Abstract
Rare genetic variants may be responsible for a significant amount of the uncharacterized genetic risk underlying many diseases. An efficient approach to characterizing the disease burden of rare variants may be to impute them into existing large datasets. It is well-known that the ability to impute a rare variant is dependent both on the array choice and number of individuals in the reference panel carrying that variant, though it is still unclear exactly how well imputation will work for rare variants. We review here the additional challenges that arise when imputing rare variants, looking at studies that have been able to impute rare variants, methods behind merging reference panels, approaches for imputing rare variants, and methods for analyzing rare variants.
Keywords: rare variants, genotype imputation, genome-wide association study
Efficiently testing for missing GWAS heritability through rare variant imputation
Genome-wide association studies (GWAS, see Glossary) genotype and test millions of genetic variants across the entire genome for association with various traits. GWAS have detected a large number of clearly replicated novel associations [1], with increasing numbers of variants found as study sample sizes grow [2]. GWAS have done particularly well at finding associations between common variants and traits, although the magnitude of associations have been much smaller than originally anticipated [3]. Genome-wide significant findings from GWAS only explain a limited proportion of the genetic variance (i.e., heritability) for many phenotypes [4,5]. This proportion is increased when considering all variants typed or tagged by GWAS arrays, although substantial heritability remains unexplained (i.e., the so-called “missing heritability”). Rare variants are one potential source of this missing heritability [6–8], although due to their rarity they may contribute very little to heritability individually. Note that in general, investigators refer to common variants as those with minor allele frequency (MAF) greater than 5%, less common variants as those with MAF between 1% and 5%, and rare variants as those with MAF less than 1%.
Next-generation sequencing is one approach to assaying rare variants. However, this remains prohibitively expensive for studying large sample sizes, which may be necessary to have sufficient statistical power for detecting rare variant associations. A much more affordable option is to type study subjects using genotyping arrays designed to measure rare variants. For example, recent GWAS arrays have been designed with increasingly rarer variation in them, such as arrays utilizing 1000 Genomes Project content in design (e.g., Illumina Omni 2.5 and 5, http://www.illumina.org, and Affymetrix Axiom arrays [9,10]), and Exome arrays of rare potentially functional content (e.g., utilizing all variants likely to be real in the Exome Sequencing Project (http://esp.gs.washington.edu).
An even more efficient approach for studying rare variants is to impute them into existing genome-wide data. Genotype imputation entails predicting genetic variants that have not been directly assayed in study subjects by matching their measured haplotypes to haplotypes from a reference panel genotyped at additional loci. That is, imputation uses complete haplotype information from a reference panel to estimate missing genetic variants in one’s study subjects. This approach began as a method to combine different GWAS arrays that genotyped different sets of variants [11]. Nowadays, imputing GWAS arrays to reference panels prior to undertaking association analyses has become a standard approach to increase statistical power and improve signal resolution [12–14]. Imputation of rare variants by leveraging information across haplotypes is becoming increasingly feasible, even when there is low pairwise correlation between rare variants and neighboring alleles.
However, a number of challenges remain for imputing rare variants. The ability to impute rare variants depends both on the GWAS array that was used and the number of reference samples used for imputation [15,16]. We will first discuss choices of reference panels used for imputation, and methods for creating a custom reference panel with additional sequence data. We then present approaches for analyzing rare genetic variants arising from imputation efforts. Finally, we examine several successful strategies and applications of rare variant imputation, making note of how the imputation was done, in particular what reference panels were used.
Overall process for imputing and analyzing rare variants
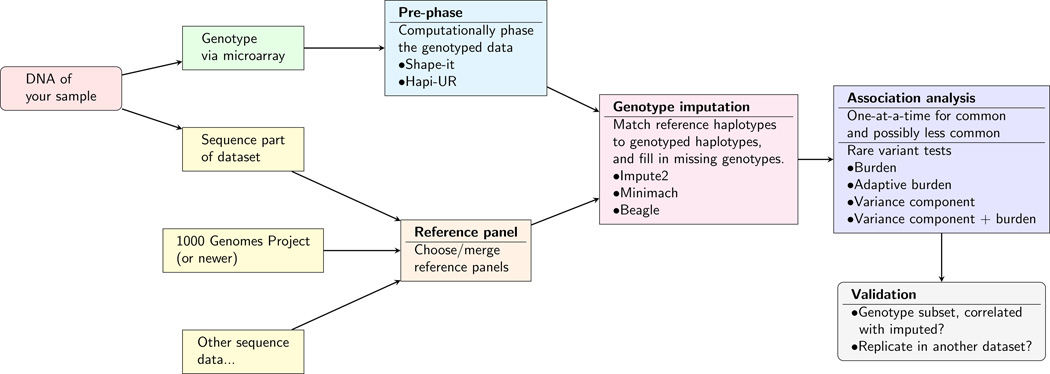
Figure 1 outlines the overall process for imputing and analyzing rare variants. The first step is choosing or creating reference panels for imputation (Box 1). With earlier imputation methods, selecting reference panels that closely matched the race/ethnicity of your sample was important. However, more recent methods are able to appropriately use ethnically diverse imputation panels, which can in turn improve imputation, especially in admixed populations, and for rare variants that may be more common in other ancestries [15].
Figure 1. Rare variant imputation process.
A flowchart describing the steps in imputing rare variants into genome-wide SNP datasets.
Box 1. Selection and development of Reference Panels.
Existing and future reference panels
Imputation reference panels have advanced rapidly in the past decade. The first set of samples used in such panels was the HapMap Phase II releases starting in 2006 [42], followed by the Phase III data in 2009 [43]. These were superceeded by the main releases of the 1000 Genomes Project. The Project began with the Pilot release, which include 60 CEU, 60 CHB and JPT, and 60 YRI individuals [44]. This was followed by the Phase I integrated release in 2012 of 1,092 individuals, combining low coverage (~4×) genome-wide data with high-coverage exomesequence data (~20–60×). In late 2014, the interim Phase III dataset with 2,504 individuals was released, more than doubling the number of rare variants (Table I). Since multiple individuals with a rare variant are needed to impute it well, the progression of larger and more comprehensive reference panels has substantially improved our ability to impute increasingly rarer genetic variants.
Sources of additional imputation reference panel information
There are various different sources for additional reference panel information. One is to use a subset of your own study samples as a reference panel (Figure 1). These individuals can be whole-genome or whole-exome sequenced for novel variants or to enrich reference panels with population-specific variants. Individuals could also be genotyped with custom or off-the shelf exome arrays, which may in addition include a GWAS backbone of a few hundred thousand SNPs with relatively high genome-wide coverage. Finally, individuals can be genotyped at a particular known locus to be used for imputation (see comments about buffers to use below in the Merging reference panels section). Note that when using your own samples to discover new rare variant content, also using them in ones rare variant association analysis can be problematic [45].
Merging reference panels
The process of merging reference panels—either just existing datasets, or an existing dataset, such as the 1000 Genomes Project, with ones own custom panel—is an area of active research. Early work focused on the situation where one reference panel contains a subset of the variants in another reference panel, and modeled them jointly (multi-panel imputation) [28]. If one has additional reference panel data on study samples and existing datasets, then it may be possible to re-call the genotypes of all of the sequenced datasets together, or at least at the loci identified in each of the panels. Another approach proposed in the newest Impute2 software (v2.3.2) is to combine reference panels by first treating them as reference panels for each other, and imputing them into each other, using the best guess allele for the haplotype [19]. The performance of this approach has not yet been fully explored.
In addition to merging entire reference panels for genome-wide imputation, it can also be useful to perform imputation on a more focused region. One might also want to follow up known association regions from GWAS, and fine map them by attempting a more thorough imputation process around a locus by using a custom more detailed and/or ethnically diverse reference panel. A window of approximately 0.5Mb past both ends of the region is probably sufficient to ensure enough of a buffer to identify haplotypes, but the exact useful size may depend on the region. It may also be useful to rephase the final reference panel if one is trying to impute a specific rare variant that was potentially not phased well in one of the reference panel groups (e.g., if a private mutation exists only in one of the groups).
Performing genotype imputation
After a reference panel has been decided upon or created, imputation proceeds in two steps (Figure 1). First, the genome-wide array data is computationally pre-phased, as the genotyping process only measures genotypes, not phase (i.e., the haplotypes). The term pre-phasing came from imputation methodology where there was a process of phasing many times for imputation, before results showed that phasing just once produced similar accuracy, but for a fraction of the computational cost [17]. The current most popular pieces of software for pre-phasing are Shapeit [17] and Hapi-ur [18].
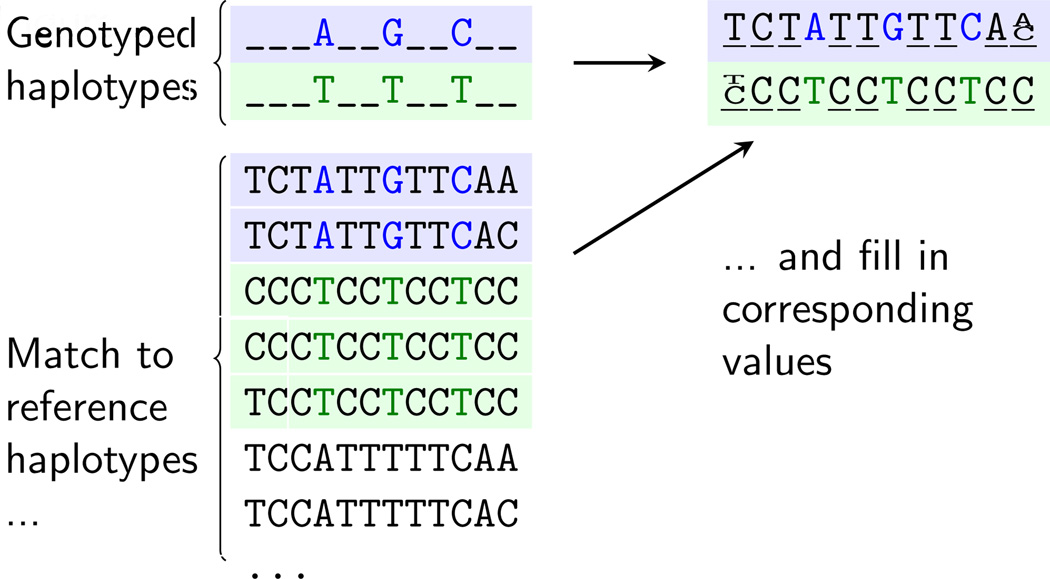
After pre-phasing, the genotypes are imputed by matching haplotype segments from the study's individuals to the reference haplotypes (which has been more densely genotyped, e.g., sequenced). This is usually done in sliding windows of 5Mb or less across the genome, with a small amount of overlap (e.g., 0.5Mb) to ensure enough of a buffer to identify the haplotypes [19]. A simplified version of this approach is depicted in Figure 2; in practice many more haplotypes are matched, resulting in a probabilistic outcome for each imputed genotype. That is, there is usually some uncertainty from this process as to which genotype an individual will have due to incomplete haplotype matching. To use the probabilities in association analyses, a simple approach is to convert them into their best guess. But it is generally better to take the uncertainty into account (discussed below). Current commonly used software for this process includes Impute2 [15,19,20], Minimac [21], and Beagle [22].
Figure 2. Genotype imputation.
Variants are imputed by matching up haplotypes from the sample to be analyzed to haplotypes in a reference panel. Note that they are generally matched to more haplotypes than shown; rare variants in particular can be limited by the sample size of the reference sample, which indicates how many haplotypes can be used in the matching process. The corresponding values are filled in probabilistically, that is, 1/3 T and 2/3 C for example for the first loci of the second genotyped haplotype. One can convert this to a “best guess” genotype, which, in the example described before, would be C.
Following imputation, the resulting common and rare variants can be tested for association with the phenotype of interest. Individually testing the association between each variant and the phenotype is the standard approach for GWAS. This approach works well if the variant is sufficiently common and/or has a strong enough effect on the phenotype. However, due to the uncommon nature of rare variants, analyzing them individually can result in imprecise estimates of effect and low power to detect associations. To address this issue one can aggregate the rare variants together for their analysis. There are a large number of different rare variant methods that analyze together rare variants, for example, in a gene (or even pathway), to increase the power to detect associations of multiple variants with phenotypes (Box 2).
Box 2. Association analysis approaches.
These methods make a range of different assumptions and the corresponding tests can be categorized as burden, adaptive burden, variance components, and hybrid [46]. The power of these methods varies considerably depending on how realistic are their assumptions. Some tests sacrifice a small amount of power when all of the assumptions are true, and behave much better in other situations. Burden tests collapse rare variants into genetic scores for testing, and include the CAST [47] and CMC [48] methods. These methods use a priori information for collapsing, such as if a variant alters the protein coding, and if it is predicted to be damaging using information such as provided in PolyPhen 2 [49], or MutationTaster [50]. They also assume that all rare variants have the same direction of effect on the phenotype (e.g., are deleterious). They are powerful if the assumptions are correct, and a large proportion of the variants are causal.
Adaptive burden tests use data-adaptive weights, thresholds, or the best of several different weights and thresholds when computing risk scores [51–54]. They are more robust than burden tests, but tend to be computationally intensive. Variance components tests evaluate the variance of genetic effects, and perform particularly well when only a small fraction of the variants are causal, and if there are both deleterious and beneficial variants [55,56]. One of the more popular such tests for rare variants is the SKAT test [55]. Finally, there are tests that try to combine the best of burden tests and variance components tests, such as SKAT-O [57]. Several of these methods have their own pieces of software and have implementations in R [58]; standalone packages that incorporate multiple rare variant tests include Plinkseq (http://atgu.mgh.harvard.edu/plinkseq), Epacts (http://genome.sph.umich.edu/wiki/EPACTS), Granvil (http://www.well.ox.ac.uk/GRANVIL) [59], Rvtests (http://genome.sph.umich.edu/wiki/Rvtests), Score-Seq (http://dlin.web.unc.edu/software/score-seq), and VAT (http://varianttools.sourceforge.net/Association/HomePage) [60].
As an example of applying these rare variant tests, Zawistowski and colleagues were the first to show that burden tests also work well with imputed dosages with their CMAT test, and provided an example applied to Psoriasis finding a novel gene SKIVL, which contained four less common variants that each trended towards significance but did not alone explain the signal [61]. Magi and colleagues provide a second successful example of this approach (discussed above in the successful rare variants examples) [27].
Imputed SNPs are analyzed in a similar manner as genotyped SNPs, but the best performing association methods account for imputation uncertainties [23]. One can do this by converting the imputed SNPs to probabilistic dosages. These are effectively the additive coding of the SNP, i.e., the number of copies an allele carried by an individual), but also incorporating the imputation uncertainty. The dosage is given by 2*Pr(genotype=AA) + 1*Pr(genotype=Aa) + 0*Pr(genotype=aa). So individuals with a dosage of 2 are extremely confident they are genotype AA, with decreasing confidence for 1.9 and 1.8, down to 1.5, where they are equally likely to be AA vs. Aa. This dosage can be analyzed using various imputation output formats (or conversion utilities to convert them) with Plink [24], with a beta version 1.9 in development that supports even more formats [25]. The other most popular software for dealing with imputed data, Snptest, can compute tests utilizing dosages, as well as allows for a few other different tests, as described in [20].
If an association is found, there are two steps to ensure that the variant has imputed correctly and the association is real. Follow-up genotyping on the variant, at least on a subset of your individuals, can confirm if the variant imputed correctly. Ideally the correlation of the imputed genotype and the actual genotype will have a correlation similar to the estimated r2 from imputation (e.g., the rinfo2 estimate) [26]. For rare variants, it may be very useful to confirm the genotype by enriching the number of individuals who are imputed to have the rare variant, though this may slightly invent inflate the true r2 because of enrichment. We recommend a rough power analysis to determine how many individuals to genotype. Second, to confirm the association is real, the variant should be tested in an independent cohort, if available.
Successful rare variant imputation: strategies and examples
A number of strategies exist for imputing rare variants. The most straightforward is to use a single existing reference panel, such as the 1000 Genomes Project data. As increasing amounts of sequence data become publicly available in centralized repositories, this is becoming a common approach. As an example, Magi and colleagues used a total of 16,179 individuals with GWAS array genotypes from the Welcome Trust Case Control Consortium (WTCCC) and imputed up to the 1000 Genomes Project Phase I data [27]. They found a total of 5,383,228 rare variants imputed with rinfo2≥0.4, and 17.3% of these imputing very well (rinfo2≥0.8). Here, rinfo2 is a quality control metric that estimates the correlation of the imputed genotype to the true genotype [20]; often those rare variants with rinfo2≥0.3 are analyzed, although this threshold is somewhat arbitrary. Using the measured and imputed data, Magi et al. used a burden test approach (discussed below) and detected genome-wide significant evidence of association between rare variants in the PRDM10 gene and coronary artery disease, as well as rare variants in 10 genes in the MHC region and Type I diabetes [16].
A second imputation strategy is a hybrid approach that combines an existing reference panel (e.g., the 1000 Genomes Project data) with additional study-specific genotyping or sequencing data. This approach allows one to obtain rare variants among individuals with the particular phenotype of interest, which could be important if one expects such individuals to be more likely to carry particular (e.g., causal) rare variants. An interesting recent example of this approach, that highlights challenges with imputation, are studies of the well-known HOXB13 G84E prostate cancer mutation [28,29]. This mutation has a MAF of 0.0017 in the 1000 Genomes Project individuals of European ancestry (the mutation is of European ancestry). A previous study reported that they were unable to impute the G84E variant using large multi-panel imputation (described below) with a large custom reference panel from 5,500 prostate cancer cases and 4,923 controls typed on their iCOGS array plus G84E, 677 cases typed on the Illumina Omni 2.5 array, and 1000 Genomes subjects [30]. However, another study found an ancestral haplotype containing the mutation [31], suggesting that imputation might be possible. By using the hybrid imputation approach and combining 1000 Genomes Project data with an enriched set of prostate cancer cases who carried the mutation, Hoffmann and colleagues were able to successfully impute the G84E mutation into a large cohort with r2=0.57 (computed from a genotyped subset) [26]. Why one study was able to impute this mutation while another was not reflects the difficulty in imputing rare variants [16]. Possible explanations here include differences in the original GWAS array [26] and the imputation approach used, as well as potential difficulty in accurately estimating r2, which is less accurate for rare variants [14]. Note also that Hoffmann et al. used the imputed data to test for a pleiotropic effect of the HOXB13 mutation across multiple different cancers [26]; this highlights the value of using imputation of discovered risk variants into cohort studies to test for cross-phenotype effects.
A third imputation strategy is to only use custom genotyping or sequencing content. As an example of this approach, Jonsson and colleagues used a custom reference panel of 1,795 whole-genome sequenced Icelanders to impute into 71,743 Icelanders with GWAS data a very rare Alzheimer's disease risk variant (MAF 0.62% cases, 0.13% controls) [32]. Another study used 2,630 whole-genome sequenced Icelanders to impute variants in 11,114 cases and 267,140 controls in Danish and Iranian samples, and discovered four low frequency variants associated with BMI and Type 2 diabetes, including one variant with MAF 0.20% and 0.65% [33]. As another example, Auer and colleagues exome sequenced 761 African Americans and used these data as a custom reference panel for imputing genetic variants in over 13,000 African Americans; they were able to impute almost 200,000 rare variants well, and identified lowfrequency coding variants (MAF 2%–3%) that may affect blood cell-associated loci [34]. Another group showed that imputing variants with MAF 0.005–0.05 was feasible by using data on 1,962 African Americans in the Women's Health Initiative (WHI) cohort who were genotyped on both the Metabochip and Affymetrix 6.0 arrays, and used this to impute genotypes on 6,459 WHI SNP Health Association Resource (SHARe) study subjects who had only been genotyped on the Affymetrix 6.0 array [35]. Li and colleagues showed that imputation performance with custom panels depends strongly on reference panel size by sequencing the exons and flanking regions of over 14,000 individuals in 202 genes and imputing into GWAS data from Affymetrix 500k or 6.0, or Illumina 550k on 8,865 of those individuals [16].
While we have outlined three strategies above, one may also try multiple different strategies or several applications of the same strategy. For example, one of the first successful examples of imputing a rare variant detected an association between a variant with MAF=0.0038 and sick sinus syndrome by repeating the imputation process for each of multiple different reference panels (HapMap, 1000 Genomes Pilot, and a set of genotyped samples) [36].
Future Challenges
There are many future challenges for imputing rare variants, summarized in Box 3. As reference panels become larger, methods to analyze them must become more computationally efficient. Alternative strategies to merge reference panels may be necessary depending on the performance of current methods. In addition, individuals usually follow very reasonable QC procedures, but the impact of these remains poorly understood. Rare variants, and in particular singletons, may even be excluded in QC procedures. In addition different sequencing technologies and lower coverage depth may make it more difficult to detect rare/low frequency variants. This may make merging of reference panels for recovering rare variants very difficult. There are also challenges with analyzing rare variants. Usually ancestry is controlled for in non-homogeneous populations by adjusting for principal components [37] or through mixed model approaches [38]. However, it is not yet known if this will be enough of an adjustment for rare variants, as they may be extremely localized [39]. In addition, most methods of rare variants tests were proposed for gene-based tests, which worked particularly well for exome sequenced and exome array genotyped individuals. As many GWAS hits have been in noncoding regions, new methods for studying non-genic regions may be useful.
Box 3. Outstanding Questions.
How do we best merge reference panels that may be on different sequencing technologies and have different coverage steps, and what is the impact of this on imputing rare variants?
When analyzing rare variants, is the adjustment for ancestry enough?
How do we best extend rare variant analysis methods, which have typically been focused on gene regions, to non-genic regions, where many GWAS hits have been found?
Concluding remarks
Imputation provides a very cost-effective means for characterizing the genetic architecture of rare variants, especially as much larger reference panels become available. We have given various examples showing the successful imputation of rare variants from current best imputation reference panels, from reference panels combining additional data with existing reference datasets, and from using exclusively independent datasets. Sources for these additional reference data include whole genome and exome sequencing, as well as exome genotyping, or even very focused genotyping. As reference panels become much larger or existing panels are merged together, it may no longer be necessary to create additional reference samples from one's own data. While imputation will never be able to recover private mutations, we expect that it will provide a means to use the increasingly large amounts of available array data (e.g., the 100,000 individuals currently available from the RPGEH project, dbGap phs000674.v1.p1, and the 500,000 individuals soon to be available from the UK biobank, http://www.ukbiobank.ac.uk/) to help characterize rare genetic variation, especially as recent work suggests discovering rare variants may require extremely large sample sizes [40,41].
Box1 Table I.
Progression of numbers of rare and common variants in different reference panels.
| Population | Na | Private: Only one |
Rare (excluding private): Private<MAF≤0.01 |
Less Common: 0.01<MAF≤0.05 |
Common: 0.05≤MAF |
|---|---|---|---|---|---|
| HapMap 2 (r22) | 210 | - | 185,748 | 514,377 | 2,321,200 |
| HapMap 3 (r2) | 1,011 | 2,126 | 27,891 | 151,744 | 1,205,633 |
| 1000 Genomes Phase I (Sep 2013) | 1,092 | 7,745 | 25,283,503 | 5,386,140 | 7,201,411 |
| 1000 Genomes Phase III (October 2014) | 2,504 | 5,614 | 67,702,634 | 5,779,865 | 7,963,730 |
N is the number of individuals (the number of haplotypes is twice that).
Trends Box.
There have been an increasing number of success stories about imputing rare variants, using different strategies such as merging reference panels.
The number of sequenced individuals available for use as reference panels is greatly increasing, allowing us to better impute rarer variation
Tests that combine a group of similar rare variants (e.g., in a gene) initially intended for sequence data are being applied to imputed data, as it also begins to capture rarer variation
Acknowledgements
This work was supported by grants R21 AG046616 and K01 DC013300 to TJH, and U01 CA127298 and R01 CA088164 to JSW from the National Institutes of Health. This work was also supported by the UCSF Goldberg-Benioff Program in Cancer Translational Biology.
Glossary Box
- Genome-wide association study (GWAS)
a study testing for associations of genetic variants and disease across the entire genome (e.g., SNPs, insertions, deletions, copy number variants, etc.)
- Genotype imputation
filling in missing genotypes that were not directly assayed by matching haplotypes to a set of reference panel haplotypes with genotypes at additional locations
- Haplotype
a collection of specific alleles in a tightly-linked (correlated) region of the genome; these are generally spatially close to each other
- Missing heritability
term used to describe that studies have not yet explained a substantial proportion of the genetic variance (i.e., heritability) for many phenotypes.
- Private mutation
A mutation that occurs in only one individual
- Phase
determining what the two haplotypes are for a set of genotypes
- Rare variant
generally a variant with frequency less than 1%. Often a SNP, but can be insertions, deletions, etc..
- Single nucleotide polymorphism (SNP)
a SNV with frequency greater than 1%
- Single nucleotide variant (SNV)
a genetic locus that contains a single nucleotide variation amongst individuals in a population; usually used for frequency less than 1%
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindquist KJ, et al. The impact of improved microarray coverage and larger sample sizes on future genome-wide association studies. Genet. Epidemiol. 2013;37:383–392. doi: 10.1002/gepi.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witte JS. Genome-Wide Association Studies and Beyond. Annu. Rev. Public Health. 2010;31:9–20. doi: 10.1146/annurev.publhealth.012809.103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witte JS, et al. The contribution of genetic variants to disease depends on the ruler. Nat. Rev. Genet. 2014;15:765–776. doi: 10.1038/nrg3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickson SP, et al. Rare Variants Create Synthetic Genome-Wide Associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazer KA, et al. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 8.Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann TJ, et al. Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann TJ, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton PR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao K, et al. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet. 2009;10:27. doi: 10.1186/1471-2156-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L, et al. The Relationship between Imputation Error and Statistical Power in Genetic Association Studies in Diverse Populations. Am. J. Hum. Genet. 2009;85:692–698. doi: 10.1016/j.ajhg.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. Genotype Imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howie B, et al. Genotype Imputation with Thousands of Genomes. G3 Genes Genomes Genet. 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, et al. Performance of Genotype Imputation for Rare Variants Identified in Exons and Flanking Regions of Genes. PLoS ONE. 2011;6:e24945. doi: 10.1371/journal.pone.0024945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaneau O, et al. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 18.Williams AL, et al. Phasing of Many Thousands of Genotyped Samples. Am. J. Hum. Genet. 2012;91:238–251. doi: 10.1016/j.ajhg.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howie BN, et al. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 21.Fuchsberger C, et al. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browning SR, Browning BL. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, et al. Genotype-Imputation Accuracy across Worldwide Human Populations. Am. J. Hum. Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4 doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann TJ, et al. Imputation of the Rare HOXB13 G84E Mutation and Cancer Risk in a Large Population-Based Cohort. PLoS Genet. 2015;11:e1004930. doi: 10.1371/journal.pgen.1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mägi R, et al. Genome-Wide Association Analysis of Imputed Rare Variants: Application to Seven Common Complex Diseases. Genet. Epidemiol. 2012 doi: 10.1002/gepi.21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewing CM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N. Engl. J. Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Cai B. G84E mutation in HOXB13 is firmly associated with prostate cancer risk: a meta-analysis. Tumor Biol. 2014;35:1177–1182. doi: 10.1007/s13277-013-1157-5. [DOI] [PubMed] [Google Scholar]
- 30.Saunders EJ, et al. Fine-Mapping the HOXB Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer. PLoS Genet. 2014;10:e1004129. doi: 10.1371/journal.pgen.1004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, et al. The G84E mutation of HOXB13 is associated with increased risk for prostate cancer: results from the REDUCE trial. Carcinogenesis. 2013;34:1260–1264. doi: 10.1093/carcin/bgt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonsson T, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 33.Steinthorsdottir V, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 34.Auer PL, et al. Imputation of Exome Sequence Variants into Population- Based Samples and Blood-Cell-Trait-Associated Loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu EY, et al. Genotype Imputation of MetabochipSNPs Using a Study-Specific Reference Panel of ~4,000 Haplotypes in African Americans From the Women’s Health Initiative. Genet. Epidemiol. 2012;36:107–117. doi: 10.1002/gepi.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm H, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat. Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 38.Loh P-R, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connor TD, et al. Fine-Scale Patterns of Population Stratification Confound Rare Variant Association Tests. PLoS ONE. 2013;8:e65834. doi: 10.1371/journal.pone.0065834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuk O, et al. Searching for missing heritability: Designing rare variant association studies. Proc. Natl. Acad. Sci. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwala V, et al. Evaluating empirical bounds on complex disease genetic architecture. Nat. Genet. 2013;45:1418–1427. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium, T. 1000 G.P. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Leal SM. Discovery of Rare Variants via Sequencing: Implications for the Design of Complex Trait Association Studies. PLoS Genet. 2009;5:e1000481. doi: 10.1371/journal.pgen.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S, et al. Rare-Variant Association Analysis: Study Designs and Statistical Tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgenthaler S, Thilly WG. A strategy to discover genes that carry multiallelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat. Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Li B, Leal SM. Methods for Detecting Associations with Rare Variants for Common Diseases: Application to Analysis of Sequence Data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarz JM, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann TJ, et al. Comprehensive Approach to Analyzing Rare Genetic Variants. PLoS ONE. 2010;5:e13584. doi: 10.1371/journal.pone.0013584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu DJ, Leal SM. A Novel Adaptive Method for the Analysis of Next-Generation Sequencing Data to Detect Complex Trait Associations with Rare Variants Due to Gene Main Effects and Interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madsen BE, Browning SR. A Groupwise Association Test for Rare Mutations Using a Weighted Sum Statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price AL, et al. Pooled Association Tests for Rare Variants in Exon-Resequencing Studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu MC, et al. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan W, et al. A powerful and adaptive association test for rare variants. Genetics. 2014;197:1081–1095. doi: 10.1534/genetics.114.165035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, et al. Optimal Unified Approach for Rare-Variant Association Testing with Application to Small-Sample Case-Control Whole-Exome Sequencing Studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team. R: A language and environment for statistical computing. 2012 [Google Scholar]
- 59.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang GT, et al. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zawistowski M, et al. Extending Rare-Variant Testing Strategies: Analysis of Noncoding Sequence and Imputed Genotypes. Am. J. Hum. Genet. 2010;87:604–617. doi: 10.1016/j.ajhg.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]