ABSTRACT
The Middle East respiratory syndrome coronavirus (MERS-CoV) causes a severe acute respiratory tract infection with a high fatality rate in humans. Coronaviruses are capable of infecting multiple species and can evolve rapidly through recombination events. Here, we report the complete genomic sequence analysis of a MERS-CoV strain imported to China from South Korea. The imported virus, provisionally named ChinaGD01, belongs to group 3 in clade B in the whole-genome phylogenetic tree and also has a similar tree topology structure in the open reading frame 1a and -b (ORF1ab) gene segment but clusters with group 5 of clade B in the tree constructed using the S gene. Genetic recombination analysis and lineage-specific single-nucleotide polymorphism (SNP) comparison suggest that the imported virus is a recombinant comprising group 3 and group 5 elements. The time-resolved phylogenetic estimation indicates that the recombination event likely occurred in the second half of 2014. Genetic recombination events between group 3 and group 5 of clade B may have implications for the transmissibility of the virus.
IMPORTANCE
The recent outbreak of MERS-CoV in South Korea has attracted global media attention due to the speed of spread and onward transmission. Here, we present the complete genome of the first imported MERS-CoV case in China and demonstrate genetic recombination events between group 3 and group 5 of clade B that may have implications for the transmissibility of MERS-CoV.
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV), first detected in the Kingdom of Saudi Arabia (KSA) in 2012, causes severe acute respiratory tract infection in humans, with a high case fatality rate (CFR) (1–4). Dromedary camels are believed to be important reservoir hosts or vectors for human infection; bats may also be implicated (5–8). As of 17 July 2015, 1,368 laboratory-confirmed cases of human infection with MERS-CoV had been reported to the World Health Organization (WHO), including at least 490 deaths, corresponding to a CFR as high as 35.45% (9). Recent MERS clusters in South Korea are thought to be the largest outbreak outside the Middle East countries (10). As of 25 July 2015, 186 laboratory-confirmed cases of MERS-CoV infection have been confirmed (including 36 deaths) in South Korea (9). A South Korean man who was a relative of some of the laboratory-confirmed cases traveled to Guangdong Province (10) and was diagnosed as the first imported MERS-CoV case in China by molecular detection of MERS-CoV (11, 12). The rapid spread of disease in South Korea raised concerns that the imported virus had evolved to become more transmissible. Here, we report a comprehensive phylogenetic analysis of the complete MERS-CoV genome sequence of the first Chinese imported case of MERS (ChinaGD01), and the results indicate its probable origin and show evidence of genetic recombination.
RESULTS
Patient and sample history.
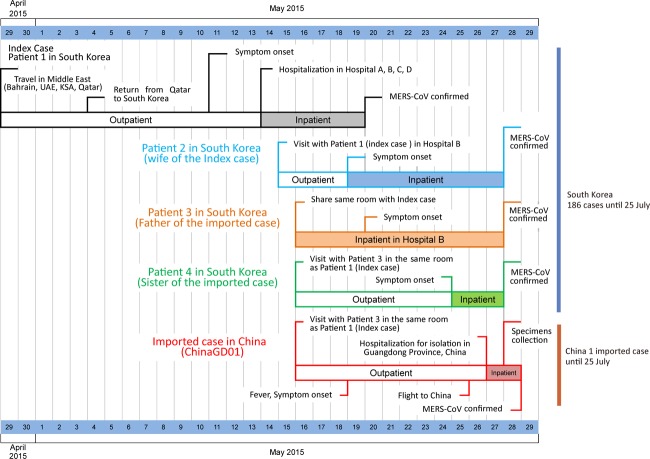
The current outbreak in South Korea and China was initiated when a 68-year-old Korean man flew back to Seoul on 4 May 2015 after a visit to four Middle East countries (Bahrain, United Arab Emirates, Saudi Arabia, and Qatar). On 26 May 2015, a 44-year-old South Korean man presented with fever to a hospital in Guangdong. He was in close contact with the index patient in South Korea on 16 May 2015 (Fig. 1), as well as a suspected second-generation patient. The timeline of the travel history, potential virus exposure, onset of disease, and diagnosis of the first imported MERS-CoV case in China are presented in Fig. 1.
FIG 1 .
Timeline of the travel history, potential virus exposure, onset of disease, and diagnosis of the first imported MERS-CoV case in China. UAE, United Arab Emirates; KSA, Kingdom of Saudi Arabia.
Characterization of genome.
With informed consent and the approval of the ethical committee of the National Institute of Viral Disease Control and Prevention, China Center for Disease Control and Prevention (CDC), nasopharyngeal swabs were collected and used for RNA extraction, followed by reverse transcription PCR and genome sequencing. Through both Sanger and Ion Torrent sequencing, the full-length virus genome (30,144 bp) of ChinaGD01 was obtained and deposited in GenBank (accession no. KT006149). Over 2,000,000 paired-end reads were quality trimmed and processed to remove human genome sequences. Nonhuman reads were assembled into contigs by CLC Genomic Workbench and aligned against representative sequences of MERS-CoV. No nucleotide insertions or deletions were observed in the genome.
The genome sequence of this virus, referred to as ChinaGD01, had high levels of nucleotide identity (99.33% to 99.79%) to previously published MERS-CoV genomes (Fig. 2), with 99.31% to 99.78% sequence identity in the open reading frame 1a and -b (ORF1ab) gene segment and 98.91% to 99.60% identity in the S gene. The E, M, and N genes had 98.93% to 100% identity with previously described MERS-CoV strains. In total, ChinaGD01 possessed 11 nonsynonymous nucleotide substitutions (Table 1), which occurred in the ORF1ab (n = 8), ORF3 (n = 1), ORF4b (n = 1), and M (n = 1) genes, respectively (Table 1). Although there were five nucleotide substitutions in the S gene, no amino acid change was discovered. Of note, in comparison with previously published MERS-CoV genomes, the ChinaGD01 genome shows 11 unique amino acid substitutions, and 8 of them were shared with the newly released South Korean strains and the latest strains prevalent in Saudi Arabia (Table 1).
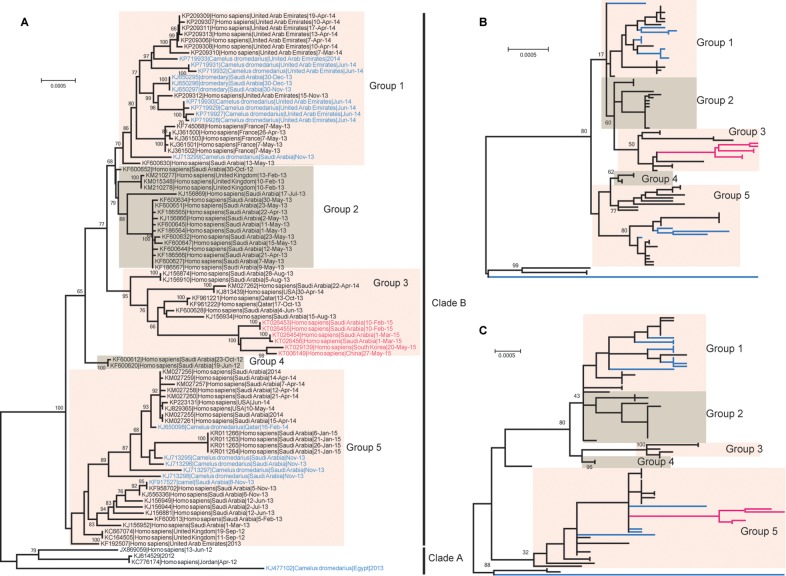
FIG 2 .
Phylogenetic relationships based on complete genomes (A), ORF1ab genes (B), and S genes (C) of MERS-CoV strains. China’s first imported MERS-CoV strain (GenBank accession no. KT006149.2), South Korea’s first MERS-CoV strain (GenBank accession no. KT029139), and the latest MERS-CoV strains prevalent in the Middle East (GenBank accession no. KT026453 to KT026456) are indicated in red. The MERS-CoV strains derived from camels are indicated in blue. All of the complete genomes were analyzed by nucleotide sequence alignment using the maximum-likelihood method implemented in the RAxML. Numbers at the nodes indicate bootstrap support for each node (percentage of 1,000 bootstrap replicates). Scale bars indicate the expected number of nucleotide substitutions per site.
TABLE 1 .
Comparison of sites of variation between gene sequences of ChinaGD01, the first South Korean strain, the latest Saudi Arabia strains, and other MERS-CoV strainsa
| Gene | Position (bp) | China |
South Korea |
Saudi Arabia |
Others |
||||
|---|---|---|---|---|---|---|---|---|---|
| Nt | Aa | Nt | Aa | Nt | Aa | Nt | Aa | ||
| ORF1ab | 472 | G | Val | G | Val | G | Val | T | Phe |
| 2496 | C | Phe | C | Phe | C | Phe | T | Phe | |
| 2930 | G | Gly | G | Gly | A | Asp | A | Asp | |
| 3706 | A | Thr | A | Thr | A | Thr | G | Ala | |
| 5895 | C | Ser | C | Ser | C | Ser | T | Ser | |
| 6357 | T | Ile | T | Ile | T | Ile | G | Met | |
| 6876 | C | Phe | C | Phe | T | Phe | T | Phe | |
| 7277 | T | Ile | T | Ile | T | Ile | C | Thr | |
| 9678 | T | Ser | T | Ser | C | Ser | C | Ser | |
| 10716 | T | Val | T | Val | C | Val | C | Val | |
| 11649 | C | Asp | C | Asp | T | Asp | T | Asp | |
| 17868 | A | Ile | A | Ile | A | Ile | G | Val | |
| 19739 | T | Ile | T | Ile | T | Ile | G | Met | |
| 20685 | A | Ser | A | Ser | G | Gly | G | Gly | |
| S | 258 | C | Val | C | Val | C | Val | T | Val |
| 1848 | C | Val | T | Val | T | Val | T | Val | |
| 2841 | C | Tyr | C | Tyr | C | Tyr | T | Tyr | |
| 3177 | C | Asp | C | Asp | T | Asp | T | Asp | |
| 3267 | T | Ala | T | Ala | C | Ala | T | Ala | |
| ORF3 | 49 | T | Phe | T | Phe | T | Phe | C | Leu |
| 183 | T | Asp | T | Asp | C | Asp | C | Asp | |
| 237 | T | Ser | T | Ser | T | Ser | A | Ser | |
| ORF4a | 258 | C | Asp | T | Asp | C | Asp | T | Asp |
| ORF4b | 17 | C | Thr | T | Met | C | Thr | T | Met |
| ORF5 | 228 | T | Leu | T | Leu | T | Leu | G | Leu |
| M | 367 | A | Ile | A | Ile | A | Ile | T | Phe |
| 438 | T | Gly | T | Gly | T | Gly | C | Gly | |
The positions of amino acid substitutions are indicated by boldface. China, imported MERS-CoV strain ChinaGD01 (GenBank accession no. KT006149); South Korea, South Korea’s first MERS-CoV strain (GenBank accession no. KT029139); Saudi Arabia, MERS-CoV strains recently identified in Saudi Arabia (GenBank accession no. KT026453 to KT026456); Others, other MERS-CoV strains detected worldwide; Nt, nucleotide; Aa, amino acid.
Phylogenetic analysis.
To further investigate the genetic relationship between ChinaGD01 and other MERS-CoV strains whose genomes are available, we performed phylogenetic analyses using the complete genome, the ORF1ab gene, and the S gene. From the whole-genome phylogeny, all available MERS-CoV strains can be clustered into two clades, the earlier clade A and the more recent clade B (Fig. 2A). ChinaGD01 fell into group 3 of clade B (Fig. 2A). Within group 3, ChinaGD01 and the South Korean and Saudi Arabian strains from 2015 were closely clustered and formed a long branch, separate from others of group 3. The nearest strain to this branch was Hafr-Al-Batin-1-2013 (GenBank accession no. KF600628), isolated in August 2013. Phylogenetic analysis of the ORF1ab gene indicated a similar topology in which ChinaGD01 and the recent MERS-CoV strains identified in South Korea were closely adjacent to Hafr-Al-Batin-1-2013 in group 3 (Fig. 2B). However, the phylogeny of the S gene differed in that the new viruses fell into group 5 and were closely related to viruses from both humans and dromedaries (Fig. 2C). These findings are consistent with recombination, a phenomenon not uncommon in coronaviruses.
Genetic recombination analysis.
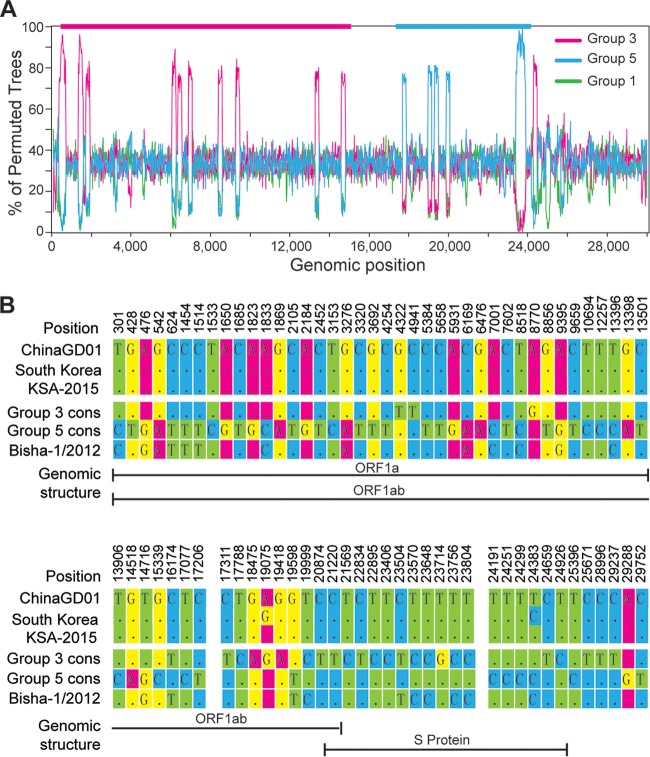
To examine whether genetic recombination has occurred in ChinaGD01, we performed bootscanning analyses. We compared ChinaGD01 with representative viruses from group 3 (Hafr-Al-Batin-1-2013; GenBank accession no. KF600628), group 5 (KSA-CAMEL-378; GenBank accession no. KJ713296), and group 1 (Abu Dhabi_UAE_9_2013; GenBank accession no. KP209312) as controls. As shown in Fig. 3A, ChinaGD01 was more similar to the group 3 strain from position 1 to 15,000 and more similar to the group 5 strain from approximately position 18,000 to 24,000. We then compared the single-nucleotide polymorphisms (SNPs) of ChinaGD01 with consensus sequences of group 3 and group 5 (Fig. 3B; see also Fig. S1 and S2 in the supplemental material). There were 78 SNPs discovered along the ChinaGD01 genome (Fig. 3B). Whereas before position 17,206, ChinaGD01’s SNP pattern is nearly identical to that of the group 3 viruses, its SNP pattern is more similar to that of group 5 viruses between positions 17,311 and 23,804. The consistency in the results of bootscanning and SNP analyses supports the hypothesis that the gene segment from approximately position 17,300 to 24,000, representing portions of the ORF1ab and S genes, reflects a recombination event (Fig. 3B).
FIG 3 .
Recombination analyses of complete MERS-CoV genomes. (A) Bootscanning analysis of MERS-CoV genome. The ChinaGD01 strain was used as a query sequence and compared with one strain from group 3 (GenBank accession no. KF600628.1), one from group 5 (GenBank accession no. KJ713296.1), and one from group 1 (GenBank accession no. KP209312.1). (B) Single-nucleotide differences between the ChinaGD01 sequence and consensus sequences of group 3 and group 5. Group 3 cons, consensus sequences of group 3 strains; group 5 cons, consensus sequences of group 5 strains; South Korea, first MERS-CoV strain (GenBank accession no. KT029139) in South Korea; KSA-2015, latest strains prevalent in Saudi Arabia (GenBank accession no. KT026453 to KT026456), Bisha-1/2012, an earlier strain used as a control.
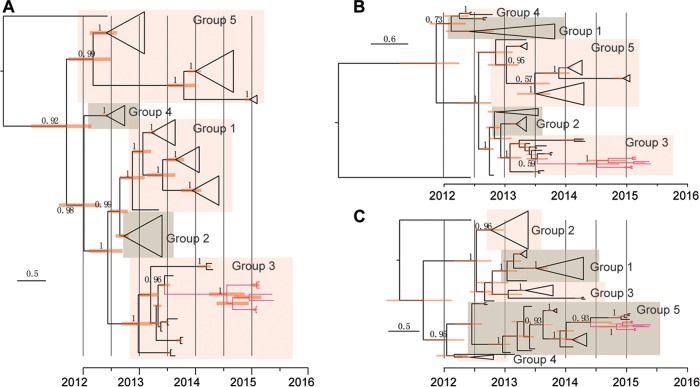
Phylogenetic analysis was further performed using BEAST with the complete genome, the nonrecombinant region (positions 1 to 17,300), and the potential recombinant region (positions 17,301 to 24,000), respectively (Fig. 4). The phylogenies revealed by the BEAST trees were consistent with those from the maximum-likelihood trees. In the trees constructed using the complete genome and the nonrecombinant region, ChinaGD01 fell within group 3; however, trees constructed using the recombinant region clustered with the group 5 sequences.
FIG 4 .
Time-resolved phylogenetic analyses of complete genomes (A), nonrecombination regions (B), and recombination regions (C) of MERS-CoV strains using BEAST. The nonrecombination region is approximately bp 1 to 17,300, and the recombination region is approximately bp 17,301 to 24,000. ChinaGD01 (GenBank accession no. KT006149), South Korea’s first MERS-CoV strain (GenBank accession no. KT029139), and the latest strains prevalent in Saudi Arabia (GenBank accession no. KT026453 to KT026456) are indicated in red.
To date the recombination event, we estimated the time to most recent common ancestor for the novel MERS-CoV from 2015. Although there was a slight difference among results from different models, the time to most recent common ancestor of the 2015 cluster was estimated to be between 0.5 and 0.7 years before the identification of the imported case in the latter months of 2014 (Table 2). Given the observation of similar recombination events in the newly released South Korean strains and the latest strains prevalent in Saudi Arabia, the travel histories of patients, and potential opportunities for virus exposure, we surmise that the recombination likely occurred in the Arabian Peninsula.
TABLE 2 .
Estimated times in years to most recent common ancestor of the 2015 cluster
| Molecular clock model | Value for indicated coalescent model [mean (95% CI)] |
|||
|---|---|---|---|---|
| Constant size | Exponential growth | Logistic growth | GMRF Bayesian skyridea | |
| Strict clock | 0.6594 (0.4259, 0.9137) | 0.6046 (0.371, 0.8562) | 0.6621 (0.4167, 0.9201) | 0.6007 (0.3611, 0.8396) |
| Exponential relaxed clock | 0.6008 (0.3586, 0.8944) | 0.5208 (0.3339, 0.7868) | 0.6037 (0.3578, 0.9059) | 0.5199 (0.3141, 0.8026) |
| Lognormal relaxed clock | 0.6394 (0.4049, 0.9247) | 0.582 (0.3684, 0.8437) | 0.641 (0.4085, 0.9167) | 0.5711 (0.3351, 0.8264) |
GMRF, Gauss Markov random fields.
DISCUSSION
Over the past 3 years, MERS-CoV infections have continued to increase, posing a serious threat to global public health. Previous studies have revealed that MERS-CoV infections are likely due to repeated introductions of MERS-CoV from dromedary camels to humans (13–15), resulting in only limited human-to-human transmission (16). However, the large number of second- and third-generation cases in South Korea raised concerns that MERS-CoV may have evolved to become more adapted to human-to-human transmission.
Our results indicate that at the whole-genome level, ChinaGD01 is >99% similar to the previously identified MERS-CoV strains. Phylogenetic analysis based on the whole-genome sequence revealed that it belongs to group 3 of clade B MERS-CoV strains and forms a separate small branch with viruses from South Korea and Saudi Arabia from 2015. Different phylogenies were observed in the trees constructed using the full-length genome and the S gene, indicating the possibility of a recombination event. Further evidence of a recombination event was obtained through bootscanning and SNP analyses. BEAST analysis revealed that it might have occurred recently, in the second half of 2014, in the Middle East.
Genetic recombination has been well established in severe acute respiratory syndrome coronavirus (SARS-CoV) (17, 18); however, there is only one report of genetic recombination in MERS-CoV (19). Dudas and Rambaut point to frequent recombination in MERS-CoV and partition the genome into two parts in which nucleotides 1 to 23,722 and nucleotides 23,723 to 30,126 have independent molecular clock rates. Based on the latest genome sequences from South Korea and the Kingdom of Saudi Arabia, our research indicated that a novel type of genetic recombination has occurred in the MERS-CoV strains prevalent in South Korea. We note that six MERS-CoV isolates from 2015 (ChinaGD01, the first MERS-CoV strain from South Korea, and the four latest strains from Saudi Arabia) had high levels of nucleotide identity (99.90% to 99.96%) and showed the same recombination signal in our analyses. We speculate that they arose from a common recombination event. However, more studies are needed to understand the relationship between genetic recombination of MERS-CoV, the biological properties it conveys, and its relevance to the recent high rate of transmission.
MATERIALS AND METHODS
Full-length genomic sequencing.
Nasopharyngeal swabs from the South Korean patient diagnosed with MERS-CoV infection were collected and used for viral RNA extraction with the QIAamp viral RNA minikit. Forty-four sets of specific primer pairs were designed and used to amplify the complete genome, followed by Sanger sequencing; meanwhile, the extracted viral RNA was also used for next-generation sequencing with the Ion Torrent PGM after random amplification.
Phylogenetic analysis.
We downloaded all (n = 92) available full-length genome sequences of MERS-CoV from GenBank and used RAxML (20) for phylogenetic analyses of the complete genome, the ORF1ab gene, and the S gene, respectively. One thousand bootstrap replicates were run. Furthermore, the Bayesian Markov chain Monte Carlo method, implemented in BEAST (21), was used to estimate the time to the most recent common ancestor. Twelve different model combinations were applied. For all the analyses, we used the general time-reversible nucleotide substitution model with gamma-distributed rate heterogeneity. Bayesian Markov chain Monte Carlo analysis was run for 50 million steps. Trees and parameters were sampled every 5,000 steps, with the first 10% removed as burn-in.
Genetic recombinant analysis.
Similarity plots and bootscanning analysis were generated by SimPlot (22); a sliding window of 200 nucleotides was used, moving in 20-nucleotide steps. Single-nucleotide-difference analysis was used to confirm the recombination event.
Nucleotide sequence accession number.
The full-length virus genome (30,144 bp) of ChinaGD01 was deposited in GenBank under accession no. KT006149.
SUPPLEMENTAL MATERIAL
Substitutions in the ORF1ab genes from all available MERS-CoV genomes. Different groups are separated by lines; only positions with substitutions are shown. Different types of substitutions are indicated using different colors, as follows: blue for synonymous, claret red for nonsynonymous, and green for untranslated region. Download
Substitutions in the indicated genes from all available MERS-CoV genomes. Different groups are separated by lines; only positions with substitutions are shown. Different types of substitutions are indicated using different colors, as follows: blue for synonymous, claret red for nonsynonymous, and green for untranslated region. Download
ACKNOWLEDGMENTS
This work was supported by funds from the Ministry of Science and Technology of China (2011CB504704), the National Health and Family Planning Commission of China (grants 2014ZX10004-001 and 2013ZX10004601), and National Natural Science Foundation of China (NSFC; grants 81100062 and 81321063). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (81321063).
We thank the clinical staff in Huizhou Central Hospital in Guangdong Province for providing the information and nasopharyngeal swab samples from this imported case.
Footnotes
Citation Wang Y, Liu D, Shi W, Lu R, Wang W, Zhao Y, Deng Y, Zhou W, Ren H, Wu J, Wang Y, Wu G, Gao GF, Tan W. 2015. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio 6(5):e01280-15. doi:10.1128/mBio.01280-15.
REFERENCES
- 1.Zumla A, Hui DS, Perlman S. 3 June 2015. Middle East respiratory syndrome. Lancet. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guery B, Poissy J, el Mansouf L, Séjourné C, Ettahar N, Lemaire X, Vuotto F, Goffard A, Behillil S, Enouf V, Caro V, Mailles A, Che D, Manuguerra JC, Mathieu D, Fontanet A, van der Werf S, MERS-CoV Study Group . 2013. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet 381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LL, Snijder EJ, Stephens GM, Woo PC, Zaki AM, Zambon M, Ziebuhr J. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, AlHakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, de Wit E, Munster VJ, Hensley LE, Zalmout IS, Kapoor A, Epstein JH, Karesh WB, Daszak P, Mohammed OB, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5(2):e00884-00814. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotten M, Watson SJ, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Al Rabeeah AA, Alhakeem RF, Assiri A, Al-Tawfiq JA, Albarrak A, Barry M, Shibl A, Alrabiah FA, Hajjar S, Balkhy HH, Flemban H, Rambaut A, Kellam P, Memish ZA. 2014. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5(1):e01062-13. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, Godeke G-J, Jonges M, Farag E, Diab A, Ghobashy H, Alhajri F, Al-Thani M, Al-Marri SA, Al Romaihi HE, Al Khal A, Bermingham A, Osterhaus AD, AlHajri MM, Koopmans MP. 2014. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis 14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization 2015. Novel coronavirus infection—update. http://www.who.int/csr/don/20-June-2015-mers-saudi-arabia/en/.
- 10.Hui DS, Perlman S, Zumla A. 2015. Spread of MERS to South Korea and China. Lancet Respir Med 3:509–510. doi: 10.1016/S2213-2600(15)00238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, Al-Rabiah FA, Al-Hajjar S, Al-Barrak A, Flemban H, Al-Nassir WN, Balkhy HH, Al-Hakeem RF, Makhdoom HQ, Zumla AI, Memish ZA. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, Van Boheemen S, Gopal R, Ballhause M, Bestebroer TM, Muth TD, Müller MA, Drexler JF, Zambon M, Osterhaus AD, Fouchier RM, Drosten C. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveillance 17(39):pii=20285 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20285. [DOI] [PubMed] [Google Scholar]
- 13.Banik G, Khandaker G, Rashid H. 2015. Middle East respiratory syndrome coronavirus “MERS-CoV”: current knowledge gaps. Paediatr Respir Rev 16:197–202. doi: 10.1016/j.prrv.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zumla A, Perlman S, McNabb SJ, Shaikh A, Heymann DL, McCloskey B, Hui DS. 2015. Middle East respiratory syndrome in the shadow of Ebola. Lancet Respir Med 3:100–102. doi: 10.1016/S2213-2600(14)70316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemida MG, Al-Naeem A, Perera RA, Chin AW, Poon LL, Peiris M. 2015. Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg Infect Dis 21:699–701. doi: 10.3201/eid2104.141949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edelstein M, Heymann DL. 2015. What needs to be done to control the spread of Middle East respiratory syndrome coronavirus? Future Virol 10:497–505. doi: 10.2217/fvl.15.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yount B, Roberts RS, Lindesmith L, Baric RS. 2006. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc Natl Acad Sci U S A 103:12546–12551. doi: 10.1073/pnas.0605438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya AE, Snijder EJ, Spaan WJ. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol 78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudas G, Rambaut A. 2015. MERS-CoV recombination: implications about the reservoir and potential for adaptation. bioRxiv doi: 10.1101/020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Substitutions in the ORF1ab genes from all available MERS-CoV genomes. Different groups are separated by lines; only positions with substitutions are shown. Different types of substitutions are indicated using different colors, as follows: blue for synonymous, claret red for nonsynonymous, and green for untranslated region. Download
Substitutions in the indicated genes from all available MERS-CoV genomes. Different groups are separated by lines; only positions with substitutions are shown. Different types of substitutions are indicated using different colors, as follows: blue for synonymous, claret red for nonsynonymous, and green for untranslated region. Download