Summary
In many species the mutation rate is higher in males than in females, a phenomenon denoted as male mutation bias. This is often observed in animals where males produce many more sperm than females produce eggs, and is thought to result from differences in the number of replication-associated mutations accumulated in each sex. Thus, studies of male mutation bias have the capacity to reveal information about the replication-dependent or replication-independent nature of different mutations. The availability of whole genome sequences for many species, as well as for multiple individuals within a species, has opened the door to studying factors, both sequence-specific and those acting on the genome globally, that affect differences in mutation rates between males and females. Here, we assess the advantages that genomic sequences provide for studies of male mutation bias and general mutation mechanisms, discuss major challenges left unresolved, and speculate about the direction of future studies.
Keywords: life history traits, male mutation bias, mutation rates
Introduction
Mutations are the primary cause of human genetic diseases. Currently, there are over 100,000 mutations associated with disease phenotypes described in the Human Gene Mutation Database (HGMD) [1]. 68% of entries are nucleotide substitutions and 31% are small insertions/deletions, with an additional approximately 300 disease-causing mutations related to repetitive elements [1]. Moreover, many mutations have been implicated in a variety of human cancers [2, 3]. Thus, it is crucial to unravel the mechanisms of mutagenesis for these common, and frequently disease-causing, mutations. The phenomenon of male mutation bias, in particular, can be utilized to address the question of whether mutations are driven by replication-dependent or by replication-independent factors.
In many species the number of germline cell divisions (and thus of DNA replications) prior to reproduction is higher in males than in females (Table 1) [4, 5]. When more mutations originate in the male germline (sperm and their precursors) than the female germline (eggs and their precursors) [6], this is referred to as a male mutation bias. One way to determine whether male mutation bias occurs is to compute α, the ratio of male-to-female mutation rates. If α equals one, there is no bias; α less than one indicates more mutations coming from the female germline, whereas α greater than one suggests a male mutation bias, also called male-driven evolution (Fig. 1). The difference in the number of germline cell divisions between males and females provides an opportunity to test the hypothesis that mutations result from errors in DNA replication. If mutations are primarily replication-driven, then α should be similar to c, the male-to-female ratio in the number of germline cell divisions (Fig. 2). If, on the other hand α is smaller than c, then the role of replication-independent factors (e.g., free radicals present in cells) in generating mutations is significant [7, 8]. Estimates of c are only available for a limited number of species (humans [9, 10], mice and rats [11], flies [12] and an estimate across birds [13]) and are highly dependent on paternal age. For instance, in humans, c is ~6 if the father’s age is, on average, 20 years (Fig. 2) [9, 10], whereas this value will increase rapidly as paternal age increases. Alternatively in rodents, c is ~2, assuming that the average age of males at reproduction in the wild is 5 months [11].
Table 1.
Estimates of α, the male-to-female mutation rate ratio, from genome-wide sequence data.
| Mammalian species | α | Reference |
|---|---|---|
| Human | ||
| Human | 20.1 | Wilson Sayres et al., 2011 [16] |
| Human–Chimpanzee | 6 | Taylor et al., 2006 [22] |
| Human | 4.7 | Presgraves and Yi, 2009 [80] |
| Chimpanzee | ||
| Chimpanzee | 6.2 | Presgraves and Yi, 2009 [80] |
| Chimpanzee | 3.6 | Wilson Sayres et al., 2011 [16] |
| Gorilla | ||
| Gorilla | 2.5 | Wilson Sayres et al., 2011 [16] |
| Gorilla | 2.1 | Presgraves and Yi, 2009 [80] |
| Orangutan | 3.5 | Wilson Sayres et al., 2011 [16] |
| Rhesus | ||
| Rhesus | 2.9 | Wilson Sayres et al., 2011 [16] |
| Rhesus–Human | 2.9 | Rhesus Macaque Genome Sequencing and Analysis Consortium [90] |
| Marmoset | 2.6 | Wilson Sayres et al., 2011 [16] |
| Tarsier | 3.0 | Wilson Sayres et al., 2011 [16] |
| Mouse lemur | 3.0 | Wilson Sayres et al., 2011 [16] |
| Bushbaby | 2.4 | Wilson Sayres et al., 2011 [16] |
| Treeshrew | 3.3 | Wilson Sayres et al., 2011 [16] |
| Mouse | ||
| Mouse | 2.2 | Wilson Sayres et al., 2011 [16] |
| Mouse–Rat | 2 | Makova, Yang and Chiaromonte, 2004 [28] Gibbs et al., 2004 [91] |
| Rat | 1.9 | Wilson Sayres et al., 2011 [16] |
| Kangaroo rat | 1.9 | Wilson Sayres et al., 2011 [16] |
| Guinea pig | 1.5 | Wilson Sayres et al., 2011 [16] |
| Squirrel | 2.4 | Wilson Sayres et al., 2011 [16] |
| Rabbit | 3.3 | Wilson Sayres et al., 2011 [16] |
| Pika | 2.0 | Wilson Sayres et al., 2011 [16] |
| Alpaca | 3.3 | Wilson Sayres et al., 2011 [16] |
| Dolphin | 3.9 | Wilson Sayres et al., 2011 [16] |
| Cow | 2.8 | Wilson Sayres et al., 2011 [16] |
| Horse | 3.5 | Wilson Sayres et al., 2011 [16] |
| Cat | 1.4 | Wilson Sayres et al., 2011 [16] |
| Dog | ||
| Dog | 2.8 | Linblad-Toh et al., 2005 [92] |
| Dog | 2.0 | Wilson Sayres et al., 2011 [16] |
| Microbat | 2.2 | Wilson Sayres et al., 2011 [16] |
| Megabat | 2.0 | Wilson Sayres et al., 2011 [16] |
| Hedgehog | 1.0 | Wilson Sayres et al., 2011 [16] |
| Shrew | 1.1 | Wilson Sayres et al., 2011 [16] |
| Elephant | 2.8 | Wilson Sayres et al., 2011 [16] |
| Rock hyrax | 2.7 | Wilson Sayres et al., 2011 [16] |
| Tenrec | 2.0 | Wilson Sayres et al., 2011 [16] |
| Armadillo | 1.6 | Wilson Sayres et al., 2011 [16] |
| Sloth | 2.0 | Wilson Sayres et al., 2011 [16] |
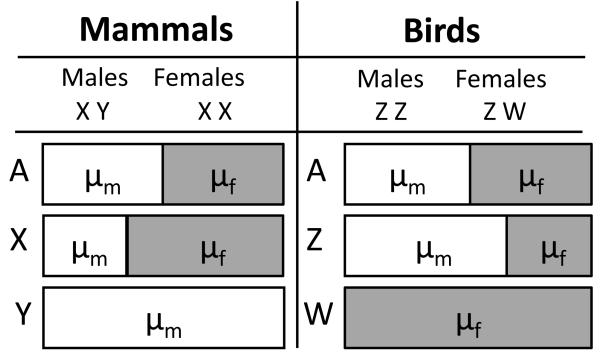
Figure 1.
The proportion of the overall mutation rate at different chromosome types attributed to the male mutation rate (μm) or the female mutation rate (μf). For both mammals (with male heterogamety) and birds (with female heterogamety), autosomes represent half μm and half μf. In mammals (XY males and XX females), the mutation rate of the heterogametic sex chromosome, the Y, is completely representative of the male mutation rate. Alternatively, in birds (ZZ males and ZW females), the heterogametic sex chromosome, the W, is entirely representative of the female mutation rate. For both mammals and birds, the homogametic sex chromosome (X and Z, respectively), represents two-thirds the mutation rate of the homogametic sex (females and males, respectively), and one-third the mutation rate of the heterogametic sex.
Figure 2.
The relative number of cumulative germline cell divisions in males and females. Until sexual maturity, the number of germline cell divisions in females (dotted red line) and males (solid blue line) is static. Then, female germ cells (eggs) experience only two divisions before reproduction while male germ cells (sperm) continue to divide throughout the remaining reproductive life of the male. Consequently the ratio of male-to-female germline cell divisions at reproduction, c, will increase as the age at reproduction of the male increases. Similarly, α, the male-to-female mutation rate ratio, is expected to increase as the average age of males at reproduction increases. The values of cumulative germline cell divisions here are modeled on human estimates, obtained from [10].
Comparisons of species with a variety of sex determination mechanisms, such as fishes [14], birds [13, 15], and mammals [16], find evidence of male mutation bias, indicating that differences in the number of germline cell divisions between males and females are important for shaping genome evolution. To date, the vast majority of mammals and birds studied have shown evidence of a male mutation bias. In mammals, with male heterogamety (XX females and XY males), estimates of α vary greatly, including a low of approximately 2 in rodents [11], an intermediate α of about 4 in Perissodactyls (horses and rhinos) [17], and even higher estimates in primates [17, 18]. Sequence comparisons between many species of birds (with ZW females and ZZ males) indicate that, as expected, male mutation bias occurs independently of male or female heterogamety, with estimates of α in birds ranging from 3.1 to 6.5 [13, 15, 19].
Not all species exhibit clear evidence for a male mutation bias. In flies, both males and females produce many gametes leading to the expectation of no male mutation bias [20]. Indeed, initial studies in Drosophila melanogaster and D. simulans estimated that there should be a slight female mutation bias when flies mated early (three days after hatching c ≈ 0.77), and a low male mutation bias when they mated later (c ≈ 1.33, 21 days after hatching), but did not observe significant differences between substitution rates on the X and autosomes, implying that, on average, flies do not exhibit sex-biased mutation rates [12]. However, a more recent genomic comparison of 205 homologous neo-X-and neo-Y-linked sequences (versus the smaller set of non-homologous loci compared previously [12]) indicated the presence of male mutation bias in D. miranda, with α ≈ 2 [21]. Given that α estimates vary greatly across mammals (see below), it is reasonable that male mutation bias may also differ between fly species, and so the aforementioned studies may not be incompatible.
Genome-wide data are particularly useful for advancing studies of male mutation bias, and, subsequently, elucidating the roles that replication-dependent and -independent mechanisms play in mutation accumulation. Indeed, the abundance of information from genome sequences allows for the study of mutation mechanism analysis of context-dependent substitutions, as well as of transposable elements, simple repeats, and insertions and deletions. Finally, genome sequences from a variety of species now make it possible to investigate branch-specific mutation rates, whereas most previous studies were only able to analyze pairwise mutation rates. Estimates of male mutation bias can vary depending on which mutation types are included in the analysis [22], which chromosomes are compared [23], and whether ancestral polymorphism for closely related species is corrected for [24]. Further, differences in life history traits may cause variations in α between species [16]. Here we will address recent advances in the study of male mutation bias brought about by the genomic era, suggest potential avenues of research, and speculate about future prospects in the field.
Utilizing comparative genomics to measure male mutation bias: An updated evolutionary approach
One way to study the differences in mutation rates between males and females in species with chromosomal sex determination is to compare the mutation rates between the two sex chromosomes, or between a sex chromosome and autosomes [25]. Usually, neutrally evolving regions of the genome (either non-coding and non-repetitive, or those that fall within ancestral interspersed repeats [22, 26]) are utilized, to avoid the confounding effects of selection. Substitution rates are used as a proxy for mutation rates. Because we know how much time each chromosome type spends in the male vs. female germline – in mammals, autosomes spend equal amounts of time in both germlines, X spends 2/3 of its time in the female germline and 1/3 in the male germline, and Y is found exclusively in the male germline (Fig. 1) – substitution rates between any two chromosome types can be compared to estimate the male-to-female mutation rate ratio, α. In species with female heterogamety, like birds, the W-specific substitution rate represents the female-specific mutation rate, while the Z spends 2/3 of its time in males and only 1/3 of its time in females, and autosomes are again split 50:50 with respect to time spent in the male and female germlines (Fig. 1). The evolutionary approach has the advantage of comparing mutation rates over a large number of sites, and it does not suffer from disease-associated ascertainment bias [27]. Moreover, although earlier studies employing this approach were affected by regional variation in substitution rates (e.g. [17]), the availability of genome-wide sequences circumvents this limitation [16, 22, 28, 29] because regional variation will be averaged out.
However, there are some drawbacks to the evolutionary approach. First, it requires at least two alignable genomes to compute evolutionary rates and at least three to polarize mutations to specific branches. In a modification of the evolutionary method, divergence in the nucleotide sequences of transposable elements on the X, Y and autosomes with a single species can be compared to evaluate the magnitude of male mutation bias [30, 31]. Another pitfall of the evolutionary method is that it may be affected by ancestral polymorphism when very closely related species are compared, which can inflate or deflate estimates of α, depending on which chromosomes are compared [24]. Furthermore, although genome-wide sequences have recently become available, allowing for global estimates of α, the complete Y chromosome has not been sequenced for most mammals (with the exception of human [32] and chimpanzee [33]), limiting, for now, the ability of researchers to explore how comparisons among different chromosome types affect estimates of male mutation bias.
Novel mutations can be identified directly
Historically, instead of being estimated from sequence changes between or within species, α was computed by inferring the parental origin (paternal versus maternal) of mutations leading to human genetic diseases (X-linked or autosomal dominant disorders). Specifically, after the origins of many mutations for a particular disorder have been inferred for a variety of individuals, α can be calculated as the ratio of the number of paternal mutations to the number of maternal mutations (reviewed in [27]). However, mutations leading to human genetic diseases usually affect gene function and, as a result, are likely to be affected by selection, confounding estimates of male mutation bias.
In contrast to observing mutations through disease associations, another way to directly study mutation rates – available now with genomic sequences – is to analyze novel mutations in families including a mother, father and one [34, 35] or several children [36]. As a pilot study of the 1,000 genomes project, whole-genome sequence data for two sets of trios were used to detect 49 and 35 de novo mutations occurring in the germlines of the two families, respectively. Initially no differentiation was made regarding parental origin of mutations [37], but a subsequent study partitioned the new mutations into those coming from the paternal or maternal germlines [35]. Curiously, results from one of the two families suggested a strong male mutation bias while the other indicated a female mutation bias [35]. Given that new mutations in the germ cells (that may be passed on to the next generation) are likely to be rare, including more than one child may greatly improve the power of a study to determine the magnitude of male mutation bias [36].
Even with the sequencing of complete genomes, the sequence of the Y chromosome is often difficult to capture because it is small, highly heterochromatic and shares many highly similar sequences with the X that can complicate alignments. However, flow sorting Y chromosomes (choosing just the Y chromosome out of the set of all autosomes and sex chromosomes) gives researchers an edge by reducing the chance of misalignment. In fact, the direct sequencing of flow-sorted Y chromosomes from two human males, with knowledge of the pedigree (relationship between individuals), resulted in a direct estimate of the mutation rate on the human Y chromosome (3.0×10−8 mutations per nucleotide per generation) [38]. A study of the autosomal mutation rate from a family including both parents and two children found that the autosomal substitution rate is approximately 1.1×10−8 mutations per nucleotide per generation [36], which is lower than the substitution rate reported for the Y chromosome [38] (as would be expected in the presence of male mutation bias), but not significantly so [36]. Because only a few mutations get passed on each generation, variation due to the small samples sizes in each study may limit the power to determine significant differences.
Using families with parents and offspring to study de novo germline mutations has the advantage of avoiding germline cell collection (which is especially invasive for females), but the challenge that each novel mutation must be validated. Single-molecule sequencing holds the promise of fast and reliable sequencing [39] of individual genomes that will make it feasible and affordable to study the male and female mutation rates directly within many families. Studies of mutation rates from families should include a large number of individuals, to account for the small number of novel mutations, and stochastic fluctuations [35]. Further, families from diverse ethnic backgrounds should be included, to take into account the effects of population structure and mating patterns. In addition to the non-uniform mating of males and females in different cultures, which might lead to variance in α estimates due to increased or decreased diversity on the X or Y chromosomes (see [40] for a more detailed discussion), individuals in some populations may have a history of reproducing earlier, on average, leading to a lower estimate of α, while individuals in others may delay reproduction, increasing the difference in germline cell divisions between males and females. In addition, one must be careful, to distinguish germline mutations from somatic mutations (which will not be passed on to the next generation) and from mutations arising during the propagation of cultured cell lines.
Estimates of male mutation bias vary across nucleotide sites
Most autosomal dominant disorders caused by nucleotide substitutions show an excess of new mutations originating in males (reviewed in [27, 41]). Male mutation bias theory assumes that the majority of new mutations are due to errors during replication, but some nucleotide substitutions are more likely to occur through replication-independent mechanisms, which may skew estimates of α. For example, a methylated cytosine followed immediately by a guanine (CpG), has an increased likelihood of being spontaneously deaminated to a uracil, which is then “repaired” as a thymine, resulting in a C-to-T transition that is independent of replication [42]. This largely replication-independent nucleotide substitution has a mutation rate that is 10-50 times higher than that at non-CpG sites [43], and is very high in the male and female germlines, effectively lowering estimates of α at CpG sites and masking the effects of male mutation bias [22].
Other, non-CpG, neighbor-dependent substitution processes are still poorly understood, but analyzing such mutations in the context of male mutation bias has a potential to determine whether they are associated with errors made during replication. For example, substitution rates in pseudogenes were found to be highest in purines surrounded by pyrimidines and in pyrimidines surrounded by purines, and slowest within purine and pyrimidine tracts [44-46]. It was hypothesized that an increase in dNTP misinsertion or transient misalignment during replication is associated with structural alterations of the DNA backbone in alternating purine-pyrimidine sequences [47], thus dNTP misinsertions in alternating purine-pyrimidine tracks should exhibit a strong male mutation bias. Another substitution type, the instability of thymines occurring in tandem (TpT) was proposed to be due to thymine photodimer formation in DNA during exposure to UV light [48]. Since exposure to UV should occur independent of replication (and likely affects germline cells only minimally), observations of thymine dimers are expected to show weak or no male mutation bias. Several models of sequence evolution have been developed to investigate neighbor-dependent substitution biases genome-wide from intraspecific [46] and interspecific comparisons [49-51]. These models hold great promise for investigating context-dependent biases within the framework and hypotheses formed by male mutation bias.
Insertions and deletions are subject to male mutation bias
Studies of male bias for indels (insertions and deletions) may provide a unique opportunity to disentangle the effects of recombination and replication on the formation of indels. Although large (>1-kb) indels, which include copy number variants, have not yet been explored genome-wide in the context of male mutation bias, small (<50-bp) indels have been studied across chromosome types, leading to some interesting conclusions [52, 53]. When replication-dependent and recombination-dependent genomic landscape predictors were analyzed, the results suggested that insertions are more often associated with recombination while deletions result mostly from errors during replication [52, 53]. Even with these different forces primarily associated with formation of insertions versus deletions, the incidence of both insertions and deletions is reduced on the X chromosome, relative to the autosomes (i.e. there is male bias for small indels), tentatively suggesting that replication may play a strong role in determining indel rates [28, 52]. Comparisons with the Y chromosome will need to be made to rule out significant effects of recombination. Future studies will also need to assess if the same patterns hold true for larger indels. Given the implied female bias of large indels in human genetic diseases (see above), this will be of great interest to evolutionary biologists and clinicians and alike. Indeed, while some small indels (< 50-bp) that cause human genetic diseases originate more frequently in males (e.g. in neurofibromatosis type 2 and Rett syndrome [54, 55]), there is evidence that some large indels (>1-kb or larger) exhibit a maternal mutation bias (e.g. the ones causing neurofibromatosis type 1 and hemophilia B [56, 57]).
Variations in microsatellite repeats are prone to replication-dependent changes
Microsatellites – repeats of short (1-6-bp) DNA motifs where the length of the repeated sequence can vary dramatically from person to person – have been implicated in a variety of human cancers [3] and are widely used in forensics, so understanding how these quickly changing sequences mutate, especially in males versus females can have far-reaching implications. Microsatellite mutation rates are predicted to occur primarily through misalignment and strand slippage of the repetitive microsatellite DNA during replication [58]. Thus, microsatellite mutations are expected to originate predominately in males, who experience a greater number of germline cell divisions, as compared with females. However, the data on the parental origin of trinucleotide microsatellite expansions leading to human neurological diseases have been contradictory. Some exhibit a clear paternal bias (e.g. dentatorubral-pallidoluysian atrophy, and spinocerebellar ataxia types 1 through 3 [59]), while others exhibit a maternal bias (e.g. myotonic dystrophy [60], FRAXE mental retardation, Freidrich’s ataxia and spinocerebellar ataxia type 8 [59]).
An evolutionary study of microsatellite mutability shows a male mutation bias where mutability is highest on the Y chromosome, intermediate on the autosomes and lowest on the X, likely due to the amount of time each chromosome type spends in the male and female germline [61]. Note that in this study [61] mutability in microsatellite repeats was observed to be significantly different between the two sex chromosomes only for mononucleotide repeats, either due to the lack of data for larger motifs or, potentially due to real differences in the importance of replication for microsatellites with different motif sizes [62]. Future studies of variation in microsatellite mutability between males and females at all motif lengths (e.g., di-, tri-, and tetranucleotide microsatellites) and compositions (e.g. A/T vs. C/G repeats for mononucleotide microsatellites) are likely to find many similarities supporting the relative importance of replication slippage in the formation of all microsatellites, but may also unveil differences that will suggest alternative processes affecting mutability. In addition, comparisons of mutability at microsatellites with mutability at other types of repetitive elements, such as minisatellites and larger-scale copy number variants, may discover some global mutation mechanisms at repetitive regions.
Strong male mutation bias is observed for some but not all transposable elements
Transposable elements (TEs) – elements that replicate and integrate themselves selfishly in our genomes – contribute significantly to both genetic diseases and cancer [63, 64], with over 65 separate human diseases caused by the de novo insertion of mobile elements [65, 66]. Male mutation bias provides a unique perspective for studying TEs, such as short and long interspersed repetitive elements (SINEs and LINEs), because it can be utilized to investigate both how and when during development TEs integrate, as well as in which germlines they integrate. Different families of transposable elements may have different preferences for integration – either into the male or female germline – or they may not show a preference. For instance, if integration occurs preferentially in the male germline, as suggested for Alus [67] and for human endogenous retroviruses [68], we expect highest, intermediate, and lowest TE densities at Y, autosomes, and X, respectively. No differences in the TE densities among chromosomal types are expected if integration occurs during early embryogenesis, as experimental evidence suggests for L1s [69] (but, others suggested integration of L1s in oocytes [70]). Largely consistent with these wet-lab observations, a recent computational study observed a strong male mutation bias for Alu elements, while no such bias was observed for L1 elements [71]. Potential male mutation bias at other interspersed repeat types (e.g. DNA transposons) has yet to be investigated. Knowing roughly when and how TEs integrate in the genome may, in the future, assist clinical tests (particularly in the egg and sperm before conception) in detecting TE-related genetic diseases.
Can life history traits account for variations in male mutation bias between species?
Estimates of male mutation bias vary greatly between mammals, even when entire genome sequences are used to compute α [16]. Life history traits, which also differ from species to species, might influence male mutation bias by affecting either the relative number of germline cell divisions or differences in DNA damage/repair between males and females. First, if the number of cell divisions in oogenesis is roughly constant across species, then species with longer generation times should also exhibit stronger male mutation bias. This is because, as in humans, and presumably all other mammals, sperm are produced continuously during a male’s reproductive life, so the total number of DNA replications accumulates with age. For instance, a sperm of a 20-year old man has completed ~160 chromosome replications, while a sperm of a 40-year old man has completed ~610 chromosome replications [9, 41]. A recent analysis of 32 mammalian genomes [16] corroborated the dominant role of generation time in determining the variation in the magnitude of male mutation bias for nucleotide substitutions, finding it explained over 30% of the variation in α estimates across mammals. Second, metabolic rate, specifically its association with oxidative damage of the DNA through reactive oxygen species, may act in a replication-independent way to affect germline mutations. Oxidative damage may increase the mutation rate in males over females if, as postulated, eggs are more efficient at avoiding such damage than sperm because the former have a denser cell membrane and the latter exist in a more reactive oxygen rich environment [72]. Consistent with these expectations, genome-wide comparisons indicate that metabolic rate is a significant positive predictor of male mutation bias; however, it loses its significance when included in a multiple regression model alongside predictors of generation time [16]. These results suggest that although metabolic rate appears to affect male mutation bias, generation time, a replication-dependent factor, is still the driving cause of differences between the male and female mutation rates. Finally, increased sperm competition, which occurs when sperm from multiple males compete to fertilize the egg(s) from a single female, is associated with increased testes size [73, 74] and sperm quantity [75], and more rounds of reproduction per unit of time [76], and thus is expected to lead to more mutations in the male germline, increasing the magnitude of male mutation bias. Although this expected positive trend was observed qualitatively for a few bird species [77], results from recent regression analyses suggest that available predictors of sperm competition are not significant in explaining variation in male mutation bias across 32 mammalian species [16]. If sperm competition can affect substitution rates on a short time scale, as suggested for flies [74], rapid changes between mating patterns may mask the effects of effects of sperm competition on male mutation bias, especially when comparing divergent species.
Male mutation bias in humans
Given the variation in estimates of male mutation bias at within and across mutation types, along with the general interest in human-specific evolution, it is unsurprising that there has been some controversy surrounding α estimates in humans, and its implications are directly relevant to understanding mutation mechanisms. Many earlier estimates were based on one or a few loci, leading to drastically different estimates (due to regional variation in mutation rates) with wide confidence intervals [7, 24, 78]. Some estimates of α in humans were much lower than 6 [78, 79], which suggested that α may be roughly constant across species, and casted doubt upon the theory that replication errors were an important mutational mechanism. However, more recent studies, accounting for ancestral polymorphism and using genome-wide data [16, 22, 80], showed that male mutation bias in humans and other primates likely reflects a replication-dependent nature of the majority of mutations, with α estimates consistent with, or sometimes even larger than, estimates of c. Future avenues of research into human-specific male mutation bias using population genetics can address whether large α values are due to reduced purifying selection, increases in the average age at reproduction, or changes in some other demographic parameters.
Factors that limit studies of male mutation bias
Several external features may affect actual and estimated values of male mutation bias. First, although the current literature supports the explanation that the male mutation bias is due to differences between the number of germline cell divisions between males and females within species, c has been measured in only a few model organisms [10, 20]. This paucity of c values hinders researchers’ ability to investigate whether α deviates from c across species, and why. Such data will need to be generated if genomic data are to be useful, especially given recent studies that suggest oocyte production may not be finished at birth (mouse ovaries likely contain some mitotically active germ cells [81]), and that the length of time needed to complete a spermatogenesis cycle may vary even between closely related species (as observed in shrews [76]). Second, if replication were the only cause of mutation rate differences between males and females, then the magnitude of male mutation bias, measured by α, should be the same whether comparing X/Y, X/A or Y/A (or alternatively Z/W, Z/A or W/A in female-heterogametic systems). However, there are differences in the estimates of α, depending on which ratio it is computed from [16, 22, 23], indicating that, unsurprisingly, replication-independent factors (e.g. recombination) might also differentially influence substitution rates on different chromosomal types. Initial studies have suggested that recombination may be mutagenic at chromosomal rearrangements and copy number variants [82], and also for nucleotide substitutions [83]. However, more research is needed because the latest genomic data from humans question the mutagenicity of recombination [37], implying that replication-independent processes other than recombination may be at play. For example, although sperm do not show increased susceptibility to induced DNA damage compared to somatic tissues [84], it not yet known whether eggs are more or less robust to such damage, which would affect estimates of α. Another possibility is that sperm accumulate replication-independent mutations as a result of the reduced DNA-repair fidelity in late spermatogenesis [85]. Third, most genomic analyses of male mutation bias study putatively neutrally evolving sequence to avoid the confounding effects of selection. But, one of the most striking conclusions of studies of mutations in sperm is the suggestion that selection may play an important role in affecting which disease-related mutations are maintained in the genome [86, 87]. However, because selection acts differently on the autosomes, and the sex chromosomes [88], it likely affects all studies of male mutation bias, even those of presumed neutral regions [16, 22, 23]. Currently there is not a systematic way to globally correct for the effects of selection, but it is an open challenge to the field.
Conclusion and future prospects
Genome-wide analyses have shown that there is, indeed a male mutation bias in many species, with estimates of α consistent with the ratio of expected germline cell divisions in males and females, suggesting this bias is largely driven by replication-dependent mutational processes. Such studies also observed variations in α across species and between mutation types. The growing abundance of completely sequenced genomes from a variety of species [89], as well as from many individuals within a species (particularly across families and populations of humans), will open up possibilities for future studies of male mutation bias. Perhaps the most exciting prospect that we only touched upon here is the ability to study mutation rates in males and females directly, from parents to offspring using sequenced families consisting of a father, mother and child (or even better, many children) [35, 36]. Further, computational studies of male mutation bias have, and will continue to inform upon the general mechanisms for a variety of mutation types including substitutions, insertions, deletions, microsatellites and transposable elements, and guide future wet-lab biochemical experiments. All of these results will illuminate how male mutation bias varies across individual human families and across species, and, when taken together with life history trait data, will allow researchers to elucidate the mechanisms that cause differences between male and female germline mutation rates. The wealth of genomic data is, unfortunately, paralleled by a dearth of similar, abundant, and high quality, references about species life history traits. This imbalance emphasizes the need for experimental advances in collecting such information, especially on the number of germline cell divisions per unit of time in both males and females. Importantly, understanding the factors affecting male mutation bias will contribute to studies of age-related diseases, and can assist in the development and timing of pre- and post-conception genetic counseling.
Acknowledgements
This study was supported by the NIH grant R01-GM072264.
REFERENCES
- 1.Stenson PD, Ball EV, Howells K, Phillips AD, et al. The Human Gene Mutation Database: providing a comprehensive central mutation database for molecular diagnostics and personalized genomics. Hum Genomics. 2009;4:69–72. doi: 10.1186/1479-7364-4-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stratton MR. Exploring the genomes of cancer cells: progress and promise. 2011;331:1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 3.Oda S, Maehara Y, Sumiyoshi Y, Sugimachi K. Microsatellite instability in cancer: what problems remain unanswered? Surgery. 2002;131:S55–62. doi: 10.1067/msy.2002.119305. [DOI] [PubMed] [Google Scholar]
- 4.Haldane J. The rate of spontaneous mutation of a human gene. J Genet. 1935;31:317–26. doi: 10.1007/BF02717892. [DOI] [PubMed] [Google Scholar]
- 5.Haldane J. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann Eugen. 1947;13:262–71. doi: 10.1111/j.1469-1809.1946.tb02367.x. [DOI] [PubMed] [Google Scholar]
- 6.Miyata T, Hayashida H, Kuma K, Mitsuyasu K, et al. Male-driven molecular evolution: a model of nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–7. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 7.Li WH, Ellsworth DL, Krushkal J, Chang BH-J, et al. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol. 1996;5:182–7. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 8.Shimmin LC, Chang BH-J, Hewett-Emmett D, Li W-H. Potential problems in estimating the male-to-female mutation rate ratio from DNA sequence data. J Mol Evol. 1993;37:160–6. doi: 10.1007/BF02407351. [DOI] [PubMed] [Google Scholar]
- 9.Hurst LD, Ellegren H. Sex biases in the mutation rate. Trends Genet. 1998;14:446–52. doi: 10.1016/s0168-9525(98)01577-7. [DOI] [PubMed] [Google Scholar]
- 10.Vogel F, Motulsky A. Human genetics: problems and applications. 3rd ed. Springer Verlag; Berlin, Heidelberg, New York: 1997. Springer Verlag, Berlin, Heidelberg, New York. [Google Scholar]
- 11.Chang BH-J, Shimmin LC, Shyue S, Hewett-Emmett D, et al. Weak male-driven molecular evolution in rodents. Proc Natl Acad Sci USA. 1994;91:827–31. doi: 10.1073/pnas.91.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer VL, Aquadro CF. Rates of DNA sequence evolution are not sex-biased in Drosophila melanogaster and D. simulans. Mol Biol Evol. 1997;14:1252–7. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- 13.Kahn NW, Quinn TW. Male-driven evolution among Eoaves? A test of the replicative division hypothesis in a heterogametic female (ZW) system. J Mol Evol. 1999;49:750–9. doi: 10.1007/pl00006597. [DOI] [PubMed] [Google Scholar]
- 14.Ellegren H, Fridolfsson A-K. Sex-specific mutation rates in salmonid fish. J Mol Evol. 2003;56:458–63. doi: 10.1007/s00239-002-2416-z. [DOI] [PubMed] [Google Scholar]
- 15.Ellegren H, Fridolfsson A-K. Male-driven evolution of DNA sequences in birds. Nat Genet. 1997;17:182–4. doi: 10.1038/ng1097-182. [DOI] [PubMed] [Google Scholar]
- 16.Wilson Sayres MA, Venditti C, Pagel M, Makova KD. Do variations in substitution rates and male mutation bias correlate with life history traits? A study of 32 mammalian genomes. Evolution. 2011 doi: 10.1111/j.1558-5646.2011.01337.x. in press, DOI: 10.1111/j.1558-5646.2011.01337.x. [DOI] [PubMed] [Google Scholar]
- 17.Goetting-Minesky MP, Makova KD. Mammalian male mutation bias; impacts of generation time and regional variation in substitution rates. J Mol Evol. 2006;63:537–44. doi: 10.1007/s00239-005-0308-8. [DOI] [PubMed] [Google Scholar]
- 18.Elango N, Lee J, Peng Z, Loh YH, et al. Evolutionary rate variation in Old World monkeys. Biol Lett. 2009;5:405–8. doi: 10.1098/rsbl.2008.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmichael AN, Fridolfsson AK, Halverson J, Ellegren H. Male-biased mutation rates revealed from Z and W chromosome-linked ATP synthase alpha-subunit (ATP5A1) sequences in birds. J Mol Evol. 2000;50:443–7. doi: 10.1007/s002390010046. [DOI] [PubMed] [Google Scholar]
- 20.Drost JB, Lee WR. Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 1995;25:48–64. doi: 10.1002/em.2850250609. [DOI] [PubMed] [Google Scholar]
- 21.Bachtrog D. Evidence for male-driven evolution in Drosophila. Mol Biol Evol. 2008;25:617–9. doi: 10.1093/molbev/msn020. [DOI] [PubMed] [Google Scholar]
- 22.Taylor J, Tyekucheva S, Zody M, Chiaromonte F, et al. Strong and weak male mutation bias at different sites in the primate genomes: Insights from the human-chimpanzee comparison. Mol Biol Evol. 2006;23:565–73. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- 23.Pink CJ, Swaminathan SK, Dunham I, Rogers J, et al. Evidence that replication-associated mutation alone does not explain between-chromosome differences in substitution rates. Genome Biol Evol. 2009;30:13–22. doi: 10.1093/gbe/evp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makova KD, Li W-H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–6. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- 25.Miyata T, Hayashida H, Kuma K, Mitsuyasu K, et al. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–7. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 26.Hardison RC, Roskin KM, Yang S, Diekhans M, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W-H, Yi S, Makova K. Male-driven evolution. Curr Opin Genet Dev. 2002;12:650–6. doi: 10.1016/s0959-437x(02)00354-4. [DOI] [PubMed] [Google Scholar]
- 28.Makova KD, Yang S, Chiaromonte F. Insertions and deletions are male biased too: a whole-genome analysis in rodents. Genome Res. 2004;14:567–73. doi: 10.1101/gr.1971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, Nusbaum C, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Erlandsson R, Wilson JF, Paabo S. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol Biol Evol. 2000;17:804–12. doi: 10.1093/oxfordjournals.molbev.a026359. [DOI] [PubMed] [Google Scholar]
- 32.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 33.Hughes JF, Skaletsky H, Pyntikova T, Graves TA, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–9. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicard P, Dawid AP, Mortera J, Lauritzen SL. Estimating mutation rates from paternity casework. Forensic Sci Int Genet. 2008;2:9–18. doi: 10.1016/j.fsigen.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Conrad DF, Keebler JE, Depristo MA, Lindsay SJ, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–4. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roach JC, Glusman G, Smit AF, Huff CD, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durbin RM, Abecasis GR, Altshuler DL, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue Y, Wang Q, Long Q, Ng BL, et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr Biol. 2009;19:1453–7. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–52. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustamante CD, Ramachandran S. Evaluating signatures of sex-specific processes in the human genome. Nat Genet. 2009;41:8–10. doi: 10.1038/ng0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crow JF. The origins, patterns and implications of human spontaneous mutation. 2000;1:40–7. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 42.Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–7. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 43.Walser JC, Furano AV. The mutational spectrum of non-CpG DNA varies with CpG content. Genome Res. 2010;20:875–82. doi: 10.1101/gr.103283.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morton BR, Oberholzer VM, Clegg MT. The influence of specific neighboring bases on substitution bias in noncoding regions of the plant chloroplast genome. J Mol Evol. 1997;45:227–31. doi: 10.1007/pl00006224. [DOI] [PubMed] [Google Scholar]
- 45.Yang YW, Chen Y, Li WH. The influence of adjacent nucleotides on the pattern of nucleotide substitution in mitochondrial introns of angiosperms. J Mol Evol. 2002;55:111–5. doi: 10.1007/s00239-001-2310-0. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Z, Boerwinkle E. Neighboring-nucleotide effects on single nucleotide polymorphisms: a study of 2.6 million polymorphisms across the human genome. Genome Res. 2002;12:1679–86. doi: 10.1101/gr.287302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hess ST, Blake JD, Blake RD. Wide variations in neighbor-dependent substitution rates. J Mol Biol. 1994;236:1022–33. doi: 10.1016/0022-2836(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 48.Arndt PF, Hwa T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics. 2005;21:2322–8. doi: 10.1093/bioinformatics/bti376. [DOI] [PubMed] [Google Scholar]
- 49.Siepel A, Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Bio Evol. 2004;21:468–88. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 50.Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. 2004;101:13994–4001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kvikstad EM, Tyekucheva S, Chiaromonte F, Makova KD. A macaque’s-eye view of human insertions and deletions: differences in mechanisms. PLoS Comput Biol. 2007;3:1772–82. doi: 10.1371/journal.pcbi.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kvikstad EM, Chiaromonte F, Makova KD. Ride the wavelet: A multiscale analysis of genomic contexts flanking small insertions and deletions. Genome Res. 2009;19:1153–64. doi: 10.1101/gr.088922.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kluwe L, Mautner V, Parry DM, Jacoby LB, et al. The parental origin of new mutations in neurofibromatosis 2. Neurogenetics. 2000;3:17–24. doi: 10.1007/s100480000088. [DOI] [PubMed] [Google Scholar]
- 55.Trappe R, Laccone F, Cobilanschi J, Meins M, et al. MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. Am J Hum Genet. 2001;68:1093–101. doi: 10.1086/320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lazaro C, Gaona A, Ainsworth P, Tenconi R, et al. Sex differences in mutational rate and mutational mechanism in the NF1 gene in neurofibromatosis type 1 patients. Hum Genet. 1996;98:696–9. doi: 10.1007/s004390050287. [DOI] [PubMed] [Google Scholar]
- 57.Sommer SS, Scaringe WA, Hill KA. Human germline mutation in the factor IX gene. Mutat Res. 2001;487:1–17. doi: 10.1016/s0921-8777(01)00108-2. [DOI] [PubMed] [Google Scholar]
- 58.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–21. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 59.Usdin K, Grabczyk E. DNA repeat expansions and human disease. Cell Mol Life Sci. 2000;57:914–31. doi: 10.1007/PL00000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tome S, Panigrahi GB, Lopez Castel A, Foiry L, et al. Maternal germline-specific effect of DNA ligase I on CTG/CAG instability. Hum Mol Genet. 2011;20:2131–43. doi: 10.1093/hmg/ddr099. [DOI] [PubMed] [Google Scholar]
- 61.Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–8. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlotterer C, Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992;20:211–5. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 64.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genetics. 2005;117:411–27. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 65.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–58. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 67.Jurka J, Kohany O, Pavlicek A, Kapitonov VV, et al. Duplication, coclustering, and selection of human Alu retrotransposons. Proc Natl Acad Sci USA. 2004;101:1268–72. doi: 10.1073/pnas.0308084100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katzourakis A, Pereira V, Tristem M. Effects of recombination rate on human endogenous retrovirus fixation and persistence. J Virol. 2007;81:10712–7. doi: 10.1128/JVI.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kano H, Godoy I, Courtney C, Vetter MR, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Georgiou I, Noutsopoulos D, Dimitriadou E, Markopoulos G, et al. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum Mol Genet. 2009;18:1221–8. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- 71.Kvikstad EM, Makova KD. The (r)evolution of SINE versus LINE distributions in primate genomes: sex chromosomes are important. Genome Res. 2010;20:600–13. doi: 10.1101/gr.099044.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Velando A, Torres R, Alonso-Alvarez C. Avoiding bad genes: oxidatively damaged DNA in germ line and mate choice. BioEssays. 2008;30:1212–9. doi: 10.1002/bies.20838. [DOI] [PubMed] [Google Scholar]
- 73.Ramm SA, Stockley P. Sperm competition and sperm length influence the rate of mammalian spermatogenesis. Biol Lett. 2010;6:219–21. doi: 10.1098/rsbl.2009.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosken DJ, Ward PI. Experimental evidence for testis size evolution via sperm competition. Ecol Lett. 2001;4:10–3. [Google Scholar]
- 75.Luepold S, Linz GM, Rivers JW, Westneat DF, et al. Sperm competition selects beyond relative testes size in birds. Evolution. 2009;63:391–402. doi: 10.1111/j.1558-5646.2008.00571.x. [DOI] [PubMed] [Google Scholar]
- 76.Parapanov R, Nussle S, Vogel P. Cycle length of spermatogenesis in shrews (Mammalia : Soricidae) with high and low metabolic rates and different mating systems. Biol Reprod. 2007;76:833–40. doi: 10.1095/biolreprod.106.058073. [DOI] [PubMed] [Google Scholar]
- 77.Bartosch-Härlid A, Berlin S, Smith NG, Møller A, et al. Life history and the male mutation bias. Evolution. 2003;57:2398–406. doi: 10.1554/03-036. [DOI] [PubMed] [Google Scholar]
- 78.Bohossian HB, Skaletsky H, Page DC. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature. 2000;406:622–5. doi: 10.1038/35020557. [DOI] [PubMed] [Google Scholar]
- 79.de Jong P, Catanese JJ, Osoegawa K, Shizuya H, et al. Initial sequencing and analysis of the human genome. Nature. 2001;412:565–6. [Google Scholar]
- 80.Presgraves DC, Yi SV. Doubts about complex speciation between humans and chimpanzees. Trends Ecol Evol. 2009;24:533–40. doi: 10.1016/j.tree.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson J, Canning J, Kaneko T, Pru JK, et al. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428:145–50. doi: 10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 82.Volker M, Backstrom N, Skinner BM, Langley EJ, et al. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 2010;20:503–11. doi: 10.1101/gr.103663.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–72. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawyer DE, Mercer BG, Wiklendt AM, Aitken RJ. Quantitative analysis of gene-specific DNA damage in human spermatozoa. Mutat Res. 2003;529:21–34. doi: 10.1016/s0027-5107(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 85.Olsen AK, Lindeman B, Wiger R, Duale N, et al. How do male germ cells handle DNA damage? Toxicol Appl Pharmacol. 2005;207:521–31. doi: 10.1016/j.taap.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 86.Yoon SR, Qin J, Glaser RL, Jabs EW, et al. The ups and downs of mutation frequencies during aging can account for the Apert syndrome paternal age effect. PLoS Genet. 2009;5:e1000558. doi: 10.1371/journal.pgen.1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi SK, Yoon SR, Calabrese P, Arnheim N. A germ-line-selective advantage rather than an increased mutation rate can explain some unexpectedly common human disease mutations. Proc Natl Acad Sci USA. 2008;105:10143–8. doi: 10.1073/pnas.0801267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans Royal Soc Biol Sci. 2000;355:1563–72. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Check Hayden E. 10,000 genomes to come. Nature. 2009;462:21. doi: 10.1038/462021a. [DOI] [PubMed] [Google Scholar]
- 90.Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 91.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 92.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]