Abstract
Circadian rhythms in mammals are driven by a feedback loop in which the transcription factor Clock–Bmal1 activates expression of Per and Cry proteins, which together form a large nuclear complex (Per complex) that represses Clock–Bmal1 activity. We found that mouse Clock–Bmal1 recruits the Ddb1–Cullin-4 ubiquitin ligase to Per, Cry, and other circadian target genes. Histone 2B mono-ubiquitination at Per genes was rhythmic and depended on Bmal1, Ddb1, and Cullin-4a. Depletion of Ddb1–Cullin-4a or independent reduction of Histone 2B mono-ubiquitination caused defective circadian feedback and reduced the association of the Per complex with DNA-bound Clock–Bmal1. Clock–Bmal1 thus covalently marks Per genes for subsequent recruitment of the Per complex. Our results reveal a chromatin-mediated signal from the positive to the negative limb of the clock that provides a licensing mechanism for circadian feedback.
INTRODUCTION
Circadian clocks are endogenous oscillators with a period close to 24 hours. In mammals, circadian clocks are found in most, if not all, tissues1,2. These distributed clocks locally regulate diverse cellular processes3-5, collectively generating coherent daily rhythms of physiology, metabolism, and behavior6-10.
The mammalian clock is built on a conserved negative feedback loop that operates as a cell-autonomous molecular oscillator1,2. At the core of the clock is the heterodimeric transcription factor Clock–Bmal1 (Bmal1 is also known as Arntl or Mop3), which acts as a positive element of the feedback loop by driving transcription of Period (Per) and Cryptochrome (Cry) genes from its E-box DNA binding sites11. The three Per and two Cry proteins, acting as negative elements, enter the nucleus and assemble into one or more protein complexes (Per complex) of >1 mDa12 that include 25-30 proteins13,14. The Per and Cry proteins of the complex interact with Clock–Bmal1, leading to suppression of Clock–Bmal1 transcriptional activity15-17. Regulated degradation of Per and Cry proteins18-20 is followed by re-activation21 of Clock–Bmal1 and the consequent initiation of a new 24-hour transcriptional cycle. Clock function is further supported by additional negative regulators of Clock–Bmal122-24, a coupled transcriptional loop involving nuclear receptors25,26, and post-transcriptional processes27,28.
Recent studies indicate that two fundamental properties of the Per complex are essential to its role in circadian negative feedback. First, upon assembly in the nucleus, the Per complex incorporates pre-existing, widely-acting transcriptional repressor complexes that serve as its effectors. This cargo includes chromatin-modifying and nucleosome-remodeling machinery that inhibits transcriptional initiation, such as the Sin3 histone deacetylase complex29, Hp1γ-Suv39h histone methyltransferase30, and the Nucleosome Remodeling and Deacetylase (NuRD) complex14, and it includes factors that inhibit transcriptional termination13, indirectly suppressing initiation. Second, the Per complex directly interacts with DNA-bound Clock–Bmal1, thereby delivering the cargo of multiple repressors to chromatin at regulatory regions of Per genes and other circadian target genes13,14,29,30. The stable association of the Per complex with Clock–Bmal1 at its E-box binding sites thus results in the targeted suppression of Clock–Bmal1-dependent transcription, a defining feature of the oscillatory mechanism and of rhythmic control of transcriptional outputs. Our recent work indicates that circadian clock negative feedback at Per genes does not rely solely on the physical interaction of the Per complex with Clock–Bmal1. One transcriptional effector of the Per complex, the NuRD complex, is initially divided between the Clock–Bmal1 activator and the nascent Per complex; it is reconstituted as an active repressor only if the Per complex successfully targets DNA-bound Clock–Bmal114. Thus at least part of the circadian negative feedback action of the Per complex is target dependent.
Compared with circadian negative feedback, the present understanding of the positive limb of the clock is fragmentary and lacks a comparable conceptual framework. During the circadian activation phase, Clock–Bmal1 has been shown to work with a number of different factors acting as transcriptional co-activators, including Cbp (p300)31, Mll132, Jarid1a33, and Trap15021. It is not known if these factors operate within a single complex to co-activate Clock–Bmal1 or if they work independently in different complexes or even in different cell-types.
With the ultimate goal of obtaining a clearer picture of Clock–Bmal1 function during the circadian activation phase, we initiated pilot experiments to optimize label-free quantitative mass spectrometry methods34 for the characterization of Bmal1 protein complexes from mouse tissues. A clue from these early pilot experiments led ultimately to the finding that the Clock–Bmal1 complex mono-ubiquitinates histones at its Per gene E-box binding sites during the circadian activation phase and that this modification is crucial not for transcriptional activation but for the subsequent binding and therefore negative feedback action of the Per complex. The results thus revealed an unanticipated mechanism for fidelity of circadian negative feedback.
RESULTS
Clock–Bmal1 recruits Ddb1–Cul4 to circadian E-box sites
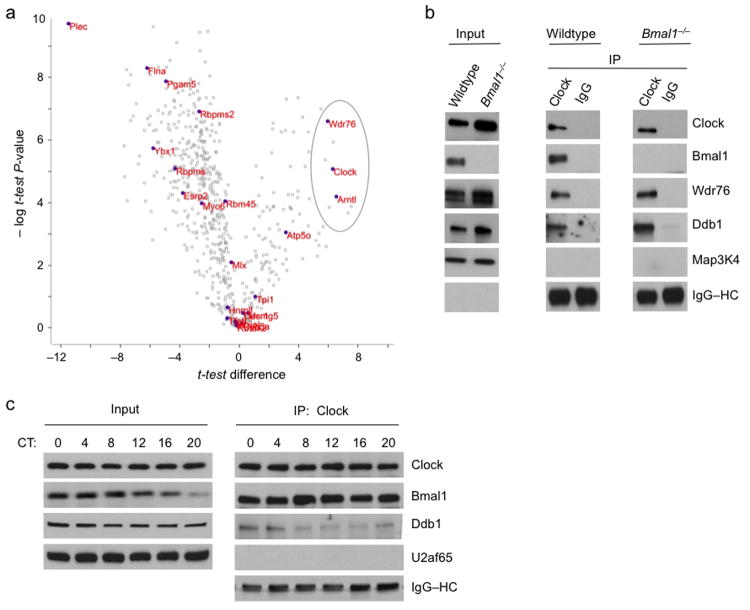
To investigate the positive limb of the circadian feedback loop, we isolated Clock–Bmal1 nuclear complexes by E-box oligonucleotide affinity purification (Supplementary Figure 1) from mouse livers harvested at circadian time 6 hours (CT6), the approximate peak of Clock–Bmal1 binding to E-box sites21,27,28 during the circadian activation phase. Although the pilot purification experiments were performed on a small scale and were not intended to be comprehensive, analysis by quantitative, label-free mass spectrometry34 identified Clock, Bmal1, and the adaptor protein Wdr76 (Wd-repeat containing protein 76) as statistically significant and specific components of an E-box-binding Clock–Bmal1 complex (Fig. 1a and Supplementary Table 1). Wdr76 is known to associate with the highly-conserved Ddb1 (DNA damage binding protein 1)–Cullin-4 (Cul4) E3 ubiquitin ligase35, which plays important roles in the DNA damage response35,36, targeted protein degradation37, and histone mono-ubiquitination36. The hint that this E3 ubiquitin ligase might be in a complex with Clock–Bmal1 seemed worth exploring because of prior work indicating connections between this pathway and the clock: Ddb1–Cul4 has been implicated in light-dependent turnover of Cry in the Drosophila circadian system38, Ddb1 was among positives in a large-scale mammalian circadian RNAi screen39, and both Ddb1 and Cul4 have been linked in a general way to mammalian Bmal1 and clock function in an analysis of a circadian protein interaction network40.
Figure 1.
Identification of Wdr76 and Ddb1 in a Clock–Bmal1 complex. (a) Volcano plot54 showing results of a modified t-test analysis of label-free mass spectrometry data comparing E-box and control oligonucleotide affinity purification from livers of wild type and Bmal1 mice (see Supplementary Figure 1). Open gray rectangles represent proteins detected, closed purple circles with accompanying names represent proteins significantly associated with the E-box sequence (FDR < 0.05, one tailed t-test of E-box vs. mutated E-box, n = 5 biological replicates), and the gray oval encompasses the three proteins significantly dependent on both the E-box sequence and the presence of Bmal1 (FDR < 0.05, one-tailed t-test of wild type vs. Bmal1–/–, n = 5 biological replicates). (b) Immunoblots of liver nuclear extract (CT6) from wild type or Bmal1–/– mice (Input) and immunoprecipitates (IP) from the extract (antibodies at top) probed with the antibodies indicated at right. Map3K4 (mitogen-activated protein kinase kinase kinase-4) served as negative control, and IgG–heavy chain (HC) served as positive control for immunoprecipitation (see Supplementary Figure 2). (c) Immunoblots of mouse liver nuclear extracts obtained across a circadian cycle (Input) and Clock immunoprecipitates from the extracts probed with the antibodies indicated at right. U2af65 (65-kD subunit of U2 small nuclear ribonucleoprotein particle auxiliary factor), negative control; IgG–HC, positive control. Uncropped images are presented in Supplementary Data Set 1.
We therefore immunoprecipitated Clock from liver nuclear extracts (CT6) obtained from wild type or Bmal1–/– mutant mice and probed for co-immunoprecipitating proteins. In both genotypes, Wdr76 and Ddb1 specifically co-immunoprecipitated with Clock (Fig. 1b), and in wild type mice Ddb1 specifically co-immunoprecipitated with Bmal1 (Supplementary Figure 2a); together the results indicate that Ddb1 associates with the Clock–Bmal1 heterodimer by virtue of a direct or indirect interaction with Clock. Similar results were obtained with lung nuclear extracts (Supplementary Figure 2b), suggesting that Wdr76 and Ddb1 are common or universal constituents of Clock–Bmal1 complexes. Although Ddb1 showed a constitutive steady-state abundance across the circadian cycle, its association with Clock–Bmal1 showed a circadian rhythm that peaked early in the circadian transcriptional activation phase, approximately CT0-4 (Fig. 1c).
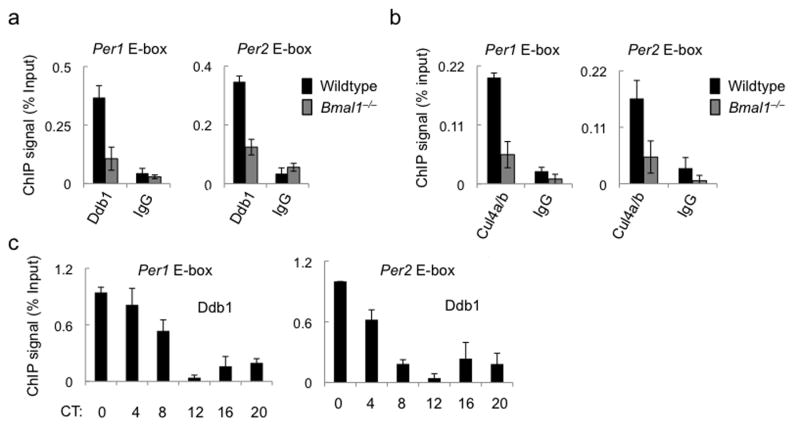
To determine if the Ddb1–Cul4 ubiquitin ligase is associated with DNA-bound Clock–Bmal1, we performed chromatin immunoprecipitation (ChIP) assays on chromatin from mouse livers obtained at CT6. We found that Ddb1 and Cul4 associated with E-box sites of the Per1 and Per2 genes and that this association was dependent on Bmal1 (Fig. 2a,b). ChIP assays performed on samples from across the circadian cycle showed that Ddb1 exhibits a circadian rhythm at Per1 and Per2 gene E-box sites (Fig. 2c), with peak occupancy around the time of maximal Clock–Bmal1–Ddb1 interaction and an overall temporal profile similar to the E-box binding cycle of Clock and Bmal121,27,28. Together these results indicate that Clock–Bmal1 interacts with the Ddb1–Cul4 ubiquitin ligase and recruits it to E-box sites of the Per1 and Per2 genes.
Figure 2.
Clock–Bmal1 recruits the E3 ubiquitin ligase Ddb1–Cul4 to circadian E-box sites. (a) ChIP assays showing Ddb1 or parallel IgG control from liver nuclear extracts (CT6) of wild type or Bmal1–/– mice at Per1 (left) and Per2 (right) gene E-box sites. (b) ChIP assays as in (a) showing Cul4a/b (the antibody does not distinguish the isoforms) or parallel IgG control at Per1 and Per2 gene E-box sites. (c) ChIP assays from wild type mouse livers across a circadian cycle (bottom) performed with Ddb1 antibody for the sites indicated at top. ChIP data are displayed as mean +/- SD of triplicate experiment; representative of three independent experiments.
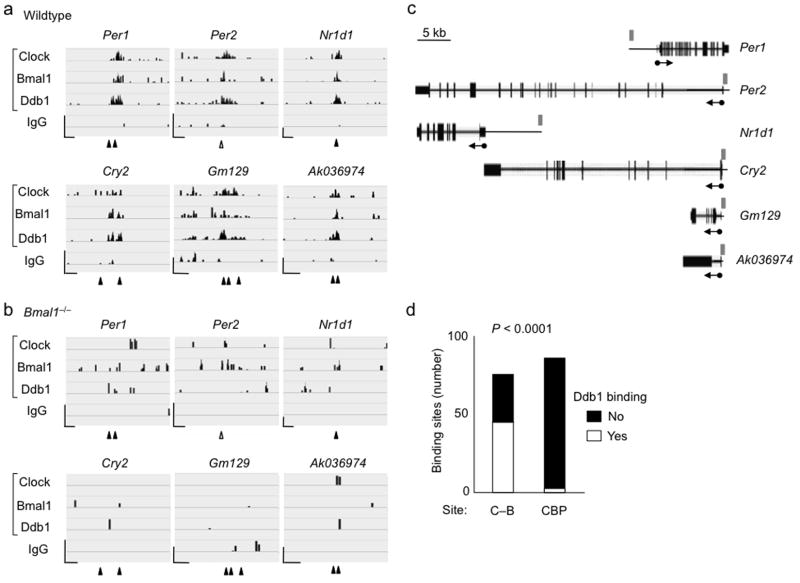
In addition, we obtained livers at CT6 from wild type and Bmal1–/– mutants, prepared chromatin, and performed ChIP-seq experiments to determine if Ddb1 is commonly co-recruited to Clock–Bmal1 binding sites and to other transcriptional activator sites throughout the genome. To identify secure Clock–Bmal1 binding sites for the analysis, we required that our top sites for Clock and Bmal1 occupancy agree with those from two other mouse liver ChIP-seq studies of Clock–Bmal1 binding sites27,28 and, additionally, that the signals be lost in our Bmal1–/– dataset (see Online Methods). This procedure generated a set of 78 validated Clock–Bmal1 sites common to three laboratories (Supplementary Table 2), of which 89% included a canonical E-box. We found that 46 of these sites were positive for Ddb1 occupancy (examples in Fig. 3a,b), likely an underestimate because the criteria for occupancy were restrictive.
Figure 3.
Clock–Bmal1 recruits Ddb1–Cul4 to many circadian target genes. (a) Six examples of ChIP-seq data alignments (Integrative Genome Viewer55) for Clock, Bmal1, and Ddb1 occupancy (as indicated) at validated Clock–Bmal1 E-box binding sites in wild type mouse liver (CT6). ChIP-Seq for IgG served as a negative control. Tick marks correspond to sequence reads along the length of the gene. Closed arrowheads, site of canonical E-box; open arrowhead, site of non-canonical E-box. Scale bars at lower left of each panel: vertical = 25 reads; horizontal = 500 bp. (b) Corresponding examples from identical experiment performed with liver chromatin from Bmal1–/– mice, displayed as in (a). Coordinates of genomic regions displayed: Per1, chr11:68906486-68910610; Per2, chr1:93354281-93357619; Nr1d1, chr11:98642602-98646726; Cry2, chr2:92272900-92275638; Gm129, chr3:95685093-95687497; AK036974, chr5:149805686-149807984. Total reads in (a) and (b), respectively: Clock, 16,103,538 and 22,043,586; Bmal1, 26,372,784 and 21,140,374; Ddb1, 15,984,254 and 19,760,087; IgG, 13,702,153 and 21,501,883. (c) Diagram showing relationship of data displayed in panels (a) and (b) to the structures of the relevant genes. Scale bar at top left. Thin boxes, untranslated regions; thick boxes, exons. Arrows denote start site and direction of transcription. Gray rectangles indicate relative positions of view shown in (a) and (b). (d) Summary of genome-wide ChIP-seq data comparing recruitment of Ddb1 to Clock–Bmal1 binding sites (C–B) and CBP binding sites, which correspond to the sites of many transcriptional activators. P-value was determined by Fisher’s exact test from a 2 × 2 contingency table (see Online Methods).
For comparison, we monitored Ddb1 occupancy at sites for Creb-binding protein (Cbp), a general co-activator that marks the binding sites of many transcriptional activators. At 85 Cbp sites in mouse liver28 that were not also Clock–Bmal1 sites (Online Methods and Supplementary Table 3), we found that only 3 were positive for Ddb1; the difference in Ddb1 association between Clock–Bmal1 sites and Cbp sites was highly significant (Fig. 3c). Ddb1 recruitment is thus a common feature of Clock–Bmal1 action across the genome but appears uncommon among other transcriptional activators.
Unexpected role of Ddb1–Cul4 in circadian negative feedback
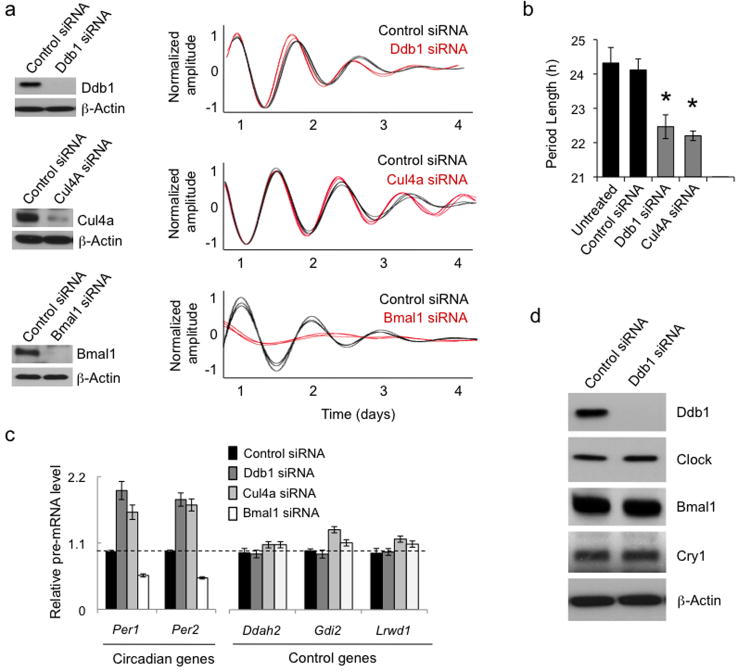
Because of its recruitment to Per genes, we examined a possible role for Ddb1–Cul4 in the core clock mechanism by introducing small interfering RNAs (siRNAs) to deplete Ddb1, Cul4a, or Bmal1 from cultured Bli23 circadian reporter fibroblasts (Fig. 4a). We then monitored real-time circadian rhythms of bioluminescence and the steady-state pre-mRNA level, a close correlate of transcription rate, of several Clock–Bmal1 circadian target genes and arbitrary control genes. Depletion of either Ddb1 or Cul4a produced the same phenotype, a significant shortening of circadian period length (Fig. 4a,b), as previously noted40. In both cases the short-period phenotype was accompanied by a substantial and specific increase in the transcription of Clock–Bmal1 target genes (Fig. 4c). In each case, the phenotype was reproduced by at least one additional, non-overlapping siRNA. Thus Ddb1 and Cul4a are important factors in the transcriptional operation of the clock. Depletion of Ddb1 had no detectable effect on the steady-state levels of Clock, Bmal1, or Cry1 (Fig. 4d), indicating that the period length and transcriptional phenotypes are unlikely to reflect a role for Ddb1–Cul4 in the turnover of the clock proteins. Depletion of the adaptor protein Wdr76 produced a marked decrease in the steady-state level of Clock and a corresponding long-period39,41 circadian phenotype (Supplementary Figure 3); the requirement of Wdr76 for Clock protein stability or expression thus precluded a detailed analysis of its likely role in the recruitment of Ddb1–Cul4. In contrast to Ddb1 or Cul4a, depletion of Bmal1 produced the expected39,41 long-period, low-amplitude circadian phenotype (Fig. 4a) and a marked decrease in Clock–Bmal1 target gene transcription (Fig. 4c). Even though Ddb1–Cul4 is recruited by Clock–Bmal1 to E-box sites during the transcriptional activation phase, the dual phenotype of shortened period length and increased target gene transcription is typical not of a Clock–Bmal1 co-activator but rather of a factor important for circadian negative feedback repression13,29,30. These results indicate that Ddb1–Cul4 unexpectedly acts somehow as a negative regulator of Clock–Bmal1 activity. The phenotypes are consistent with a role in promoting the negative feedback action of the Per complex.
Figure 4.
Ddb1 and Cul4a play a role in circadian clock feedback repression. (a) Left panels: western blots showing the effect of indicated siRNA or scrambled control siRNA on the steady-state abundance of the corresponding endogenous protein in cultured circadian reporter Bli fibroblasts. β-Actin, loading control. Right panels: circadian oscillations of bioluminescence in synchronized reporter fibroblasts after delivery of control siRNA (black) or indicated specific siRNA (red). Traces from three independent cultures are shown for each. (b) Circadian periods of fibroblasts in (a) (mean +/- SD; n = 3 cultures for each; *P < 0.0005, t-test, two-tailed). A period length could not be measured for Bmal1 siRNA traces. (c) Activation of Clock–Bmal1 circadian target genes by depletion of Ddb1 or Cul4a. Quantitative RT-PCR assays showing the steady-state abundance of the indicated pre-mRNAs in mouse fibroblasts after introduction of the indicated siRNAs. Data are normalized to control condition (dashed line). Shown are mean +/- SD of triplicate experiment; representative of 3 experiments. (d) Immunoblot showing the effect of control siRNA or Ddb1 siRNA on the steady-state abundance of the proteins indicated at right. β-Actin, loading control. Uncropped images can be found in Supplementary Data Set 1.
Ddb1–Cul4 promotes H2B mono-ubiquitination at E-box sites
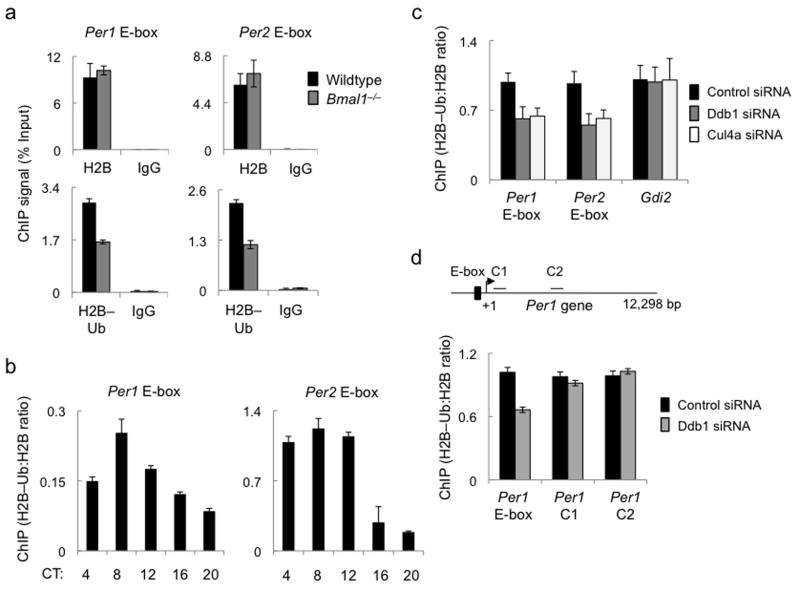
Since a role in regulated Clock–Bmal1 turnover seemed unlikely, we next examined whether the recruitment of a Ddb1–Cul4 complex by Clock–Bmal1 to Per E-box sites promotes local mono-ubiquitination of histones, which are known mono-ubiquitination substrates for Ddb1–Cul4 in vitro36. In ChIP assays with the available antibodies, we detected specific signals at Per gene E-box sites for mono-ubiquitinated H2B (at Lysine 120; H2B–Ub) but none for mono-ubiquitinated H2A, suggesting that this antibody is problematic for ChIP. To determine whether the local H2B–Ub depended upon functional Clock–Bmal1 complexes (as does recruitment of Ddb1–Cul4 to the sites, Fig. 2a), we compared H2B and H2B–Ub at the Per E-box sites in chromatin from livers of wild type and Bmal1–/– mutant mice. We found that total H2B at Per1 and Per2 gene E-box sites (CT6) was comparable in wild type and Bmal1–/– mice, but H2B–Ub was substantially reduced at both sites in the Bmal1–/– mutants (Fig. 5a). In wild type mice, H2B–Ub at both Per1 and Per2 E-box sites showed a circadian oscillation (Fig. 5b), peaking in the activation phase shortly after the peak of the interaction of Ddb1 with Clock–Bmal1 and its association with the E-box (as shown in Figs. 1 and 2). We found a similar circadian oscillation of H2B–Ub at an E-box site of the Dbp gene (Supplementary Figure 4), a well-known circadian target of Clock–Bmal142.
Figure 5.
Ddb1 and Cul4a promote H2B mono-ubiquitination at circadian E-box sites. (a) Top panels: ChIP assays showing total H2B or parallel IgG control from liver nuclear extracts (CT6) of wild type (black) or Bmal1–/– mice (gray) at Per1 (left) and Per2 (right) E-box sites. Bottom panels: ChIP assays (as in top panels) showing H2B–Ub or parallel IgG control. (b) ChIP assays from mouse liver nuclear extracts showing H2B–Ub (normalized to total H2B) across the circadian cycle at the Per1 (left) and Per2 (right) E-box sites (see Supplementary Figure 4). (c) ChIP assays from mouse fibroblasts showing H2B–Ub (normalized to total H2B) at Per E-box sites or an arbitrary control gene promoter (marked at bottom) after introduction of indicated siRNAs. (d) Top: diagram of mouse Per1 gene showing positions of E-box and control sites C1 and C2 within the gene. Arrow and +1 mark transcription start site. Bottom: ChIP assays from mouse fibroblasts showing H2B–Ub (normalized to total H2B) at the indicated Per gene sites (marked at bottom) after introduction of control siRNA or Ddb1 siRNA. ChIP data are displayed as mean +/- SD of triplicate experiment; representative of 3 independent experiments.
Similar to the finding in Bmal1–/– mice, depletion of Ddb1 or Cul4a from unsynchronized mouse fibroblasts caused a reduction of mean levels of H2B–Ub at Per1 and Per2 gene E-box sites, but it had no detectable effect on H2B–Ub at the promoter of an irrelevant control gene (Fig. 5c). Compared with the Per1 E-box site, depletion of Ddb1 had little evident effect on H2B–Ub at arbitrary sites within the body of the Per1 gene (Fig. 5d). Together these results indicate that the recruitment of Ddb1–Cul4 by Clock–Bmal1 to Per1 and Per2 gene E-box sites promotes local mono-ubiquitination of H2B (and possibly other histones). The residual H2B–Ub observed in the absence of Bmal1 or after depletion of Ddb1–Cul4 suggests that other factors, such as global histone ubiquitin ligases43,44, generate a baseline level of H2B–Ub at the sites. The circadian rhythm of H2B–Ub at E-box sites implies the action of at least one opposing histone de-ubiquitinase.
Independent reduction of H2B–Ub phenocopies Ddb1–Cul4a
Is the histone mono-ubiquitination activity of Ddb1–Cul4 associated with Clock–Bmal1 the basis for the circadian phenotypes of Ddb1 and Cul4a, or do the phenotypes reflect some other function of Ddb1–Cul4? To answer this question, we used depletion of Rnf20, a global E3 ligase for H2B mono-ubiquitination43,44, as a method for independently reducing H2B–Ub at E-box site (as well as other sites). Rnf20 is unrelated to Ddb1–Cul4, is specific for H2B44 (as opposed to other histones), and is not implicated in circadian clock function. As would be expected, we found no detectable interaction between Rnf20 and Clock or Bmal1 (Supplementary Figure 2a), nor did we detect stable recruitment of Rnf20 to Per gene E-box sites (Supplementary Figure 4b).
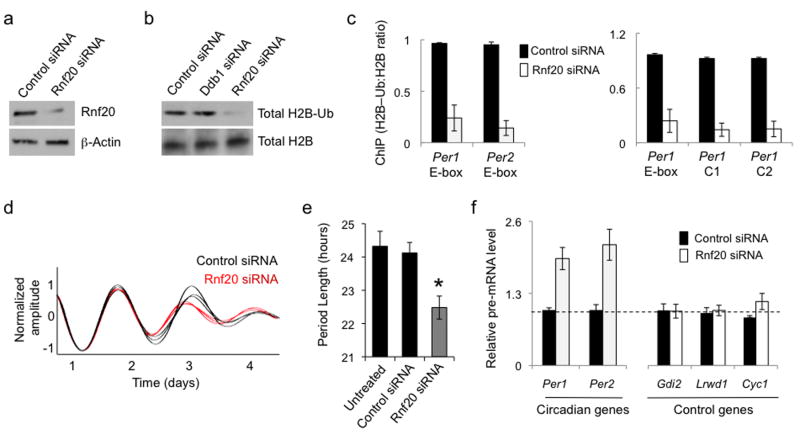
As expected from its global action, depletion of Rnf20 from circadian reporter fibroblasts (Fig. 6a) reduced total H2B–Ub (Fig. 6b), whereas depletion of Ddb1 had no detectable effect on total H2B–Ub (Fig. 6b), as previously noted36. Depletion of Rnf20 substantially reduced H2B–Ub at Per1 and Per2 E-box sites; it also reduced H2B–Ub at sites within the Per1 gene (Fig. 6c), also as would be expected from its global action.
Figure 6.
Independent reduction of H2B–Ub mimics circadian phenotypes of Ddb1 and Cul4a. (a) Immunoblots showing the effect of control siRNA or Rnf20 siRNA on the steady-state abundance of Rnf20, which globally mono-ubiquitinates H2B, in mouse fibrobloasts. β-Actin, loading control. (b) Immunoblots showing the effect of control siRNA or Rnf20 siRNA on the abundance of total H2B–Ub or H2B in mouse fibrobloasts. Uncropped images for (a) and (b) can be found in Supplementary Data Set 1. (c) Left: ChIP assays from mouse fibroblasts showing H2B–Ub (normalized to total H2B) at Per E-box sites (marked at bottom) after introduction of control siRNA (black) or Rnf20 siRNA (white). Right: ChIP assays from mouse fibroblasts showing H2B–Ub (normalized to total H2B) at Per1 gene sites (marked at bottom, as in Fig. 4d), after introduction of the indicated siRNAs. ChIP data are displayed as mean +/- SD of triplicate experiment; representative of 3 independent experiments. (d) Circadian oscillations of bioluminescence in synchronized reporter fibroblasts after delivery of control siRNA (black) or Rnf20 siRNA (red). Traces from three independent cultures are shown for each. (e) Circadian periods of fibroblasts in (d) (mean +/- SD; N = 3 cultures for each; *P < 0.005, t-test, two-tailed). (f) Quantitative RT-PCR assays showing the steady-state abundance of the indicated pre-mRNAs in mouse fibroblasts after introduction of control siRNA (black) or Rnf20 siRNA (white). Data are normalized to control (dashed line). Shown are mean +/- SD of triplicate experiment; representative of 3 experiments.
If the circadian phenotypes of Ddb1 and Cul4a reflect their targeted H2B mono-ubiquitination activity, rather than some other function, then depletion of Rnf20 should produce the same circadian phenotypes because loss of Rnf20 reduces H2B–Ub at E-box sites, even though Rnf20 is not part of the Ddb1–Cul4 pathway and has no apparent link to the clock mechanism. As with Ddb1 and Cul4, depletion of Rnf20 caused a significant shortening of circadian period length (Fig. 6d,e), and it produced a robust increase in Clock–Bmal1 target gene transcription without detectably effecting the transcription of irrelevant control genes (Fig. 6f). Thus independent reduction of H2B–Ub closely mimicked the dual circadian phenotypes, indicating that H2B mono-ubiquitination, and not the ubiquitination of other protein substrates or even other histones, is the activity of Ddb1–Cul4 that is relevant for its function in the clock.
H2B–Ub promotes binding of the Per complex at Per E-box sites
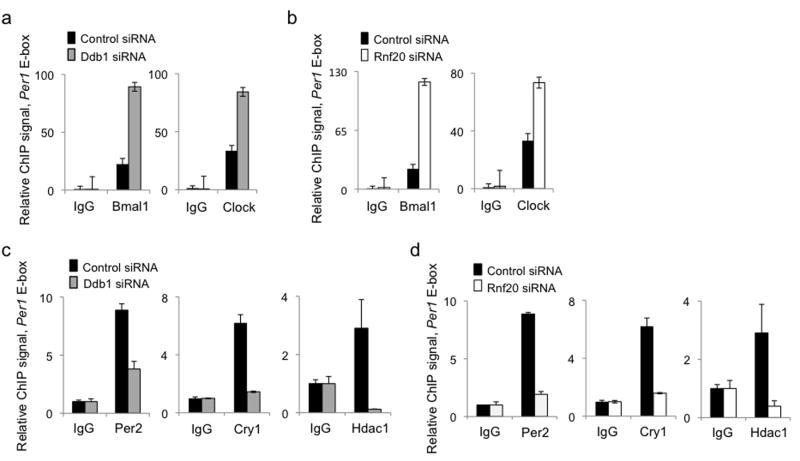
How might Clock–Bmal1-dependent histone mono-ubiquitination during the activation phase promote repression of Clock–Bmal1 activity? One possible explanation for this finding is that histone mono-ubiquitination by the Clock–Bmal1 complex is important for the subsequent interaction of the Per complex with Clock–Bmal1 and adjacent chromatin. To test this hypothesis, we depleted Ddb1 from unsynchronized fibroblasts and performed ChIP assays to monitor the mean binding of Clock–Bmal1 to Per gene E-box sites and the association of the Per complex with Clock–Bmal1 at those sites. Depletion of Ddb1 produced a substantial increase in the binding of Clock and Bmal1 to Per1 (Fig. 7a) and Per2 (Supplementary Figure 5a) E-box sites; the same results were observed after depletion of Rnf20 (Fig. 7b and Supplementary Figure 5b). Despite the large increase in Clock–Bmal1 occupancy, depletion of Ddb1 caused at the same time a marked reduction in the Per complex associated with Clock–Bmal1 (as monitored by core proteins Per2 and Cry1 and effector protein Hdac1) at Per1 and Per2 E-box sites (Fig. 7c and Supplementary Figure 5c, respectively). A similar large reduction in the Per complex associated with Clock–Bmal1 at the E-box sites was observed after depletion of Rnf20 (Fig. 7d and Supplementary Figure 5d), indicating that it is the histone mono-ubiquitination activity of Ddb1–Cul4, and not some other function, that promotes the stable association of the Per complex with E-box-bound Clock–Bmal1. These results indicate that the Clock–Bmal1 complex, by virtue of its constituent Ddb1–Cul4 histone ubiquitin ligase, covalently modifies the chromatin of Per genes so as 1) to decrease its own affinity for the E-box site and 2) to promote the stable binding and therefore the negative feedback action of the Per complex.
Figure 7.
H2B–Ub reduces Clock–Bmal1 and stabilizes the Per complex at Per gene E-box sites. (a) ChIP assays from mouse fibroblasts showing Bmal1 and IgG control (left) or Clock and IgG control (right) at Per1 E-box site after introduction of control siRNA (black) or Ddb1 siRNA (gray). (b) ChIP assays as in (a) after introduction of control siRNA (black) or Rnf20 siRNA (white). (c) ChIP assays from mouse fibroblasts showing Per complex proteins Per2 (left), Cry1 (middle), and Hdac1 (right) at Per1 E-box site after introduction of control siRNA (black) or Ddb1 siRNA (gray). (d) ChIP assays as in (c) after introduction of control siRNA (black) or Rnf20 siRNA (white). Shown in (a-d) are mean +/- SD of triplicate experiment; each representative of 3 experiments.
DISCUSSION
The results described here reveal that Clock–Bmal1 marks Per genes for subsequent feedback repression. The findings provide an example of active communication from the positive to the negative limb of the clock and provide a mechanism for fidelity of circadian clock transcriptional feedback different from that we previously described14. Our work supports a model in which the DNA-bound Clock–Bmal1 complex includes the Ddb1–Cul4 ubiquitin ligase, which mono-ubiquitinates H2B and possibly other histones on neighboring nucleosomes. This enzymatic action is crucial for the subsequent stable interaction of the Per complex with DNA-bound Clock–Bmal1 at Per genes. We postulate that this accumulation of H2B–Ub at Per E-box sites creates a chromatin structure favoring a stable interaction of the Per complex with both DNA-bound Clock–Bmal1 and adjacent chromatin; non-specific effector proteins in the Per complex can then modify the chromatin so as to repress transcription29,30 (Fig. 8). The negative feedback action of the Per complex at Per genes thus requires molecular recognition of two independent signals at the same site, a specific protein interaction interface on DNA-bound Clock–Bmal1 and a high level of H2B–Ub on adjacent chromatin. This “coincidence detection” feature of the feedback loop likely acts as a safeguard, licensing Per genes for negative feedback action. Our evidence that recruitment of Ddb1 to E-box sites is a common feature of Clock–Bmal1 action suggests that the dual signal requirement might also operate outside the core feedback loop, perhaps restricting the opportunities for off-target repression of non-circadian genes by the Per complex—as our data shows, other transcriptional activators are unlikely to recruit Ddb1–Cul4, and non-target sites with high H2B mono-ubiquitination would lack bound Clock–Bmal1.
Figure 8.
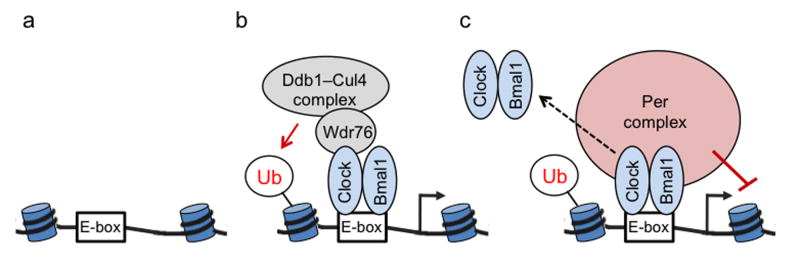
Model: Clock–Bmal1 licenses target genes for circadian feedback repression by the PER complex. (a) Inactive Clock–Bmal1 target gene after PER proteins are degraded but before Clock–Bmal1 is activated. Depicted are nucleosomes with wound DNA adjacent to Clock–Bmal1 E-box binding site. (b) At initiation of cycle, Clock–Bmal1 binds to E-box site and activates target gene transcription; Ddb1–Cul4 complex, associated with Clock via adaptor Wdr76, mono-ubiquitinates (Ub) H2B on adjacent nucleosome. (c) Accumulation of H2B–Ub at the site causes a change in chromatin conformation. Several hours later, when the PER complex is assembled at the onset of the circadian repression phase, the local H2B–Ub-dependent chromatin conformation promotes stable association of the PER complex with DNA-bound Clock–Bmal1 and adjacent chromatin, which it modifies to repress transcription. Local H2B–Ub also promotes Clock–Bmal1 dissociation from the E-box site (dashed arrow), either directly or as a consequence of its facilitation of PER complex binding and action.
Unlike other histone post-translational modifications, such as acetylation, phosphorylation, or methylation, mono-ubiquitination of H2B introduces a moiety nearly as large as the histone itself, changing the physical properties of chromatin and altering transcription, nucleosome dynamics, and chromatin compaction45,46. Studies manipulating global H2B E3 ubiquitin ligases, such Rnf20 and Rnf40, have shown that H2B–Ub can be associated with either transcriptional activation or repression, depending on the specific gene or the location of H2B–Ub in relation to gene structure46. In addition to the global factors, several E3 ubiquitin ligases that are likely to be selective regulators have been linked to H2B mono-ubiquitination, including Brca147 and Mdm248.
At least some of the transcriptional effects of H2B mono-ubiquitination are thought to result from trans-histone crosstalk between H2B–Ub and Histone 3 methylation or other histone marks, although a consensus has not yet emerged regarding the underlying mechanisms45,46,49. Accumulating evidence also suggests that at least some of the transcriptional effects of H2B mono-ubiquitination result not from direct physical effects on chromatin structure, as initially conceived, but rather from the recruitment of specific H2B–Ub “reader” proteins that regulate transcription46.
Our findings suggest that at least one component of the Per complex acts as a specific H2B–Ub reader, recognizing either H2B–Ub itself or some feature of an altered chromatin conformation resulting from the presence of H2B–Ub. Unlike the H2B–Ub reader proteins identified in a recent affinity screen50, we envision that the Per complex acts as a weak H2B–Ub reader, with an affinity inadequate for stable interaction with H2B–Ub-enriched chromatin without additional binding energy from an interaction with DNA-bound Clock–Bmal1.
H2B–Ub appears to act in the mammalian circadian clock as a selective, dynamically-regulated signal for specificity superimposed on a baseline of globally-regulated H2B–Ub. Ddb1 and Cul4, components of the relevant E3 ubiquitin ligase, and H2B–Ub itself are highly conserved, present in animals43, yeast51, and plants52. A recent study in Arabidopsis found that genetic disruption of H2B mono-ubiquitination most severely affected the transcription of dynamically-regulated loci, including light-induced genes and cyclically-transcribed circadian clock genes53. H2B mono-ubiquitination thus appears to be a genomic regulatory mechanism with an ancient connection to the dynamic regulation of transcription.
ONLINE METHODS
Mice and tissue collection
Mice (C57BL/6; 12-15 weeks of age; both sexes) on a standard diet (Pico Lab rodent diet 5053) were entrained to a 12-12-h light-dark cycle for at least 2 weeks and then transferred to constant darkness. Euthanasia was performed under infrared light, and tissues were collected under room light and frozen immediately in liquid nitrogen. All animal procedures were performed in accordance with the protocol approved by the Harvard Medical School Standing Committee on Animals.
Isolation of nuclei and preparation of nuclear extracts
Nuclei were isolated from mouse liver or lung as described56,57 with a few modifications. Briefly, homogenized tissue was incubated in ice-cold hypotonic buffer (10 mM HEPES, 7.9 pH, 10 mM KCl, 1.5 mM MgCl2, 1mM DTT, EDTA free protease inhibitors from Roche and phosphatase inhibitors from Sigma) for 10 min prior to lysis by Dounce. Nuclei were spun down and washed with isotonic buffer (10 mM HEPES, 7.9 pH, 150 mM NaCl, 1.5 mM MgCl2, 1mM DTT, protease inhibitors and phosphatase inhibitors). Nuclei were lysed with either hypertonic buffer (10 mM Tris-HCl, 7.4 pH, 400 mM NaCl, 1.5 mM MgCl2, 1mM DTT, 10% glycerol, protease inhibitors and phosphatase inhibitors) or detergent lysis buffer (100 mM Tris-HCl, 7.4 pH, 0.5% Igepal-CA 630 from Sigma, 150 mM NaCl, 1.5 mM MgCl2, 1mM DTT, 10% glycerol, protease inhibitors and phosphatase inhibitors). After incubation for 30 min on ice, insoluble material was removed by centrifugation at 21,000 x g for 10 minutes at 4°C. Nuclear extract was diluted to ~150 mM NaCl (final concentration) by addition of dilution buffer (10 mM Tris-HCl, 7.4 pH, 1.5 mM MgCl2, 1mM DTT, 10% glycerol, protease inhibitors and phosphatase inhibitors).
DNA Affinity Precipitation
Sense and anti-sense biotin-labeled single stranded DNA oligonucleotides containing either wild type or scrambled E-box sequences (see below) were heated to 98° C for 10min and allowed to anneal in Annealing Buffer (10 mM Tris-HCl, 50 mM NaCl, 1.5 mM MgCl2, 0.01% Tween-20) for at least 1 hour at room temperature. Fifty microliters of streptavidin conjugated magnetic beads (M270, Life Technologies) were incubated with 4 μM double stranded DNA for at least 20 minutes at room temperature to bind DNA to beads. The magnetic beads with conjugated DNA were then washed three times with annealing buffer, and incubated with roughly 2.5 mg of liver nuclear extract for at least 1 hour at 4° C. Beads were then washed at least three times for 5 minutes in Wash Buffer (10 mM Tris-HCl, 300 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.1% Igepal CA 630, protease inhibitors and phosphatase inhibitors). All proteins were eluted from beads by incubation with SDS-PAGE loading buffer at 98° C for 5 minutes. All experiments were performed in quadruplicate or quintuplicate.
DNA Affinity Precipitation Oligonucleotides
Widtype E-boxes
Sense: Biotin-5’-CAGTATTTAGCCACGTGACAGTGTAAGCACACGTGGGCCCTCA AGTCCACGTGCAGGGA-3’
Anti-sense: TCCCTGCACGTGGACTTGAGGGCCCACGTGTGCTTACACTGTCACGTGG CTAAATACTG -3’
Mutated E-boxes
Sense: Biotin-5’-CAGTATTTAGCCTGAGCACAGTGTAAGCACTGAGCGGCCCTCA AGTCCTGAGCCAGGGA-3’
Anti-sense: 5-TCCCTGGCTCAGGACTTGAGGGCCGCTCAGTGCTTACACTGTGC TCAGGCTAAATACTG -3’
LC-MS/MS Analysis
Samples from each oligonucleotide affinity-purification were separated by 1D gel electrophoresis followed by LC-MS/MS analysis of two fractions as previously described58. Briefly, peptides were desalted on StageTips and analysed on a nanoflow HPLC system (Thermo Fisher Scientific) connected to a hybrid LTQ-Orbitrap XL (Thermo Fisher Scientific), equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). Peptides were separated by reversed phase chromatography using in-house-made C18 microcolumns with a diameter of 75 μm packed with ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) in 4 hours LC gradient from 3% to 75% acetonitrile in 0.5% acetic acid and directly electrosprayed into the mass spectrometer. The LTQ-Orbitrap XL was operated with a Top10 MS/MS spectra acquisition method in the linear ion trap per MS full scan.
MS spectrum and data analysis
Raw MS files were processed with MaxQuant59, a freely available software suite (http://www.maxquant.org). Peak list files were searched by the ANDROMEDA engine, incorporated into the MaxQuant framework60, against the decoy UniProt database containing forward and reverse sequences. Initial maximum precursor and fragment mass deviations were set to 7 ppm and 0.5 Da, respectively, but MaxQuant achieved sub-ppm mass accuracy for the majority of peptide precursors. The search included variable modifications for oxidation of methionine, protein N-terminal acetylation and carbamidomethylation. Peptides with at least seven amino acids were considered for identification, specifying as enzyme trypsin allowing N-terminal cleavage to proline. The false discovery rate, determined by searching a reverse database, was set at 0.01 for both peptides and proteins. Identification across different replicates and adjacent fractions was achieved by enabling matching between runs option in MaxQuant within a time window of 2 minutes. Protein quantification was done using the label-free quantification algorithm within the MaxQuant software suite59,60 . Only proteins identified with at least two unique peptides and two quantification events were considered.
Immunoblotting
Extracts were resolved by SDS–PAGE (BioRad) and blotted onto PVDF (Millipore) by wet transfer. The buffer was PBS-0.05% Tween-20 with or without 5 mg/ml skim milk (EMD). In cases where proteins were detected after immunoprecipitation, the secondary antibody used was Clean-Blot IP Detection Reagent (Pierce). Signals were detected by enhanced chemiluminescence (G.E.).
Co-immunoprecipitation
Co-immunoprecipitation was performed as described with a minor modifications23. Briefly, magnetic beads conjugated to Protein A/G (Life Technologies) were incubated with primary antibody, and then incubated with protein extracts at 4° C. Protein complexes were eluted from beads with 0.2 M Glycine, pH 2.5.
Chromatin immunoprecipitation and quantitative RT-PCR
ChIP-seq
DNA from ChiP experiments was sequenced and aligned by the Center for Cancer Computational Biology at the Dana-Farber Cancer Institute. Genome aligned (Bowtie61) sequence reads were viewed using the Inetgrated Genome Viewer (IGV) software55. Briefly, to measure depth and breadth of mouse genome (mm9) aligned sequence reads mapping to published Clock, Bmal1, and binding sites27,28 we used the coverageBed software of the BEDTools suite62 located on the Harvard Medical school Orchestra computing cluster. Enrichment of Clock or Bmal1 protein at a specific genomic site was considered positive if at least 10 reads could be detected, if the fold enrichment over IgG control was 2 or greater (normalized to total reads) and if the fold enrichment over Bmal1–/– mutant control was 3 or greater. Of these secure Clock–Bmal1 binding sites, Ddb1 was considered enriched if at least 10 counts could be detected and if the fold enrichment over IgG was 2 or greater (normalized to total reads). The signal was considered Bmal1-dependent if fold enrichment over the Bmal1–/– mutant control was 3 or greater.
siRNAs and real-time monitoring of circadian oscillations
Bli cells23 (generated in this laboratory; not tested for mycopolasma) were plated at 250,000 cells/well in 6-well tissue culture plates. Transfections were performed using 90 pmol siRNA (Life Technoogies) per well in RNAi Max lipofectamine (Life Technologies). For analysis of clock gene transcription, unsynchronized cells were collected 48-72 h after transfection for determining silencing efficiency by Western Blot and RNA extraction as described7,29. In our experience, the most robust results for pre-mRNA analysis are obtained when performing the experiment in unsynchronized cells, in which all circadian phases are sampled simultaneously. This method results in a loss of sensitivity (i.e., only some of the cells are at a phase appropriate for detection of a difference in a given pre-mRNA), but it more than compensates by avoiding the need for precise sampling of the same phases from different cultures, particularly problematic (and liable to false-positive results) if depletion of the protein alters the circadian period compared with controls. The pre-mRNA assay in unsynchronized cells represents an average value for expression over the entire circadian cycle.
For real-time bioluminescence circadian reporter assays, transfected cells were synchronized and analyzed as described23. siRNA’s were purchased from Life Technologies: Control- 4390843; Wdr76- s10958; Ddb1- s64879; Cul4a- s97421; Rnf20- s99518; Bmal1- s62620. Non-overlapping siRNAs that confirmed the phenotypes: Ddb1- s64880; Cul4a- s97422; Rnf20- s99519).
Antibodies
For the antibodies below, data validating their use in mice can be found on the manufacturers’ websites, except for those marked by citation of a reference providing the validation data. For western blots (WB), antibodies were diluted according to the supplier’s instructions, except for those with dilutions indicated below. The following antibodies and control IgGs were obtained from the indicated suppliers: Abcam - anti-Clock (ab3517: ChIP, IP, WB); anti-Bmal1 (ab3350: ChIP, IP); anti-Bmal1 (Weitz Lab: ChIP, WB); anti-Hdac1 (ab7028: ChIP); anti-Ddb1 (ab109027: ChIP, WB); anti-Cul4a (ab72548: ChIP, WB); anti-U2AF65 (ab37530: WB); anti-Wdr76 (ab108149: WB); anti-Rnf20 (ab32629: WB); anti-beta-Actin (ab6276: WB); anti-Map3k4 (ab186125: WB); anti-Cry1 (ab104736: WB); ADI- anti-PER2 (PER21-A: ChIP)63; anti-CRY1 (CRY11-A: ChIP)64; Cell Signaling - mouse IgG1 (G3A1: ChIP, IP); anti-H2B–Ub (5546: ChIP, WB); Sigma-Aldrich rabbit IgG (I5006: ChIP, IP); Bethyl- anti-Clock (A302-618A5: WB); Millipore - normal mouse IgG (12-371: ChIP, IP); normal rabbit IgG (PP64: ChIP, IP); GE Healthcare - rabbit or mouse HRP-conjugated secondary antibodies.
siRNAs
Control siRNA
Forward: 5’-rArArUrUrCrUrCrCrGrArArArCrGrUrGrUrCrGtt-3’
Reverse: 5’-rUrUrCrGrArCrArCrGrUrUrUrCrGrGrArGrArAga-3’
Wdr76 siRNA:
Forward: 5’-CrGrUrGrGrArUrGrCrUrUrArUrArCrUrGrArAtt-3’
Reverse: 5’-rUrUrCrArGrUrArUrArArGrCrArUrCrCrArCrGga-3’
Ddb1 siRNA:
Forward: 5’-rGrCrCrUrGrUrArUrCrUrUrGrGrArGrUrArUrAtt-3’
Reverse: 5’-rUrArUrArCrUrCrCrArArGrArUrArCrArGrGrCrAat-3’
Rnf20 siRNA:
Forward: 5’-rGrUrGrGrArUrCrUrCrUrUrArUrCrCrCrGrGrAtt-3’
Reverse: 5’-rUrCrCrGrGrGrArUrArArGrArGrArUrCrCrArCtt-3’
Cul4a siRNA:
Forward: 5’-rGrCrArCrGrUrGrGrArCrUrCrArArArGrUrUrAtt-3’
Reverse: 5’-rUrArArCrUrUrUrGrArGrUrCrCrArCrGrUrGrCtt-3’
Bmal1 siRNA:
Forward: 5’-rGrCrArUrCrGrArUrArUrGrArUrArGrArUrArAtt-3’
Reverse: 5’-rUrUrArUrCrUrArUrCrArUrArUrCrGrArUrGrCct-3’
Quantitative RT-PCR primers
-
Per1 pre-mRNA :
Forward: 5’-ATTCTGAGGGTGTATCTGCCGCTT-3’
Reverse: 5’-TAAGGAATCACCACACCACCACCA-3’
-
Per2 pre-mRNA :
Forward: 5’-CCAAGTGACGGGCCGAGCAA-3’
Reverse: 5’-CCGAGCCGCCGGTTACGTAA-3’
-
Lrwd1 pre-mRNA:
Forward: 5’-AGGGAAGATGGGCCACAGAAATGA-3’
Reverse: 5’-GGAATGGGCACAATCAAAGCGTGA-3’
-
Cyc1 pre-mRNA:
Forward: 5’-AGCATTCTCCATTTGCCCTCCAGA-3’
Reverse: 5’-TCTGAATGAACGCCCATGTCTTCC-3’
-
Ddah2 pre-mRNA:
Forward: 5’-AAGGTTGATGGAGTGCGTAAAGCC-3’
Reverse: 5’-AGTCTCCCAAACTCTGCTTCCCTT-3’
-
Gdi2 pre-mRNA:
Forward: 5’-AGCATTCTCCATTTGCCCTCCAGA-3’
Reverse: 5’-TCTGAATGAACGCCCATGTCTTCC-3’
ChIP quantitative PCR primers
-
Per1 Promoter:
Forward: 5’-ATCCTGATCGCATTGGCTGACTGA-3’
Reverse: 5’-TCTCTTCCTGGCATCTGATTGGCT-3’
-
Per1-C1:
Forward: 5’-AGCCAGGACCCAGAAAGAACTCAT-3’
Reverse: 5’-AACTCACTCACCCTGAACCTGCTT-3’
-
Per1-C2:
Forward: 5’-TCCCATTGTGAGTTAGGCAGAGCA-3’
Reverse: 5’-AGCCAGAGAAGGGCACAGTTACAT-3’
-
Per2 Promoter:
Forward: 5’-AAGAGCGCGCAGCATCTTCATT-3’
Reverse: 5’-ATTGGTCGGAGTGCCACCTCATTT-3’
-
Gdi2 Promoter:
Forward: 5’-TTACAGGCAAGCTGGGCCTTAGTT-3’
Reverse: 5’-TTATCTGCGAGAGACAACGCCACA-3’
Sample size and conditions
Sample sizes were not predetermined by statistical methods, and experiments were not randomized or conducted blind to conditions.
Supplementary Material
Acknowledgments
We thank Ming Liu for mouse colony management and genotyping. This work was supported by an award from the G. Harold & Leila Y. Mathers Charitable Foundation (C.J.W.), a grant from the National Institute of General Medical Sciences (C.J.W.), a U.S. National Institutes of Health Training Grant in Fundamental Neurobiology (A.G.T.; H.A.D.), a Dean’s Postdoctoral Fellowship at Harvard Medical School (A.G.T.), and European Community's Seventh Framework Program Marie Curie Fellowship (M.S.R.).
Footnotes
AUTHOR CONTRIBUTIONS
A.T.G., M.S.R., and H.A.D. performed experiments, A.T.G., M.S.R., H.A.D., M.M., and C.J.W. designed experiments and analyzed data, and C.J.W. oversaw the project.
ACCESSION CODES (NCBI accession numbers pending)
References
- 1.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–9. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 3.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 4.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 5.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 6.Durgan DJ, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–69. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- 7.Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–41. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–31. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–4. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 12.Brown SA, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–6. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 13.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 14.Kim JY, Kwak PB, Weitz CJ. Specificity in circadian clock negative feedback from targeted reconstitution of the NuRD corepressor. Mol Cell. 2014;56:1–11. doi: 10.1016/j.molcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Sangoram AM, et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK–BMAL1-induced transcription. Neuron. 1998;21:1101–13. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 16.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 17.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–71. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 18.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and Period gene expression. Cell. 2007;129:1011–23. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian Period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 20.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–4. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 21.Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK–BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci USA. 2013;110:16021–6. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao WN, et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat Cell Biol. 2007;9:268–75. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 23.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327:463–6. doi: 10.1126/science.1180067. [DOI] [PubMed] [Google Scholar]
- 24.Anafi RC, Lee Y, Sato TK, Venkataraman A, Ramanathan C, Kavakli IH, Hughes ME, Baggs JE, Growe J, Liu AC, Kim J, Hogenesch JB. Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol 15. 12(4):e1001840. doi: 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solt LA, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–8. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–9. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol. 2014;21:126–132. doi: 10.1038/nsmb.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosoda H, et al. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain. 2009;2:34. doi: 10.1186/1756-6606-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–21. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK–BMAL1 and influences the circadian clock. Science. 2011;333:1881–5. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell. 2013;49:68–78. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–83. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB1-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–94. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Iovine B, Iannella ML, Bevilacqua MA. Damage-specific DNA binding protein 1 (DDB1): a protein with a wide range of functions. Int J Biochem Cell Biol. 2011;43:1664–7. doi: 10.1016/j.biocel.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Ozturk N, VanVickle-Chavez SJ, Akileswaran L, Van Gelder RN, Sancar A. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1–CUL4-ROC1 E3 ligase complex. Proc Natl Acad Sci USA. 2013;110:4980–5. doi: 10.1073/pnas.1303234110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallach T, Schellenberg K, Maier B, Kalathur RK, Porras P, Wanker EE, Futschik ME, Kramer A. Dynamic circadian protein-protein interaction networks predict temporal organization of cellular functions. PLoS Genet. 2013;9:e1003398. doi: 10.1371/journal.pgen.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–18. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripperger JA, Schibler U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 43.Fuchs G, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–73. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage repsonse, and cancer. Front Oncol. 2012;2 doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright DE, Wang CY, Kao CE. Histone ubiquitylation and chromatin dynamics. Front Biosci (Landmark Ed) 2012;17:1051–78. doi: 10.2741/3973. [DOI] [PubMed] [Google Scholar]
- 46.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim Biophys Acta. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Thakar A, Parvin J, Zlatanova J. BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J Biomol Struct Dyn. 2010;27:399–406. doi: 10.1080/07391102.2010.10507326. [DOI] [PubMed] [Google Scholar]
- 48.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–63. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Shema-Yaacoby E, et al. Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep. 2013;4:601–8. doi: 10.1016/j.celrep.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 51.Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–29. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seo KI, Lee JH, Nezames CD, Zhong S, Song E, Byun MO, Deng XW. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell. 2014;26:695–711. doi: 10.1105/tpc.113.119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 2012;8(7):e1002825. doi: 10.1371/journal.pgen.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W. Volcano plots in analyzing differential expressions with mRNA microarrays. J Bioinform Comput Biol. 2012;10:231003. doi: 10.1142/S0219720012310038. [DOI] [PubMed] [Google Scholar]
- 55.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotech. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carey MF, Peterson CL, Smale ST. Dignam and Roeder nuclear extract preparation. Cold Spring Harb Protoc. 2009;12 doi: 10.1101/pdb.prot5330. pdb.prot5330. [DOI] [PubMed] [Google Scholar]
- 58.Butter F, Kappei D, Buchholz F, Vermeulen M, Mann M. A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 2010;11:305–311. doi: 10.1038/embor.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 60.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 61.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory- efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR, Hall IM. BEDtools: a flexible suite of utilities comparingt genomic features. Bioinformat. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishii K, et al. Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: a real-time analysis. Neurosci Lett. 2006;401:44–8. doi: 10.1016/j.neulet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 64.Lee C, et al. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–67. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.