Abstract
Cooperative interactions between RNA and vesicle membranes on the prebiotic earth may have led to the emergence of primitive cells. The membrane surface offers a potential platform for the catalysis of reactions involving RNA, but this scenario relies upon the existence of a simple mechanism by which RNA could become associated with protocell membranes. Here, we show that electrostatic interactions provided by short, basic, amphipathic peptides can be harnessed to drive RNA binding to both zwitterionic phospholipid and anionic fatty acid membranes. We show that the association of cationic molecules with phospholipid vesicles can enhance the local positive charge on a membrane and attract RNA polynucleotides. This phenomenon can be reproduced with amphipathic peptides as short as three amino acids. Finally, we show that peptides can cross bilayer membranes to localize encapsulated RNA. This mechanism of polynucleotide confinement could have been important for primitive cellular evolution.
Keywords: origin of life, peptides, protocells, RNA, vesicles
The emergence of a protocell capable of Darwinian evolution is expected to require two key components: a self-replicating membrane and a self-replicating nucleic acid.[1] RNA, due to its ability to store genetic information and fold into catalytic structures, is hypothesized to be the ancestral nucleic acid in primitive cells.[2] A major question in the design and study of protocells is how membranes could promote reactions involving RNA in the absence of highly evolved macromolecular catalysts. Membranes provide an obvious structural role in a protocell, spatially localizing RNA by passively trapping the nucleic acids in the lumen of vesicles.[2b] In principle, membranes could also play a catalytic role by co-localizing reactants on their surface.[3] The localization of short polynucleotides to membranes could promote a variety of catalytic processes of RNA, ranging from faster nonenzymatic template copying to faster assembly of oligonucleotides into multi-component ribozymes. Determining mechanisms for RNA–membrane association is a fundamental step towards the goal of demonstrating increased RNA reactivity in the presence of vesicle membranes.
Polynucleotides can be localized to vesicle membranes through a variety of approaches ranging from hydrophobic modification of the polynucleotide[4] to selections for membrane-associating complexes[5] to increasing electrostatic interactions between the membrane and polynucleotide by the use of cationic lipids.[6] We sought to explore a prebiotically plausible means by which unmodified RNA strands, irrespective of sequence, could localize to both anionic and neutral membranes.
One prebiotically accessible mechanism for RNA–membrane association is by electrostatic interactions with cationic, membrane-bound small molecules or peptides. Peptides are of particular interest since amino acids are readily synthesized by a variety of potentially prebiotic routes[7] suggesting that peptides were likely to have been present in any protocellular system.[8] Membrane associating peptides are also found in current biological systems. For example, cell penetrating peptides (CPPs) permeate and antimicrobial peptides (AMPs) associate with bilayer membranes, respectively.[9] Several CPPs have also demonstrated the ability to non-covalently complex with oligonucleotides and transport them past cellular membranes,[10] suggesting that with some sequence modification, peptides might be altered to reside at the membrane along with their nucleic acid cargo. Both CPPs and AMPs are generally amphipathic, basic, and less than 30 amino acids in length.[11] We reasoned that short peptides (equal to or shorter than seven amino acids) that are sufficiently hydrophobic to associate with membranes and sufficiently cationic to interact electrostatically with the phosphate groups on RNA might localize short RNA oligomers to a vesicle surface.
In order to determine the physical factors required for RNA–membrane localization, we first analyzed a model amphipathic cationic molecule, 2-undecylimidazole (Figure 1 a). We expected that the hydrophobic undecyl alkyl chain would lead to membrane binding, while the positive charge on the imidazole moiety might attract negatively charged RNA oligonucleotides to the vesicle surface. We began our investigation with 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-choline (POPC) vesicles as a model membrane system. As expected the zeta potential (a measure of surface charge) of POPC vesicles becomes progressively more positive with an increasing mole fraction of undecylimidazole in the membrane (Figure 1 b). In contrast, a less hydrophobic imidazole derivative, 2-methylimidazole, does not change the zeta potential of vesicles, indicating that membrane association of the cationic molecule is essential to increase the charge on the vesicle surface.
Figure 1.
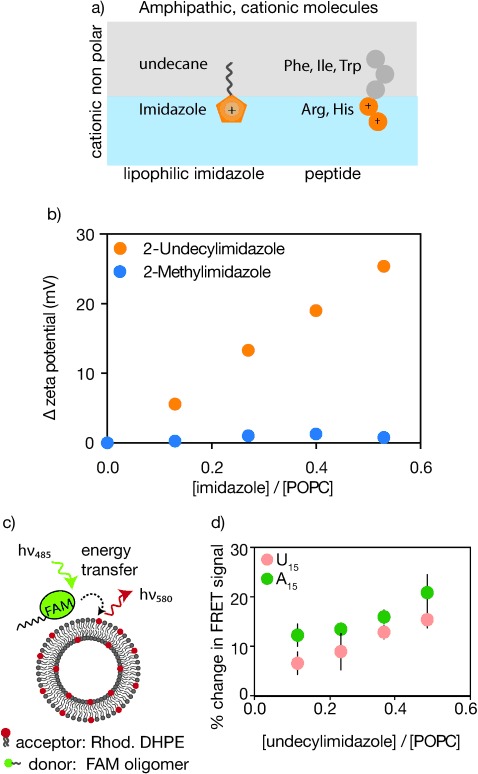
RNA localization with a model amphipathic, cationic molecule. a) Design of RNA-localizing molecules that include both nonpolar and cationic regions. b) The change in zeta potential is reported for POPC samples prepared with 2-undecylimidazole (yellow) or incubated with 2-methylimidazole (blue) with respect to control POPC samples that had no imidazole derivatives added. c) Schematic of the FRET assay used to assess RNA localization to vesicle membranes: The FRET efficiency between a FAM-labeled oligomer and a rhodamine-labeled lipid in vesicle membranes is measured. A positive change in FRET indicates an increase in membrane–RNA localization with respect to control samples. d) RNA (5′-FAM-U15 and 5′-FAM-A15) shows increasing localization to POPC membranes that contain increased amounts of undecylimidazole. n=3, error bars represent standard deviation.
We then measured RNA localization to the outside of vesicles using a fluorescence resonance energy transfer (FRET) assay. This assay reports the association of RNA with the vesicle surface by measuring the FRET efficiency between a carboxyfluorescein (FAM)-labeled RNA oligomer and a rhodamine-labeled phospholipid in small (100 nm), empty, unilamellar POPC vesicles (Figure 1 c). We validated the FRET method by using a secondary gel filtration assay to confirm RNA–membrane association (Figure S1-2). The FRET assay was designed to be maximally sensitive to increases or decreases in RNA–membrane association. A positive or negative change in FRET indicates increased or decreased membrane association, respectively, with respect to control samples. An increase in FRET signal could be due to an increased number of RNA molecules associating with the membrane, stronger RNA–membrane binding, or both.
Using this FRET method, we found that RNA oligonucleotides become increasingly localized to vesicle membranes in the presence of increasing concentrations of undecylimidazole (Figure 1 d). RNA–membrane association is not dependent on the identity of the oligonucleotides, as both poly(uracil), 5′-FAM-U15, and poly(adenosine), 5′-FAM-A15, 15-mer oligomers demonstrated similar levels of localization to the surface of POPC vesicles. In support of electrostatically mediated binding, we found that the extent of RNA–membrane association was tunable by adjusting the ionic strength and the pH around the pKa of the imidazole group (ca. pH 8; Figure S3). Increasing the pH from 8 to 9 reduced RNA localization to POPC membranes, as observed through the FRET assay. We measured a corresponding reduction in the zeta potential of vesicles that contain undecylimidazole as the pH was increased from 8 to 9 (Figure S4), showing that decreased protonation of the cationic molecule decreases the surface charge of the vesicles to which they are bound.
Having shown by indirect fluorescence methods that undecylimidazole can localize RNA oligonucleotides to membranes, we then attempted to directly visualize RNA–membrane association by microscopy. We prepared giant POPC vesicles (GUVs, 5–25 μm) with an encapsulated FAM-U15 oligomer. Because undecylimidazole has poor solubility in aqueous solutions, we were unable to simply add undecylimidazole to these giant POPC vesicles and observe localization of the encapsulated RNA. We therefore prepared small unilamellar POPC vesicles (SUVs) that contained undecylimidazole. Such small, neutral sonicated vesicles can be unstable and tend to aggregate or fuse with available membranes[12] bringing their cargo in close proximity to the surface of GUVs. Since undecylimidazole is a single-chain amphiphile, it should be able to exchange between vesicle membranes and flip-flop across the phospholipid bilayer to access the interior of the giant phospholipid vesicles.[13] Indeed, we observed that undecylimidazole-containing SUVs rapidly associated with giant POPC vesicles (Figure 2 b). After 1 hour, we observed that the encapsulated RNA had become localized to the vesicle membrane, including internal membranes within the vesicle, indicating the undecylimidazole could flip across the bilayer and cause encapsulated RNA to localize to the inner membrane leaflets (Figure 2 c, Figure S5).
Figure 2.
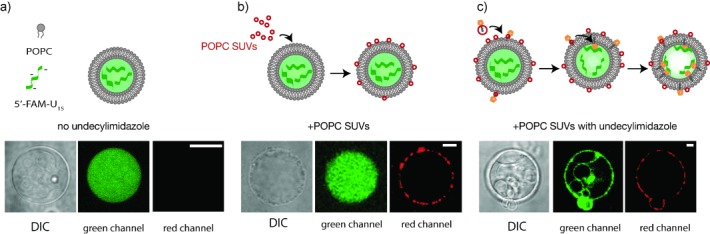
Microscopy of encapsulated RNA localization to POPC membranes with 2-undecylimidazole. Confocal images of 5′-FAM-U15 RNA (green) association with giant POPC vesicles membranes in the presence of 2-undecylimidazole. Differential interference contrast (DIC) microscopy images are shown for each vesicle. a) RNA appears uniformly distributed in the interior of POPC GUVs. b) The addition of SUVs containing a rhodamine-labeled lipid (red) leads to SUV aggregation and association with the giant vesicle membranes, but RNA (green) remains uniformly encapsulated in the vesicle interior. c) The addition of SUVs containing a rhodamine-labeled lipid (red) and 40 mol % 2-undecylimidazole leads to SUV association with vesicle membranes and RNA (green) localizes to the vesicle surface. The scale bar is 20 μm.
Encouraged by the ability of undecylimdazole to mediate RNA–membrane association, we proceeded to design a series of short cationic and lipophilic peptides to see if simple peptides could exhibit the same effect. We chose to work primarily with arginine as the cationic residue since its ability to associate with membranes and RNA alike is well documented.[11a, 14] We also examined histidine, which has an imidazole functionality similar to the model undecylimidazole molecule. Finally, we chose phenylalanine, isoleucine, and tryptophan as the nonpolar residues, because of their hydrophobicity and/or tendency to reside at the interface of a bilayer membrane.
We first screened the resulting set of peptides for their propensity to insert into phospholipid membranes. All peptides containing hydrophobic amino acid side chains inserted into POPC membranes, as detected through an increase in membrane surface area and leakage of an encapsulated dye (Figure S6). We next screened the same peptides for their ability to localize RNA to the surface of phospholipid vesicles, using the same FRET assay as used above to demonstrate undecylimidazole-mediated RNA–membrane association. We measured the change in FRET efficiency between donor-labeled RNA and acceptor-labeled POPC vesicles in response to the addition of 1 mm of peptide at a 0.13 molar peptide:lipid ratio (Figure 3, gray bars). Several peptides were identified that caused a greater than 5 % increase in FRET efficiency from control samples, including RF2, R3F2, R5F2, R3F3, R3I3, R5W2, and R3W3, suggesting these peptides increased RNA association to POPC membranes.
Figure 3.
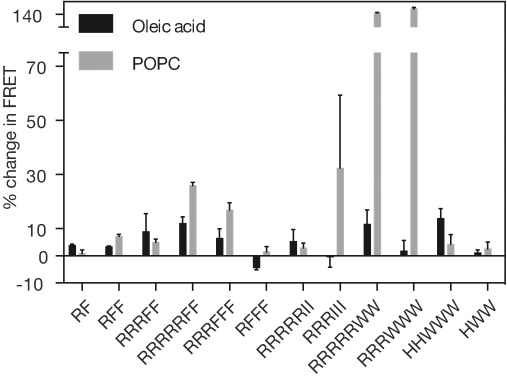
Peptide-induced RNA–membrane association. A FRET assay reports RNA localization (5′-FAM-U15) to POPC and oleic acid membranes (7.5 mm) 10 h after the addition of 1 mm of various peptides to the vesicle solution at pH 8. Data is reported as a percentage change from control samples that lack peptide. n=4, error bars represent the standard error of the mean.
Because early protocell membranes are thought to have assembled from single-chain amphiphiles such as fatty acids,[8] we also asked whether amphipathic, cationic peptides could localize RNA to such anionic membranes. Several peptides caused a greater than 5 % increase in FRET from control samples, including R3F2, R5F2, R3F3, R5I2, R5W2, and H2W3 (Figure 3, black bars). Overall, the extent of RNA association with oleic acid membranes was less than that observed with POPC membranes, which could be due to electrostatic repulsion between the anionic oleic acid amphiphiles and RNA polynucleotides, and/or due to shielding of the cationic peptide by the anionic oleate. The identification of peptides that localize RNA to fatty acid vesicles, however, indicates that repulsive interactions between oligonucleotides and anionic amphiphiles can be overcome to enable RNA binding to potentially prebiotic membranes.
In general, increasing both the number of positively charged amino acid residues and the number of hydrophobic residues resulted in increased localization of RNA to vesicle membranes. With peptides that have phenylalanine as the hydrophobic amino acid, there appears to be a minimum number of arginine residues necessary to enable localization to oleic acid membranes: neither RF, nor RF2 nor RF3 cause strong RNA–membrane association, while R3F2 does localize RNA to oleic acid membranes, and R5F2 causes even stronger localization. In most cases, binding to POPC membranes appeared to be stronger than binding to oleate membranes. For example, both R3I3 and R3W3 appeared to cause strong association with POPC membranes but not to oleate membranes.
To further confirm that cationic hydrophobic peptides could mediate the association of RNA with vesicle membranes, we used fluorescence microscopy to directly observe the nature of the interaction. We investigated the peptide R3F3, because according to the FRET assay, it localized RNA to both POPC and oleic acid membranes. We also investigated the peptide R3W3, because it induced the largest increase in FRET signal with POPC membranes. We prepared giant, micron-scale vesicles from POPC or a blend of oleic acid (90 %) and POPC (10 %). In our initial experiments, we premixed an AlexaFluor647-labeled RNA strand (a random sequence 15-mer) with either peptide and then added the mixture to the outside of giant POPC or oleic acid/POPC membranes. The R3F3 peptide caused visible and relatively homogeneous binding of RNA to the outside of at least some POPC vesicles (Figure 4). In contrast, the R3W3 peptide appeared to condense the RNA into large aggregates that then associated with the POPC vesicles (Figure 4, Figure S7). Both the R3F3 and the R3W3 peptides caused RNA localization in punctate aggregates around the 90 % oleic acid membranes.
Figure 4.
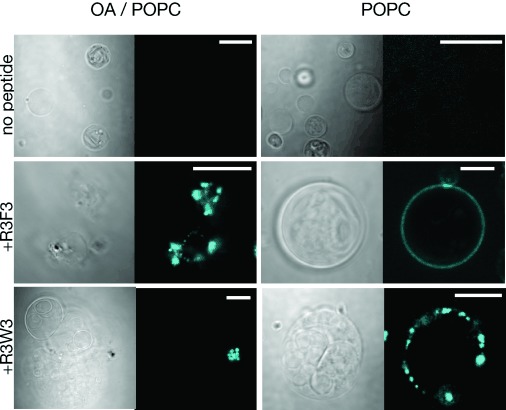
Microscopy of peptide-induced RNA–membrane association. Confocal images show RNA localization (5′-AlexaFluor647-labeled 15-mer, cyan) to the outside of oleic acid/POPC (90 %/10 %) and pure POPC membranes in the presence of R3F3 and R3W3 peptides. Control samples had no peptide added. For each image, the left panel shows the DIC image and the right panel shows AlexaFluor647 fluorescence. The scale bar is 20 μm.
The large RNA aggregates that we observed are likely to contain both peptide and lipid. Lee et al. observed through microscopy the aggregation of penetratin peptides on giant vesicles once a critical concentration ratio of peptide:lipid was reached.[15] As peptide aggregation in a membrane has often been found to be dependent on the molar ratio of peptide to lipid,[16] some of the variability in our observations may be due to variation in total lipid concentration between batches of our giant vesicles. For example, the R3F3 peptide in some cases induced uniform localization of RNA to the outside of POPC vesicles (Figure 4, panel 2), but in other cases led to the formation of punctate aggregates surrounding the vesicles (Figure S7).
Given that the pre-mixing of peptide and RNA, followed by addition to vesicles led primarily to the formation of large membrane-associated aggregates (with the partial exception of R3F3), we decided to examine the effect of adding either R3F3 or R3W3 to giant vesicles containing encapsulated RNA oligonucleotides. In these experiments, we wished to assess the ability of the peptide to first bind to vesicles and then flip to the interior face of the membrane where it could potentially localize encapsulated RNA. After an overnight incubation with R3F3, we again observed that encapsulated RNA had become localized in a relatively uniform manner to the membranes of POPC vesicles. Because the RNA was originally uniformly dispersed in the interior of the vesicle, this result indicates that the R3F3 peptide can cross the membrane and then localize encapsulated RNA to the membrane surface (Figure 5). Somewhat surprisingly, the tryptophan containing R3W3 peptide once again induced the formation of large RNA-lipid peptide aggregates that appeared to cluster around the perimeter of some of the giant vesicles.
Figure 5.
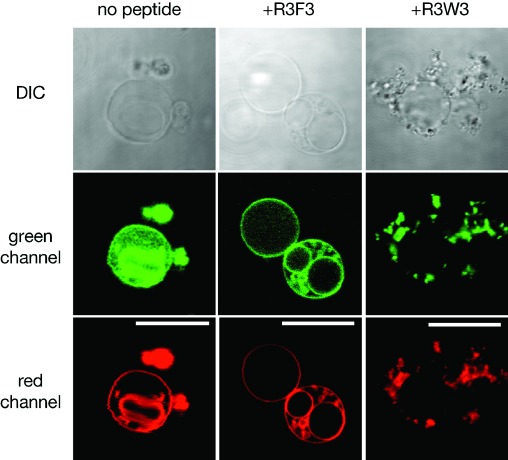
Microscopy of encapsulated RNA localization to POPC membranes with peptides. Confocal images show that RNA (5′-FAM-U15, green) encapsulated in POPC vesicles (containing a rhodamine-labeled lipid, red) becomes localized to the membrane of certain vesicles after an overnight incubation with R3F3 and R3W3 peptides. The scale bar is 20 μm.
In summary, we have found that the small molecule undecylimidazole, and a series of short peptides have the ability to localize RNA to POPC membranes. Our results show that different peptides and membrane amphiphiles cause differential RNA–membrane interactions. While R3F3 can induce a more uniform RNA–membrane association, the R3W3 peptide leads to the formation of RNA aggregates on POPC vesicles, and both the R3F3 and the R3W3 peptides form large aggregates with RNA in the presence of oleate vesicles. These different types of RNA sequestration could ultimately alter RNA reactivity or the strength of RNA–membrane binding in yet unknown ways. The physical association of RNA with vesicle membranes may have been an important initial step in accelerating prebiotically relevant reactions,[17] including RNA replication and ribozyme assembly, and may therefore have played an important role in early RNA-based protocells. Our study motivates the continued search for small molecules and simple peptides that could mediate the association of RNA oligonucleotides with model protocell membranes. In addition, our work addresses a potentially prebiotic route through which RNA could become associated with primitive membranes and opens the door to studying membrane-localized prebiotic reactions with RNA catalysts or substrates.
Acknowledgments
This work was supported in part by grants from the NASA Exobiology Program (grnat number NNX07AJ09G) and from the Simons Foundation (grant number 290363) to J.W.S. J.W.S. is an Investigator of the Howard Hughes Medical Institute (HHMI). N.P.K. is supported by an appointment to the NASA Postdoctoral Program, administered by Oak Ridge Associated Universities through a contract with NASA. I.T.H. is supported by an NSF Graduate Fellowship. We thank Albert Fahrenbach, Victor Lelyveld, Tony Jia, and all other members of the Szostak lab for helpful comments on the manuscript.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
References
- 1.Meierhenrich UJ, Filippi JJ, Meinert C, Vierling P, Dworkin JP. Angew. Chem. Int. Ed. 2010;49:3738. doi: 10.1002/anie.200905465. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010;122:3826. [Google Scholar]
- 2a.Orgel LE. Crit. Rev. Biochem. Mol. Biol. 2004;39:99. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 2b.Szostak JW, Bartel DP, Luisi PL. Nature. 2001;409:387. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 3a.Walde P, Umakoshi H, Stano P, Mavelli F. Chem. Commun. 2014;50:10177. doi: 10.1039/c4cc02812k. [DOI] [PubMed] [Google Scholar]
- 3b.Rajamani S, Vlassov A, Benner S, Coombs A, Olasagasti F, Deamer D. Origins Life Evol. Biospheres. 2008;38:57. doi: 10.1007/s11084-007-9113-2. [DOI] [PubMed] [Google Scholar]
- 4.Pfeiffer I, Höök F. J. Am. Chem. Soc. 2004;126:10224. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]
- 5.Vlassov A, Khvorova A, Yarus M. Proc. Natl. Acad. Sci. USA. 2001;98:7706. doi: 10.1073/pnas.141041098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Muñoz-Úbeda M, Misra SK, Barrán-Berdón AL, Aicart-Ramos C, Sierra MB, Biswas J, Kondaiah P, Junquera E, Bhattacharya S, Aicart E. J. Am. Chem. Soc. 2011;133:18014. doi: 10.1021/ja204693f. [DOI] [PubMed] [Google Scholar]
- 6b.Thomas CF, Luisi PL. J. Phys. Chem. B. 2005;109:14544. doi: 10.1021/jp0512210. [DOI] [PubMed] [Google Scholar]
- 7.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. Nat. Chem. 2015;7:301. doi: 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen IA, Walde P. Cold Spring Harbor Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a002170. a002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Henriques ST, Melo MN, Castanho MA. Biochem. J. 2006;399:1. doi: 10.1042/BJ20061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Wheaten SA, Ablan FD, Spaller BL, Trieu JM, Almeida PF. J. Am. Chem. Soc. 2013;135:16517. doi: 10.1021/ja407451c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margus H, Padari K, Pooga M. Mol. Ther. 2012;20:525. doi: 10.1038/mt.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Bechara C, Sagan S. FEBS Lett. 2013;587:1693. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 11b.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GC. J. Am. Chem. Soc. 2011;133:6720. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu PS, Tin GW, Baldeschwieler JD, Shen TY, Ponpipom MM. Proc. Natl. Acad. Sci. USA. 1981;78:6211. doi: 10.1073/pnas.78.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Thomas RM, Baici A, Werder M, Schulthess G, Hauser H. Biochemistry. 2002;41:1591. doi: 10.1021/bi011555p. [DOI] [PubMed] [Google Scholar]
- 13b.Kamp F, Hamilton JA, Kamp F, Westerhoff HV, Hamilton JA. Biochemistry. 1993;32:11074. doi: 10.1021/bi00092a017. [DOI] [PubMed] [Google Scholar]
- 14a.Herce HD, Garcia AE, Cardoso MC. J. Am. Chem. Soc. 2014;136:17459. doi: 10.1021/ja507790z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b.Alhakamy NA, Berkland CJ. Mol. Pharm. 2013;10:1940. doi: 10.1021/mp3007117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CC, Sun Y, Huang HW. Biophys. J. 2010;98:2236. doi: 10.1016/j.bpj.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen FY, Lee MT, Huang HW. Biophys. J. 2002;82:908. doi: 10.1016/S0006-3495(02)75452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Black RA, Blosser MC, Stottrup BL, Tavakley R, Deamer DW, Keller SL. Proc. Natl. Acad. Sci. USA. 2013;110:13272. doi: 10.1073/pnas.1300963110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17b.Damer B, Deamer D. Life. 2015;5:872. doi: 10.3390/life5010872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miscellaneous_information


