ABSTRACT
With over 3.5 billion people at risk and approximately 390 million human infections per year, dengue virus (DENV) disease strains health care resources worldwide. Previously, we and others established models for DENV pathogenesis in mice that completely lack subunits of the receptors (Ifnar and Ifngr) for type I and type II interferon (IFN) signaling; however, the utility of these models is limited by the pleotropic effect of these cytokines on innate and adaptive immune system development and function. Here, we demonstrate that the specific deletion of Ifnar expression on subsets of murine myeloid cells (LysM Cre+ Ifnarflox/flox [denoted as Ifnarf/f herein]) resulted in enhanced DENV replication in vivo. The administration of subneutralizing amounts of cross-reactive anti-DENV monoclonal antibodies to LysM Cre+ Ifnarf/f mice prior to infection with DENV serotype 2 or 3 resulted in antibody-dependent enhancement (ADE) of infection with many of the characteristics associated with severe DENV disease in humans, including plasma leakage, hypercytokinemia, liver injury, hemoconcentration, and thrombocytopenia. Notably, the pathogenesis of severe DENV-2 or DENV-3 infection in LysM Cre+ Ifnarf/f mice was blocked by pre- or postexposure administration of a bispecific dual-affinity retargeting molecule (DART) or an optimized RIG-I receptor agonist that stimulates innate immune responses. Our findings establish a more immunocompetent animal model of ADE of infection with multiple DENV serotypes in which disease is inhibited by treatment with broad-spectrum antibody derivatives or innate immune stimulatory agents.
IMPORTANCE
Although dengue virus (DENV) infects hundreds of millions of people annually and results in morbidity and mortality on a global scale, there are no approved antiviral treatments or vaccines. Part of the difficulty in evaluating therapeutic candidates is the lack of small animal models that are permissive to DENV and recapitulate the clinical features of severe human disease. Using animals lacking the type I interferon receptor only on myeloid cell subsets, we developed a more immunocompetent mouse model of severe DENV infection with characteristics of the human disease, including vascular leakage, hemoconcentration, thrombocytopenia, and liver injury. Using this model, we demonstrate that pathogenesis by two different DENV serotypes is inhibited by therapeutic administration of a genetically modified antibody or a RIG-I receptor agonist that stimulates innate immunity.
INTRODUCTION
Dengue virus (DENV) is a mosquito-transmitted, enveloped, positive-sense RNA virus and member of the flavivirus genus of the Flaviviridae family, which includes several other viruses (e.g., West Nile virus [WNV], Japanese encephalitis virus [JEV], and yellow fever virus [YFV]) that cause disease globally. Infection by any of four serologically distinct viruses (DENV serotype 1 [DENV-1], DENV-2, DENV-3, and DENV-4) causes dengue fever (DF), an acute self-limiting febrile illness, or severe dengue, which manifests as a potentially fatal hemorrhagic fever and vascular leakage syndrome. Epidemiological studies suggest that two populations are at highest risk for severe dengue infection: infants born to dengue-immune mothers who are infected for the first time (infant dengue hemorrhagic fever) and children or adults who experience a second infection with a different DENV serotype (1–3).
DENV has a 10.7-kb, positive-sense RNA genome with 5′ and 3′ untranslated regions flanking a polyprotein that encodes three structural (C, prM/M, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. The E protein is comprised of three domains, I (E-DI), II (E-DII), and III (E-DIII), with E-DII and E-DIII containing the fusion peptide and putative viral receptor binding site(s), respectively (reviewed in references 4 and 5). Among the structural proteins, prM and E are primary antigenic targets of the humoral immune response (6–9). The most potently neutralizing antibodies target sites on the lateral ridge and A strand of E-DIII (10–16), quaternary epitopes on adjacent E proteins near the E-DI-DII hinge region (17–20), amino acids near the bc loop of E-DII (21), and a conserved epitope at the E dimer interface (22).
One hypothesis as to why certain individuals are more vulnerable to severe DENV infection is that preexisting, poorly neutralizing antibodies acquired passively (infants) or after primary infection (children and adults) facilitate virus entry into Fcγ receptor (FcγR)-bearing target cells, thereby increasing viral replication, cytokine levels, inflammation, and ultimately, disease severity (reviewed in reference 23). Experimental evidence in mice supports this idea. Initial studies showed that passive transfer of subneutralizing concentrations of monoclonal antibody (MAb) or polyclonal antibody (PAb) enhanced infection and disease caused by DENV-2 in 129/Sv mice deficient in both alpha/beta interferon (IFN-α/β) receptor (Ifnar) and IFN-γ receptor (Ifngr) (known as AG129) (24–26). Subsequent reports extended these findings to other DENV serotypes in AG129 mice (DENV-1 [19], DENV-3 [27], and DENV-4 [13, 28, 29]) or Ifnar−/− C57BL/6 mice. Ifnar−/− mice in either the 129/Sv or C57BL/6 background develop a severe DENV-like disease when infected with very high DENV-2 doses or in the presence of enhancing anti-DENV antibodies (25, 30–33).
The utility of these highly immunocompromised mice to provide a mechanistic understanding of DENV pathogenesis and disease remains controversial. The use of laboratory or mouse-adapted DENV-2 strains has been required to induce mortality or neuroinvasive disease (34), and the latter is not commonly observed in DENV-infected humans. Studies with DENV-2 indicate that mice with deficiencies in innate immunity are needed to study DENV pathogenesis because the viral NS3 and NS5 proteins induce degradation of human but not mouse STING and STAT2, respectively (35–38); STING and STAT2 are key components of the IFN induction and signaling pathway. Thus, DENV generally does not replicate to high titers or cause clinical signs of disease in wild-type (WT) mice, in part because DENV nonstructural proteins fail to antagonize host innate immune responses efficiently. We recently demonstrated that WNV infection of the more immunocompetent LysM Cre+ Ifnarflox/flox (denoted as Ifnarf/f herein) or CD11c Cre+ Ifnarf/f mice, which lack Ifnar expression only on subsets of myeloid cells, resulted in a sepsis-like syndrome that shared features of DENV disease in humans (39). Another group recently used the LysM Cre+ Ifnarf/f and CD11c Cre+ Ifnarf/f mice to generate a model of DENV-2 infection and study adaptive immunity after immunization with a candidate vaccine (40). Here, we administered enhancing amounts of anti-E and anti-prM MAbs to LysM Cre+ Ifnarf/f mice prior to infection with either DENV-2 or DENV-3 and generated an antibody-dependent enhancement (ADE) of infection model of disease that shared many characteristics of severe dengue in humans. We used this model to establish the therapeutic efficacy of an antibody-based bispecific dual-affinity retargeting molecule (DART) that targets epitopes on the A strand of E-DIII and the fusion loop in E-DII and a novel sequence-optimized RIG-I receptor agonist that stimulates antiviral innate immunity. These studies provide the first demonstration of severe ADE-mediated DENV infection in LysM Cre+ Ifnarf/f mice and illustrate the utility of this model to evaluate antibody- and innate immune-based antiviral therapies to limit DENV pathogenesis.
RESULTS
Dengue virus infection in LysM Cre+ Ifnarf/f mice.
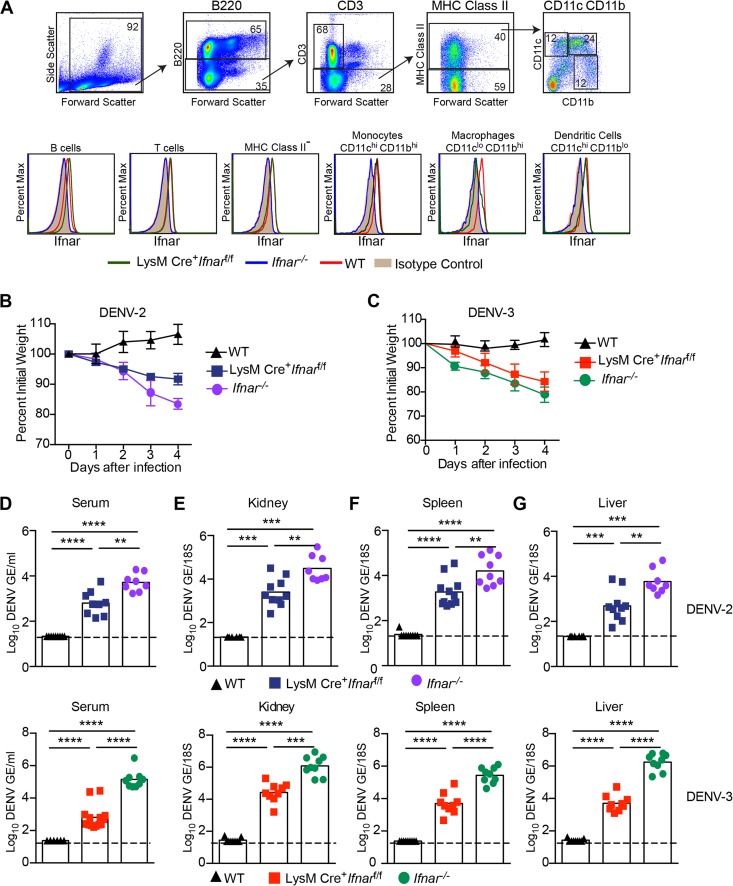
Prior analysis established that myeloid cells are targets for human DENV infection in vivo (41). The relevance of Ifnar−/− mice in studying DENV pathogenesis has been questioned because of the central role of IFN signaling in priming innate and adaptive immune responses (42–44). In an attempt to study DENV pathogenesis in a more immunocompetent animal, we used LysM Cre+ Ifnarf/f mice that conditionally delete Ifnar only in subsets of myeloid cells. In splenocytes, flow cytometric analysis revealed substantially reduced Ifnar expression on the surface of CD11bhi CD11clo macrophages from LysM Cre+ Ifnarf/f mice compared to the results for WT mice. In contrast, the levels of Ifnar expression on B220+ B cells, CD3+ T cells, CD11bhi CD11chi monocytes, and CD11blo CD11chi dendritic cells from LysM Cre+ Ifnarf/f mice were comparable to the levels seen in WT mice (Fig. 1A).
FIG 1 .
Dengue virus infection of LysM Cre+ Ifnarf/f mice. (A) Ifnar expression on splenic B cells, T cells, class II MHC-negative cells, monocytes, dendritic cells, and macrophages from WT, LysM Cre+ Ifnarf/f, and Ifnar−/− mice was measured using flow cytometry. Cells were gated (example in top panels is from DENV-2-infected LysM Cre+ Ifnarf/f mice) using the following markers: B cells (B220+), T cells (B220− + CD3+), class II MHC-negative cells (B220− CD3− class II MHC−), monocytes (B220− CD3− class II MHC+ CD11bhi CD11chi), dendritic cells (B220− CD3− class II MHC+ CD11blo CD11chi), and macrophages (B220− CD3− class II MHC+ CD11bhi CD11clo). Ifnar expression was measured using the MAR1-5A3 MAb. Data are representative of 4 mice from two independent experiments. (B and C) Weight loss of 4-week-old WT, LysM Cre+ Ifnarf/f, and Ifnar−/− mice after intravenous injection of 106 FFU of DENV-2 (D2S20) (B) or 107 FFU of DENV-3 (C0360/94) (C). Weight loss differences between WT and LysM Cre+ Ifnarf/f or Ifnar−/− mice were statistically significant on days 2, 3, and 4 after DENV-2 or DENV-3 infection in three independent experiments with 8 to 12 mice per group. (D to G) Viral burdens in LysM Cre+ Ifnarf/f, WT, and Ifnar−/− mice after DENV-2 (top) or DENV-3 (bottom) infection. Four-week-old mice were infected intravenously with 106 FFU of DENV-2 (D2S20) or 107 FFU of DENV-3 (C0360/94). Levels of viral RNA in (D) serum, (E) kidney, (F) spleen, and (G) liver samples harvested 4 days after infection were determined using qRT-PCR. Data are shown as log10 DENV genome equivalents (GE) per 18S ribosomal RNA of tissue or per milliliter of serum for 8 to 9 mice per group, from three independent experiments. The dotted line represents the limit of sensitivity of the assay. Asterisks indicate values that are statistically significant (*, P < 0.05, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) as determined by using the Mann-Whitney test.
Initially, we compared morbidity in DENV-infected WT, LysM Cre+ Ifnarf/f, and Ifnar−/− mice. Four-week-old mice lacking Ifnar expression on all cells or only on myeloid cell subsets rapidly developed disease within days of DENV-2 (strain D2S20, 1 × 106 focus-forming units [FFU]) or DENV-3 (strain C0360/94, 1 × 107 FFU) infection, as reflected by the development of weight loss (Fig. 1B and C) ruffled fur, and hunched appearance (data not shown). In comparison, and as expected, WT mice infected with DENV-2 or DENV-3 did not develop signs of disease.
To define the basis for the DENV-induced disease in LysM Cre+ Ifnarf/f mice, we assessed viral burdens. WT, LysM Cre+ Ifnarf/f, and Ifnar−/− mice were inoculated intravenously with 1 × 106 FFU of DENV-2 or 1 × 107 FFU of DENV-3, and the levels of viral RNA in the serum, kidney, spleen, and liver were determined at 4 days after infection using quantitative reverse transcription-PCR (qRT-PCR). DENV-2 or DENV-3 replication was markedly enhanced (Fig. 1D to G) in LysM Cre+ Ifnarf/f mice compared to the levels of replication in WT mice. These values approached but did not attain those observed in Ifnar−/− mice, which lack Ifnar signaling in all cell types.
Antibody-enhanced DENV disease in LysM Cre+ Ifnarf/f mice.
Given that LysM Cre+ Ifnarf/f mice sustained infection and disease after DENV inoculation, we evaluated whether more severe disease would be observed if enhancing and cross-reactive anti-prM (2H2) and anti-E (4G2) MAbs were administered. Indeed, more severe disease was observed in LysM Cre+ Ifnarf/f mice that were treated with enhancing MAbs than in those treated with an isotype control anti-Chikungunya virus MAb (CHK-152) (45) prior to infection with 1 × 106 FFU of DENV-2 (Fig. 2A). However, no mortality was seen with the 1 × 106 FFU dose of DENV-2, even in the presence of enhancing MAbs (data not shown). With a higher dose of DENV-2 (1 × 107 FFU), we did observe increased mortality under ADE conditions (55 versus 8 percent mortality; P < 0.006) (Fig. 2B). As DENV infection in humans is predominantly a disease that does not cause lethality, we performed subsequent experiments using the lower 1 × 106 FFU dose of DENV-2.
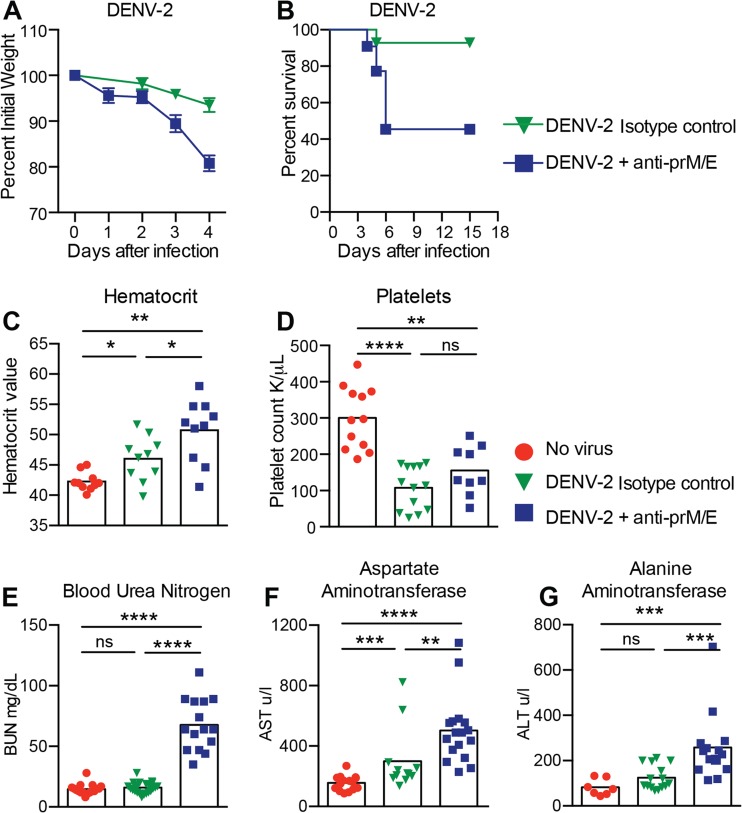
FIG 2 .
Weight loss, survival, hematology, and blood chemistry of DENV-2-infected LysM Cre+ Ifnarf/f mice under ADE conditions. (A and B) LysM Cre+ Ifnarf/f mice received passively transferred isotype control MAb (30 µg of CHK-152) or enhancing MAbs (15 µg of anti-prM MAb [2H2] and 15 µg anti-E MAb [4G2]) and then infected a day later with 106 (A) or 107 (B) FFU of DENV-2 (strain D2S20) via an intravenous route. Weight change and mortality were monitored; the data reflect the results from three to four independent experiments with 3 to 8 mice per group per experiment. (C to G) LysM Cre+ Ifnarf/f mice were either not infected, infected with DENV-2 (106 FFU), or administered enhancing anti-prM and anti-E MAbs (as in the experiment whose results are shown in panel A) one day before DENV-2 (106 FFU) infection. Whole blood was collected at day 4 after infection and evaluated. Hematocrit (C) and platelet levels (D) were analyzed from whole blood. Clinical chemistry was performed on serum and included measurement of blood urea nitrogen (BUN) (E), aspartate amino transferase (AST) (F), and alanine aminotransferase (ALT) (G) levels. Three independent experiments were completed, with 3 to 5 mice per group per experiment. Statistically significant differences between individual groups were determined by using the Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant).
To define the relevance of the LysM Cre+ Ifnarf/f mouse model of DENV-2 infection, we assessed the impact of ADE on laboratory parameters that are linked to severe DENV disease in humans. Hemoconcentration (shown by elevated hematocrit [HCT]), which reflects changes in vascular permeability, was observed in DENV-infected LysM Cre+ Ifnarf/f mice, and this phenotype was exacerbated in the presence of enhancing antibodies (HCT was 42 ± 2 for no virus, 46 ± 4 for DENV-2 alone, and 51 ± 5 for DENV-2 plus anti-prM and anti-E MAbs) (Fig. 2C). Thrombocytopenia (low platelet [PLT] counts) was also detected in DENV-infected LysM Cre+ Ifnarf/f mice, although ADE did not alter its magnitude (PLT count was 300 ± 85 for no virus, 108 ± 59 for DENV-2 alone, and 155 ± 67 for DENV-2 plus anti-prM and anti-E MAbs) (Fig. 2D). An elevated blood urea nitrogen (BUN) level, which reflects renal damage, was measured only in DENV-infected LysM Cre+ Ifnarf/f mice treated with enhancing antibodies (BUN was 15 ± 4 for no virus, 16 ± 4 for DENV-2 alone, and 68 ± 21 for DENV-2 plus anti-prM and anti-E MAbs) (Fig. 2E). Liver injury, as manifested by elevated aspartate aminotransferase (AST) or alanine aminotransferase (ALT) levels in serum, was also observed in DENV-infected LysM Cre+ Ifnarf/f mice, with greater damage present under conditions of ADE (AST and ALT were 156 ± 49 and 82 ± 36, respectively, for no virus, 300 ± 208 and 125 ± 53 for DENV-2 alone, and 503 ± 223 and 258 ± 149 for DENV-2 plus anti-prM and anti-E MAbs) (Fig. 2F and G). Overall, enhancing antibodies in the context of DENV infection of LysM Cre+ Ifnarf/f mice worsened many of the laboratory parameters that are seen during cases of severe DENV in humans (46). In comparison, in Ifnar−/− mice, the administration of enhancing antibodies resulted in an increased hematocrit (44 ± 3 for DENV-2 alone and 53 ± 9 for DENV-2 plus anti-prM and anti-E MAbs) but did not alter AST or ALT levels or platelet counts beyond that seen with virus alone (data not shown).
To determine the basis for the exacerbated clinical disease in LysM Cre+ Ifnarf/f mice, we assessed the effects of enhancing antibodies on the DENV-2 viral burden. At day 4 after infection, we observed higher levels of DENV RNA in the sera of infected LysM Cre+ Ifnarf/f than in the sera of WT mice, with further increases when animals were pretreated with enhancing MAbs (Fig. 3A). Analogous results were observed in other tissues after DENV infection and antibody treatment, including the kidney, spleen, and liver (Fig. 3B to D).
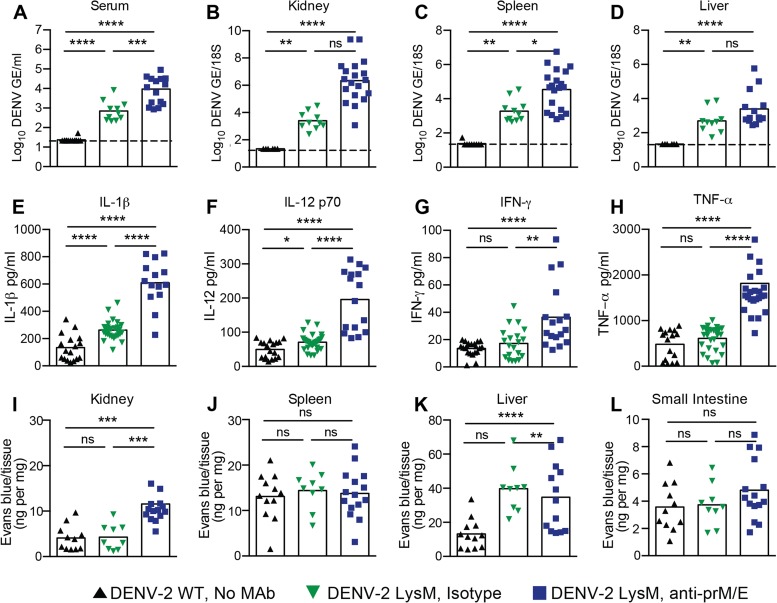
FIG 3 .
Viral burdens, cytokines, and vascular permeability following DENV-2 infection of LysM Cre+ Ifnarf/f mice under ADE conditions. WT and LysM Cre+ Ifnarf/f mice were infected with DENV-2 (106 FFU) via an intravenous route. Some of the LysM Cre+ Ifnarf/f mice were pretreated with an isotype control MAb (CHK-152) or enhancing amounts of anti-prM and anti-E MAbs. (A to D) Levels of viral RNA in serum (A), kidney (B), spleen (C), and liver (D) samples harvested 4 days after infection were determined using qRT-PCR. Data are shown as log10 DENV genome equivalents (GE) per 18S ribosomal RNA of tissue or per milliliter of serum from 11 to 16 mice per condition. The dotted line represents the limit of sensitivity of the assay. Asterisks indicate values that are statistically significant by the Mann-Whitney test compared to the values for isotype control MAb-treated mice. (E to H) Proinflammatory cytokine analysis. Cytokines IL-1β (E), IL-12p70 (F), IFN-γ (G), and TNF-α (H) were measured at day 4 after infection using a Bioplex instrument; the data reflect the results for 9 to 12 mice per group. Data are pooled from three to four independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [Mann-Whitney test]). (I to L) Vascular permeability. Four days after infection, Evans blue dye was administered intravenously 20 min prior to sacrifice and tissue harvest. Levels of Evans blue (ng/mg tissue) in kidney (I), spleen (J), liver (K), and small intestine (L) were determined. Data are shown as ng of Evans blue per mg of tissue for 9 to 12 mice per group. Data are pooled from three to four independent experiments (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001 [Mann-Whitney test]).
We evaluated cytokine levels in serum and vascular leakage in tissues, which are two features of severe DENV infection in humans (23, 47). Elevated levels of interleukin 1β (IL-1β) and IL-12 were observed in LysM Cre+ Ifnarf/f mice compared to the levels in WT mice after DENV-2 infection (Fig. 3E and F). Moreover, in the setting of ADE, the levels of several other proinflammatory cytokines were substantially higher in LysM Cre+ Ifnarf/f mice than they were in the setting of DENV infection alone (Fig. 3G and H; Table 1). Since previous studies in DENV-infected AG129 mice demonstrated increased vascular permeability in the liver (24, 25, 27), as measured by extravasation of Evans blue dye, we performed similar studies in LysM Cre+ Ifnarf/f mice (Fig. 3I to L). Leakage of Evans blue dye into the kidney and liver was observed at day 4 after DENV infection in the context of ADE in LysM Cre+ Ifnarf/f mice, which parallels the enhanced capillary permeability seen during severe DENV disease in humans (48). However, the increased dye extravasation was tissue restricted and was not observed in the spleen or small intestine of these mice.
TABLE 1 .
Cytokine levels in sera of DENV-2-infected WT and LysM Cre+ Ifnarf/f mice under ADE conditions
| Mouse treatment |
Amt of cytokine [pg/ml (mean ± SD)] in sera of mice with indicated genotype and treatmenta |
||
|---|---|---|---|
| WT mice, no MAb |
LysM Cre+ Ifnarf/f mice, isotype MAb (P value vs WT) |
LysM Cre+ Ifnarf/f mice, prM and E MAbs (P value vs WT; P value vs isotype MAb) |
|
| IL-1α | 5 ± 1 | 16 ± 2 (<0.0001) | 20 ± 4 (<0.0001; NS) |
| IL-1β | 134 ± 23 | 262 ± 13 (<0.0001) | 609 ± 45 (<0.0001; <0.0001) |
| IL-2 | 24 ± 1 | 42 ± 5 (0.01) | 48 ± 6 (0.01; NS) |
| IL-3 | 23 ± 1 | 62 ± 9 (0.01) | 93 ± 3 (<0.0001; 0.01) |
| IL-4 | 16 ± 2 | 47 ± 5 (<0.0001) | 50 ± 8 (0.001; NS) |
| IL-5 | 24 ± 4 | 89 ± 10 (<0.0001) | 100 ± 11 (<0.0001; NS) |
| IL-6 | 35 ± 7 | 86 ± 17 (0.01) | 88 ± 5 (<0.0001; NS) |
| IL-10 | 52 ± 6 | 62 ± 6 (NS) | 80 ± 10 (NS; NS) |
| IL-12(p40) | 59 ± 6 | 79 ± 12 (NS) | 215 ± 49 (<0.0001; 0.00) |
| IL-12(p70) | 50 ± 6 | 71 ± 4 (0.01) | 196 ± 22 (<0.0001; <0.0001) |
| IL-13 | 28 ± 3 | 44 ± 5 (0.04) | 51 ± 7 (0.0026; NS) |
| IL-17 | 18 ± 1 | 40 ± 6 (0.02) | 60 ± 4 (<0.0001; 0.007) |
| Eotaxin | 592 ± 35 | 595 ± 101 (NS) | 1,092 ± 165 (NS; 0.008) |
| G-CSF | 110 ± 10 | 322 ± 73 (0.0004) | 3,081 ± 745 (<0.0001; <0.0001) |
| GM-CSF | 64 ± 4 | 76 ± 15 (NS) | 214 ± 7 (<0.0001; <0.0001) |
| IFN-γ | 14 ± 1 | 17 ± 3 (NS) | 36 ± 6 (<0.0001; 0.002) |
| KC | 48 ± 7 | 111 ± 11 (<0.0001) | 1,171 ± 307 (<0.0001; <0.0001) |
| MCP-1 | 219 ± 34 | 712 ± 92 (<0.0001) | 1,973 ± 349 (<0.0001; <0.0001) |
| MIP-1α | 5 ± 0 | 8 ± 1 (NS) | 25 ± 1 (<0.0001; <0.0001) |
| MIP-1β | 41 ± 9 | 74 ± 6 (0.0014) | 135 ± 7 (<0.0001; <0.0001) |
| RANTES | 7 ± 1 | 20 ± 8 (<0.0001) | 323 ± 113 (<0.0001; <0.0001) |
| TNF-α | 480 ± 81 | 609 ± 50 (NS) | 1,815 ± 221 (<0.0001; <0.0001) |
Mice were pretreated with the indicated MAb(s) (none, isotype control anti-CHK-152, or enhancing anti-prM and anti-E MAbs) one day prior to infection with DENV-2. On day 4 after infection, sera were harvested and processed for cytokines and chemokines as described in Materials and Methods. Data from 9 to 12 mice per group in three or four independent experiments were pooled. Statistical significance was assessed using the Mann-Whitney test. NS, not significant.
To corroborate these findings, we performed analogous experiments with enhancing MAbs in DENV-3-infected LysM Cre+ Ifnarf/f mice using a non-mouse-adapted DENV-3 human isolate (strain C0360/94) (27). Although lethal infection was not observed in this model at a dose of 1 × 107 FFU, some mortality (~33%) was seen at a dose of 3 × 107 FFU in A129 mice (data not shown). More severe DENV-3-induced disease occurred in LysM Cre+ Ifnarf/f but not WT mice when enhancing amounts of anti-E MAb were present, as determined by greater weight loss (Fig. 4A; also data not shown). The addition of the anti-prM MAb to the anti-E MAb did not worsen clinical disease further after DENV-3 infection of LysM Cre+ Ifnarf/f mice (data not shown), and thus, it was not included in subsequent experiments.
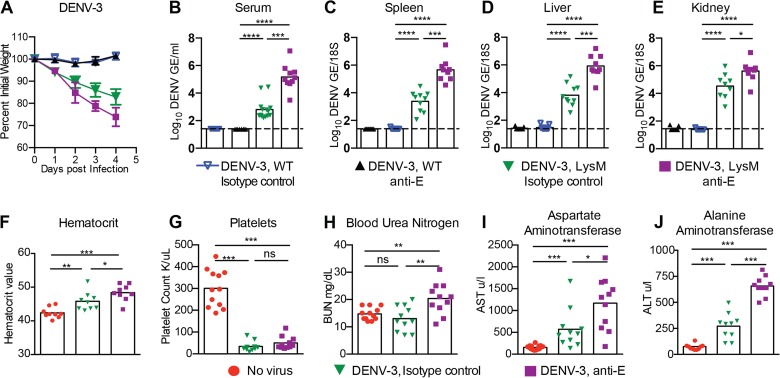
FIG 4 .
Weight loss, viral burdens, hematology, and blood chemistry for DENV-3-infected WT and LysM Cre+ Ifnarf/f mice under ADE conditions. WT or LysM Cre+ Ifnarf/f mice received passively transferred isotype control MAb (10 µg of CHK-152) or enhancing MAb (10 µg of anti-E MAb [4G2]) and were infected a day later with 107 FFU of DENV-3 (strain C0360/94) via an intravenous route. (A) Weight change was monitored daily in four independent experiments with 4 to 5 mice per group per experiment. (B to E) Levels of viral RNA in serum (B), spleen (C), liver (D), and kidney (E) samples harvested 4 days after infection were determined using qRT-PCR. Data are shown as log10 DENV genome equivalents (GE) per 18S of tissue or per milliliter of serum from 9 to 12 mice per condition. The dotted line represents the limit of sensitivity of the assay. Asterisks indicate values that are statistically significant compared to the values for isotype control MAb-treated animals. (F to J) LysM Cre+ Ifnarf/f mice were either not infected (naive, no virus) or were administered isotype control (anti-CHK 152) or enhancing anti-E MAb one day before DENV-3 infection. Whole blood was collected at day 4 after infection, and hematocrit (F) and platelet counts (G) were analyzed. Clinical chemistry analysis was performed on serum and included measurement of blood urea nitrogen (BUN) (H), aspartate amino transferase (AST) (I), and alanine aminotransferase (ALT) (J) levels. Three independent experiments were completed with 3 to 5 mice per group per experiment. Statistically significant differences between individual groups were determined by using the Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Virologic and clinical laboratory studies were performed after DENV-3 infection in WT and LysM Cre+ Ifnarf/f mice. Higher viral burdens at day 4 after DENV-3 infection were observed in LysM Cre+ Ifnarf/f mice only when enhancing anti-E MAb was administered (Fig. 4B to E; also data not shown). Hemoconcentration, lower platelet levels, enhanced liver enzymes (elevated AST and ALT), and renal injury (elevated BUN) were also observed in LysM Cre+ Ifnarf/f mice after DENV-3 infection in the presence of enhancing antibodies (Fig. 4F to J). Thus, the LysM Cre+ Ifnarf/f mouse model shows enhanced disease in the setting of ADE of two different DENV serotypes.
A bispecific tetravalent Fc-DART protects against DENV infection in LysM Cre+ Ifnarf/f mice.
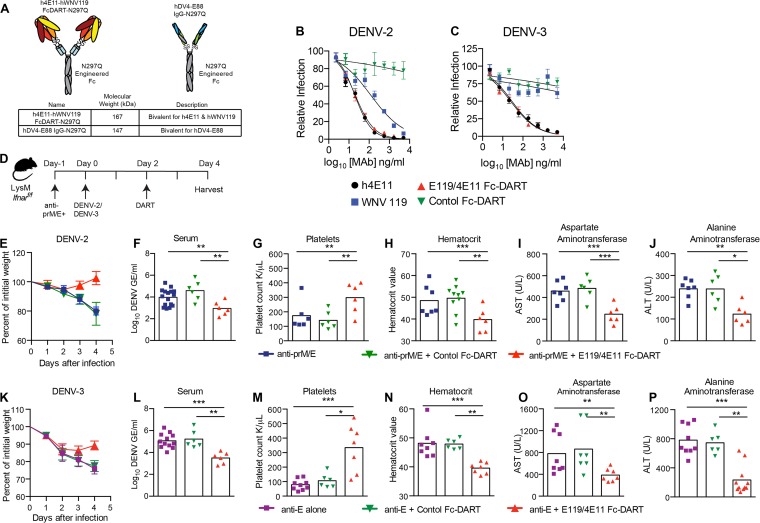
Previously, we generated an antibody-variable-region-based bispecific Ig-DART against DENV that targeted two distinct epitopes (49). That DART used WNV-E60, a cross-reactive neutralizing MAb that binds the fusion loop on DII, and 4E11, a group-specific neutralizing MAb that binds the A-strand epitope on DIII. Our subsequent studies revealed that a second DII fusion loop antibody, WNV-E119 (50), had superior inhibitory activity against other DENV serotypes (13; also data not shown). Accordingly, we generated a new bispecific tetravalent DART that contained the VH and VL regions of humanized WNV-E119 (hE119) and humanized 4E11 (h4E11) (Fig. 5A) (51, 52). This Fc-DART was engineered with the human IgG Fc constant regions (CH1, CH2, and CH3) containing a point mutation (N297Q) that prevents FcγR engagement and ADE (26). A nonbinding, bivalent DENV-4 type-specific IgG was developed as a negative control (Fig. 5A).
The initial studies confirmed the in vitro binding activity of N297Q h4E11 plus hWNV-E119A against recombinant E protein from prototype strains of all four DENV serotypes (data not shown). We next investigated the neutralization potential of the Fc-DART against the viruses we used for the in vivo studies and compared it to that of the parental h4E11 and hWNV-E119 MAbs. The h4E11 MAb efficiently neutralized DENV-2 (50% effective concentration [EC50] of 20 ng/ml) and DENV-3 (EC50 of 29 ng/ml) (Fig. 5B and C). In comparison, the hWNV-E119 MAb neutralized DENV-2 (EC50 of 120 ng/ml) but not DENV-3 strain C0360/94. The h4E11–hWNV-E119A N297Q bispecific Fc-DART showed a neutralization profile that was consistent with the inhibitory activity of h4E11.
FIG 5 .
Protection of LysM Cre+ Ifnarf/f mice from antibody-enhanced disease using an Fc-DART. (A) Fc-DART diagram and description. (B and C) Neutralization data. DENV-2 (B) or DENV-3 (C) was mixed with the indicated MAbs or Fc-DART prior to performing a focus reduction neutralization test. The data are representative of three independent experiments performed in triplicate. (D) Scheme of Fc-DART therapeutic studies. LysM Cre+ Ifnarf/f mice were treated with 15 µg each of anti-E and anti-prM MAbs or 10 µg of anti-E MAb. One day, later, mice were infected with 106 FFU of DENV-2 D2S20 or 107 FFU of DENV-3 C0360/94 by intravenous injection. Two days later, animals were administered 25 µg of anti-DENV Fc-DART (h4E11–WNV-hE119 N297Q) or control antibody (DENV-4 hE88 N297Q). Weight loss was monitored (E and K), and serum titers (F and L), platelet counts (G and M), hematocrit (H and N), and AST (I and O) and ALT (J and P) levels in blood and serum samples obtained 4 days after infection were analyzed. Two independent experiments were completed, with 3 to 4 mice per group per experiment. Statistically significant differences between individual groups were determined by using the Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To assess the utility of the LysM Cre+ Ifnarf/f mouse model of ADE and DENV pathogenesis, we performed protection studies using the bispecific N297Q Fc-DART. Initially, prophylaxis studies were performed against DENV-2. One 25-µg dose of the bispecific h4E11–hWNV-E119 N297Q Fc-DART administered one day prior to DENV-2 inoculation and enhancing treatment with anti-E plus anti-prM MAb completely protected LysM Cre+ Ifnarf/f mice against weight loss, viremia, thrombocytopenia, hemoconcentration, and liver injury (data not shown). Subsequently, LysM Cre+ Ifnarf/f mice were administered a single 25-µg dose of the bispecific N297Q Fc-DART 48 h after infection with DENV-2 or DENV-3 (Fig. 5D). The h4E11–hWNV-E119 Fc-DART was active therapeutically, as judged by increased weight gain, reduced viral burden, and improved laboratory test values at 4 days after infection in the setting of ADE for either DENV-2 (Fig. 5E to J) and DENV-3 (Fig. 5K to P).
A RIG-I receptor agonist protects against severe DENV infection in LysM Cre+ Ifnarf/f but not Ifnar−/− mice.
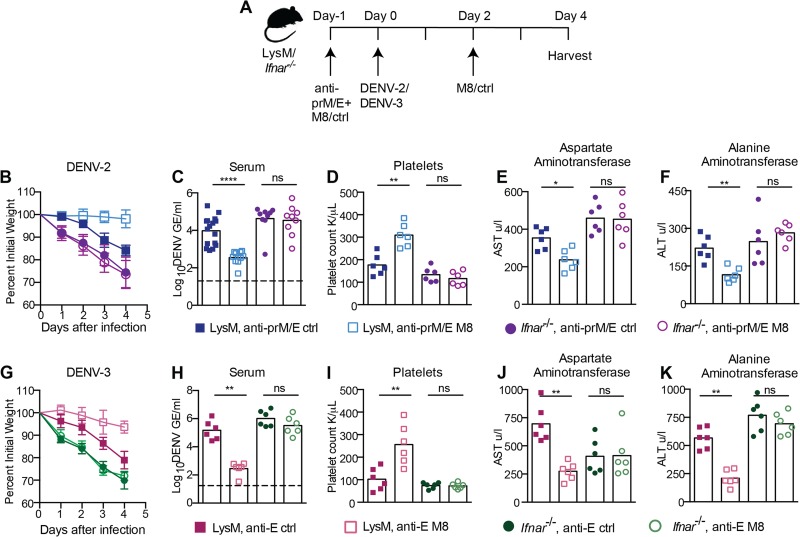
One of the limitations of using Ifnar−/− or AG129 mice is that the complete absence of IFNAR signaling does not permit the evaluation of anti-DENV therapies that act by modulating the host innate immune response. Recently, members of our group developed a 99-nucleotide, uridine-rich hairpin 5′-ppp RNA (termed M8) RIG-I agonist that can be delivered in vivo to stimulate protective antiviral responses against alphaviruses and orthomyxoviruses (53). We tested the utility of the LysM Cre+ Ifnarf/f model by administering M8 5′-ppp RNA in parallel to LysM Cre+ Ifnarf/f and Ifnar−/− mice prior to and after the addition of enhancing anti-prM and -E MAbs and DENV-2 (Fig. 6A). Protection, as judged by the absence of weight loss, was observed only in LysM Cre+ Ifnarf/f and not in Ifnar−/− mice (Fig. 6B). Associated with this clinical improvement, at day 4, we observed decreased viremia (Fig. 6C), normalized platelet counts (Fig. 6D), and decreased liver injury (Fig. 6E and F) in M8-treated LysM Cre+ Ifnarf/f mice but not in M8-treated Ifnar−/− mice. An analogous improvement in clinical and laboratory parameters was observed in M8 5′-ppp RNA-pretreated LysM Cre+ Ifnarf/f mice but not Ifnar−/− mice infected with DENV-3 in the context of ADE (Fig. 6G to K). These experiments confirm that 5′-ppp RNA uses Ifnar signaling in cells other than macrophages to inhibit DENV infection in vivo, and thus, establish an animal model for testing immunomodulatory agents against DENV.
FIG 6 .
Pretreatment with a RIG-I receptor agonist protects LysM Cre+ Ifnarf/f but not Ifnar−/− mice against DENV-2 and DENV-3 infection and disease. (A) Scheme of treatment and infection. LysM Cre+ Ifnarf/f or Ifnar−/− mice were treated with 5 µg of 5′-ppp M8 RNA or control RNA immediately before injection with 15 µg of anti-prM MAb (DENV-2 only) and anti-E MAb (DENV-2 and DENV-3). Animals were infected one day later with 106 FFU of DENV-2 D2S20 (B to F) or 107 FFU of DENV-3 C0360/94 (G to K). A second dose of 5′-ppp M8 RNA or control RNA was given 2 days after DENV infection. (B and G) Weight loss was monitored for 4 days. Laboratory parameters, including viremia (C and H), platelet counts (D and I), and levels of liver enzymes AST (E and J) and ALT (F and K) in serum were monitored as described in the legend to Fig. 5. Two to three independent experiments were performed, with 3 to 5 mice per group per experiment. Statistically significant differences between individual groups were determined by using the Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
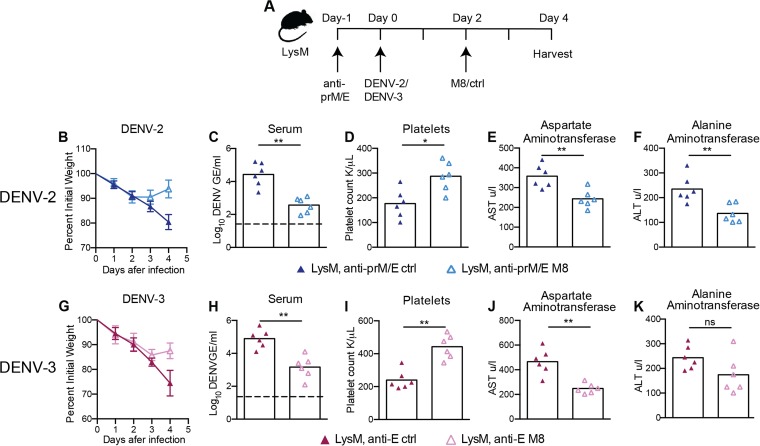
As a follow-up to these studies, we demonstrated the postexposure therapeutic efficacy of M8 5′-ppp RNA against DENV-2 and DENV-3 infection in LysM Cre+ Ifnarf/f mice when the RIG-I agonist was administered 2 days after infection in the setting of ADE (Fig. 7A). Therapeutic administration of M8 5′-ppp RNA after infection with DENV-2 or DENV-3 resulted in improved weight gain in the M8-treated group, with a significant difference in weight at 4 days postinfection compared to the results for the control (Fig. 7B and G). Concurrently, there was a reduction in viremia (Fig. 7C and H), a normalization of platelet levels (Fig. 7D and I), and an improvement in liver function tests (Fig. 7E, F, J, and K) associated with M8 treatment. These results suggest that RIG-I activation is a potential therapeutic target during the course of DENV infection.
FIG 7 .
Postexposure therapy with a RIG-I receptor agonist controls DENV-2 and DENV-3 infection and disease in LysM Cre+ Ifnarf/f mice. (A) Scheme of treatment and infection. LysM Cre+ Ifnarf/f mice were injected with 15 µg of anti-prM MAb (DENV-2 only) and anti-E MAb (DENV-2 and DENV-3) and infected one day later with 106 FFU of DENV-2 D2S20 (B to F) or 107 FFU of DENV-3 C0360/94 (G to K). Two days after DENV infection, mice were treated with 5 µg of 5′-ppp M8 RNA or control RNA. Weight loss was monitored for 4 days (B and G). Laboratory parameters, including viremia (C and H), platelet counts (D and I), and levels of liver enzymes AST (E and J) and ALT (F and K) in serum were monitored as in described in the legend to Fig. 5. Two independent experiments were performed, with 3 mice per group per experiment. Statistically significant differences between individual groups were determined by using the Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
DENV continues to emerge globally, with an estimated 390 million infections per year (54). Infection by the four distinct serotypes can cause several clinical syndromes, ranging from the debilitating DF to life-threatening shock syndrome. Severe DENV disease most often is associated with a second infection with a heterologous DENV serotype, due to the presence of preexisting and nonneutralizing, cross-reactive antibodies and/or T cells. Although live-attenuated tetravalent prophylactic vaccines are in advanced clinical evaluation (55–58), there is still a need for therapeutics that can be utilized in DENV-infected individuals. Preclinical testing has been hampered by the lack of small animal models that support DENV replication and pathogenesis. Most studies have used highly immunocompromised mice lacking intact type I and type II IFN signaling pathways (59). Here, using LysM Cre+ Ifnarf/f mice that delete Ifnar expression only on a subset of target myeloid cells, we established a more immunocompetent model of ADE, which shares many of the same clinical features of severe DENV in humans. ADE of DENV-2 or DENV-3 infection in LysM Cre+ Ifnarf/f mice resulted in plasma leakage, elevated levels of proinflammatory and vasoactive cytokines in blood, liver injury, hemoconcentration, and thrombocytopenia. We used this more immunocompetent model to establish the therapeutic activity against DENV of a bispecific antibody-based Fc-DART and a RIG-I receptor immunomodulatory agonist.
A prior study showed that LysM Cre+ Ifnarf/f mice were vulnerable to DENV-2 infection (40). Rather than try to augment DENV disease using enhancing antibodies as was done in our study, this group showed that an Ifnar deficiency on CD11c+ dendritic cells or LysM+ macrophages resulted in complete lethality after infection with a 10-fold-higher dose of DENV-2. The mice lacking Ifnar expression only on myeloid cell subsets developed a robust DENV-specific CD8+ T cell response, compared to a weak response in Ifnar−/− mice, and sustained a protective immune response to a candidate subunit vaccine. Our experiments confirm and extend these findings. First, in the direct DENV-2 infection model in LysM Cre+ Ifnarf/f mice, we observed increased viral infection, elevated levels of cytokines, and altered laboratory parameters without significant mortality, which is characteristic of primary DENV infection in humans. These results confirm that protection of mice from DENV infection depends on type I IFN signaling in LysM-expressing myeloid cells. Second, we showed greater DENV infection and disease in LysM Cre+ Ifnarf/f mice when preexisting enhancing anti-prM and anti-E antibodies were present. This led to the development of a vascular leakage syndrome, which parallels that seen in severe DENV (60). Third, we extended our findings in LysM Cre+ Ifnarf/f mice to a second DENV serotype, using a nonadapted DENV-3 isolate.
A criticism of existing Ifnar-deficient mouse models of DENV is that the type I IFN receptor signaling pathway is required for optimal antigen-specific T and B cell priming, expansion, and memory formation (40). The LysM Cre+ Ifnarf/f mouse model overcomes these limitations and more closely recapitulates several aspects of human disease. The deletion of Ifnar expression on LysM+ mouse myeloid cells overcomes the species immune antagonism issues and enables higher levels of DENV infection and inflammatory responses in myeloid cells, which are the natural cellular targets in human disease (41). Although LysM Cre+ Ifnarf/f mice supported increased DENV replication, showed signs of illness, and developed thrombocytopenia during primary infection, they did not develop hemoconcentration, vascular leakage, or lethality when virus alone was administered. In comparison, when enhancing antibodies against prM and E were provided, the viral burden was greater, and this was associated with more severe disease, including death, as is seen in the most severe cases of dengue in humans.
Another limitation of DENV infection experiments in Ifnar−/− or AG129 mice is that most agonists that modulate innate immune responses cannot be evaluated. There is increasing interest in the development of molecules that can stimulate IRF3 and/or type I IFN responses as means of controlling virus infection. Such agents may be important particularly in the context of DENV, which can inhibit IRF3 and IFN induction by virtue of its ability to antagonize STING-dependent responses (37, 38). However, such agents would not induce robust antiviral responses in mice lacking type I IFN responses in all cells. Indeed, 5′-ppp RNA moieties that activate RIG-I had no therapeutic effect against DENV in Ifnar−/− mice. In comparison, administration of the M8 5′-ppp RNA pathogen-associated molecular pattern protected LysM Cre+ Ifnarf/f mice from severe DENV disease. These experiments establish the utility of the conditional LysM Cre+ Ifnarf/f mouse model for testing possible novel immunomodulatory therapies against DENV.
We previously generated a bispecific tetravalent Ig-DART (E60 and 4E11) and showed protective activity against DENV in AG129 mice (49). In the current study, we used a different cross-reactive DII fusion loop MAb (E119) and a sequence-optimized version of a group-specific MAb (4E11) (52) to create a new bispecific tetravalent Fc-DART with an N297Q modified Fc region that cannot promote ADE in vitro or in vivo. This Fc-DART showed marked protective activity against DENV-2 and DENV-3 in LysM Cre+ Ifnarf/f mice and prevented clinical disease and laboratory parameter abnormalities. These experiments confirm the utility of Fc-modified-antibody-based therapeutics against DENV (24, 26).
In summary, we established LysM Cre+ Ifnarf/f mice as a more immunocompetent model of antibody-enhanced DENV infection in vivo. We demonstrated the utility of this model by administering antibody- or innate immune-based therapeutic agents against two different DENV serotypes, including a non-mouse-adapted DENV-3 strain. Future studies are planned to evaluate the pathogenesis of the remaining two DENV serotypes (DENV-1 and DENV-4) and assess whether disease occurs in LysM Cre+ Ifnarf/f mice using circulating, contemporary, and other non-mouse-adapted isolates.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381-01). Dissections and injections were performed under anesthesia that was induced with ketamine hydrochloride and xylazine.
Viruses and cells.
DENV-2 strain D2S20 is a mouse-adapted strain and has been described previously (33). The DENV-3 strain is a non-mouse-adapted DENV-3 Thai human isolate (strain C0360/94) (27). All viruses were passaged in C6/36 Aedes albopictus cells and ultracentrifuged (30,000 RPM for 3 h in an SW32 rotor [110,500 × g]) through a 25% glycerol cushion. Pelleted virus was resuspended in 10 mM Tris, pH 8.0, 150 mM NaCl, and 1 mM EDTA, and stored at −80°C. The titers of virus stocks were determined by a focus-forming assay on Vero cells (49).
Mice.
WT C57BL/6 mice were purchased commercially (Jackson Laboratories). Ifnar−/− mice (61) were backcrossed for 10 generations onto the C57BL/6 background. LysM Cre+ Ifnarf/f mice were obtained from R. Schreiber (St. Louis, MO) and U. Kalinke (Hannover, Germany). The Ifnarf/f (62) and LysM Cre+ mice were backcrossed using speed congenic analysis to 99% C57BL/6 as judged by microsatellite analysis. Mice (4 to 5 weeks old) were inoculated intravenously with DENV-2 (D2S20) or DENV-3 (C0360/94).
Analysis of Ifnar expression.
Blood was obtained by intracardiac heart puncture, and spleens were recovered. Live cells were stained with MAbs specific for CD11c, CD3, class II major histocompatibility complex (MHC), CD11b, B220, and Ifnar (BioLegend) to define cell types and determine Ifnar expression. All samples were processed on an LSRII or Fortessa flow cytometer (BD Biosciences). The resulting data were analyzed using FlowJo (Treestar).
Fc-DART generation.
To humanize WNV-E119 antibody (50), the complementarity-determining regions (CDRs) from VH and VL were grafted onto the most homologous human germline antibody framework sequences, VH3-48/VH3-23 and Vκ-A30/Vκ-L15, respectively. For humanization of the 4E11 antibody (52), the CDRs from VH and VL were grafted onto the human germline antibody framework sequences VH1-46 and Vκ-B3, respectively. Necessary framework back mutations were engineered to rescue the antibody binding affinity. The Fc-DART was produced from plasmids that coexpressed two polypeptide chains: chain 1, with the humanized VL domain of WNV-E119 linked to the humanized VH domain of 4E11, followed by a K coil sequence, and chain 2, with the humanized VL of 4E11 linked to the humanized VH of WNV-E119, followed by an E coil sequence and the CH2 and CH3 of the human γ1 constant region, containing an N297Q point mutation that abolishes C1q and FcγR interactions (63). The oppositely charged E coil and K coil promote the heterodimerization of chain 1 and chain 2, and this assembly is stabilized further by an interchain disulfide bond within the E-coil/K-coil domain. The Fc-DART was expressed in CHO-S cells and purified from supernatants by serial protein A affinity and Superdex 200 size exclusion chromatography to generate purified recombinant material.
Animal treatments. (i) ADE studies.
To induce ADE, mice were treated via the intravenous route with the flavivirus cross-reactive antibody 4G2 (64) (10 µg/mouse) for DENV-3 or with the combination of dengue complex-specific anti-prM antibody 2H2 (64) and 4G2 (15 µg of each MAb per mouse) for DENV-2 one day prior to infection.
(ii) Fc-DART therapy.
Mice were treated via an intraperitoneal route with 25 µg/mouse of h4E11–WNV-hE119 N297Q Fc-DART or negative-control recombinant (type-specific DENV-4 hE88 N297Q [13]) IgG either one day prior to infection or at 48 h after infection.
(iii) RIG-I agonist treatment.
Mice were treated with a single dose (5 µg/mouse) of a 99-nucleotide, uridine-rich hairpin 5′-ppp RNA termed M8 (53, 65) or a synthetic RNA control lacking 5′-ppp control combined with jetPEI transfection reagent (Polyplus) via intravenous injection before and after infection.
qRT-PCR analysis of viral burden.
Fluorogenic quantitative RT-PCR (qRT-PCR) was used to measure viral genome copy number. Total RNA from organs or serum of infected animals was isolated using the Qiagen RNeasy kit or the Qiagen viral RNA isolation kit. DENV viral RNA was determined using the pan-DENV primer probe set (66), which recognizes a conserved sequence in the 3′ untranslated region of all DENV serotypes. Viral copy number was determined using a defined positive single-stranded RNA generated in vitro using T7 polymerase containing the DENV target sequences.
Neutralization assay.
Focus-forming reduction neutralization assays (FRNTs) were performed as described previously (49) to determine the 50% inhibitory concentrations of MAbs and Fc-DARTs against the virus isolates used to infect mice. Infected foci were enumerated by counting using a CTL-Immunospot S6 (Cellular Technology Limited).
Clinical hematology and chemistry analysis.
At specified times after DENV infection of mice, blood was collected by intracardiac heart puncture. Blood for clinical hematology analysis was collected into EDTA-coated tubes (Becton Dickinson), and analyzed using a ProCyte DX machine (IDEXX). Serum was collected for clinical chemistry analysis of alanine transaminase (ALT), aspartate amino transferase (AST), and blood urea nitrogen (BUN) using the ProCyte DX machine.
Cytokine and chemokine measurements.
The BioPlex Pro assay (Bio-Rad) was performed according to the manufacturer’s protocol on serum isolated at day 4 postinfection. The cytokine screen included IL-1α, IL-1β, IL-2, IL-3 IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-17, Eotaxin, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, KC, monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), MIP-1β, RANTES (CCL5), and tumor necrosis factor alpha (TNF-α).
Vascular permeability analysis.
Two hundred microliters of sterile filtered Evans blue solution (0.5% [wt/vol]) in phosphate-buffered saline (PBS) was injected intravenously. After 20 min, mice were sacrificed and perfused with 40 ml of PBS. Livers, spleens, small intestines, and kidneys were collected into tubes containing formamide (Fisher Scientific), homogenized, weighed, and incubated overnight at 25°C. The following day, samples were centrifuged at 14,000 × g and 100 µl of supernatant was quantified at 610 nm against a standard curve generated by serial dilutions of a stock Evans blue solution.
Statistical analysis.
All data were analyzed using Prism software (GraphPad, San Diego, CA). Kaplan-Meier survival curves were analyzed by the log rank test. Differences in results for viral burden, clinical chemistry and hematology, cytokine levels, and Evans blue staining were analyzed by the Mann-Whitney test.
ACKNOWLEDGMENTS
We gratefully acknowledge Ulrich Kalinke and Robert Schreiber for providing the Ifnarf/f and LysM Cre+ Ifnarf/f mice and William Tang and William Eddy for technical assistance in some of the experiments.
This work was supported by the Burroughs Wellcome Fund (M.S.D.), NIH grants R01 AI077955 (M.S.D.), R01 AI108861 (J.H.), U54 AI057517 (the Southeastern Regional Center of Excellence for Emerging Infectious and Biodefense), and R56 A1085063 (S.S.). V.V.S. was supported by a T32 postdoctoral fellowship (NIAID AI 7536-13).
Footnotes
Citation Pinto AK, Brien JD, Lam C-YK, Johnson S, Chiang C, Hiscott J, Sarathy VV, Barrett AD, Shresta S, Diamond MS. 2015. Defining new therapeutics using a more immunocompetent mouse model of antibody-enhanced dengue virus infection. mBio 6(5):e01316-15. doi:10.1128/mBio.01316-15.
REFERENCES
- 1.Halstead SB, Nimmannitya S, Cohen SN. 1970. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J Biol Med 42:311–328. [PMC free article] [PubMed] [Google Scholar]
- 2.Kliks SC, Nimmanitya S, Nisalak A, Burke DS. 1988. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am J Trop Med Hyg 38:411–419. [DOI] [PubMed] [Google Scholar]
- 3.Chau TN, Quyen NT, Thuy TT, Tuan NM, Hoang DM, Dung NT, Lien LB, Quy NT, Hieu NT, Hieu LT, Hien TT, Hung NT, Farrar J, Simmons CP. 2008. Dengue in Vietnamese infants—results of infection-enhancement assays correlate with age-related disease epidemiology, and cellular immune responses correlate with disease severity. J Infect Dis 198:516–524. doi: 10.1086/590117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4:229–238. doi: 10.1016/j.chom.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond MS, Pierson TC. 2015. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 162(3):488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brien JD, Tsai WY, Wang WK, Halstead S, Kliks S, Diamond MS, Baric R, Lanzavecchia A, Sallusto F, de Silva AM. 2011. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis 5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE Jr.. 2014. Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol 88:12233–12241. doi: 10.1128/JVI.00247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, Diamond MS. 2010. Genotype specific neutralization and protection by antibodies against dengue virus type 3. J Virol 84:10630–10643. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. 2010. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol 84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O’Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. 2010. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog 6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. 2013. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol 87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gromowski GD, Barrett AD. 2007. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366:349–360. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 15.Gromowski GD, Barrett ND, Barrett AD. 2008. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 virus. J Virol 82:8828–8837. doi: 10.1128/JVI.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, de Silva AM. 2010. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog 6:e1000821. doi: 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE Jr., de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med 4:139ra183. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- 19.Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, Crowe JE Jr., Lok SM. 2014. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med 6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, Lok SM. 2015. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun 6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, Pfaff JM, Xiang X, Doranz BJ, de Silva AM, Austin SK, Sukupolvi-Petty S, Diamond MS, Crowe JE Jr.. 2013. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. mBio 4(6):e00873-13. doi: 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Desprès P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA. 2015. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 23.Guzman MG, Alvarez M, Halstead SB. 2013. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch Virol 158:1445–1459. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 24.Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E. 2010. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog 6:e1000790. doi: 10.1371/journal.ppat.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zellweger RM, Prestwood TR, Shresta S. 2010. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe 7:128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams KL, Sukupolvi-Petty S, Beltramello M, Johnson S, Sallusto F, Lanzavecchia A, Diamond MS, Harris E. 2013. Therapeutic efficacy of antibodies lacking Fcγ receptor against lethal dengue virus infection is due to neutralizing potency and blocking of enhancing antibodies. PLoS Pathog 9:e1003157. doi: 10.1371/journal.ppat.1003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarathy VV, White M, Li L, Gorder SR, Pyles RB, Campbell GA, Milligan GN, Bourne N, Barrett AD. 2015. A lethal murine infection model for dengue virus 3 in AG129 mice deficient in type I and II interferon receptors leads to systemic disease. J Virol 89:1254–1266. doi: 10.1128/JVI.01320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs J, Chu H, O’Day P, Pyles R, Bourne N, Das SC, Milligan GN, Barrett AD, Partidos CD, Osorio JE. 2014. Investigating the efficacy of monovalent and tetravalent dengue vaccine formulations against DENV-4 challenge in AG129 mice. Vaccine 32:6537–6543. doi: 10.1016/j.vaccine.2014.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milligan GN, Sarathy VV, Infante E, Li L, Campbell GA, Beatty PR, Harris E, Barrett AD, Bourne N. 2015. A dengue virus type 4 model of disseminated lethal infection in AG129 mice. PLoS One 10:e0125476. doi: 10.1371/journal.pone.0125476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry ST, Prestwood TR, Lada SM, Benedict CA, Shresta S. 2009. Cardif-mediated signaling controls the initial innate response to dengue virus in vivo. J Virol 83:8276–8281. doi: 10.1128/JVI.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, Beatty R, Harris E. 2012. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J Gen Virol 93:2152–2157. doi: 10.1099/vir.0.045088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, Lada SM, Shresta S. 2012. Gamma interferon (IFN-γ) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-α/β receptor-deficient mice. J Virol 86:12561–12570. doi: 10.1128/JVI.06743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makhluf H, Buck MD, King K, Perry ST, Henn MR, Shresta S. 2013. Tracking the evolution of dengue virus strains D2S10 and D2S20 by 454 pyrosequencing. PLoS One 8:e54220. doi: 10.1371/journal.pone.0054220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zellweger RM, Shresta S. 2014. Mouse models to study dengue virus immunology and pathogenesis. Front Immunol 5:151. doi: 10.3389/fimmu.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashour J, Laurent-Rolle M, Shi PY, García-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, García-Sastre A. 2010. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe 8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto AK, Ramos HJ, Wu X, Aggarwal S, Shrestha B, Gorman M, Kim KY, Suthar MS, Atkinson JP, Gale M Jr., Diamond MS. 2014. Deficient IFN signaling by myeloid cells leads to MAVS-dependent virus-induced sepsis. PLoS Pathog 10:e1004086. doi: 10.1371/journal.ppat.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Züst R, Toh YX, Valdés I, Cerny D, Heinrich J, Hermida L, Marcos E, Guillén G, Kalinke U, Shi PY, Fink K. 2014. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: implications for a new mouse model to test dengue vaccines. J Virol 88:7276–7285. doi: 10.1128/JVI.03827-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. 2008. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology 376:429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prchal M, Pilz A, Simma O, Lingnau K, von Gabain A, Strobl B, Müller M, Decker T. 2009. Type I interferons as mediators of immune adjuvants for T- and B cell-dependent acquired immunity. Vaccine 27(Suppl 6):G17–G20. doi: 10.1016/j.vaccine.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Crouse J, Kalinke U, Oxenius A. 2015. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 45.Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, Smit JM, Fremont DH, Pierson TC, Heise MT, Diamond MS. 2013. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog 9:e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rathakrishnan A, Klekamp B, Wang SM, Komarasamy TV, Natkunam SK, Sathar J, Azizan A, Sanchez-Anguiano A, Manikam R, Sekaran SD. 2014. Clinical and immunological markers of dengue progression in a study cohort from a hyperendemic area in Malaysia. PLoS One 9:e92021. doi: 10.1371/journal.pone.0092021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathakrishnan A, Wang SM, Hu Y, Khan AM, Ponnampalavanar S, Lum LC, Manikam R, Sekaran SD. 2012. Cytokine expression profile of dengue patients at different phases of illness. PLoS One 7:e52215. doi: 10.1371/journal.pone.0052215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aye KS, Charngkaew K, Win N, Wai KZ, Moe K, Punyadee N, Thiemmeca S, Suttitheptumrong A, Sukpanichnant S, Prida M, Halstead SB. 2014. Pathologic highlights of dengue hemorrhagic fever in 13 autopsy cases from Myanmar. Hum Pathol 45:1221–1233. doi: 10.1016/j.humpath.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 49.Brien JD, Sukupolvi-Petty S, Williams KL, Lam CY, Schmid MA, Johnson S, Harris E, Diamond MS. 2013. Protection by immunoglobulin dual-affinity retargeting antibodies against dengue virus. J Virol 87:7747–7753. doi: 10.1128/JVI.00327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J Virol 80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thullier P, Demangel C, Bedouelle H, Mégret F, Jouan A, Deubel V, Mazié JC, Lafaye P. 2001. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J Gen Virol 82:1885–1892. [DOI] [PubMed] [Google Scholar]
- 52.Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. 2012. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure 20:303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Chiang C, Beljanski V, Yin K, Olagnier D, Ben Yebdri F, Steel C, Goulet ML, DeFilippis VR, Streblow DN, Haddad EK, Trautmann L, Ross T, Lin R, Hiscott J. 2015. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J Virol 89:8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osorio JE, Velez ID, Thomson C, Lopez L, Jimenez A, Haller AA, Silengo S, Scott J, Boroughs KL, Stovall JL, Luy BE, Arguello J, Beatty ME, Santangelo J, Gordon GS, Huang CY, Stinchcomb DT. 2014. Safety and immunogenicity of a recombinant live attenuated tetravalent dengue vaccine (DENVax) in flavivirus-naive healthy adults in Colombia: a randomised, placebo-controlled, phase 1 study. Lancet Infect Dis 14:830–838. doi: 10.1016/S1473-3099(14)70811-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon IK, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A. 2014. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384:1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 57.Villar L, Dayan GH, Arredondo-García JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramírez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortés Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F. 2015. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med 372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- 58.Kirkpatrick BD, Durbin AP, Pierce KK, Carmolli MP, Tibery CM, Grier PL, Hynes N, Diehl SA, Elwood D, Jarvis AP, Sabundayo BP, Lyon CE, Larsson CJ, Jo M, Lovchik JM, Luke CJ, Walsh MC, Fraser EA, Subbarao K, Whitehead SS. 2015. Robust and balanced immune responses to all 4 dengue virus serotypes following administration of a single dose of a live attenuated tetravalent dengue vaccine to healthy, flavivirus-naive adults. J Infect Dis 212:702–710. doi: 10.1093/infdis/jiv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shresta S, Sharar KL, Prigozhin DM, Beatty PR, Harris E. 2006. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J Virol 80:10208–10217. doi: 10.1128/JVI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guzman MG, Harris E. 2015. Dengue. Lancet 385:453–465. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- 61.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 62.Le Bon A, Durand V, Kamphuis E, Thompson C, Bulfone-Paus S, Rossmann C, Kalinke U, Tough DF. 2006. Direct stimulation of T cells by type I IFN enhances the CD8+ T cell response during cross-priming. J Immunol 176:4682–4689. doi: 10.4049/jimmunol.176.8.4682. [DOI] [PubMed] [Google Scholar]
- 63.Tao MH, Morrison SL. 1989. Studies of aglycosylated chimeric mouse-human IgG. Role of carbohydrate in the structure and effector functions mediated by the human IgG constant region. J Immunol 143:2595–2601. [PubMed] [Google Scholar]
- 64.Brandt WE, McCown JM, Gentry MK, Russell PK. 1982. Infection enhancement of dengue type 2 virus in the U-937 human monocyte cell line by antibodies to flavivirus cross-reactive determinants. Infect Immun 36:1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. 2013. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog 9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gurukumar KR, Priyadarshini D, Patil JA, Bhagat A, Singh A, Shah PS, Cecilia D. 2009. Development of real time PCR for detection and quantitation of dengue viruses. Virol J 6:10. doi: 10.1186/1743-422X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]