelves; purple, N shores; cyan, south (S) shelves; orange, S shores; gray, remaining CpGs not related to CpG islands, shores, or shelves. Download
Gene expression in professional phagocytes in LTBI and TB. (A) Variance decomposition of the principal component (PC) analysis (PCA) of the full data set. Variance (black line) in the data set was broken down into known sources of variance within each component of PCA, illustrating the majority of variance being explained in the first component, of which cell type explained most of the variance. (B) Heat map showing correlation coefficients between the samples, from low (red) to high (yellow). Samples were clustered using hierarchical clustering. Sample code: for the first digit, T represents TB and C represents LTBI, for the second-to-last digit, N represents granulocytes and M represents monocytes, and for the last digit, M represents male and F represents female. (C) PCA of gene expression data set illustrating major sources of variance in PC1, PC2, and PC5. Green, LTBI; yellow, TB; spheres, monocytes; cubes, granulocytes. Download
Overall analysis of microRNA (miR) microarray data. (A) Variance decomposition of the PCA of the full data set. Variance (black line) in the methylation data set is broken down into the known sources of variance within each component of PCA, illustrating the majority of variance being explained in the first two components, of which gender and cell type explains most of the components. (B) Heat map showing correlation coefficients between the samples, from low (red) to high (yellow). Samples were clustered using hierarchical clustering. Sample code: for the first digit, T represents TB and C represents LTBI, for the second-to-last digit, N represents granulocytes and M represents monocytes, and for the last digit, M represents male and F represents female. (C) PCA of methylation data sets. Green, LTBI; yellow, TBA; spheres, monocytes; cubes, granulocytes. Download
miR-146a-5p data illustrating (A) significant differential expression between both monocytes and neutrophils of LTBI and TB active (TBA), measured in a microarray for two different probes. Blue and red are the respective probes (B). These results were interrogated using a qRT-PCR TaqMan assay of miR-146a-5p in monocytes from these individuals and more from the same cohort. CD14+ (n = 10), TB active (n = 10). RNU6B was used as the reference gene. P = 0.055, Wilcoxon paired test. Download
Functional enrichment of hypomethylated CpGs in blood transcriptional modules (BTMs), compared with total genetic background. Enrichment represents the proportion of CpGs associated with the given module in the set of hypomethylated sites divided by the overall proportion of CpGs associated with the given module. The q value is the P value from hypergeometric test corrected for multiple testing.
Differential analysis of Infinium450K results reporting DNA methylation status of CpGs in monocytes and granulocytes. Shown are results from a Wilcoxon paired rank test. The q values represent P values after correction for multiple testing using the Benjamini-Hochberg procedure.
Most significant 10% of results depicting global differential expression between LTBI and TB participants’ monocytes or granulocytes alone and monocytes and granulocytes combined. Probe names are the probe IDs from the Agilent human 8-by-60,000 array; the gene name and systematic name are probe names mapped onto assembly GRCh38.p2 (Ensembl database). Log fold change (LFC), difference between LTBI and TB calculated with limma, with the q value representing the P value after Bonferroni correction. LFC < 0 indicates overexpression in TB. Combined results represent the limma test for differential expression in both monocytes and granulocytes. A full table of results is available on request. Colored text marks q values that are <0.05.
Blood transcriptional module (BTM) enrichment analysis of genes differentially expressed between monocytes and granulocytes. “Title” represents the title of the blood transcription module according to references 17 and 18. The prefix DC represents BTMs according to Chaussabel et al. (18), and the prefix LI represents BTMs according to Li et al. (17). N1, number of genes in a module; AUC, area under the curve. The q value represents the adjusted P value after correction for multiple testing (Benjamini-Hochberg). Only modules with a functional annotation are shown. Other abbreviations: BAFF, B-cell activating factor; IFN, interferon; IRF2, interferon regulatory factor 2; LPS, lipopolysaccharide; MHC, major histocompatibility complex; NK, natural killer; TF, transcription factor; TLR, Toll-like receptor.
Results depicting global differential expression in miRs between LTBI and TB participants’ monocytes or granulocytes alone and monocytes and granulocytes combined. The probe name represents probe IDs from Agilent human 8-by-60,000 miR array, and the gene name represents the probe names mapped onto miRs according to assembly GRCh38.p2 (Ensembl database). LFC, difference between LTBI and TB calculated with limma, with the q value representing the P value after Bonferroni correction. LFC < 0 indicates overexpression in TB. Combined results represent the limma test for differential expression in both monocytes and granulocytes. A full table of results is available on request. Colored text marks q values that are <0.05.
Results depicting translational differences between LTBI and TB participants’ monocytes or granulocytes alone and monocytes and granulocytes combined. The gene name and systematic name represent the probe IDs mapped onto human genome assembly GRCh38.p2 (Ensembl database). LFC, log fold change between LTBI and TB with limma, with the q value representing the P value after Bonferroni correction. LFC < 0 indicates overexpression in TB. Combined results represent the limma test for differential expression in both monocytes and granulocytes. A full table of results is available on request. Colored text marks q values that are <0.05.
ACKNOWLEDGMENTS
We thank all study participants for participating in the study, Christoph Bock for critical reading of the manuscript, Laura Lozza, Silvana Gromoeller, and Joachim Schmidt for help, the nursing staff for collecting samples, Robert Golinski for logistical assistance, and Mary Louise Grossman for excellent help in preparation of the manuscript.
This project has received funding from the European Union’s Research and Innovation Programme Horizon 2020 (grant no. 643381), the European Union’s Seventh Framework Programmes ADITEC (FP7/2007-2013, grant no. 280873) and SysteMTb (HEALTH-F3-2009-241587), the Innovative Medicines Initiative Joint Undertaking “Biomarkers for Enhanced Vaccine Safety” project BioVacSafe (IMI JU grant no. 115308), the The European and Developing Countries Clinical Trials Partnership (EDCTP) project “African European Tuberculosis Consortium” (AE-TBC), and SystemsX.ch (2013/154).
M.M.E., H.L., R.A., G.W., and S.H.E.K. designed the research, M.M.E., M.I., A.G.L., C.W., K.S., E.C., H.-J.M. and M.S. performed the research, M.M.E., J.W., and W.E.W. analyzed the data, and M.M.E., S.H.E.K., J.W., P.S., and H.L. wrote the article.
Footnotes
Citation Esterhuyse MM, Weiner J, 3rd, Caron E, Loxton AG, Iannaccone M, Wagman C, Saikali P, Stanley K, Wolski WE, Mollenkopf H-J, Schick M, Aebersold R, Linhart H, Walzl G, Kaufmann SHE. 2015. Epigenetics and proteomics join transcriptomics in the quest for tuberculosis biomarkers. mBio 6(5):e01187-15. doi:10.1128/mBio.01187-15.
REFERENCES
- 1.WHO 2014. Global tuberculosis report 2014. WHO Press, Geneva, Switzerland. [Google Scholar]
- 2.Abubakar I, Zignol M, Falzon D, Raviglione M, Ditiu L, Masham S, Adetifa I, Ford N, Cox H, Lawn SD, Marais BJ, McHugh TD, Mwaba P, Bates M, Lipman M, Zijenah L, Logan S, McNerney R, Zumla A, Sarda K, Nahid P, Hoelscher M, Pletschette M, Memish ZA, Kim P, Hafner R, Cole S, Migliori GB, Maeurer M, Schito M, Zumla A. 2013. Drug-resistant tuberculosis: time for visionary political leadership. Lancet Infect Dis 13:529–539. doi: 10.1016/S1473-3099(13)70030-6. [DOI] [PubMed] [Google Scholar]
- 3.Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. 2011. Immunological biomarkers of tuberculosis. Nat Rev Immunol 11:343–354. doi: 10.1038/nri2960. [DOI] [PubMed] [Google Scholar]
- 4.Ottenhoff TH, Kaufmann SH. 2012. Vaccines against tuberculosis: where are we and where do we need to go? PLoS Pathog 8:e1002607. doi: 10.1371/journal.ppat.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebeis SL, Kalman D. 2009. Aligning antimicrobial drug discovery with complex and redundant host-pathogen interactions. Cell Host Microbe 5:114–122. doi: 10.1016/j.chom.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Romieu I, Samet JM, Smith KR, Bruce N. 2002. Outdoor air pollution and acute respiratory infections among children in developing countries. J Occup Environ Med 44:640–649. doi: 10.1097/00043764-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Marcos A, Nova E, Montero A. 2003. Changes in the immune system are conditioned by nutrition. Eur J Clin Nutr 57(Suppl 1):S66–S69. doi: 10.1038/sj.ejcn.1601819. [DOI] [PubMed] [Google Scholar]
- 8.Kant S, Gupta H, Ahluwalia S. 2015. Significance of nutrition in pulmonary tuberculosis. Crit Rev Food Sci Nutr 55:955–963. doi: 10.1080/10408398.2012.679500. [DOI] [PubMed] [Google Scholar]
- 9.Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA, Sharifi N, Janssen-Megens EM, Ter Huurne M, Mandoli A, van Schaik T, Ng A, Burden F, Downes K, Frontini M, Kumar V, Giamarellos-Bourboulis EJ, Ouwehand WH, van der Meer JW, Joosten LA, Wijmenga C, Martens JH, Xavier RJ, Logie C, Netea MG, Stunnenberg HG. 2014. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, Xavier RJ, van der Meer JW, van Crevel R, Netea MG. 2012. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A 109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maertzdorf J, Kaufmann SH, Weiner J III. 2015. Toward a unified biosignature for tuberculosis. Cold Spring Harb Perspect Med 5:a018531. doi: 10.1101/cshperspect.a018531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blankley S, Berry MP, Graham CM, Bloom CI, Lipman M, O’Garra A. 2014. The application of transcriptional blood signatures to enhance our understanding of the host response to infection: the example of tuberculosis. Philos Trans R Soc Lond B Biol Sci 369:20130427. doi: 10.1098/rstb.2013.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad S. 2011. Pathogenesis, immunology, and diagnosis of latent Mycobacterium tuberculosis infection. Clin Dev Immunol 2011:814943. doi: 10.1155/2011/814943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilbauer M, Rayner TF, Clark C, Coffey AJ, Joyce CJ, Palta P, Palotie A, Lyons PA, Smith KG. 2013. Genome-wide methylation analyses of primary human leukocyte subsets identifies functionally important cell-type-specific hypomethylated regions. Blood 122:e52–e60. doi: 10.1182/blood-2013-05-503201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. 2011. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood 117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 16.Bock C. 2012. Analysing and interpreting DNA methylation data. Nat Rev Genet 13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, Kasturi S, Carlone GM, Quinn C, Chaussabel D, Palucka AK, Mulligan MJ, Ahmed R, Stephens DS, Nakaya HI, Pulendran B. 2014. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, Bennett L, Allantaz F, Mejias A, Ardura M, Kaizer E, Monnet L, Allman W, Randall H, Johnson D, Lanier A, Punaro M, Wittkowski KM, White P, Fay J, Klintmalm G, Ramilo O, Palucka AK, Banchereau J, Pascual V. 2008. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, Söderhäll C, Scheynius A, Kere J. 2012. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baena A, Porcelli SA. 2009. Evasion and subversion of antigen presentation by Mycobacterium tuberculosis. Tissue Antigens 74:189–204. doi: 10.1111/j.1399-0039.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding CV, Boom WH. 2010. Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat Rev Microbiol 8:296–307. doi: 10.1038/nrmicro2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgado JC, Baena A, Thim S, Goldfeld AE. 2006. Aspartic acid homozygosity at codon 57 of HLA-DQ beta is associated with susceptibility to pulmonary tuberculosis in Cambodia. J Immunol 176:1090–1097. doi: 10.4049/jimmunol.176.2.1090. [DOI] [PubMed] [Google Scholar]
- 23.Theng SS, Wang W, Mah WC, Chan C, Zhuo J, Gao Y, Qin H, Lim L, Chong SS, Song J, Lee CG. 2014. Disruption of FAT10-MAD2 binding inhibits tumor progression. Proc Natl Acad Sci U S A 111:E5282–E5291. doi: 10.1073/pnas.1403383111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodridge JP, Burian A, Lee N, Geraghty DE. 2013. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol 191:3553–3562. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maertzdorf J, Weiner J III, Mollenkopf HJ, Bauer T, Prasse A, Muller-Quernheim J, Kaufmann SH. 2012. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A 109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cliff JM, Lee JS, Constantinou N, Cho JE, Clark TG, Ronacher K, King EC, Lukey PT, Duncan K, Van Helden PD, Walzl G, Dockrell HM. 2013. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis 207:18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 27.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.So AY, Zhao JL, Baltimore D. 2013. The Yin and Yang of microRNAs: leukemia and immunity. Immunol Rev 253:129–145. doi: 10.1111/imr.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spinelli SV, Diaz A, D’Attilio L, Marchesini MM, Bogue C, Bay ML, Bottasso OA. 2013. Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol Immunol 53:265–269. doi: 10.1016/j.molimm.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. 2009. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum 60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 31.Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. 2009. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One 4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Morgan M, Hutchison K, Calhoun VD. 2010. A study of the influence of sex on genome wide methylation. PLoS One 5:e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. 2011. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 6:623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaghlool SB, Al-Shafai M, Al Muftah WA, Kumar P, Falchi M, Suhre K. 2015. Association of DNA methylation with age, gender, and smoking in an Arab population. Clin Epigenetics 7:6. doi: 10.1186/s13148-014-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 36.Smyth GK. 2005. Limma: linear models for microarray data, p 397–420. In Gentelmen R, Carey V, Dudoit S, Irizarry R, Huber W (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer Verlag, New York, NY. [Google Scholar]
- 37.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 38.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. 2005. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol 1:2005.0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- 40.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 41.Chaussabel D, Pascual V, Banchereau J. 2010. Assessing the human immune system through blood transcriptomics. BMC Biol 8:84. doi: 10.1186/1741-7007-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Density distribution of methylation data in monocytes (A) and granulocytes (B). Horizontal axis, methylation, where 0 indicates no methylation at the locus and 1 indicates full methylation. Dark gray bars depict data from TB, white bars depict data from LTBI, and light gray reflects overlap of both phenotypic states. (C) Stratification of differentially methylated CpGs in the filtered set between LTBI and TB (green slice of pie); (D) stratification of CpGs differentially methylated between monocytes and granulocytes (green slice of pie). Square box, stratification of differentially methylated CpGs; red, CpG islands; green, north (N) shelves; purple, N shores; cyan, south (S) shelves; orange, S shores; gray, remaining CpGs not related to CpG islands, shores, or shelves. Download
Gene expression in professional phagocytes in LTBI and TB. (A) Variance decomposition of the principal component (PC) analysis (PCA) of the full data set. Variance (black line) in the data set was broken down into known sources of variance within each component of PCA, illustrating the majority of variance being explained in the first component, of which cell type explained most of the variance. (B) Heat map showing correlation coefficients between the samples, from low (red) to high (yellow). Samples were clustered using hierarchical clustering. Sample code: for the first digit, T represents TB and C represents LTBI, for the second-to-last digit, N represents granulocytes and M represents monocytes, and for the last digit, M represents male and F represents female. (C) PCA of gene expression data set illustrating major sources of variance in PC1, PC2, and PC5. Green, LTBI; yellow, TB; spheres, monocytes; cubes, granulocytes. Download
Overall analysis of microRNA (miR) microarray data. (A) Variance decomposition of the PCA of the full data set. Variance (black line) in the methylation data set is broken down into the known sources of variance within each component of PCA, illustrating the majority of variance being explained in the first two components, of which gender and cell type explains most of the components. (B) Heat map showing correlation coefficients between the samples, from low (red) to high (yellow). Samples were clustered using hierarchical clustering. Sample code: for the first digit, T represents TB and C represents LTBI, for the second-to-last digit, N represents granulocytes and M represents monocytes, and for the last digit, M represents male and F represents female. (C) PCA of methylation data sets. Green, LTBI; yellow, TBA; spheres, monocytes; cubes, granulocytes. Download
miR-146a-5p data illustrating (A) significant differential expression between both monocytes and neutrophils of LTBI and TB active (TBA), measured in a microarray for two different probes. Blue and red are the respective probes (B). These results were interrogated using a qRT-PCR TaqMan assay of miR-146a-5p in monocytes from these individuals and more from the same cohort. CD14+ (n = 10), TB active (n = 10). RNU6B was used as the reference gene. P = 0.055, Wilcoxon paired test. Download
Functional enrichment of hypomethylated CpGs in blood transcriptional modules (BTMs), compared with total genetic background. Enrichment represents the proportion of CpGs associated with the given module in the set of hypomethylated sites divided by the overall proportion of CpGs associated with the given module. The q value is the P value from hypergeometric test corrected for multiple testing.
Differential analysis of Infinium450K results reporting DNA methylation status of CpGs in monocytes and granulocytes. Shown are results from a Wilcoxon paired rank test. The q values represent P values after correction for multiple testing using the Benjamini-Hochberg procedure.
Most significant 10% of results depicting global differential expression between LTBI and TB participants’ monocytes or granulocytes alone and monocytes and granulocytes combined. Probe names are the probe IDs from the Agilent human 8-by-60,000 array; the gene name and systematic name are probe names mapped onto assembly GRCh38.p2 (Ensembl database). Log fold change (LFC), difference between LTBI and TB calculated with limma, with the q value representing the P value after Bonferroni correction. LFC < 0 indicates overexpression in TB. Combined results represent the limma test for differential expression in both monocytes and granulocytes. A full table of results is available on request. Colored text marks q values that are <0.05.
Blood transcriptional module (BTM) enrichment analysis of genes differentially expressed between monocytes and granulocytes. “Title” represents the title of the blood transcription module according to references 17 and 18. The prefix DC represents BTMs according to Chaussabel et al. (18), and the prefix LI represents BTMs according to Li et al. (17). N1, number of genes in a module; AUC, area under the curve. The q value represents the adjusted P value after correction for multiple testing (Benjamini-Hochberg). Only modules with a functional annotation are shown. Other abbreviations: BAFF, B-cell activating factor; IFN, interferon; IRF2, interferon regulatory factor 2; LPS, lipopolysaccharide; MHC, major histocompatibility complex; NK, natural killer; TF, transcription factor; TLR, Toll-like receptor.
Results depicting global differential expression in miRs between LTBI and TB participants’ monocytes or granulocytes alone and monocytes and granulocytes combined. The probe name represents probe IDs from Agilent human 8-by-60,000 miR array, and the gene name represents the probe names mapped onto miRs according to assembly GRCh38.p2 (Ensembl database). LFC, difference between LTBI and TB calculated with limma, with the q value representing the P value after Bonferroni correction. LFC < 0 indicates overexpression in TB. Combined results represent the limma test for differential expression in both monocytes and granulocytes. A full table of results is available on request. Colored text marks q values that are <0.05.
Results depicting translational differences between LTBI and TB participants’ monocytes or granulocytes alone and monocytes and granulocytes combined. The gene name and systematic name represent the probe IDs mapped onto human genome assembly GRCh38.p2 (Ensembl database). LFC, log fold change between LTBI and TB with limma, with the q value representing the P value after Bonferroni correction. LFC < 0 indicates overexpression in TB. Combined results represent the limma test for differential expression in both monocytes and granulocytes. A full table of results is available on request. Colored text marks q values that are <0.05.