Abstract
Among the three phases of mRNA translation - initiation, elongation, and termination - initiation has traditionally been considered to be rate-limiting and thus the focus of regulation. Emerging evidence, however, demonstrates that control of ribosome translocation (polypeptide elongation) can also be regulatory, and indeed exerts a profound influence on development, neurologic disease, and cell stress. The correspondence of mRNA codon usage and the relative abundance of their cognate tRNAs is equally important for mediating the rate of polypeptide elongation. Here, we discuss recent results showing that ribosome pausing is a widely used mechanism for controlling translation and as a result, biological transitions in health and disease.
Introduction
Since translational control became a distinct field of study, the term ‘control’ for many investigators was synonymous with initiation, the first and most complicated phase of protein synthesis. Initiation includes formation of the 43S pre-initiation complex, its association with the mRNA 5′ terminal 7mG cap in coordination with the eIF4F (eIF4A, eIF4G, eIF4E) complex, scanning of the 40S ribosomal subunit to the initiation AUG codon, and joining of the 60S subunit to form the 80S monosome (Hinnebusch 2014).
Cells generally contain a dearth of the cap-binding factor eIF4E (Mamane et al 2004), and its interaction with eIF4G and hence its ability to recruit the translational apparatus is widely regulated by different classes of protein factors, including the eIF4E binding proteins (4EBPs) (Richter and Sonenberg 2006). Additionally, scanning of the 40S ribosomal subunit along the mRNA can be impeded by interacting proteins or secondary structure in the 5′UTR. Given these distinct control points, tradition has dictated that initiation would be rate-limiting for protein synthesis. Moreover, it makes intuitive sense that the first step in translation would be the most likely to be regulated. However, emerging evidence indicates that polypeptide elongation (ribosome transit) can also be regulatory and indeed may be critical for controlling early development, neural function, and cancer etiology. Here, we review salient observations pointing to an important role for regulated ribosome translocation in diverse biological contexts.
Translational elongation at a glance
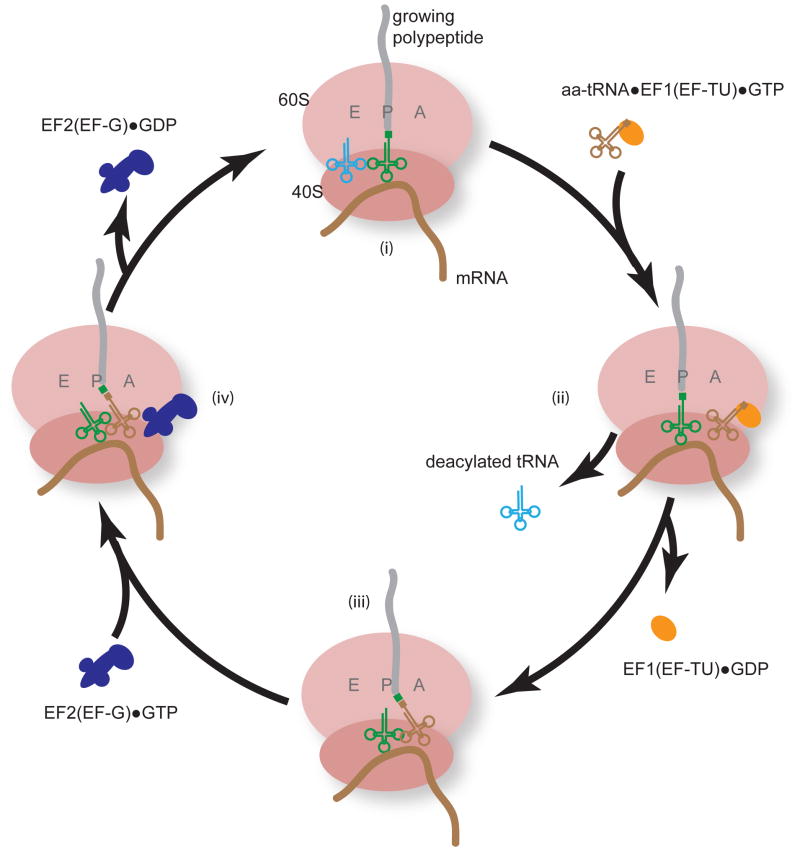
Translational initiation involves dozens of individual polypeptides and given its complexity, it is not surprising that distinct sub-steps can be regulated. In comparison, translational elongation is relatively simple. In concert with elongation factor EF-1/EF-TU and GTP, an aminoacylated tRNA enters the A-site of the ribosome; cognate tRNA - mRNA codon recognition then stimulates GTP hydrolysis and eviction of EF-1/EF-TU from the A-site. Concomitantly, the ribosome undergoes a conformational shift, stimulating contact between the 3′ ends of the aminoacylated tRNA in the A-site and the tRNA bearing the polypeptide chain in the P-site. When the two tRNAs shift position (A to P and P to E site) peptide bond formation occurs as the polypeptide is now transferred to the aminoacylated tRNA, extending the protein by one amino acid. A second elongation factor, EF-2/EF-G then enters the A-site, hydrolyzing GTP and resetting the ribosome to a conformation competent to receive the next aminoacylated tRNA in the A-site. The process repeats itself over and over again (Figure 1).
Figure 1.
Translational elongation at a glance. Shown are the four basic steps of translational elongation. The ribosome has three major tRNA pockets, the A, P, and E sites. The first step of polypeptide elongation is the recognition and accommodation of the cognate tRNA, as directed by a mRNA codon, within the ribosomal A-site (i). The cognate tRNA is brought into the ribosome as a complex with elongation factor 1 (EF-TU in bacteria) and GTP. Recognition of the cognate tRNA catalyzes the hydrolysis of GTP and the eviction of EF1 from the A-site (ii). At this point, the deacylated tRNA in the E-site is also thought to be evicted. The A-site tRNA and the P-site tRNA move into close proximity for the peptidyl transfer reaction where the growing polypeptide chain is added to the amino acid on the A-site tRNA (iii). Elongation factor 2 (EF-G in bacteria) then enters the A-site and completes ribosome translocation by moving the A-site tRNA to the P-site, and the P-site deacylated tRNA into the E-site (iv). The process then repeats itself over and over again.
Despite the “simplicity” of elongation, regulation can and does occur. Indeed, for decades we have known that ribosomes stall after reading only the first 5–30 codons of mRNAs encoding secreted proteins (Siegel and Walter 1988; Halic et al 2004). This activity requires the signal recognition particle (SRP), which binds the N-terminus of the nascent polypeptide and simultaneously inserts itself into the ribosome A-site (Halic et al 2004). Docking of SRP in the ribosomal A-site blocks further tRNA entry, arresting elongation until the ribosome/mRNA complex is localized to the endoplasmic reticulum. The lesson from SRP function is that A-site occlusion is a viable and potent means to arrest translational elongation. Thus any factor (protein or RNA) that can interact in or near the A-site has the potential to stall elongation by blocking tRNA entry.
In this regard, there are many known factors that do interact with the A-site - all for distinct reasons. The release factors, eRF1 and eRF3, the elongation factor EF-G, the ribosome recycling factors/mRNA decay factors DOM34 and HBS1, and the mRNA decay factor SKI7 are all thought to interact at the A-site. Thus the A-site is a busy place and a potential target for mRNA-specific regulation. A-site occlusion could easily be achieved by a message-specific regulator provided it has sequence specific binding properties for its mRNA transcript and a motif capable of A-site docking.
The SRP example demonstrates, in clear molecular terms, how elongation can be regulated. Importantly, the literature contains a number of less clear, but still tantalizing glimpses of where modulating ribosome translocation might be a driving force in regulation. In several of these cases, regulated elongation is inferred from observations that repressed mRNA co-sediments with polysomes in sucrose gradients and/or that this co-sedimentation is resistant to puromycin treatment, which causes release of translating ribosomes. Nevertheless, accumulating evidence hints to a broad influence of translational elongation on the control of gene expression.
Control of elongation during early development
Mechanisms of translational control during the early development of model organisms are often recapitulated in adult mammalian tissues. Consider, for example, the case of masked (i.e., repressed) mRNA in the oocytes (eggs) of sea urchins and frogs. As the oocytes prepare for fertilization of during early embryogenesis, this mRNA is massively mobilized onto polysomes, which coincides with a substantial decrease in ribosome translocation time (Brandis and Raff 1978; Richter et al 1983). These and other observations of this era now seem archaic because for the most part they pre-dated one’s ability to assess the time required for a ribosome to transit any particular mRNA. Even so, they illustrate the point that polypeptide elongation rates can be regulated by cellular transitions.
Masked mRNA is also a hallmark of Drosophila development. Here, the translation of nanos mRNA, which encodes a posterior pole determinant, is regulated both spatially and temporally. Although nanos mRNA translation is controlled in multiple ways, one is by ribosome stalling (Clark et al 2000; Andrews et al 2011). Nanos mRNA co-sediments with polyribosomes in sucrose gradients even though no Nanos protein is detected; yet, when the polysomes were added to an in vitro ribosome run-off system, the stalled polysomes resumed their transit and produced Nanos protein, showing that they were paused rather than immobilized in an inactive form. This scenario is somewhat similar to that observed with oskar mRNA, another posterior pole determinant in Drosophila. Oskar mRNA also co-sediments with polysomes even though little Oskar protein is detected (Braat et al 2004). Moreover, when added to an in vitro translation system derived from ovaries, puromycin, an antibiotic that acts on translating ribosomes by mimicking tRNA and causing premature polypeptide release and ribosome dissociation, caused only a partial shift of the sedimentation of oskar mRNA to lighter fractions of sucrose gradients, suggesting that it is associated with stalled ribosomes.
Micro RNAs, which have profoundly changed our notion of how biological processes are regulated, were discovered during examination of C. elegans larval development (Lee et al 1993; Wightman et al 1993). In spite of the huge number of studies that have analyzed miRNA activity, the mechanism(s) by which they silence mRNA expression remains somewhat enigmatic, perhaps because they repress translation a number of different ways. That mRNAs silenced by miRNAs are often eventually destroyed is beyond doubt, but the step(s) at which the silencing occurs is seemingly manifold. Olsen and Ambros (1999) noted that although C. elegans lin-4 miRNA inhibited Lin-14 mRNA translation, the message appeared to be stable and co-sedimented with polysomes in sucrose gradients. This observation gave rise to the hypothesis that miRNAs repress translation by stalling ribosomes. Using cell lines, several labs subsequently found miRNA-inhibited mRNAs that also co-sedimented with polysomes (Nottrott et al 2006; Maroney et al 2006) and one proposed that miRNAs promote pre-mature drop-off of translating ribosomes (Petersen et al 2006). Although ribosome profiles derived from developing zebrafish embryos showed that at least miR-430 did not induce ribosome drop-off (Bazzini et al 2012), it remains an open question as to the extent to which miRNAs can promote post-initiation mRNA silencing.
Synaptic plasticity in neurons
Neuronal processes, particularly dendrites, have long been known to harbor mRNAs whose translation is critical for synaptic plasticity, the underlying cellular basis of learning and memory (Kang and Shuman 1996; Martin et al, 1997). The regulation of dendritic mRNA translation must be considered in conjunction with cellular localization as mRNAs are transported from cell bodies into dendrites on molecular motors in a mostly silent form. The mRNAs are then activated in response to synaptic activity (Kanai et al 2004). Early work found that a substantial portion of neuronal mRNAs reside in granules that sediment in sucrose gradients to fractions much heavier than polysomes. Electron microscopy revealed these granules to be composed of densely packed polysomes, but because they lack the initiation factors eIF4E and eIF4G, they were thought to represent stalled ribosomes. Membrane depolarization of neurons by KCl stimulated translation partially dispersed the aggregates, suggesting that the stalled polysomes resumed translation (Krichevsky and Kosik 2001).
In a contemporaneous study, Scheetz et al (2000) stimulated synaptoneurosomes (a biochemical preparation of pre- and post-synaptic compartments) isolated from rat brain with the neurotransmitter N-methyl-D-aspartate (NMDA) and observed an increase in eEF2 phosphorylation, which would inactivate the enzyme and thus slow ribosome translocation. These investigators found a concurrent increase in the synthesis of the critical synaptic protein alpha calcium/calmodulin protein kinase II (αCaMKII) and hypothesized that inhibition of elongation of some mRNAs allows for the elevated translation of other mRNAs by mechanisms involving enhanced initiation. This hypothesis suggests that phospho-eEF2 could discriminate among mRNAs, which might be accomplished by, for example, spatial segregation of some components of the translational apparatus.
A link between eEF2 phosphorylation and synaptic activity was also observed by Sutton et al (2007), who found that eEF2’s enzymatic activity can be toggled by the type of neurotransmission to which a neuron is subjected. eEF2 is mostly nonphosphorylated when neurotransmission is evoked by action potentials. On the other hand, miniature synaptic transmission, which is spontaneous in nature and independent of action potentials, results in eEF2 phosphorylation. As might be expected, protein synthesis is up or down regulated depending on the state of eEF2 phosphorylation. For this reason, Sutton et al (2007) proposed that eEF2 is a sensor that links synaptic activity to local (i.e., dendritic) control of polypeptide elongation.
More contemporary studies are consistent with these findings. Using an indirect in vivo ribosome run-off assay in neurons, Graber et al (2013) found that mRNAs in dendrites are associated with stalled polysomes that can be reactivated by stimulation of metabotropic glutamate receptors, which induces long term depression (LTD), a protein synthesis-dependent form of synaptic plasticity. Buxbaum et al (2015) used three color fluorescence in situ hybridization (FISH) and single molecule detection technology to explore actin mRNA masking/unmasking dynamics in neuronal dendrites. Their results buttress the interpretation that quiescent mRNAs in neurons associate with stalled ribosomes and that induction of long tem potentiation (LTP), another form of synaptic plasticity, activates the stalled ribosomes to complete translation.
Ribosome stalling in Fragile X Syndrome
Fragile X Syndrome is the most common form of inherited intellectual disability and most frequent monogenic cause of autism. The syndrome is caused by a CGG repeat expansion in the Fmr1 gene, which causes its transcriptional inactivation. Fmr1 encodes FMRP, an RNA binding protein that represses translation. In the absence of FMRP, protein synthesis in the brain is excessive and it is commonly thought that this leads to synaptic dysfunction and other anomalies associated with this disease.
Several studies have shown that FMRP co-sediments with polysomes, suggesting that it inhibits translation by stalling ribosomes (Feng et al 1997; Corbin et al 1997; Stefani et al; 2004). In a groundbreaking study by Darnell et al (2011), FMRP was found to crosslink to nearly 1000 mRNAs in the brain in an experiment where UV irradiation to covalently link RNAs and proteins is followed by immunoprecipitation and deep sequencing (this procedure is referred to as “CLIP”). Surprisingly, most of the sites in mRNA to which FMRP was crosslinked were distributed in coding regions in a cis element-independent manner. Moreover, extensive analysis of the associated mRNAs showed them to co-sediment with polysomes, but to be largely resistant to puromycin treatment (because puromycin causes dissociation of transiting, but not static ribosomes, one infers that ribosomes do not move, or move only very slowing, on FMRP-bound mRNAs). By contrast, mRNAs not crosslinked to FMRP dissociated from polysomes following puromycin treatment and sedimented with the nontranslating ribonucleoprotein (RNP) fractions of sucrose gradients. Therefore, a synthesis of these two observations –that FMRP binds to coding regions and that the ribosomes on such mRNAs do not transit – strongly implies that FMRP stalls ribosomes.
In a number of cases, mouse models have shown that the excessive protein synthesis in the Fragile X brain can be restored to normal levels if a second gene is deleted, which leads rescue of many disease phenotypes (Dolen et al 2007; Bhattacharya et al 2012; Udagawa et al 2013; Gross et al 2015). For example, Udagawa et al focused on CPEB1, which generally (although not necessarily exclusively) stimulates translation in the brain, and co-localizes and co-immunoprecipitates with FMRP. The binding site for CPEB1 is present in about 30% of the FMRP-associated mRNAs, and thus has the potential to regulate their expression. These authors proposed that translational homeostasis in the brain might be restored if the CPEB1 gene as well as Fmr1 were disrupted. Indeed, not only was the excessive protein synthesis restored to normal levels in FMRP/CPEB double knockout mice, but so too were a variety of pathophysiologies associated with Fragile X. Moreover, Udagawa et al found that ribosome transit time in the Fragile X brain was ~45% faster than in the wild type brain, which was rescued to normal in the FMRP/CPEB1 double knockout brain. How the loss of CPEB1 slows ribosome translocation in the FMRP-deficient brain is unknown, but irrespective of the mechanism involved, these data directly demonstrate that FMRP controls polypeptide elongation and that Fragile X may be a disease, at least in part, of accelerated ribosome translocation.
Because the ribosome has intrinsic helicase-like activity and is capable of removing protein/RNA complexes as it moves along the mRNA (Takyar et al 2005), FMRP acting as a simple roadblock to stall ribosomes, while tenable as a model, seems too simplistic. An intriguing structural study of Drosophila FMRP showed that it binds directly to the ribosome via an interaction with ribosomal protein L5 (Chen et al 2014). The two KH (hnRNP K homology) domains of FMRP interact with the ribosome while the single RGG (arginine-glycine-glycine) box could associate directly with RNA, suggesting that FMRP may act as a bridge between the two to impede ribosome movement (Figure 2). However, such a configuration does not predict whether the specificity of FMRP binding is imparted by its association with the mRNA or the ribosome.
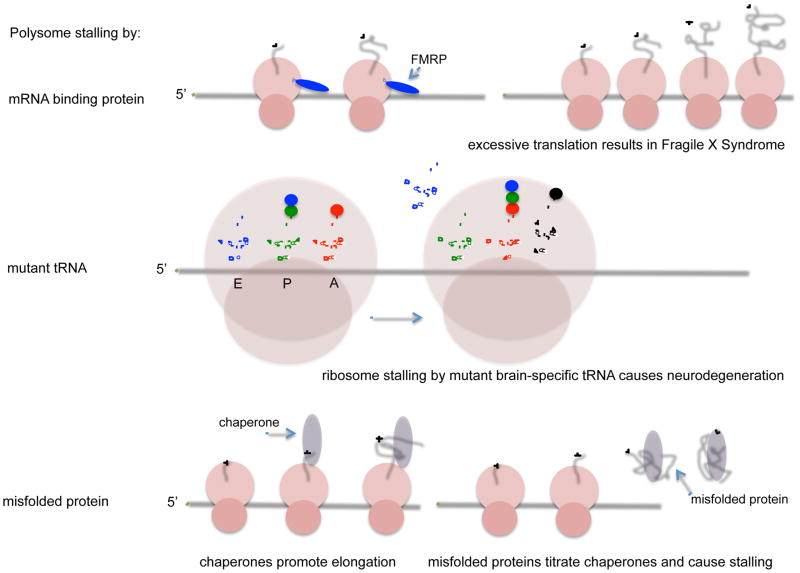
Figure 2.
Three examples of regulated polypeptide elongation. FMRP is proposed to bind both the ribosome and the engaged mRNA to impede ribosome translocation. FMRP is not produced when the Fmr1 gene is inactivated, which results in an elevated rate of polypeptide elongation and the Fragile X syndrome (top). In the brain, a mutated tissue-specific tRNA causes ribosome stalling at its corresponding codon. Neurodegeneration occurs if GTPBP2 is also mutated (middle). During proteotoxic stress, the chaperone HSC70, which normally binds the nascent peptide as it emerges from the ribosome, is titrated by misfolded proteins and causes ribosome stalling after reading about 50 codons (bottom).
One extant question arising from the above discussion is whether FMRP-mediated translational repression is permanent or reversible. At least one form of synaptic plasticity, LTD, induces FMRP phosphorylation and ubiquitin-mediated destruction (Nalavadi et al 2012; Huang et al 2015), which one surmises would remove the block to ribosome translocation and allow polypeptide elongation to proceed.
Ribosome stalling in neurodegeneration
The brain contains the most complex mixture of mRNAs of any adult tissue, and therefore it is not surprising that translational control is becoming a hallmark of neural development and function. One stunning observation on translational control at the level of ribosome translocation came from a mutagenesis screen for neurological disorders in mice where one line in particular displayed profound brain degeneration, ataxia, and death by about two months of age (Ishimura et al 2014). Mapping the mutation by crosses to congenic mouse strains combined with SNP analysis showed that the mutagen (N-ethyl-N-nitrosourea) produced a point mutation in a splice site of the Gtpbp2 gene, which encodes a protein with homology to GTPases involved in translation, particularly the ribosome recycling factors HBS1 and eRF3. These two proteins interact with the ribosome release factors Dom34 (yeast nomenclature) and eRF1, and GTPBP2 does indeed co-immunoprecipitate with Pelota, the mammalian homologue of Dom34. However, neurodegeneration was manifest only in the commonly used C57BL/6J background, suggesting that a second modifier gene specific to this strain was necessary to produce the phenotype. The modifier gene was found to encode an arginine tRNA isodecoder (isodecoder tRNAs share the same anticodon, but have changes elsewhere in the molecule). This arginine tRNAUCU, which contains a single C to U mutation in the T-stem loop that is likely to result in RNA misfolding, is CNS-specific; in contrast, GTPBP2 is widely-expressed. Ishimura et al hypothesized that the mutant tRNAArgUCU would cause the ribosome to stall at AGA codons. Indeed, ribosome profiling (Ingolia et al 2009) revealed a particularly high number of reads at AGA codons in cerebellar material containing the mutant tRNA, indicating strong ribosome stalling at these sites. Such strong stalling was not evident in cerebellar tissue from animals with wild type tRNAArgUCU (Ishimura et al 2014). Thus, ribosome stalling at AGA codons caused by the mutant tRNA may allow mutant GTPBP2 to recruit Pelota and promote premature polypeptide release, resulting in neurodegeneration (Figure 2) (Ishimura et al 2014; Darnell 2014). tRNA mis-charging (Lee et al 2006) or mis-folding appears to have particularly profound consequences for CNS function, suggesting that disruption of tRNA activity could contribute to other CNS pathologies as well.
Polysome pausing by mis-folded proteins
Cellular stress has long been known to cause reduced translation and promote the formation of stress granules and processing bodies (p-bodies) where mRNAs have been hypothesized to undergo silencing and decay, respectively (Anderson and Kedersha 2009; Decker and Parker 2012). Proteotoxicity can be one source of cellular stress and can be simulated by feeding cells with amino acid analogs that result in misfolded proteins. Liu et al (2013) found that culturing cells in L-azetdine-2-carboxylic acid (AZC), a proline analog, caused rapid turnover of newly synthesized protein, and when AZC treatment was combined with the proteasome inhibitor MG132, protein synthesis substantially decreased. This treatment did not induce rapid stress granule formation or eIF2α phosphorylation, which are common stress-induced events (Proud 2005), nor did it alter other parameters normally associated with a block at initiation. Instead, AZC and MG132 caused polypeptide elongation to slow. Ribosome profiling showed that the stress promoted ribosome stalling within ~50 codons of the initiation AUG, enough to encode a polypeptide partially buried in the exit tunnel of the ribosome and partially exposed to the cellular milieu. Liu et al hypothesized that because the chaperone HSC70 binds and helps fold nascent peptides as they emerge from the ribosome, it would naturally facilitate ribosome transit. During proteotoxic stress, however, the chaperone may be titrated by misfolded protein and thus cause ribosome stalling and reduced translation (Figure 2).
Heat shock, perhaps of the most common form of stress, has been known for decades to inhibit protein synthesis at least partly at the level of elongation (Ballinger and Pardue 1983). Using ribosome profiling, Shalgi et al (2013) found that heat stress induces widespread stalling at about codon 65. Similar to the data presented by Liu et al, this study found that HSP70 was responsible for the ribosomal stall. Although Shalgi et al did not determined how the chaperone induces stalling during heat stress, they suggest it may involve HSP70 association with the emergent nascent peptide, that it somehow clogs the peptide exit tunnel, or that it mitigates elongation factor activity.
An additional mode by which heat shock proteins modulate elongation comes out of structural analysis of yeast ribosomes (Zhang et al 2014). A ribosome-associated complex (RAC), which is composed of HSP40 and HSP70, binds both ribosomal subunits through a single long alpha-helix. As a consequence, a necessary rotation between the subunits during translation is limited and thus ribosome translocation is reduced. Zhang et al propose that RAC somehow responds the folding needs of the nascent peptide as it emerges from the exit tunnel and binds the ribosome to reduce translocation such that the peptide can assume a productive tertiary structure.
Caloric restriction or nutrient deprivation also induces cell stress. Although a number of proteins can sense that nutrients are in short supply, the major one is mTORC1 (mTOR complex 1). When amino acid levels are low, this kinase is inactivated, which leads to a number of downstream dephosphorylation events including that of 4EBP1. The non-phosphorylated form of this protein disrupts the ability of eIF4E to bind eIF4G and recruit factors necessary to initiate translation. A second nutrient sensor is AMP kinase (AMPK). In response to amino acid starvation, it down-regulates protein synthesis and other high ATP-demanding processes in an effort to conserve energy. One way AMPK accomplishes this task is by activating eEF2 kinase (eEF2K), which reduces elongation by phosphorylating eEF2 (Leprivier et al, 2013). Through this kind of energy conservation, AMPK-mediated eEF2 phosphorylation promotes cell survival in the face of nutrient restriction.
Perhaps the most unusual but well-defined example of stress-regulated elongation is yeast Hac1 mRNA, which encodes a transcription factor involved in the unfolded protein response (UPR). As the 5′end of Hac1 mRNA emerges, it associates with ribosomes and begins to be translated. When the 3′UTR enters the cytoplasm, an intron contained within it base pairs with the 5′UTR, thereby forming a closed loop that stalls the ribosomes. In response to ER stress, the 3′ UTR intron is removed by the nuclease Ire1p and the RNA ends are unconventionally spliced by tRNA ligase, which consequently allows the ribosomes to continue to catalyze polypeptide elongation (Chapman and Walter 1997; Ruegsegger et al 2001).
Regulated elongation during oncogenic transformation
The AMPK-eEF2 kinase pathway mediates tumorigenesis as well as caloric restriction-induced stress. When tumor cells with low levels of eEF2K are starved, polypeptide elongation remains robust and energy is consumed at a high rate, thereby causing the cells to undergo apoptosis. However, some tumor cells can adapt to starvation conditions by up-regulating the AMPK-eEF2 pathway, which inhibits polypeptide elongation, conserves energy, and promotes cell survival.
mTOR is a master regulator of cell physiology and can induce many types of cancer through phosphorylation of 4EBP1 and other substrates. As noted previously, 4EBP1 inhibits initiation by competitively binding eIF4E to the exclusion of eIF4G. When 4EBP1 is phosphorylated, it dissociates from eIF4E, which allows eIF4E, eIF4G, and eIF4A to form the eIF4F initiation complex. As a consequence of these events, mRNAs with particularly long and complex 5′ UTRs are preferentially translated because eIF4A is an RNA helicase that unwinds RNA secondary structure. Many mRNAs encoding oncogenes or growth-promoting factors have complex 5′ UTRs, and are up-regulated by mTOR and promote cellular transformation (Pelletier et al 2015). mTOR can also modulate polypeptide elongation to facilitate cell proliferation and cancer etiology. In intestinal cancer caused by mutations in adenomatous polyposis coli (APC), a tumor suppressor, mTORC1 can modulate translation via phosphorylation of 4EBP1 or S6 kinase (S6K), the latter of which is an upstream inactivator of eEF2K. Thus, at least for this cancer type, tumor growth is regulated by an axis of mTORC1-S6K-eEF2K-eEF2, which culminates in decreased elongation rates that are sensitive to the levels of S6K, but not 4EBP1 (Faller et al 2015).
Regulation by tRNAs
In the previous examples, we have seen that mRNA-specific regulation might be achieved by turning translational elongation on or off. In addition to these specialized cases, some evidence hints that the control of translation elongation might broadly occur and be critical for gene regulation. Unlike the aforementioned binary switches, ribosome translocation rates may subtly influence the expression of all messages. If translocation rates are mRNA-specific, then elongation would play a pivotal role in the synthesis, folding, and perhaps function of all proteins. The hypothesis that each mRNA has a distinct elongation rate is based on the notion of supply and demand: supply of functional tRNAs and demand by expressed codons. In this light, tRNAs are implicated as critical regulators of the expressed transcriptome.
In 1968 Francis Crick referred to the degeneracy of the genetic code as a “frozen accident” based on the required 64 combinations needed to code for 20 amino acids (Crick, 1968). Since then, a prevailing zeitgeist has been that synonymous codon substitutions are silent, having no bearing on gene function. Antithetically, a growing body of literature suggests that synonymous codons are differentially recognized by the translational apparatus. This concept has been referred to as codon optimality (Reis, et al., 2004; Novoa, et al., 2012; Pechmann and Frydman, 2013; Kri, et al., 2014). Codon optimality should not be confused with codon usage or bias. Codon usage/bias is the overrepresentation of certain codons within the genome and is the result of numerous selective pressures including translational elongation rate, translational accuracy, splicing, and 5′UTR structure (Akashi, 1994; Parmley et al., 2006; Drummond and Wilke, 2008; Gu et al., 2010). The term codon optimality has been introduced in an attempt to specifically define the differential recognition of codons by the translational apparatus and is a property that is distinct from the usage of codons within the genome. For instance, commonly occurring codons can be classified as optimal or non-optimal, while uncommon codons can also be optimal or non-optimal with respect to their influence on translational elongation rate (Presynak et al., 2015).
Conceptually, codon optimality reflects the balance between the supply of charged tRNA molecules and their demand imposed by the concentration of codons engaged in translation (Figure 3). Thus the ribosome decodes some codons quickly because their cognate tRNAs are abundant, while other codons are read more slowly because their tRNA concentrations are more limiting (Tuller et al., 2010; Novoa, et al., 2012). In addition, codon optimality is based somewhat on the accuracy of tRNA anticodon/codon interactions, which can influence decoding rate (Akashi, 1994; Drummond and Wilke, 2008). The theory that each codon is read by the ribosome at subtly distinct rates would predict that the kinetics of protein synthesis are determined by the primary sequence of every gene (Presnyak et al., 2015). Thus, the overall elongation rate is the sum of each codon’s infinitesimally small effect on ribosome translocation. Recent support for this hypothesis has come several labs demonstrating that codon optimality is a powerful determinant of both mRNA translation elongation and mRNA stability, which are tightly coupled events (Pechmann and Frydman, 2013; Presnyak et al., 2015).
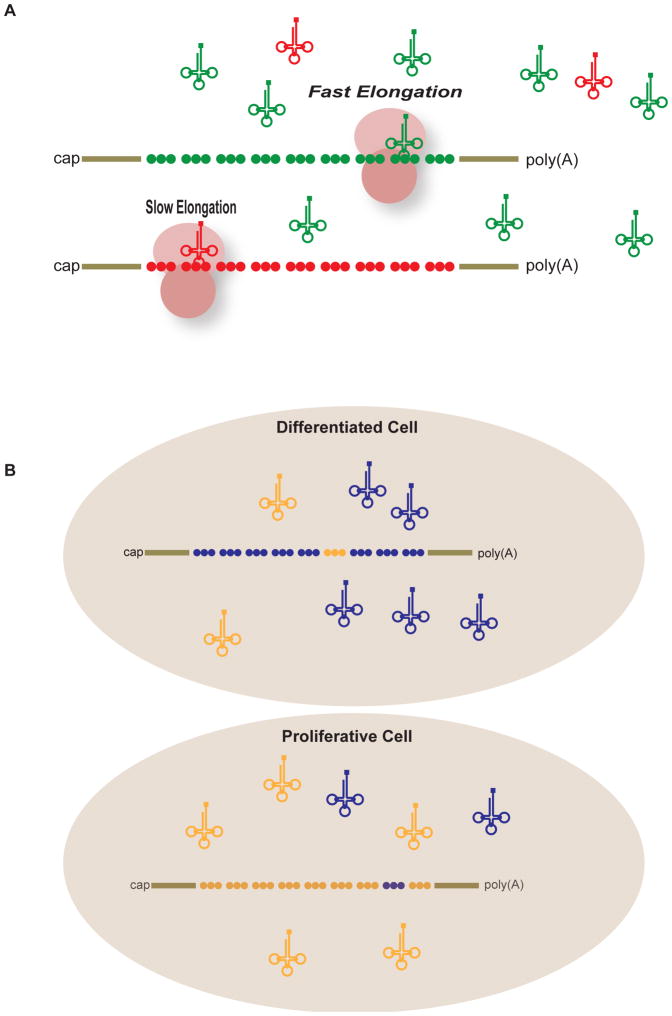
Figure 3.
(A) Illustration of codon optimality showing two hypothetical mRNAs. The green circles represent optimal codons while the red circles represent non-optimal codons. Designation of codons as optimal or non-optimal is a function of the concentration of tRNA in the cell. The green tRNA concentrations are high while the red tRNA concentrations are low; this difference impacts the speed of elongation. (B) Differentiated and proliferative cells have varied concentrations of tRNAs that are tailored to the decoding requirements of the expressed mRNAs. In this example, differentiated cells have an abundance of certain types of tRNAs (depicted in blue) because the mRNAs they express require similarly high levels of the corresponding codons. Conversely, proliferating cells contain different sets of tRNAs (in orange) to match the codons in mRNAs enriched in these cells (see Gingold et al, 2014).
If codon content dictates translational elongation rate, then perhaps it can be regulated by changing the functional concentration of tRNAs. If tRNA pools change in response to stress, environmental conditions, or other biological cues then so would the rate at which a codon is read, and thus ultimately ribosome translocation rates. Recent studies demonstrate tRNA levels and modifications indeed fluctuate in response to biological cues. First, Gingold et al. (2014) demonstrated that tRNA pools fluctuate in over 470 tumor samples when compared to quiescent cells. Specifically, a subset of tRNAs are induced in proliferating cells and repressed in quiescent cells. Moreover, a distinct subset of tRNAs are repressed in proliferating cells that are active in quiescent cells. Importantly, the tRNAs that are induced during proliferation often have anticodons corresponding to codons enriched in cell-autonomous genes. In contrast, tRNAs induced in differentiated cells carry anticodons for codons enriched in mRNAs for cell adhesion, cell-junction assembly, toll-like receptor signaling and extracellular matrix genes. In a study in which mouse embryonic development was examined, Schmitt et al (2014) demonstrated that tRNA expression patterns were controlled to generate an anticodon pool that corresponds to the codon demand by mRNAs (Schmitt et al. 2014). Thus the patterns of tRNA expression match codon usage within the expressed transcriptome (Figure 3).
Stress can also alter the level of functional tRNAs within a cell. Specifically, reprogramming of tRNA modifications occurs in cells exposed to different conditions. Chan et al. (2011) demonstrated that exposure of cells to hydrogen peroxide results in an increase in the amount of tRNALeu(CAA) containing 5-methylcytosine (m5C) at the wobble position. This increase in m5C causes selective translation of mRNAs enriched in the TTG codon. A nutrient-driven tRNA modification has also been observed in Drosophila. Here the bioavailability of queuine (a modified base) affects the levels of queuosine-modified tRNAs (Zaborske et al., 2014). Queuine is scavenged by eukaryotes from the tRNAs of bacteria and absorbed in the gut where, at least in flies, it alters translation profiles. Together, these data suggest tRNA pools can and do fluctuate in response to biological cues. If these concepts occur more broadly, then it places tRNAs as important and under-appreciated regulators of mRNA post-transcriptional regulation.
Why regulate at elongation?
The regulation of translational elongation might afford several advantages to an mRNA and the cell. First, loading an mRNA onto polyribosomes and then stalling the polysomes in response to the presence or absence of a stimulus would allow instantaneous production of new polypeptides once the stimulus was changed. This rapid response might be especially important in situations where, as in neurons, immediate protein synthesis is needed in response to synaptic stimulation. Lodging mRNA on translationally quiescent polyribosomes might also serve a protective function, limiting access by nucleases. In the cell, RNAs tend to be degraded when not associated with protein factors, therefore unless there are active events reorganizing the mRNA out of translation and into a translation-repressed ribonucleoprotein complex (e.g. stress granules or maternal mRNA storage granules) polyribosomes might serve a protective role.
Basal regulation of elongation rate by tRNAs might in fact be a primary driver of protein levels within the cell. It was recently observed that tightly coordinated optimal codon content occurs in genes encoding proteins with common physiological function (Presnyak et al 2015). This finding suggests that there is evolutionary pressure towards certain synonymous codon usage to coordinate gene expression at the level of protein synthesis and mRNA decay. The coordination of protein complexes through coordinate codon-dependent elongation rates would provide an elegant mechanism to ensure a consistent stoichiometric relationship between all members of a given complex. Because this coordination is based on codon choice, changing tRNA levels and/or modifications would provide a simple yet sophisticated means to uniformly regulate an entire physiological process by changing ribosome elongation rates.
The Road Ahead
The advent of ribosome profiling (Ingolia et al 2009), which displays stalled ribosomes on an mRNA-specific and codon-specific basis, has firmly placed elongation on the map of important gene regulatory mechanisms. Although in most cases the salient molecular details for stalling are just beginning to come into focus, perhaps some general principles may be involved. One is exemplified by FMRP in which a protein acts on a group of mRNAs to stall ribosome translocation. Certainly, intrinsic mRNA sequence gives specificity to the stalling, but is there also something intrinsic to the ribosome that allows it to be stalled? For example, we know the ribosome has trouble with lysine AAA codons (Koutmou et al. 2015), and that polyproline in the exit tunnel can also slow elongation (Gutierrez et al. 2013). Thus, certain ribosome characteristics could be exploited by an mRNA to slow elongation and consolidation of this event could occur through a second factor such as FMRP. Extrapolation of the data from several studies on FMRP suggests that this could be the case and that both message and ribosome contribute to the stalling.
A second general principle centers on tRNA. The astonishing study of Ishimua et al (2014) shows that the brain contains a tRNA that is absent from other tissues. When this tRNA has a single base change, ribosomes stall at its cognate mRNA codon. This stall results in CNS-specific pathology in instances where GTPBP2 is also mutated. Is tRNA specificity widespread and if so, how does it influence health and disease? A corollary of tRNA tissue-specificity is ‘optimality’ – the matchup between codon prevalence and relative of abundance of the tRNA bearing the anticodon. If this ratio becomes skewed during development or times of stress, it is easy to see how it could alter ribosome translocation and result in a biological transition.
Therapeutics aimed at the translational landscape have, for the most part, concentrated on initiation (e.g., Pelletier et al 2015). Importantly, however, just this year a novel multiple-stage antimalarial agent was discovered whose target is Elongation Factor 2 (Baragaña et al., 2015). Thus, we wonder whether the elongation phase of protein synthesis is an equally promising target for therapeutic intervention to ameliorate disease.
Acknowledgments
We thank Drs. Lori Lorenz, Botao Liu, and Sophie Martin for comments on the manuscript. Work from the authors’ laboratories was supported by grants from the National Institutes of Health (R01 GM46779, R01 NS079415, U54 082013 to JDR and R01 GM080465 to JC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Andrews S, Snowflack DR, Clark IE, Gavis ER. Multiple mechanisms collaborate to repress nanos translation in the Drosophila ovary and embryo. RNA. 2011;17:967–977. doi: 10.1261/rna.2478611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger DG, Pardue ML. The control of protein synthesis during heat shock in Drosophila cells involves altered polypeptide elongation rates. Cell. 1983;33:103–113. doi: 10.1016/0092-8674(83)90339-2. [DOI] [PubMed] [Google Scholar]
- Baragaña B, Hallyburton I, Lee MCS, Norcross NR, Grimaldi R, Otto TD, et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522(7556):315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat AK, Yan N, Arn E, Harrison D, Macdonald PM. Localization-dependent oskar protein accumulation; control after the initiation of translation. Dev Cell. 2004;7:125–131. doi: 10.1016/j.devcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Brandis JW, Raff RA. Translation of oogenetic mRNA in sea urchin eggs and early embryos. Demonstration of a change in translational efficiency following fertilization. Dev Biol. 1978;67:99–113. doi: 10.1016/0012-1606(78)90303-2. [DOI] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nature Communications. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr Biol. 2000;10:1311–1314. doi: 10.1016/s0960-9822(00)00754-5. [DOI] [PubMed] [Google Scholar]
- Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Darnell JC. Ribosome rescue and neurodegeneration. Science. 2014;345:378–379. doi: 10.1126/science.1257193. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell R. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, Dudek KM, Casey HA, Scopelliti A, Cordero JB, Vidal M, Pende M, Ryazanov AG, Sonenberg N, Meyuhas O, Hall MN, Bushell M, Willis AE, Sansom OJ. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Graber TE, Hébert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, Sossin WS. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci USA. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Computational Biology. 2010;6(2):1–8. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Huang J, Ikeuchi Y, Malumbres M, Bonni A. A Cdh1-APC/FMRP ubiquitin signaling link drives mGluR-dependent synaptic plasticity in the mammalian brain. Neuron. 2015;86:726–739. doi: 10.1016/j.neuron.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife. 2015;4 doi: 10.7554/eLife.05534. http://doi.org/10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kri KOA, Copi T, Gabaldón T, Lehner B, Supek F. Inferring gene function from evolutionary change in signatures of translation efficiency. Genome Biol. 2014;15:R44. doi: 10.1186/gb-2014-15-3-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–584. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler L, Sonenberg N. eIF4E – from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012;32:2582–257. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Parmley JL, Chamary JV, Hurst LD. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Molecular Biology and Evolution. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucl Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Richter JD, Wasserman WJ, Smith LD. The mechanism for increased protein synthesis during Xenopus oocyte maturation. Dev Biol. 1982;89:159–167. doi: 10.1016/0012-1606(82)90304-9. [DOI] [PubMed] [Google Scholar]
- Rüegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- Schmitt BM, Rudolph KLM, Karagianni P, Fonseca NA, White RJ, Talianidis I, et al. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Research. 2014;24:1797–1807. doi: 10.1101/gr.176784.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P. Each of the activities of the signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, Lorenz LJ, Bassell GJ, Akbarian S, Chattarji S, Klann E, Richter JD. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zaborske JM, DuMont VLB, Wallace EWJ, Pan T, Aquadro CF, Drummond DA. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biology. 2014;12:e1002015. doi: 10.1371/journal.pbio.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, Gao N. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat Struct Mol Biol. 2014;21:1042–1046. doi: 10.1038/nsmb.2908. [DOI] [PubMed] [Google Scholar]