FIG 4 .
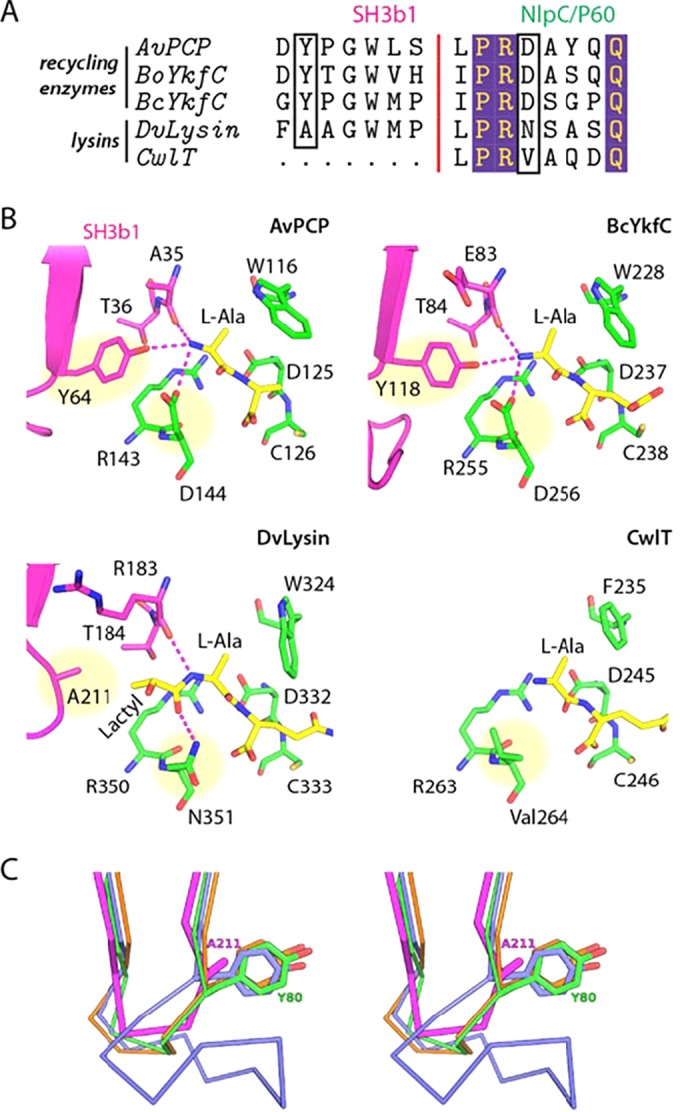
Residues at the SH3b1-NlpC/P60 domain interface affect substrate specificity in cell wall recycling enzymes and lysins. (A) Sequence alignment of the S3 and S2 subsites in CwlT, DvLysin, YkfCs, and AvPCP. The residues that we propose are the key determinants of substrate specificity for cell wall lysins and recycling enzymes are highlighted by black boxes. (B) Close-up views of the S3 and S2 subsites in lysins and recycling enzymes. Residues are colored by domain (magenta, SH3b1; green, NlpC/P60), while substrates (modeled or observed) are colored in yellow. The key residues indicated in panel A are highlighted by yellow circles. Hydrogen bonds are shown as dashed lines. (C) Comparison of the distal loops of DvLysin (magenta), BoYkfC (green), BcYkfC (blue), and AvPCP (orange), shown in stereoview. Side chains at the position equivalent to Ala211 of DvLysin are shown as sticks.
