Abstract
Cleavage of the cell-cell adhesion molecule, PTPμ, occurs in human glioblastoma multiforme brain tumor tissue and glioma cell lines. PTPμ cleavage is linked to increased cell motility and growth factor independent survival of glioma cells in vitro. Previously, PTPμ was shown to be cleaved by furin in the golgi to generate membrane associated E- (extracellular) and P- (phosphatase) subunits, and by ADAMs and the gamma secretase complex at the plasma membrane. We also identified the presence of additional extracellular and intracellular PTPμ fragments in brain tumors. We set out to biochemically analyze PTPμ cleavage in cancer cells. We determined that, in addition to the furin-processed form of PTPμ, a pool of 200 kDa full-length PTPμ exists at the plasma membrane that is cleaved directly by ADAM to generate a larger shed form of the PTPμ extracellular segment. Notably, in glioma cells, full-length PTPμ is also subject to calpain cleavage, which generates novel PTPμ fragments not found in other immortalized cells. We also observed glycosylation and phosphorylation differences in the cancer cells. Our data suggest that an additional serine protease also contributes to PTPμ shedding in glioma cells. We hypothesize that a “protease storm” occurs in cancer cells whereby multiple proteases converge to reduce the presence of cell-cell adhesion molecules at the plasma membrane and to generate protein fragments with unique biological functions. As a consequence, the “protease storm” could promote the migration and invasion of tumor cells.
Keywords: Proteolysis, Shedding, PTPmu, receptor protein tyrosine phosphatase, protein tyrosine phosphatase, ADAM, Calpain, Furin, Serine Protease, glioma
Introduction
Proteolysis of the cell-cell adhesion molecule PTPμ is implicated in regulating the motility and growth of an aggressive type of brain tumor, glioblastoma multiforme (GBM) [Burgoyne et al., 2009a; Burgoyne et al., 2009b]. PTPμ is a type I transmembrane protein with cell adhesion molecule motifs in its extracellular segment, a single pass transmembrane domain and an intracellular segment with tyrosine phosphatase activity [Brady-Kalnay and Tonks, 1993; Gebbink et al., 1991; Gebbink et al., 1993a]. PTPμ is a member of both the immunoglobulin superfamily of cell-cell adhesion molecules and the receptor protein tyrosine phosphatase (RPTP) IIb subfamily [Brady-Kalnay and Tonks, 1995]. PTPμ is produced as a single polypeptide chain that is glycosylated to produce a mature protein with a molecular weight (MW) of 200 kDa. PTPμ is processed by a furin-like protease in the golgi to yield two non-covalently associated fragments, the extracellular (E) subunit (100 kDa MW), and the intracellular phosphatase (P) subunit (100 kDa MW, Fig. 1A) [Brady-Kalnay and Tonks, 1994; Campan et al., 1996; Gebbink et al., 1995]. Both the 200 kDa and the furin-processed forms are present at the cell surface [Burgoyne et al., 2009b; Gebbink et al., 1995].
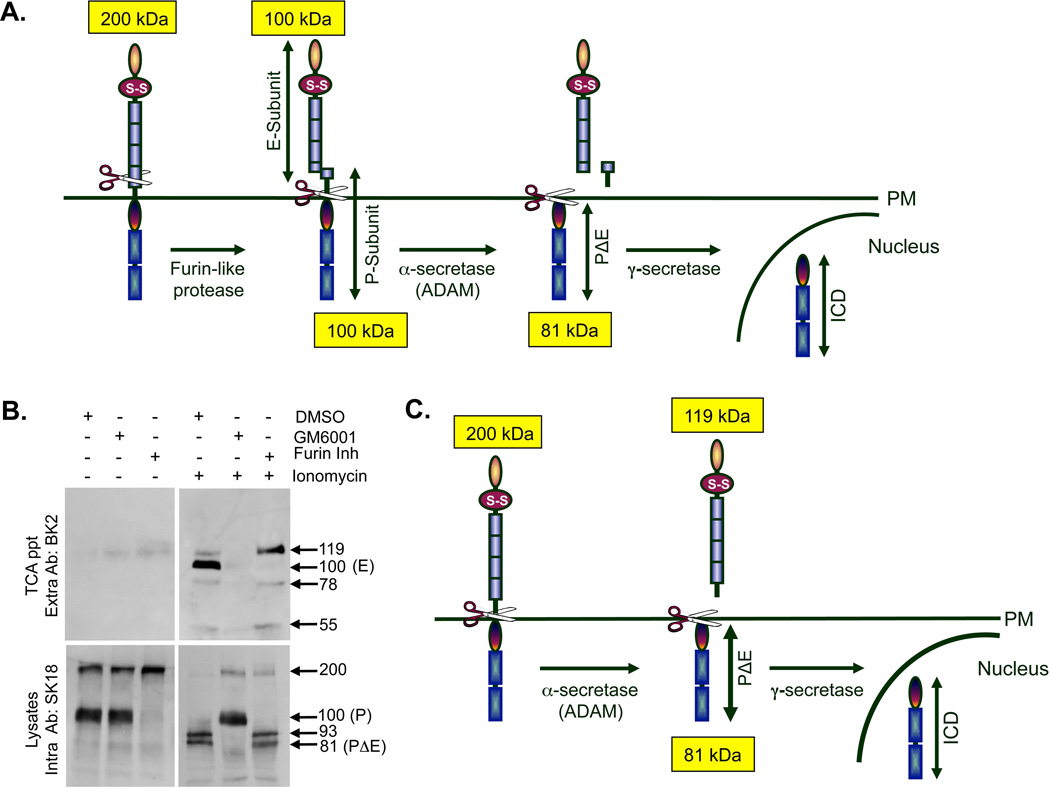
Figure 1. PTPμ is proteolytically processed by a Notch-like paradigm in Mv 1 Lu cells.
Schematic of PTPμ proteolytic processing at the plasma membrane. Full-length (200 kDa) PTPμ is sequentially processed by a furin-like protease into two tightly associated fragments, the E-subunit (100 kDa) and the P-subunit (100 kDa). PTPμ is further processed by an α-secretase/ADAM to shed the E-subunit, resulting in a membrane tethered PΔE fragment (81 kDa) consisting of the entire intracellular segment and a few residues of the remaining extracellular segment. The PΔE fragment is a substrate for γ-secretase, which cleaves PTPμ within the transmembrane domain to release an intracellular fragment (ICD) capable of translocating to the nucleus (A). Mv 1 Lu cells were treated with ionomycin, after a 17–24 hour pre-incubation with DMSO, GM6001 or furin Inhibitor. Shed proteins were recovered from the culture supernatant by TCA precipitation and cell lyates were made as described in the Methods. Protein samples were resolved by SDS-PAGE and PTPμ was detected by immunoblotting with either the BK2 extracellular antibody or the SK18 intracellular antibody (B). Numbers indicate molecular weights in kDa. In this study we demonstrate that full-length PTPμ at the plasma membrane is a substrate for α-secretase/ADAM-induced ectodomain shedding in the absence of furin processing; producing a shed fragment of 119 kDa, and containing most of the extracellular segment (C).
PTPμ is expressed in the lung, heart, vasculature and nervous system [Brady-Kalnay et al., 1995; Burden-Gulley and Brady-Kalnay, 1999; Ensslen et al., 2003; Gebbink et al., 1991; Campan et al., 1996]. PTPμ associates with members of the cadherin family of cell adhesion molecules [Brady-Kalnay et al., 1998; Brady-Kalnay et al., 1995; Sui et al., 2005] as well as with various regulators of the actin cytoskeleton, including RACK1 [Mourton et al., 2001] IQGAP1 [Phillips-Mason et al., 2006], and p120 catenin [Zondag et al., 2000]. PTPμ is stabilized at the plasma membrane at high cell density [Brady-Kalnay et al., 1995; Gebbink et al., 1995], where it mediates cell-cell adhesion [Brady-Kalnay et al., 1993; Gebbink et al., 1993b].
Full-length and furin-processed cell surface forms of PTPμ are absent in GBM tissue and glioma cell lines [Burgoyne et al., 2009a; Burgoyne et al., 2009b]. The loss of cell surface PTPμ observed in GBM is due to A Disintegrin And Metalloproteases (ADAM) metalloprotease or matrix metalloprotease (MMP)-mediated ectodomain shedding following furin processing (Fig. 1A). Ectodomain shedding results in a membrane associated fragment, PΔE [Burgoyne et al., 2009b]. γ-secretase then cleaves PΔE to generate the membrane-free intracellular domain (ICD) of PTPμ (Fig. 1A).
By using ADAM and γ-secretase inhibitors as well as short hairpin (sh)RNA knockdown of PTPμ, it was demonstrated that the PΔE and the ICD fragments promoted migration and growth factor independent survival of glioma cells, suggesting that cleaved PTPμ functions as an oncogene [Burgoyne et al., 2009b]. Conversely, reduction of full-length PTPμ using shRNA knockdown in the non-migratory glioma U-87 MG cell line induced migration and invasion, suggesting that full-length, plasma membrane associated PTPμ functions as a tumor suppressor [Burgoyne et al., 2009a].
Proteolysis of full-length cell-cell adhesion molecules is often observed in cancers [Craig and Brady-Kalnay, 2011a; Craig and Brady-Kalnay, 2011b]. The high level of proteases in the tumor microenvironment combined with the presence of activating stimuli, such as constitutive growth factor signaling, induce shedding via calcium influx and Protein Kinase C activation [Reiss et al., 2006; Rucci et al., 2011]. Just as has been observed with PTPμ proteolysis, when other cell-cell adhesion molecules are cleaved and released from the plasma membrane they no longer act as cell-cell adhesion molecules and instead induce cancer cell motility and invasion. This may lead to the loss of contact inhibition observed in cancer [Craig and Brady-Kalnay, 2011a]. Exploiting the presence of novel fragments generated from the cleavage of cell-cell adhesion molecules has been proposed as a means of diagnosing and treating cancers [Craig and Brady-Kalnay, 2011b].
Among the proteases observed to proteolyze cell-cell adhesion molecules are metalloproteases (MMPs), ADAMs, caspases, rhomboid proteases and calpains [Craig and Brady-Kalnay, 2011a; Craig and Brady-Kalnay, 2011b; David and Rajasekaran, 2012; Gil-Henn et al., 2001; Reiss et al., 2006]. MMPs and ADAMs are known as sheddases, as MMPs are found in the extracellular space while ADAMs are transmembrane proteases. Both MMPs and ADAMs proteolyze proteins outside of the cell membrane, resulting in the release or “shedding” of the protein’s extracellular segment [Reiss et al., 2006]. Rhomboid proteases cut adhesion molecules in eukaryotic parasites, such as Toxoplasma and Plasmodium species [Urban and Dickey, 2011] as well as the mammalian ephrin B cell surface guidance protein [Pascall and Brown, 2004]. Rhomboid proteases are serine proteases that cut within the plasma membrane [Urban and Dickey, 2011], thus also generating shed extracellular protein fragments.
Calpains are calcium-dependent cytoplasmic cysteine proteases that proteolyze cytoskeletal-associated proteins, phosphatases and cell adhesion molecules, including β-integrin and receptor PTPs [Chakraborti et al., 2012; Gil-Henn et al., 2001]. Because calpains are intracellular, when they cleave transmembrane proteins it does not result in shedding of the extracellular fragment from cell membranes. Instead calpain cleavage results in the generation of unique, membrane disassociated, cytosolic fragments.
In this study of PTPμ proteolysis, we demonstrate that additional PTPμ fragments exist in glioma cell lines besides the full-length (200 kDa), P (100 kDa), E (100 kDa), PΔE (81 kDa), and ICD (78 kDa) fragments previously identified [Burgoyne et al., 2009a; Burgoyne et al., 2009b]. In order to identify the additional cleavage products and analyze any related post-translational modifications to the PTPμ protein, we conducted biochemical analyses in the Mv 1 Lu immortalized, non-transformed cell line that expresses high levels of PTPμ and in which PTPμ has been well characterized. In this study, the Mv 1 Lu cell line simulated “normal” cells. We compared the Mv 1 Lu results to those obtained in the LN-229 human glioma cell line in which full-length PTPμ is lost due to proteolysis. PTPμ was exogenously expressed in LN-229 cells. Then, proteolysis was preferentially induced with ionomycin stimulation, which promotes calcium influx and is analogous to constitutive growth factor activation observed in tumor cells. We determined that although some of the same processing occurs in the immortalized and the glioma cell lines following ionomycin stimulation, additional post-translational modifications including differential glycosylation and phosphorylation occur in the tumor cell line. Importantly, we determined that the ADAM protease cleaves full-length PTPμ directly to generate a larger shed extracellular fragment. Furthermore, we determined that the calcium activated protease calpain cleaves at three different sites within the PTPμ cytoplasmic domain only in glioma cells to generate distinct PTPμ fragments. Finally, we demonstrated that simultaneous inhibition of furin, ADAM, calpain and another serine protease is required to block proteolysis of PTPμ in glioma cells. Together these data suggest that distinct proteolytic cascades occur in tumor cells to generate novel PTPμ fragments. The insights gained from this study reinforce the theory of a “protease storm” occurring in cancer cells which proteolyzes cell-cell adhesion molecules such as PTPμ to promote tumorigenesis by reducing adhesion and generating biologically active fragments that can function in new, potentially oncogenic, ways.
Materials and Methods
Cells and Lentiviral Infection
LN-229 human glioma cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco’s modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (HyClone, Logan, UT) at 37°C, 5% CO2. Mv 1 Lu mink cells were obtained from ATCC and maintained in DMEM supplemented with 10% fetal bovine serum at 37°C, 5% CO2. Where indicated, LN-229 and Mv 1 Lu cells were infected with lentiviral particles to express exogenous full-length PTPμ as previously described [Burgoyne et al., 2009b].
Lentiviral shRNA constructs to ADAM 10 (TRCN 0000006672), ADAM 17 (TRCN0000294262) and a PLKO vector control were purchased from Sigma-Aldrich (St. Louis, MO) and used to make lentiviral particles which were used to infect cells as previously described [Burgoyne et al., 2009a].
Chemical Reagents and Antibodies
The following chemicals were purchased from EMD Millipore (San Diego, CA) and used at the concentrations indicated in parenthesis: ionomycin (5 µM), furin inhibitor I (30 µM), GM6001 (25 µM), DAPT (1 µM) and proprotein convertase inhibitor (PPCI, 25 µM). Calpain inhibitor I (ALLN) was purchased from Sigma-Aldrich (St. Louis, MO) and used at 20 µM. The serine protease inhibitors 3,4-Dicholoroisocoumarian (DCI), N-p-tosyl-L-phenylalanine ketone (TPCK) and aprotinin were purchased from Sigma and used at 100 µM, 25 µM and 10µg/ml, respectively. All inhibitors were made up in DMSO with the exception of calpain inhibitor I, which was made up in methanol. A methanol control behaved similarly to DMSO and was not included in the figures (data not shown). The SK18 monoclonal antibody, directed to the intracellular domain, and the BK2 monoclonal antibody, directed to the MAM domain of PTPμ, have been described previously [Brady-Kalnay et al., 1993; Brady-Kalnay and Tonks, 1994]. Polyclonal antibodies to ADAM 10 and ADAM 17 were obtained from Calbiochem and Millipore, respectively. A monoclonal antibody to vinculin was obtained from Sigma-Aldrich.
Precipitation of secreted proteins from the tissue culture media
Mv 1 Lu and LN-229 cells were plated in 100 mm dishes. Two days after plating, the cells were washed twice with basal DMEM (serum-free without further additions) and the media was replaced with basal DMEM overnight. The following day, cells were either untreated or treated with 5 µM ionomycin for 30 min. The culture supernatant was collected and centrifuged to pellet any floating cells. Culture supernatants were incubated on ice with 20% trichloroacetic acid (TCA) for 1–2 hours to precipitate out all proteins. Precipitated protein was then recovered by centrifugation at 16,000 rpm for 15 min. Protein pellets were washed once with ice cold acetone, briefly allowed to dry, resuspended in 2X SDS sample buffer with the addition of 1N NaOH to neutralize the acid and heated at 95°C for 5 min. TCA precipitated proteins were analyzed by immunoblot as described below. For deglycosylation studies, TCA precipitated proteins were incubated with PNGase F or Protein Deglycosylation Mix, both purchased from New England Biolabs (NEB, Ipswich, MA). Deglycoslation of TCA precipitated protein was carried out according to the protocols supplied by NEB.
Immunoprecipitations and Immunoblotting
Mv 1 Lu and LN-229 cells were plated and treated with or without ionomycin as described above. Cellular proteins were isolated in RIPA buffer (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 0.5% deoxycholate, 1% Triton X-100, 0.1% SDS, 2 mM benzamidine, 10 µg/ml aprotinin and leupeptin, 2 µg/ml pepstatin, 40 mM β-glycerol phosphate, 1 mM sodium orthovanadate and 1 mM sodium fluoride). After brief sonication, protein lysates were cleared by centrifugation at 10,000 rpm for 5 min. Protein concentration was determined using a bicinchoninic acid protein assay (Pierce, Rockford, IL). 4X SDS sample buffer was added to the lysates and heated at 95°C for 5 min. Proteins were resolved on low cross-linking 8% SDS-PAGE gels and immunoblotted with PTPμ antibodies BK2 or SK18. For immunoprecipitations, protein lysates were made in RIPA buffer without phosphatase inhibitors (β-glycerol phosphate, sodium orthovanadate and sodium fluoride) to perform dephosphorylation reactions. Equal amounts (400 µg) of protein were added to protein A/G agarose (Roche Applied Science, Indianapolis, IN) pre-loaded with SK18 monoclonal antibody or mouse IgG control antibody. Immunoprecipitated proteins were washed and either untreated or treated with calf intestinal phosphatase (CIP, New England Biolabs) for 30 min at 37°C or treated with PNGase F for 30 min at room temperature. After enzyme treatments, 4X SDS sample buffer was added to the samples and heated at 95°C for 5 min. Immunoprecipitated proteins were resolved on 8% SDS-PAGE gels and immunoblotted with the PTPμ antibody SK18.
Sequence analysis
To determine the approximate expected molecular weight of PTPμ extracellular fragments, we needed to take into consideration the fact that PTPμ is a glycosylated protein. Using NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) we identified 12 potential N-linked glycosylation sites in PTPμ. Using NetOGlyc 4.0 (http://www.cbs.dtu.dk/services/NetOGlyc/) we identified one potential O-linked glycosylation site in PTPμ.
To determine sites of calpain cleavage in PTPμ protein, the online cleavage site predictor program at CaMPDB (calpain.org) was employed. PTPμ amino acid sequence was input with FASTA formatting, and the location of potential calpain cleavage sites were identified.
Results
PTPμ processing in Mv 1 Lu immortalized cells
In our previous studies of PTPμ cleavage, we analyzed intracellular PTPμ fragments, but did not analyze extracellular shed forms of PTPμ [Burgoyne et al., 2009b]. In unstimulated Mv 1 Lu cells, very little shed PTPμ is TCA precipitated from culture media (Fig. 1B). This was expected since very little shedding should occur in cells in the absence of a stimulus. To induce shedding experimentally in vitro, cells can be treated with phorbol esters (PMA or TPA) or the calcium ionophore, ionomycin [Reiss et al., 2006] These methods are commonly used to mimic constitutive growth factor stimulation by activating PKCs and inducing calcium release from intracellular stores. We tested the ability of both PMA and ionomycin to induce shedding of PTPμ in Mv 1 Lu cells. Full-length PTPμ and PTPμ fragments were identified using the PTPμ-specific antibodies BK2 and SK18, directed against the extracellular segment and the intracellular segment of PTPμ, respectively [Brady-Kalnay et al., 1993; Brady-Kalnay and Tonks, 1994]. As detected by immunoblot using the BK2 antibody, stimulation of Mv 1 Lu cells with ionomycin, but not PMA (data not shown), resulted in the shedding of PTPμ extracellular bands with MW of 119, 100, 78 and 55 kDa (Fig. 1B), demonstrating that ionomycin induced shedding of four different PTPμ fragments. The predominant bands observed were the 119 and 100 kDa bands, whereas the lower MW bands were fainter and more difficult to observe consistently.
We hypothesized that if ADAM cleaved full-length PTPμ directly, the only difference in ectodomain shedding between furin-processed and the full-length forms of cell surface PTPμ would be the size of the shed fragments. In the furin-processed case, the shed fragment is predicted to be 100 kDa E-subunit (Fig. 1A). However, if full-length PTPμ is processed by ADAM, the shed fragment is predicted to be 119 kDa (Fig. 1C). This is because the ADAM shed fragment from full-length PTPμ contains additional amino acid residues that are retained in the P-subunit if PTPμ is first cut by furin (Figs. 1A and C). In the presence of the furin inhibitor, we observed a block of the 100 kDa band, demonstrating that this band is the previously identified E-subunit of PTPμ (Fig. 1B + ionomycin + furin inh). We also observed the predicted 119 kDa band, demonstrating that full-length PTPμ exists at the plasma membrane and is processed by ADAM (Fig. 1B + ionomycin + DMSO). Since furin inhibition increases the full-length precursor pool (Fig. 1B, lysates + furin inh), more 119 kDa fragment could be generated when furin is inhibited. We observe an increase in the 119 kDa band (Fig. 1B TCA ppt + ionomycin + furin inh), which confirms that full-length PTPμ is a direct substrate for ADAM cleavage (Fig. 1C).
The broad spectrum ADAM inhibitor GM6001 eliminated nearly all PTPμ in the TCA precipitates indicating that all shedding in Mv 1 Lu cells is ADAM-dependent (Fig. 1B + ionomycin + GM6001), as predicted.
Using the SK18 PTPμ antibody directed against the intracellular segment of PTPμ, we evaluated PTPμ expression in Mv 1 Lu immortalized cell lysate preparations in unstimulated cells. We observed the full-length cell surface form of PTPμ (200 kDa) in addition to the P-subunit (100 kDa) in the cell lysates (Fig. 1B, lysates + DMSO). By treating the cells with the furin inhibitor, we confirmed earlier findings that the P-subunit is generated by cleavage of the 200 kDa protein into 2–100 kDa fragments, P- and E- (Fig. 1B lysate + furin inh only lane; [Brady-Kalnay and Tonks, 1994; Gebbink et al., 1995]).
Following ionomycin stimulation, smaller fragments with MWs of 93 kDa and 81 kDa are generated in the cell lysates (Fig. 1B lysate + ionomycin). These intracellular fragments are generated by ADAM cleavage from both full-length PTPμ and furin processed PTPμ, as use of GM6001 results in an increase in the presence of both the 200 kDa full-length and 100 kDa P-subunit band and the disappearance of the 93 and 81 kDa (PΔE) bands (Fig. 1B lysate + ionomycin + GM6001). Furin inhibition stabilizes some 200 kDa PTPμ but does not affect the generation of the 93 and 81 kDa bands, again confirming that these bands are produced as a result of ADAM, not furin, cleavage (Fig. 1B + ionomycin + furin inh). The identity of the 93 kDa band is unclear since it is bigger than PΔE but smaller than the P-subunit. Furthermore, neither dephosphorylation nor deglycosylation alters the size of this band (data not shown).
Based upon these findings, we observe that there is one pool of cell surface PTPμ that is furin processed (Fig. 1A) and one that remains full-length in the immortalized Mv 1 Lu cell line (Fig. 1C). Ionomycin induces ADAM-dependent shedding of PTPμ from both full-length and furin-cleaved PTPμ. The ratio of full-length to furin-processed (based on the amount of P subunit detected by an intracellular antibody) is 1:2.7 in cellular lysates in the absence of ionomycin stimulation in Mv 1 Lu cells. In the presence of the furin-inhibitor, there is a shift to full-length protein in the absence of ionomycin. In the presence of ionomycin, both the full-length and furin-processed forms are shed in approximately the same ratio as they are present on the cell surface (1:2.3 compared to 1:2.7 based upon the detection of the E-subunit in TCA precipitations and immunoblotting with an extracellular antibody). Therefore, we conclude that the two proteolytic pathways are relatively balanced. Furthermore, ADAM cleavage generates the intracellular 81 kDa PΔE fragment, which in turn is processed by γ-secretase (Figs. 1A and C) [Burgoyne et al., 2009b].
PTPμ glycosylation
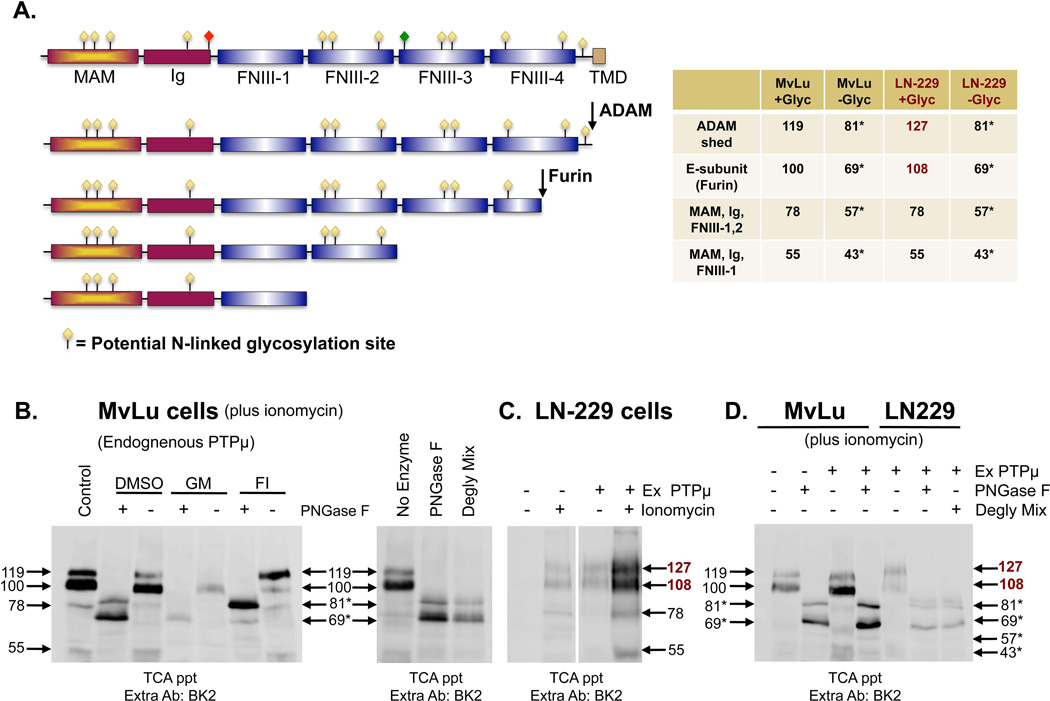
Based on the 1452 amino acids present in PTPμ, we predict that PTPμ should migrate at a MW of 165, yet full-length cell surface PTPμ migrates at 200 kDa. Likewise, the shed PTPμ fragments should be 69 and 81 kDa based on the sites of furin and ADAM cleavage, but they migrate at 100 and 119 kDa, respectively (Figs. 2A and B). N-linked glycosylation could be responsible for this difference as both the full-length and furin-processed forms of PTPμ are susceptible to glycosylation as they are trafficked to the cell surface via the ER and Golgi of Mv 1 Lu cells [Burgoyne et al., 2009b; Gebbink et al., 1995]. To determine whether glycosylation is the reason for the difference in the predicted MW and observed MW of PTPμ and its fragments, we used NetNGlyc 1.0 (See Materials and Methods) to identify potential N-linked glycosylation sites. There are 12 potential N-linked glycosylation sites in PTPμ (Fig. 2A). Based on the size shift observed when full-length PTPμ is treated with PNGase F [Gebbink et al., 1995], we calculated an increase in size of 3.16 kDa per N-linked glycan if all sites are occupied. Using the amino acid sequence and the 12 potential N-linked glycosylation sites within the PTPμ extracellular segment, we predicted the size of the PTPμ fragment shed in response to ionomycin-treatment to be 119 kDa for the ADAM cleaved fragment (Fig. 2A). In the absence of N-linked glycosylation, the size of the ADAM cleaved fragment is predicted to be 81 kDa (119 kDa minus 3.16 kDa per glycan × 12 glycosylation sites = 81). Using a similar calculation, we predicted the MW of all of the PTPμ fragments identified in Figure 1B based on their predicted glycosylation patterns (Fig. 2A). The predicted MWs of 119, 100, 78 and 55 are consistent with the observed MWs (Fig. 2B, left panel, control) indicating that most of the potential glycosylation sites are utilized. Deglycosylation of the TCA precipitated proteins with PNGase F to remove all N-linked oligosaccharides confirmed our predictions (Fig. 2B, left panel + PNGase F). Two major bands are observed on the immunoblots from TCA precipitations of ionomycin stimulated cells treated with PNGase F (DMSO + PNGase F), one at 81 kDa and one at 69 kDa (Fig. 2B, left panel, DMSO + PNGase F and right panel).
Figure 2. PTPμ is differentially glycosylated in Mv 1 Lu and LN-229 cells.
Extracellular fragments of PTPμ generated by furin and ADAM cleavage and the predicted PTPμ domains included in the 78 kDa and 55 kDa fragments, induced by ionomycin, are drawn based on size and antibody recognition. Meprin/PTPμ/A5 (MAM), immunoglobulin (Ig), fibronectin type III repeat (FNIII) and transmembrane (TMD) domains are indicated. Potential N-linked glycosylation sites based on NetNGlyc 1.0 are represented by yellow diamonds. Differential glycosylation was observed to “mark” the RPTP PTPkappa for proteolysis. Based on sequence homology, a similar site is present in PTPμ and is indicated by the red diamond. An O-linked glycosylation site predicted by NetOGlyc 1.0 exists in PTPμ as marked by a green diamond. The numbers indicate the observed sizes of the fragments when they are completely glycosylated or deglycosylated (asterisks). Numbers in burgundy represent the sizes of fragments observed in LN-229 cells that differ from those seen with Mv 1 Lu (A). Mv 1 Lu cells were treated with ionomycin, after a 17–24 hour pre-incubation with DMSO, GM6001 or furin inhibitor. Shed proteins were recovered from the culture supernatant by TCA precipitation. Protein samples were divided in half; one half was untreated, one half was subjected to deglycosylation by PNGase F. Proteins were resolved by SDS-PAGE and PTPμ fragments were detected using the BK2 antibody (B). TCA-precipitated shed proteins from Mv 1 Lu cell culture supernatant treated with ionomycin, was divided and either untreated, subjected to deglycosylation by PNGase F or to deglycosylation with a Deglycosylation Mix. Proteins were resolved by SDS-PAGE and PTPμ fragments were detected using the BK2 antibody (B, right panel). Parental LN-229 cells or LN-229 cells expressing exogenous PTPμ were either untreated or treated with ionomycin (C). Shed proteins were recovered from the culture supernatant by TCA precipitation and resolved by SDS-PAGE then detected by the BK2 antibody (C). Parental Mv 1 Lu cells, Mv 1 Lu cells expressing exogenous PTPμ and LN-229 cells expressing exogenous PTPμ were treated with ionomycin (D). Shed proteins were recovered from the culture supernatant by TCA precipitation and were subjected to deglycosylation by PNGase F or by a Deglycosylation Mix. Protein samples were resolved by SDS-PAGE and shed PTPμ detected by immunoblotting with BK2 (D). Numbers indicate molecular weights in kDa.
The identity of the 69 kDa band was confirmed as the deglycosylated E-subunit by subjecting TCA precipitates from cells treated with furin inhibitor (FI) with PNGase F. In the + PNGase F + FI lane, very little of the 69 kDa band is observed while the 81 kDa remains, demonstrating that the 69 kDa fragment is furin-dependent (Fig. 2B, left panel). The identity of the 81 kDa band as the deglycosylated 119 kDa fragment was confirmed when we deglycosylated the TCA precipitates from cells treated with GM6001. In the + PNGase F + GM6001 lane, only a faint 69 kDa band is observed while the 81 kDa band is eliminated (Fig. 2B, left panel). The deglycosylated forms of the 78 and 55 kDa bands, which should migrate at 57 and 43 kDa, are difficult to detect on immunoblots (Fig. 2B). These two lower MW deglycosylated bands were slightly increased when exogenous PTPμ was expressed in ionomycin stimulated Mv 1 Lu cells (Fig. 2D).
Treatment of the TCA precipitates from culture media of ionomycin stimulated cells with a deglycosylation mix of enzymes that removes all N-linked, simple O-linked and some complex O-linked glycans, generated the same pattern shift in the PTPμ immunopositive bands, from 119 kDa → 81 kDa and 100 kDa → 69 kDa as treatment with PNGase F alone did (Fig. 2B, right panel). We conclude from this data that in Mv 1 Lu cells most, if not all, of PTPμ glycosylation is N-linked.
PTPμ shedding in glioma cells
Previous studies demonstrated the presence of a shed 55 kDa PTPμ fragment in human GBM tumor tissue [Burden-Gulley et al., 2010]. We wanted to evaluate whether any shed fragments of PTPμ are observed in human glioma cells in vitro. To do this, we treated LN-229 glioma cells with ionomycin to induce PTPμ shedding. LN-229 cells contain PTPμ mRNA, but because PTPμ is constitutively processed in these cells [Burgoyne et al., 2009b], there is very little endogenous PTPμ protein (Fig. 2C). Therefore, exogenous PTPμ was expressed in LN229 cells (hereafter abbreviated Ex PTPμ) to conduct the subsequent experiments. All of the bands observed in Ex PTPμ cells were also present on darker exposures of immunoblots from endogenous PTPμ in LN-229 cells (Fig. 3A).
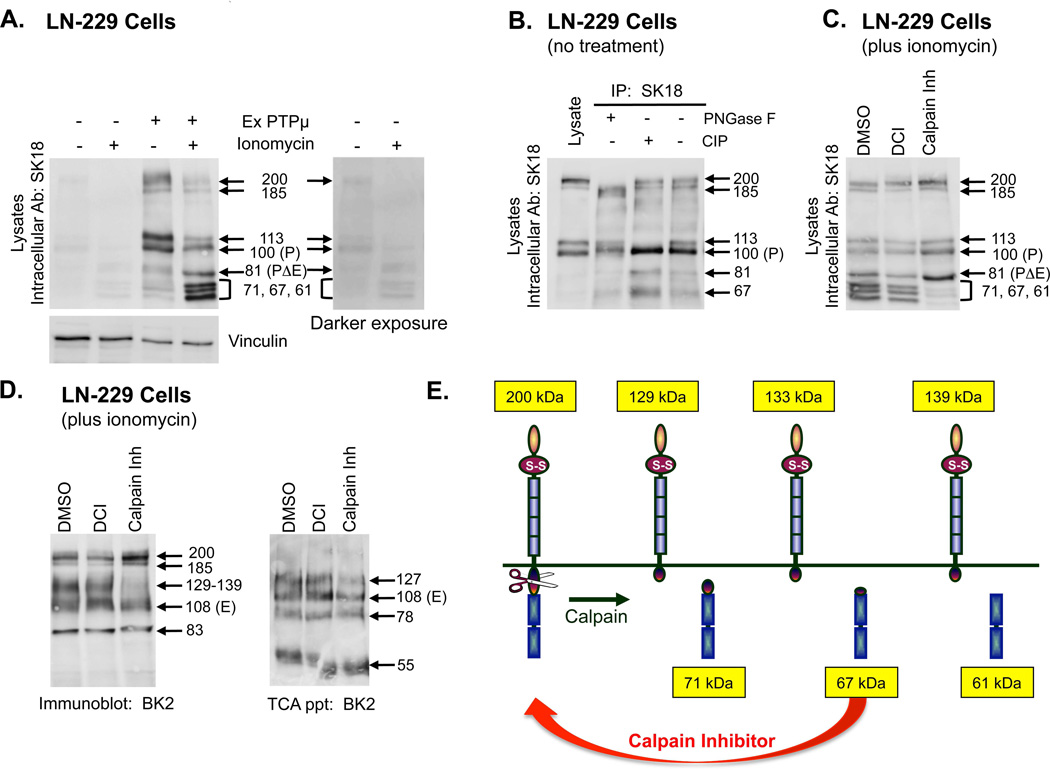
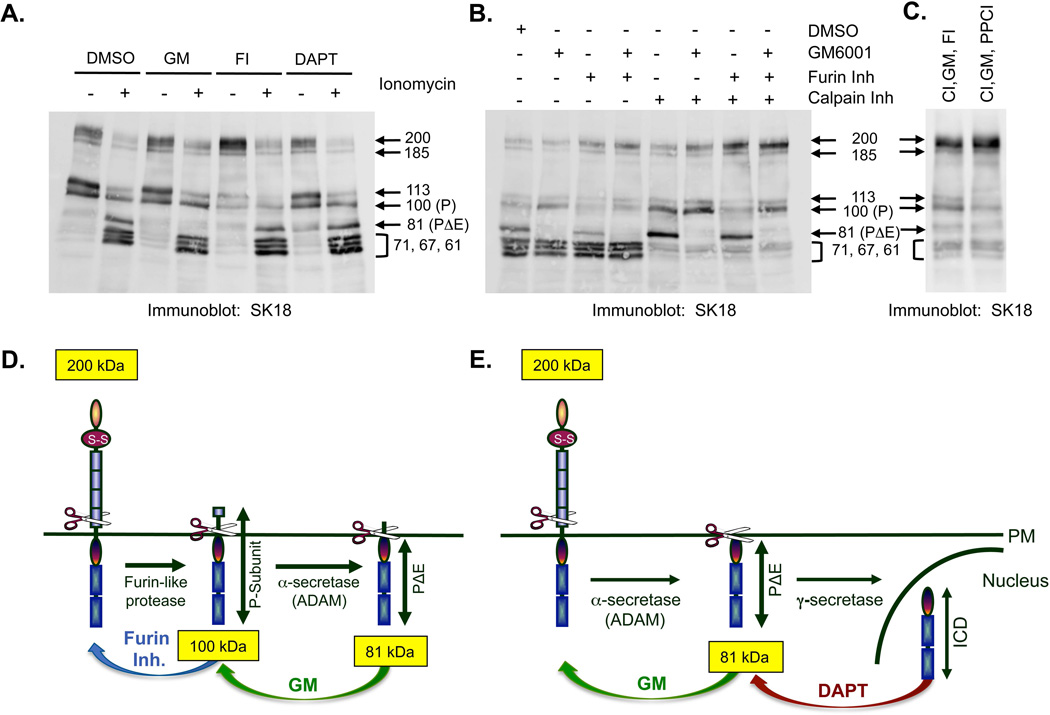
Figure 3. PTPμ is a substrate for calpain cleavage in LN-229 cells.
Parental LN-229 cells or LN-229 cells expressing exogenous PTPμ were either untreated or treated with ionomycin for 30 minutes. Cellular protein lysates were made and proteins were resolved by SDS-PAGE as described in the Methods. Cell associated PTPμ was detected by SK18 (A). An antibody to vinculin was put on the SK18 immunoblot to show relative protein loading. A darker exposure of PTPμ expression in parental LN-229 cells plus and minus ionomycin is also shown (A). Protein lysates were made from untreated LN-229 cells expressing exogenous PTPμ and used for immunoprecipitations (IPs) with SK18. SK18 IPs were untreated or treated with CIP or PNGase F as described in the Methods and then resolved by SDS-PAGE. PTPμ full-length or intracellular fragments were detected with SK18 (B). LN-229 cells expressing exogenous PTPμ were treated with ionomycin after a 30 min pre-incubation with DMSO, DCI or calpain inhibitor. Cell-associated intracellular PTPμ was detected with SK18 in cellular protein lysates (C). Cell-associated extracellular fragments were detected with BK2 (D, left panel). Shed proteins were recovered from culture supernatant by TCA precipitation and resolved by SDS-PAGE and detected with BK2 (D, right panel). Numbers indicate molecular weights in kDa. A model of calpain cleavage of full-length PTPμ illustrates the observed products (E).
In the Ex PTPμ LN-229 cells, ionomycin induced the shedding of four PTPμ fragments with MWs of 127, 108, 78 and 55 kDa (Fig. 2C), similar in size to those observed in Mv 1 Lu cells (119, 100, 78, and 55 kDa, Fig. 2B). The shed PTPμ fragments from LN-229 cells appeared as smears on the immunoblots instead of discrete bands as they did in Mv 1 Lu cell precipitations. The smears suggest differential glycosylation of PTPμ may occur in LN-229 cells to yield protein fragments that migrate at slightly different rates. The 78 and 55 kDa bands migrate at the same MW as they do in Mv 1 Lu cells (Fig. 2D, compare + Ex PTPμ lanes). The presence of the 55 kDa band is in agreement with earlier observations of a shed PTPμ fragment of this size in human GBM tumors [Burden-Gulley et al., 2010].
Deglycosylation of TCA precipitated extracellular fragments from ionomycin stimulated Ex PTPμ LN-229 cells yielded expected bands at 81 kDa (deglycosylated 127 kDa fragment), and 69 kDa (deglycosylated 108 kDa band, Fig. 2D). These bands are identical to those observed in Mv 1 Lu cells (Fig. 2B). An 88 kDa band not detected in Mv 1 Lu cells was also generated following PNGase F or Deglycosylation Mix treatment of Ex PTPμ LN-229 cells (Fig. 2D). This may represent a modification that is not susceptible to these enzymes, such as specific types of O-linked glycosylation (Fig. 2D). An O-linked glycosylation site predicted by NetOGlyc 1.0 exists in PTPμ as marked by a green diamond (Fig. 2A). To summarize, these data suggest there are four major forms of shed PTPμ ectodomains in the Ex PTPμ LN-229 glioma cells, some of which migrate differently from immortalized cells due to differences in glycosylation.
PTPμ processing in glioma cells
Evaluation of PTPμ expression in the unstimulated Ex PTPμ cell lysates with SK18 shows a slightly different pattern of immunopositive bands than that observed in the Mv 1 Lu cells (Fig. 3A compared to Fig. 1B), with two sets of doublet bands, one at 200 and 185 kDa and another at 113 and 100 kDa (Fig. 3A + ExPTPμ).
Treatment of various cell lines with PMA causes a shift from the 100 kDa P-subunit band of PTPμ to the 113 kDa band, suggesting the P-subunit is phosphorylated in response to PMA, presumably due to PKC activation (unpublished data). To test if phosphorylation was responsible for producing the doublets observed, we treated PTPμ immunoprecipitates from Ex PTPμ LN-229 cells with calf alkaline intestinal phosphatase (CIP) to remove any phosphorylation. CIP treatment did convert the 113 kDa band to the 100 kDa band (Fig. 3B + CIP), supporting the hypothesis that phosphorylation increases the molecular weight of the 100 kDa P-subunit to 113 kDa. CIP treatment had no effect on the doublet that migrates in the range of full-length PTPμ, however (Fig. 3B + CIP).
It has been suggested that the lower band of the higher MW doublet (approximately 185 kDa) is an incompletely glycosylated form of cell surface PTPμ [Gebbink et al., 1995]. We treated immunoprecipitated PTPμ with PNGase F to determine if glycosylation could account for the difference in size between the 185 and 200 kDa. Treatment with PNGase F generated a faster migrating band, suggesting both bands are differentially glycosylated forms of cell surface PTPμ (Fig. 3B + PNGase F).
Following ionomycin stimulation of Ex PTPμ LN-229 cells, the 81 kDa (PΔE) fragment of PTPμ is generated, just as in ionomycin stimulated Mv 1 Lu cells (Fig. 3A + ionomycin + ExPTPμ). Ionomycin treatment of Ex PTPμ LN-229 cells also generated a triplet of bands previously unobserved in other cell types with the approximate MWs of 71, 67 and 61 kDa (Fig. 3A). Because ionomycin increases intracellular calcium levels, we wanted to evaluate whether the calcium-dependent cysteine protease, calpain, could be generating the additional PTPμ fragments. To test this, we used the CaMPDB tool [duVerle et al., 2010] to identify potential calpain cleavage sites. Based on the top scores obtained with CaMPDB and the sizes of the calpain triplet observed, we predict calpain cleaves PTPμ at amino acids 825, 855 and 911 to generate fragments of 71, 67 and 61 kDa, respectively (Fig. 3E). We confirmed that these bands were generated by calpain cleavage by pre-incubating cells with a calpain inhibitor prior to ionomycin stimulation and observing the loss of the three bands (Fig. 3C). We next evaluated the membrane-associated forms of PTPμ in glioma cell lysates using the BK2 antibody. If calpain preferred full-length PTPμ as a substrate, we predicted the calpain generated plasma membrane associated forms would have MWs of approximately 129, 133, to 139 kDa (ex. 200 kDa full-length PTPμ - 71 kDa fragment = 129 kDa; Fig. 3E). Following ionomycin treatment, we observed a smear of bands ranging from 129–139 kDa in cell lysates with the BK2 antibody (Fig. 3D, left panel). These bands were inhibited by the addition of a calpain inhibitor (Fig. 3D, left panel). Calpain inhibition had no effect on the ionomycin-induced shedding of 127, 108, 78, and 55 kDa bands of PTPμ (Fig. 3D, right panel). Treatment with a serine protease inhibitor, DCI, did not change the cleavage or shedding of PTPμ in response to ionomycin (Figs. 3C and D), supporting the conclusion that calpain generates the cell-associated extracellular segment containing forms of PTPμ.
A band with the approximate MW of 83 kDa is also detected in immunoblots of cell lysates using the BK2 antibody (Fig. 3D, left panel). It is not the 78 kDa shed fragment observed earlier (Fig. 1B), as the 83 kDa band is observed in cell lysates, not TCA precipitates of culture media. The 83 kDa band represents a membrane associated extracellular fragment that exists only in the presence of ionomycin (Fig. 4A, Lysates, Extra Ab BK2 blot, + ionomycin). It is a discrete band, not like the “fuzzy” glycosylated bands observed in TCA precipitates, and it remains unchanged with the addition of any of the inhibitors (Figs. 3D and 4A, Lysates, Extra Ab: BK2). Therefore, the mechanism whereby it is generated is currently unknown.
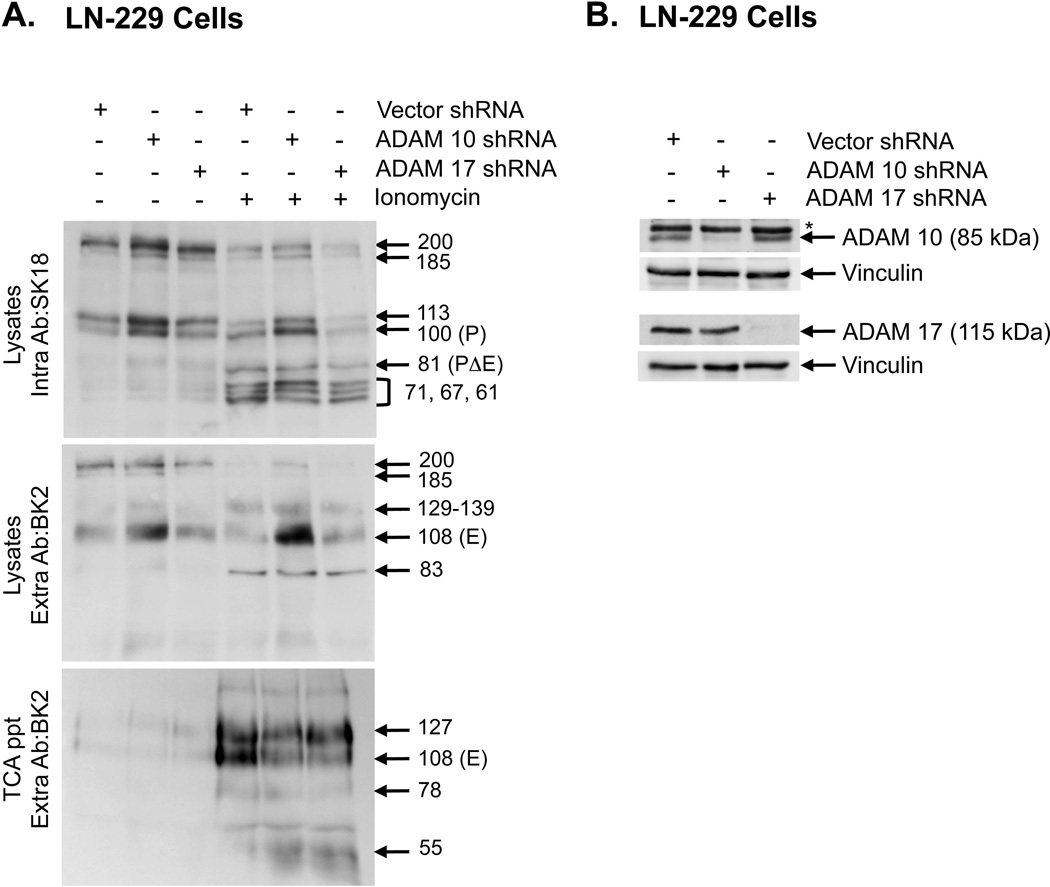
Figure 4. ADAM 10 regulates PTPμ shedding.
Lentiviral shRNA constructs to ADAM 10, ADAM 17 and vector control were used to make lentiviral particles and infect LN-229 cells. Infected cells were either untreated or treated with ionomycin for 30 minutes. Cellular protein lysates were made and proteins were resolved by SDS-PAGE. Cell associated, full-length or intracellular fragments of PTPμ were detected in lysates with SK18 (top panel). Cell-associated extracellular fragments were detected in lysates by BK2 (middle panel). Shed proteins were recovered from the culture supernatant by TCA precipitation and resolved by SDS-PAGE were detected by the BK2 antibody (bottom panel) (A). Lysates from cells infected with lentiviral particles as described above, were resolved by SDS-PAGE and immublotted with polyclonal antibodies to either ADAM 10 or ADAM 17. The immunoblots were reprobed with a monoclonal antibody to vinculin to control for protein loading. The ADAM 10 iummunoblot shows a reduction in the 85 kDa precursor of ADAM 10 recognized by this antibody. The asteriks on the ADAM 10 immunoblot denotes a non-specific band. The ADAM 17 immunoblot shows a reduction in the 115 kDa band consistent with the expected molecular weight of ADAM 17 (B). Numbers indicate molecular weights in kDa.
Down regulation of ADAM 10 by shRNA stabilizes PTPμ
The site of ADAM cleavage in PTPμ was determined based on the ADAM 10 site identified in RPTPκ, a closely related type IIb protein tyrosine phosphatase [Anders et al., 2006]. The ADAM 10 cleavage site in RPTPκ is between amino acids 746 and 747 (KQ/TD), which results in the shedding of all but 6 amino acid residues of the extracellular segment of RPTPκ. PTPμ has the same amino acid sequence N-terminal to its transmembrane domain. We hypothesized that ADAM 10 would cleave PTPμ between amino acids 736 and 737 (KQ/TD) (Fig. 2A). To determine whether ADAM 10 was the α-secretase involved in PTPμ shedding, we infected Ex PTPμ LN-229 cells with lentiviral particles containing shRNA to either ADAM 10 or ADAM 17, another α-secretase implicated in the shedding of cell-cell adhesion molecules [Dang et al., 2011]. Both ADAM 10 and ADAM 17 protein levels were significantly reduced in LN-229 cells infected with shRNA containing lentivirus as determined by immunoblot (Fig. 4B). In the absence or presence of ionomycin, cells containing ADAM 10 shRNA stabilized cell-associated full-length, E- and P-subunits of PTPμ as expected, compared to cells containing ADAM 17 shRNA or vector shRNA (Fig. 4A). ADAM 10 shRNA had only a minor effect on the total amount of PΔE generated (Fig. 4A, top panel) whereas the GM6001 inhibitor was effective in altering PΔE levels. As discussed later, there may be additional proteases involved in generating a band of similar molecular weight. ADAM 10 shRNA had only a small effect on the amount of PTPμ shed (Fig. 4A, bottom panel) suggesting multiple proteases are involved in mediating the shedding of PTPμ.
PΔE is generated by ADAM cleavage in Mv 1 Lu cells (Fig. 1B). It is further proteolyzed by γ-secretase to generate ICD (Fig. 1C) [Burgoyne et al., 2009b]. We next tested whether the PΔE fragment observed in the Ex PTPμ LN-229 cell lysates was also sensitive to GM6001 inhibition and stabilized with the γ-secretase inhibitor DAPT. In the absence of ionomycin stimulation more PΔE is observed with DAPT treatment than in DMSO only lanes (Fig. 5A). This suggests that DAPT stabilizes PΔE. When cells are pre-incubated with GM6001, the PΔE band is eliminated (Fig. 5A), suggesting it is dependent upon ADAM activity to be generated. These data demonstrate that the 81 kDa band produced in response to ionomycin stimulation in Ex PTPμ LN-229 cells is PΔE. As expected, pre-incubation with the furin inhibitor did not affect the levels of PΔE in cells treated with or without ionomycin (Fig. 5A). In addition to the inhibitors discussed above, we used inhibitors of caspase, uPA and cysteine proteases (E-64). None of these inhibitors affected the cleavage of PTPμ (data not shown). As previously published [Burgoyne et al., 2009b], the ICD fragment is not detected unless stabilized by proteasome inhibitors (data not shown).
Figure 5. Inhibition of proteolysis in LN-229 cells.
Proposed PTPμ cleavage mechanisms in LN-229 cells with furin and ADAM (A) or calpain (B). LN-229 cells expressing exogenous PTPμ were either untreated or treated with ionomycin with or without a 17–24 hour pre-incubation with DMSO, GM6001 (25µM), furin inhibitor (30µM) or DAPT (1µM) (A). Cell-associated PTPμ in protein lysates resolved by SDS-PAGE was detected with SK18 (A). In B, LN-229 cells expressing exogenous PTPμ were treated with ionomycin after a 17–24 hour pre-incubation with DMSO, GM6001, furin Inhibitor or a 30 minute pre-incubation with calpain inhibitor (20µM), as single inhibitor treatments or in combination. Cell-associated PTPμ in protein lysates resolved by SDS-PAGE was detected with SK18 (B). LN-229 cells expressing exogenous PTPμ were treated with ionomycin after a pre-incubation with a combination of calpain inhibitor (CI), GM6001 (GM) and furin inhibitor (FI) or a combination of calpain inhibitor (CI), GM6001 (GM) and proprotein convertase inhibitor (PPCI, 25µM) (C). Cell-associated PTPμ in protein lysates resolved by SDS-PAGE was detected with SK18 (C). Numbers indicate molecular weights in kDa. PTPμ is a substrate for both furin and ADAM. The furin inhibitor stabilizes full-length PTPμ, and the ADAM inhibitor, GM6001, stabilizes the P-subunit (D). We determined full-length PTPμ is a direct substrate for ADAM cleavage. PΔE is generated in the presence of the furin inhibitor and is sensitive to the ADAM inhibitor, GM6001. PΔE is a substrate for constitutive cleavage by γ-secretase. Cleavage of PΔE by γ-secretase generates a labile ICD fragment of 78 kDa. The γ-secretase inhibitor, DAPT, can stabilize PΔE in the absence of ionomycin (E).
There are clearly multiple cleavage events going on in glioma cells. To determine which PTPμ proteins are cleaved to yield the PΔE and the calpain fragments, we used protease inhibitors singly or in combination to identify which fragments were eliminated or stabilized in cell lysates by the treatments (Fig. 5B). In this way, we could determine the precursors for fragment generation, i.e. the protease’s substrates. Using the SK18 antibody, we observed that GM6001 stabilizes the P-subunit (Fig. 5B + GM6001). This suggests that the P subunit is a substrate for ADAM cleavage as in Mv 1 Lu cells. Alternatively, ADAM inhibition may increase the full-length PTPμ pool for subsequent furin processing to generate more P subunit. In fact, combining the furin inhibitor with GM6001 increases the 200 kDa PTPμ but yields very little P or PΔE (Fig. 5B + GM6001 + furin inh). In the presence of the calpain inhibitor, both P and PΔE are stabilized (Fig. 5B + calpain inh). This suggests that if full-length PTPμ is increased by inhibition of calpain cleavage, it immediately becomes a substrate for furin and ADAM cleavage to generate those fragments, and conversely, if furin and ADAM are inhibited, calpain can cleave P and PΔE. As expected, when GM6001 is added in addition to the calpain inhibitor, no PΔE or calpain fragments are generated, and the 200 kDa full-length PTPμ and the 100 kDa P-subunit are stabilized (Fig. 5B + GM6001 + calpain inh). When the furin inhibitor is added in addition to the calpain inhibitor, we observe more 200 kDa PTPμ and more PΔE (Fig. 5B + furin inh + calpain inh). This is likely due to a shift in the precursor pool to the full-length form of PTPμ, which is then cleaved by ADAM. When GM6001, the furin inhibitor and the calpain inhibitor are all added together, full-length 200 kDa PTPμ and 100 kDa P-subunit are stabilized (Fig. 5B + GM6001 + furin inh + calpain inh). We hypothesize that the P-subunit is still generated because the furin inhibitor may not function efficiently when all three inhibitors are added together. To test this, we used another inhibitor of furin-like proteases (PPCI) combined with the calpain inhibitor and GM6001. Addition of the PPCI inhibitor resulted in the stabilization of the 200 kDa PTPμ but very little 100 kDa P-subunit (Fig. 5C), suggesting that in LN-229 cells, the P-subunit is also generated by furin cleavage. Moreover, the combination of calpain inhibitor, GM6001 and PPCl blocks nearly all the cleavage of PTPμ based on immunoblot with an intracellular antibody (Fig. 5C).
We now have a general model of PTPμ proteolysis in human glioma cells; the furin processed PTPμ (represented as the 100 kDa P-subunit in SK18 immunoblots) is present at the cell membrane, and is cleaved by ADAM to generate PΔE (Fig. 5D), just as in Mv 1 Lu cells (Fig. 1A). In addition, ADAM directly cleaves cell-surface full-length PTPμ to generate PΔE, which is stabilized by DAPT (Fig. 5E). γ-secretase cleavage then generates ICD (Fig. 5E).
Generation of novel shed and cell surface associated PTPμ fragments in glioma cells
To identify the substrate for calpain cleavage, we evaluated cell lysates with the BK2 antibody to examine which form of cell surface associated PTPμ is stabilized when the calpain fragments are not generated. As we discussed earlier, when we stimulate Ex PTPμ LN-229 cells with ionomycin, we observe membrane-associated fragments of approximately 129, 133, and 139 kDa MWs using the BK2 PTPμ extracellular antibody on immunoblots (Fig. 3D, left panel and Fig, 6A + DMSO). These bands correspond in MW to the extracellular counterparts of the intracellular calpain triplet we observe: e.g. 200 kDa full-length - 129 kDa membrane associated fragment = 71 kDa intracellular fragment, the exact size of the intracellular calpain fragment generated (see Fig. 3E). When the furin inhibitor alone was added to the cells, the abundance of the 129–139 kDa bands increased, whereas the amount of the 100 kDa E-subunit decreased (Fig. 6A + furin inh). Since we know furin inhibition stabilizes the full-length 200 kDa form of PTPμ (Fig. 5A), we infer that the 200 kDa full-length PTPμ must be the substrate for the calpain-generated fragments. The addition of GM6001 did not affect the production of the 100 kDa E-subunit or of the 129–139 kDa bands, suggesting that ADAM cleavage is not involved in the calpain cleavage cascade (Fig. 6A + GM6001).
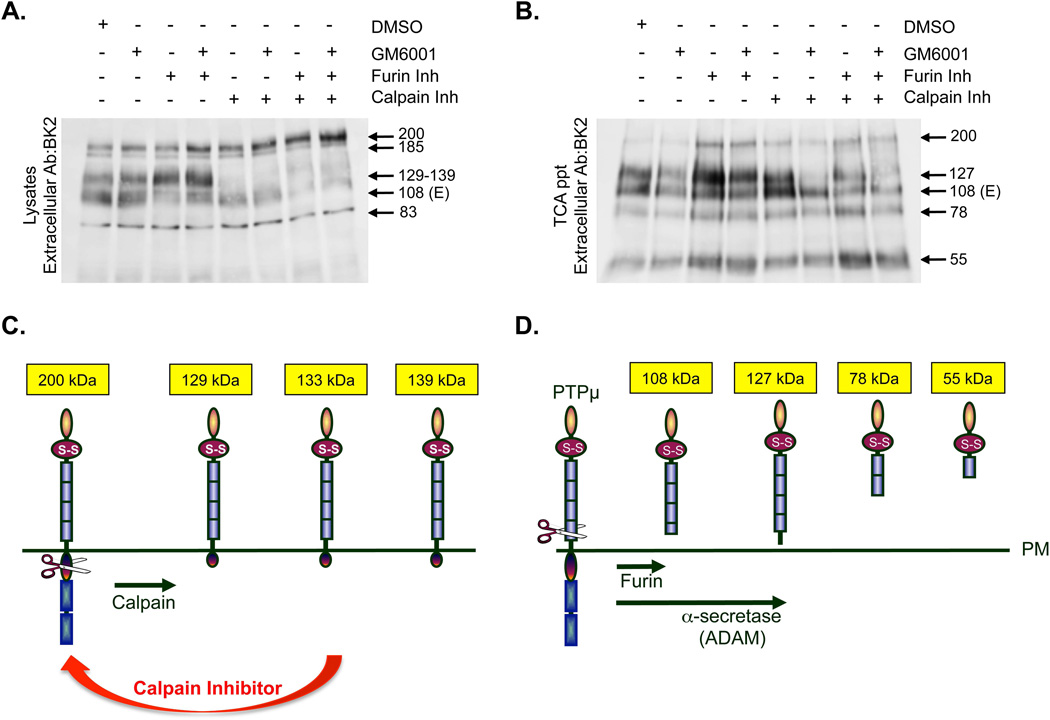
Figure 6. Multiple enzymes are involved in generating truncated membrane associated or ectodomain shed forms of PTPμ in LN-229 cells.
LN-229 cells expressing exogenous PTPμ were treated with ionomycin after a 17–24 hour pre-incubation with DMSO, GM6001, furin inhibitor or a 30 minute pre-incubation with calpain inhibitor as single inhibitor treatments or in combination. Cellular protein lysates were made as described and resolved by SDS-PAGE. Cell-associated PTPμ was detected with the extracellular anitibody BK2 (A). Cells were treated as described above and the shed proteins were recovered from the culture supernatant by TCA precipitation. Shed proteins were resolved by SDS-PAGE and detected by the BK2 antibody (B). Numbers indicate molecular weights in kDa. Schematic of PTPμ membrane associated fragments generated by calpain cleavage induced by ionomycin (C). Calpain is predicted to cleave PTPμ at residues 825, 855 and 911. This diagram depicts the cell-associated extracellular fragments of PTPμ that would result from calpain cleavage and their predicted size (C). Schematic of the potential mechanisms used to generate the observed extracellular PTPμ fragments shed in response to ionomycin. PTPμ is a substrate for furin and ADAM. ADAM cleavage of furin-processed PTPμ generates a shed 100 kDa fragment. ADAM cleavage of full-length PTPμ generates a shed 127 kDa fragment in glioma cells. We also observe shed fragments of 78 and 55 kDa. These bands could arise either from full-length, furin-processed or ADAM shed PTPμ (D).
When we examine TCA precipitates from ExPTPμ LN-229 cells treated with ionomycin, we observe four PTPμ fragments: 127 kDa (directly ADAM shed), 108 kDa (E-subunit), 78 kDa and 55 kDa (Figs. 2C and 6B). The shedding of the four bands was partially inhibited by GM6001 treatment alone (Fig. 6B). We consistently saw a decrease in the 127 kDa band when GM6001 was added in combination with the calpain inhibitor (Fig. 6B). The calpain inhibitor did not block shedding directly (Fig. 6B). The calpain and furin inhibitors affect the amount of full-length PTPμ that is present at the cell surface (Figs. 5B and 6A). Since full-length PTPμ is a substrate for direct ADAM cleavage, altering the amount of full-length changes the substrate pool available for shedding. The 127 kDa band is generated by direct ADAM cleavage of full-length PTPμ based upon cleavage site, observed MW and inhibitor studies (Figs. 2C and 6B). Figures 5 and 6 suggest that the calpain inhibitor stabilizes full-length PTPμ at the cell surface, which is then a substrate for ADAM cleavage. Therefore, the combination of the ADAM and calpain inhibitor would block shedding of the 127 kDa band but not effect the 108 kDa shed E-subunit. The shedding of the extracellular fragments observed in the TCA precipitates was further blocked with the combination of GM6001, calpain inhibitor and furin inhibitor, however, this still did not eliminate the presence of all of the shed forms of PTPμ (Fig. 6B + GM6001 + furin inh + calpain inh) further implicating another protease. A diagram summarizes the predicted domain structure of the four shed forms of PTPμ (Fig. 6D).
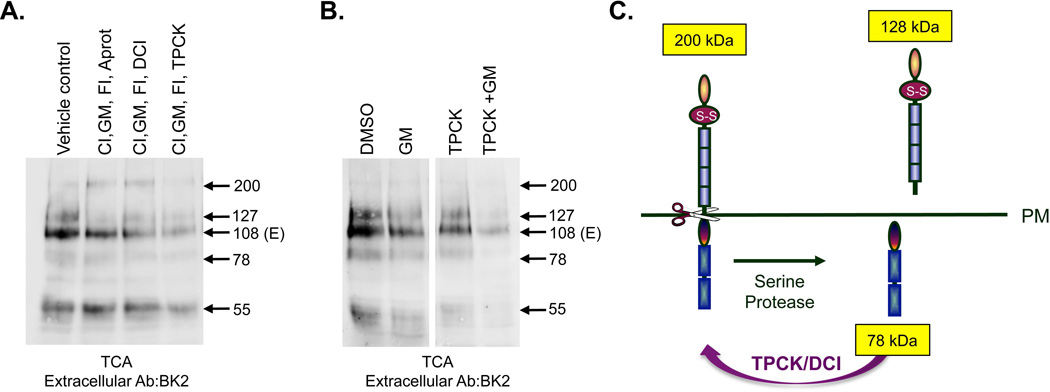
Since we were unable to fully block shedding, even with the combination of three inhibitors (Fig. 6B), we determined another protease was involved in PTPμ cleavage in glioma cells. We tested whether inclusion of serine protease inhibitors would have any effect on PTPμ shedding. Addition of aprotinin to the triple combination had almost no effect on the shedding of PTPμ whereas the addition of DCI or TPCK further reduced PTPμ shedding (Fig. 7A). DCI and TPCK are used as inhibitors of rhomboid proteases since they are most active on that class of serine proteases [Urban et al., 2001; reviewed in Ha et al., 2013 and Urban and Dickey, 2011]. Intrigued by the effect TPCK addition had on PTPμ shedding we wanted to investigate whether TPCK in combination with GM6001 could fully block PTPμ shedding. Since the shedding of PTPμ is an independent event from other cleavage events, we reasoned that shedding potentially could be blocked by the addition of two inhibitors: one targeting ADAMs and one targeting serine proteases. Figure 7B shows that GM6001 and TPCK blocked nearly all PTPμ shedding when added in combination, while each showed a partial effect when added alone. We consistently observe better reduction of shedding with GM6001 and TPCK in combination than with any of the other serine protease inhibitors. These data suggest that both an ADAM and a serine protease contribute to PTPμ shedding in glioma cells. The fact that DCI and TPCK can block the shedding of PTPμ suggests the involvement of a rhomboid protease, however, we cannot rule out other serine proteases. Based on the fact that rhomboid proteases typically cleave close to the outer leaflet of the plasma membrane, rhomboid cleavage of PTPμ should generate a shed extracellular form of PTPμ with a MW of approximately 128 kDa and an intracellular form of PTPμ with a MW of approximately 78 kDa (Fig. 7C). The ADAM cleaved 127 kDa band and the 81 kDa PΔE band may obscure these fragments on BK2 and SK18 immunoblots, respectively.
Figure 7. The combination of ADAM and serine protease inhibitors are required to block PTPμ cleavage.
LN-229 cells expressing exogenous PTPμ were treated with ionomycin after a pre-incubation for 24 hours with GM6001 (GM), furin inhibitor (FI) combined with a 30 minutes pre-incubation with calpain inhibitor (CI) and Aprotinin, DCI, or TPCK. The vehicle control includes amounts of DMSO and methanol equal to the volume used for inhibitor delivery. Cell lysates were made and resolved by SDS-PAGE. Shed proteins were recovered from the culture supernatant by TCA precipitation, resolved by SDS-PAGE and detected by the BK2 antibody (A). Numbers indicate molecular weights in kDa. LN-229 cells expressing exogenous PTPμ were treated with ionomycin after a pre-incubation for 24 hours with DSMO or GM6001 (GM) combined with a 30 minute pre-incubation with TPCK. Cell lysates were analyzed for shed PTPμ as described above. (B). A model of serine protease cleavage of full-length PTPμ illustrates the observed products of a 128 kDa shed extracellular fragment and a 78 kDa intracellular fragment (C).
Discussion
This study adds new information regarding furin and ADAM processing of PTPμ in both immortalized but non-transformed cell lines (ie. “normal” cells) and in cancer cells. PTPμ exists at the cell surface in two forms: a full-length 200 kDa form and a furin-processed form composed of two 100 kDa non-covalently associated subunits, the E- and P-subunits. The P-subunit can exist in a phosphorylated 113 kDa and an unphosphorylated 100 kDa form. The E-subunit is glycosylated. These pools of PTPμ can both be further processed by ADAM to generate two shed fragments: a 119 kDa form when the full-length 200 kDa PTPμ is the substrate and a 100 kDa shed fragment when the furin-processed form of PTPμ is the substrate in immortalized cells. ADAM proteolysis generates the membrane associated 81 kDa PΔE fragment. Further proteolysis by the γ-secretase complex generates a membrane-freed 78 kDa fragment that traffics into the nucleus [Burgoyne et al., 2009b].
The model for PTPμ proteolysis in glioma cells includes furin, ADAM10, and γ-secretase processing as described above but additional cleavage occurs as discussed extensively below. In glioma cells, calpain cleavage of cell-surface PTPμ produces three membrane associated extracellular forms of PTPμ and three membrane-freed cytoplasmic fragments. A serine protease, such as a rhomboid protease, proteolyzes cell-surface full-length PTPμ to generate novel shed and cytoplasmic PTPμ fragments. In addition, we demonstrated that differential glycosylation of PTPμ occurs in cancer cells that may contribute to the additional cleavage events observed.
Inducers of transmembrane protein shedding
Proteolysis and receptor shedding is generally upregulated in cancer due to constitutive growth factor signaling, and may be a mechanism used by cells to lose contact inhibition [Craig and Brady-Kalnay, 2011a]. Major proteases involved in receptor shedding include ADAM and the rhomboid proteases [Craig and Brady-Kalnay, 2011a; Craig and Brady-Kalnay, 2011b; David and Rajasekaran, 2012; Gil-Henn et al., 2001; Reiss et al., 2006]. PMA and ionomycin are used in vitro to induce receptor shedding and to mimic constitutive growth factor signaling observed in vivo. The mechanism by which PMA or ionomycin induce shedding is not clear, but is proposed to be via the induction of post-translational modifications of the cytoplasmic segments of transmembrane protein substrates [Dang et al., 2011]. Intracellular modifications, such as phosphorylation, are translated into conformational changes in the extracellular segment or changes in protein stability, allowing access to proteases. Moreover, the stimuli used to induce shedding appear to direct which ADAMs are activated [Dang et al., 2011].
Based upon our previous study, MMP/ADAMs were predicted to be the major inducers of transmembrane shedding [Burgoyne et al., 2009b]. ADAM 10 and ADAM 17 can be activated by calcium ionophores such as ionomycin [Horiuchi et al., 2007; Le Gall et al., 2009]. While ADAM 10 shRNA did stabilize full-length PTPμ as well as the E- and P-subunits, it did not inhibit PΔE generation (Fig. 4A, top panel), whereas the broad spectrum inhibitor for ADAMs and MMPs, GM6001, was able to block PΔE production. This implies that ADAM 10 is only one of the proteases involved in PTPμ shedding. We saw no major changes in any of the PTPμ fragments in the presence of ADAM 17 shRNA, suggesting this α-secretase is not involved.
Post-translational events segregating pools and/or inducing conformational effects
PTPμ processing in glioma cells may be regulated by localization, glycosylation and/or phosphorylation. For example, in this study, we determined the 100 kDa furin-processed P-subunit of PTPμ is phosphorylated in the LN-229 cells to produce a fragment that migrates at 113 kDa. This modification could potentially “mark” a pool of PTPμ for cleavage by a particular protease. Furthermore, the existence of multiple pools of PTPμ at the cell surface is a curiosity. Are both forms of cell-surface full-length PTPμ present at the same location? We hypothesize that in addition to phosphorylation, the localization within membrane microdomains regulates which proteases cleave PTPμ.
Glycosylation differences may also “mark” certain pools of PTPμ for cleavage. Glycosylation can regulate proper folding, transport and stability of proteins. Aberrant glycosylation is associated with disease states including cancer. An increase in β(1,6)-branching on N-linked glycans was observed in malignant cells, catalyzed by N-aceytlglucosaminyl transferase V (GnT-V). RPTPκ is a target for GnT-V in colon cancer cells [Kim et al., 2006], and differential glycosylation was observed to “mark” RPTPκ for proteolysis (indicated in Fig. 2A by a red diamond). Additional studies suggest that aberrant glycosylation of RPTPκ by GnT-V facilitates its cleavage by furin [Wang et al., 2009]. Glycosylation reduces RPTPκ‘s phosphatase activity towards EGFR, resulting in cell migration. Based on sequence homology to RPTPκ, PTPμ also has a site for potential modification by GnT-V. O-linked glycosylation regulates Notch cleavage and activity by regulating Notch ligand binding [Okajima et al., 2003]. Notably, an O-linked glycosylation site exists in PTPμ’s extracellular domain as marked by a green diamond in Figure 2A.
PTPμ proteolysis results in five shed adhesive forms of PTPμ
Earlier studies identified the existence of a shed 55 kDa PTPμ fragment in human GBM tumor samples, present at two-fold higher levels in the center and edge of the tumors than in adjacent normal cortex [Burden-Gulley et al., 2010]. This finding complimented an earlier study that demonstrated PTPμ cleavage occurs preferentially in GBM tissue and generates novel cytoplasmic fragments of PTPμ [Burgoyne et al., 2009b]. Based on size and antibody recognition, the 55 kDa shed PTPμ fragment likely corresponds to the MAM, Ig and first fibronectin III repeat of PTPμ [Burden-Gulley et al., 2010], and as such would be capable of binding homophilically to any form of PTPμ containing the homophilic binding site. Because of this homophilic binding capability, fluorescently-tagged PTPμ probes were devised to bind to and detect the shed 55 kDa fragment in tumor tissue [Burden-Gulley et al., 2010]. These probes, termed SBK probes, are very effective at labeling tumor cells and highlighting the boundary of GBM tumor tissue sections in vitro and labeling flank tumors of glioma cells in vivo [Burden-Gulley et al., 2010]. In addition, the SBK probes can cross the blood brain barrier and infiltrate a GBM tumor to label virtually all GFP-positive glioma cells in a xenograft GBM tumor model even when those cells have migrated 4mm away from the main tumor mass [Burden-Gulley et al., 2010].
In the current study, we identified five shed fragments of PTPμ released from glioma cells, including the 55 kDa form. Based on their size, our data suggests two of these fragments, the 127 and 108, are released by ADAM processing of either the full-length form of PTPμ or the furin-processed form of PTPμ, respectively. In addition, a serine protease such as a rhomboid could generate shed fragments of a similar size. We are unable to discern the size difference of ADAM or serine protease shed fragments by immunoblot but we show that both ADAM and serine protease inhibitors are needed to fully block PTPμ shedding. The protease(s) that generate the 78 and 55 kDa fragments must be regulated by one of the identified proteases as the combinations of inhibitors block their shedding although no individual protease did. The proteases involved may include an MMP or another serine protease. Given that we now know there are four additional shed fragments of PTPμ, it is possible that the SBK probes are actually detecting any of the five shed PTPμ fragments, since the homophilic binding site resides in the N-terminal part of the protein, the immunoglobulin domain [Brady-Kalnay and Tonks, 1994].
Calpain cleavage generates three novel cell surface forms of PTPμ that lack catalytic activity and three novel catalytically active cytoplasmic fragments of PTPμ
Three novel cell-surface forms of PTPμ generated by calpain cleavage were also identified in this study. The calpain cleaved cell-surface fragments (139, 133, and 129 kDa in size) are likely functional adhesion molecules, as only the extracellular domain of PTPμ is required for homophilic cell-cell adhesion [Brady-Kalnay et al., 1993; Gebbink et al., 1993b]. Unlike the full-length 200 kDa and the furin-processed forms of PTPμ, however, these cell-surface forms of PTPμ lack catalytic phosphatase domains, and would therefore be unable to signal intracellularly in response to adhesion. Calpain cleavage of cell-surface full-length PTPμ also generates three membrane-released phosphatase domain-containing fragments that are detectable in cell lysate preparations, with MWs of 71, 67, and 61 kDa (Fig. 3).
Calpain cleavage has never been demonstrated for type IIb RPTPs. However, the type IV RPTPs, RPTPα and PTPɛ, are cleaved by calpain to generate cytoplasmic fragments in primary cortical neurons following stimulation with CaCl2 [Gil-Henn et al., 2001]. Calpain cleavage of RPTPα generates a cytoplasmic PTPα fragment which is unable to act on the membrane-localized substrates of RPTPα, including Src and the Kv2.1 potassium channel [Gil-Henn et al., 2001]. Cleavage, therefore, can have major biological effects by relocating the catalytic domains of RPTPs away from substrates at the plasma membrane towards another set of substrates at different subcellular locations [Phillips-Mason et al., 2011].
Calpains have been implicated in tumor progression as their expression and activity is up-regulated by oncoproteins including v-Src, v-Myc, v-Jun, v-Fos and k-Ras [Carragher et al., 2004; Carragher et al., 2002]. A role for calpain has been established in tumor cell migration and invasion [Leloup, 2011; Storr et al., 2011], in glioma cell invasion in vitro [Jang et al., 2010] and in organotypic mouse brain slices [Lal et al., 2012]. Calpains also regulate MMP expression [Jang et al., 2010; Popp et al., 2003] and activity [Lal et al., 2012], which contribute to invasion via remodeling of the cell surface and extracellular matrix, and by proteolyzing actin associated proteins [Lal et al., 2012].
Membrane-free phosphatase domains that are no longer regulated by adhesion
Four membrane-released cytoplasmic fragments of PTPμ are generated by proteolysis: ICD [Burgoyne et al., 2009b], and the 71, 67, and 61 kDa calpain fragments. These fragments have the catalytically active membrane proximal phosphatase domain, as well as the catalytically inactive membrane distal phosphatase domain [Gebbink et al., 1993a]. They also have varying amounts of the juxtamembrane region of PTPμ (see Fig. 3E). The membrane distal phosphatase domain and the juxtamembrane region both regulate tyrosine phosphatase activity of the membrane proximal domain [Aricescu et al., 2001; Brady-Kalnay and Tonks, 1993; Feiken et al., 2000; Gebbink et al., 1993a]. Given that some fragments have more or less of the juxtamembrane domain, it is likely that the fragments would bind to different substrates, have alternative protein-protein interactions and/or could be regulated differentially by protein folding.
Our data combined with earlier studies suggest that the membrane-released cytoplasmic fragments have different levels of stability but may share the same localization and function within cells. A proteosome inhibitor was needed to stabilize the 78 kDa ICD fragment in GBM tissue samples [Burgoyne et al., 2009b], but was not required to visualize the calpain-generated fragments in this study. The stability of the calpain-generated fragments may result in longer biological activity than the labile ICD fragment. Finally, a wedge inhibitor peptide that binds to PTPμ near the first phosphatase domain and inhibits PTPμ phosphatase activity decreased migration of LN-229 cells in a migration assay [Burgoyne et al., 2009b]. This function was ascribed to the ICD fragment, as parental LN-229 glioma cells only have proteolytic cytoplasmic fragments and not full-length PTPμ [Burgoyne et al., 2009b]. Given the findings of the current study, the wedge peptide may have inhibited the phosphatase activity of some or all of the cytosolic fragments identified in this manuscript, which may all regulate glioma cell migration.
Our studies suggest that proteolysis of cell-cell adhesion molecules is even more complex than previously demonstrated in cancer cells and that a “protease storm” ensures a loss of contact inhibition. Our data suggest that individual protease inhibitors are unlikely to be effective cancer chemotherapeutics. Instead, a cocktail of protease inhibitors may be required to block the cleavage of cell-cell adhesion molecules to prevent the loss of contact inhibition observed in cancer.
Acknowledgments
The authors would like to thank Michael Howell for laboratory assistance. This research was supported by NIH grant R01-NS063971 and the Tabitha Yee-May Lou Endowment Fund for Brain Cancer Research.
List of nonstandard abbreviations
- GBM
glioblastoma multiforme
- PTPμ
receptor protein tyrosine phosphatase mu
References
- Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, Haass C, Ullrich A. Furin-, ADAM 10-, and gamma-Secretase-Mediated Cleavage of a Receptor Tyrosine Phosphatase and Regulation of beta-Catenin's Transcriptional Activity. Mol Cell Biol. 2006;26:3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Fulga TA, Cismasiu V, Goody RS, Szedlacsek SE. Intramolecular interactions in protein tyrosine phosphatase RPTPmu: kinetic evidence. Biochem Biophys Res Commun. 2001;280:319–327. doi: 10.1006/bbrc.2000.4094. [DOI] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Flint AJ, Tonks NK. Homophilic binding of PTP mu, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J Cell Biol. 1993;122:961–972. doi: 10.1083/jcb.122.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Mourton T, Nixon JP, Pietz GE, Kinch M, Chen H, Brackenbury R, Rimm DL, Del Vecchio RL, Tonks NK. Dynamic interaction of PTPmu with multiple cadherins in vivo. J Cell Biol. 1998;141:287–296. doi: 10.1083/jcb.141.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Rimm DL, Tonks NK. Receptor protein tyrosine phosphatase PTPmu associates with cadherins and catenins in vivo. J Cell Biol. 1995;130:977–986. doi: 10.1083/jcb.130.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Tonks NK. Purification and characterization of the human protein tyrosine phosphatase, PTP mu, from a baculovirus expression system. Mol Cell Biochem. 1993;127–128:131–141. doi: 10.1007/BF01076764. [DOI] [PubMed] [Google Scholar]
- Brady-Kalnay SM, Tonks NK. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTP mu. J Biol Chem. 1994;269:28472–28477. [PubMed] [Google Scholar]
- Brady-Kalnay SM, Tonks NK. Protein tyrosine phosphatases as adhesion receptors. Curr Opin Cell Biol. 1995;7:650–657. doi: 10.1016/0955-0674(95)80106-5. [DOI] [PubMed] [Google Scholar]
- Burden-Gulley SM, Brady-Kalnay SM. PTPmu regulates N-cadherin-dependent neurite outgrowth. J Cell Biol. 1999;144:1323–1336. doi: 10.1083/jcb.144.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, Sloan AE, Miller RH, Basilion JP, Brady-Kalnay SM. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12:305–316. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, Robinson S, Sloan AE, Vogelbaum MA, Miller RH, Brady-Kalnay SM. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009a;11:767–778. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, Brady-Kalnay SM. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009b;69:6960–6968. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campan M, Yoshizumi M, Seidah NG, Lee ME, Bianchi C, Haber E. Increased proteolytic processing of protein tyrosine phosphatase mu in confluent vascular endothelial cells: the role of PC5, a member of the subtilisin family. Biochemistry. 1996;35:3797–3802. doi: 10.1021/bi952552d. [DOI] [PubMed] [Google Scholar]
- Carragher NO, Fonseca BD, Frame MC. Calpain activity is generally elevated during transformation but has oncogene-specific biological functions. Neoplasia. 2004;6:53–73. doi: 10.1016/s1476-5586(04)80053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher NO, Westhoff MA, Riley D, Potter DA, Dutt P, Elce JS, Greer PA, Frame MC. v-Src-induced modulation of the calpain-calpastatin proteolytic system regulates transformation. Mol Cell Biol. 2002;22:257–269. doi: 10.1128/MCB.22.1.257-269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti S, Alam MN, Paik D, Shaikh S, Chakraborti T. Implications of calpains in health and diseases. Indian J Biochem Biophys. 2012;49:316–328. [PubMed] [Google Scholar]
- Craig SE, Brady-Kalnay SM. Cancer cells cut homophilic cell adhesion molecules and run. Cancer Res. 2011a;71:303–309. doi: 10.1158/0008-5472.CAN-10-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SE, Brady-Kalnay SM. Tumor-Derived Extracellular Fragments of Receptor Protein Tyrosine Phosphatases (RPTPs) as Cancer Molecular Diagnostic Tools. Anticancer Agents Med Chem. 2011b;11:133–140. doi: 10.2174/187152011794941244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang M, Dubbin K, D'Aiello A, Hartmann M, Lodish H, Herrlich A. Epidermal growth factor (EGF) ligand release by substrate-specific a disintegrin and metalloproteases (ADAMs) involves different protein kinase C (PKC) isoenzymes depending on the stimulus. J Biol Chem. 2011;286:17704–17713. doi: 10.1074/jbc.M110.187823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JM, Rajasekaran AK. Dishonorable discharge: the oncogenic roles of cleaved E-cadherin fragments. Cancer Res. 2012;72:2917–2923. doi: 10.1158/0008-5472.CAN-11-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- duVerle D, Takigawa I, Ono Y, Sorimachi H, Mamitsuka H. CaMPDB: a resource for calpain and modulatory proteolysis. Genome Inform. 2010;22:202–213. [PubMed] [Google Scholar]
- Ensslen SE, Rosdahl JA, Brady-Kalnay SM. The receptor protein tyrosine phosphatase mu, PTPmu, regulates histogenesis of the chick retina. Dev Biol. 2003;264:106–118. doi: 10.1016/j.ydbio.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Feiken E, van Etten I, Gebbink MF, Moolenaar WH, Zondag GC. Intramolecular interactions between the juxtamembrane domain and phosphatase domains of receptor protein-tyrosine phosphatase RPTPmu. Regulation of catalytic activity. J Biol Chem. 2000;275:15350–15356. doi: 10.1074/jbc.275.20.15350. [DOI] [PubMed] [Google Scholar]
- Gebbink MF, van Etten I, Hateboer G, Suijkerbuijk R, Beijersbergen RL, Geurts van Kessel A, Moolenaar WH. Cloning, expression and chromosomal localization of a new putative receptor-like protein tyrosine phosphatase. FEBS Lett. 1991;290:123–130. doi: 10.1016/0014-5793(91)81241-y. [DOI] [PubMed] [Google Scholar]
- Gebbink MF, Verheijen MH, Zondag GC, van Etten I, Moolenaar WH. Purification and characterization of the cytoplasmic domain of human receptor-like protein tyrosine phosphatase RPTP mu. Biochemistry. 1993a;32:13516–13522. doi: 10.1021/bi00212a017. [DOI] [PubMed] [Google Scholar]
- Gebbink MF, Zondag GC, Koningstein GM, Feiken E, Wubbolts RW, Moolenaar WH. Cell surface expression of receptor protein tyrosine phosphatase RPTP mu is regulated by cell-cell contact. J Cell Biol. 1995;131:251–260. doi: 10.1083/jcb.131.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbink MF, Zondag GC, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993b;268:16101–16104. [PubMed] [Google Scholar]
- Gil-Henn H, Volohonsky G, Elson A. Regulation of protein-tyrosine phosphatases alpha and epsilon by calpain-mediated proteolytic cleavage. J Biol Chem. 2001;276:31772–31779. doi: 10.1074/jbc.M103395200. [DOI] [PubMed] [Google Scholar]
- Ha Y, Akiyama Y, Xue Y. Structure and mechanism of rhomboid protease. J Biol Chem. 2013;288:15430–15436. doi: 10.1074/jbc.R112.422378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Lal S, Greenwood JA. Calpain 2 is required for glioblastoma cell invasion: regulation of matrix metalloproteinase 2. Neurochem Res. 2010;35:1796–1804. doi: 10.1007/s11064-010-0246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kang HY, Kim JY, Oh S, Kim CH, Ryu CJ, Miyoshi E, Taniguchi N, Ko JH. Identification of target proteins of N-acetylglucosaminyl transferase V in human colon cancer and implications of protein tyrosine phosphatase kappa in enhanced cancer cell migration. Proteomics. 2006;6:1187–1191. doi: 10.1002/pmic.200500400. [DOI] [PubMed] [Google Scholar]
- Lal S, La Du J, Tanguay RL, Greenwood JA. Calpain 2 is required for the invasion of glioblastoma cells in the zebrafish brain microenvironment. J Neurosci Res. 2012;90:769–781. doi: 10.1002/jnr.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall SM, Bobe P, Reiss K, Horiuchi K, Niu XD, Lundell D, Gibb DR, Conrad D, Saftig P, Blobel CP. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, L-selectin, and tumor necrosis factor alpha. Mol Biol Cell. 2009;20:1785–1794. doi: 10.1091/mbc.E08-11-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup LaW A. Calpains as potential anti-cancer targets. Expert Opin Ther Targets. 2011;15:309–323. doi: 10.1517/14728222.2011.553611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourton T, Hellberg CB, Burden-Gulley SM, Hinman J, Rhee A, Brady-Kalnay SM. The PTPmu protein-tyrosine phosphatase binds and recruits the scaffolding protein RACK1 to cell-cell contacts. J Biol Chem. 2001;276:14896–14901. doi: 10.1074/jbc.M010823200. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J Biol Chem. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- Pascall JC, Brown KD. Intramembrane cleavage of ephrinB3 by the human rhomboid family protease, RHBDL2. Biochem Biophys Res Commun. 2004;317:244–252. doi: 10.1016/j.bbrc.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Phillips-Mason PJ, Craig SE, Brady-Kalnay SM. Should I stay or should I go? Shedding of RPTPs in cancer cells switches signals from stabilizing cell-cell adhesion to driving cell migration. Cell Adh Migr. 2011;5:298–305. doi: 10.4161/cam.5.4.16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Mason PJ, Gates TJ, Major DL, Sacks DB, Brady-Kalnay SM. The receptor protein-tyrosine phosphatase PTPmu interacts with IQGAP1. J Biol Chem. 2006;281:4903–4910. doi: 10.1074/jbc.M506414200. [DOI] [PubMed] [Google Scholar]
- Popp O, Heidinger M, Ruiz-Heinrich L, Ries C, Jochum M, Gil-Parrado S. The calpastatin-derived calpain inhibitor CP1B reduces mRNA expression of matrix metalloproteinase-2 and −9 and invasion by leukemic THP-1 cells. Biol Chem. 2003;384:951–958. doi: 10.1515/BC.2003.107. [DOI] [PubMed] [Google Scholar]
- Reiss K, Ludwig A, Saftig P. Breaking up the tie: disintegrin-like metalloproteinases as regulators of cell migration in inflammation and invasion. Pharmacol Ther. 2006;111:985–1006. doi: 10.1016/j.pharmthera.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Rucci N, Sanità P, Angelucci A. Roles of metalloproteases in metastatic niche. Current Molecular Medicine. 2011;11:609–622. doi: 10.2174/156652411797536705. [DOI] [PubMed] [Google Scholar]
- Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- Sui XF, Kiser TD, Hyun SW, Angelini DJ, Del Vecchio RL, Young BA, Hasday JD, Romer LH, Passaniti A, Tonks NK, Goldblum SE. Receptor protein tyrosine phosphatase micro regulates the paracellular pathway in human lung microvascular endothelia. Am J Pathol. 2005;166:1247–1258. doi: 10.1016/s0002-9440(10)62343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Dickey SW. The rhomboid protease family: a decade of progress on function and mechanism. Genome Biol. 2011;12:231. doi: 10.1186/gb-2011-12-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Wang C, Yang Y, Yang Z, Liu M, Li Z, Sun L, Mei C, Chen H, Chen L, Wang L, Zha X. EGF-mediated migration signaling activated by N-acetylglucosaminyltransferase-V via receptor protein tyrosine phosphatase kappa. Arch Biochem Biophys. 2009;486:64–72. doi: 10.1016/j.abb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Zondag GC, Reynolds AB, Moolenaar WH. Receptor protein-tyrosine phosphatase RPTPmu binds to and dephosphorylates the catenin p120(ctn) J Biol Chem. 2000;275:11264–11259. doi: 10.1074/jbc.275.15.11264. [DOI] [PubMed] [Google Scholar]