Abstract
Genomic instability can initiate cancer, augment progression, and influence the overall prognosis of the affected patient. Genomic instability arises from many different pathways, such as telomere damage, centrosome amplification, epigenetic modifications, and DNA damage from endogenous and exogenous sources, and can be perpetuating, or limiting, through the induction of mutations or aneuploidy, both enabling and catastrophic. Many cancer treatments induce DNA damage to impair cell division on a global scale but it is accepted that personalized treatments, those that are tailored to the particular patient and type of cancer, must also be developed. In this review, we detail the mechanisms from which genomic instability arises and can lead to cancer, as well as treatments and measures that prevent genomic instability or take advantage of the cellular defects caused by genomic instability. In particular, we identify and discuss five priority targets against genomic instability: (1) prevention of DNA damage; (2) enhancement of DNA repair; (3) targeting deficient DNA repair; (4) impairing centrosome clustering; and, (5) inhibition of telomerase activity. Moreover, we highlight vitamin D and B, selenium, carotenoids, PARP inhibitors, resveratrol, and isothiocyanates as priority approaches against genomic instability. The prioritized target sites and approaches were cross validated to identify potential synergistic effects on a number of important areas of cancer biology.
Keywords: Genomic instability, Cancer therapy, Cancer prevention, DNA damage, Nutraceutical
1. Cellular mechanisms that prevent or promote genomic instability
Genomic instability plays critical roles in both cancer initiation and progression. This instability can manifest itself genetically on several different levels, ranging from simple deoxyribonucleic acid (DNA) sequence changes to structural and numerical abnormalities at the chromosomal level. This section will briefly outline the mechanisms that maintain the stability of nuclear and mitochondrial DNA and how these mechanisms may become corrupted in cancer cells.
1.1. Telomeres foster chromosomal stability and can inhibit or promote malignant transformation
The chromosome stabilizing role of intact telomeres was recognized as early as the 1930s from independent research by McClintock [1] and Muller [2] and more recent work has further strengthened the connection between telomere dysfunction and chromosomal instability (CIN) [3], [4]. Telomeres, which are located at the ends of each chromosome, consist of approximately 5–10 kbp of specialized, tandem repeat, noncoding DNA complexed with a variety of telomere associated proteins [5], [6]. These elements create a protective cap that prevents the recognition of the chromosomal termini as DNA double strand breaks (DSBs) and their consequent aberrant repair via nonhomologous end joining (NHEJ) or homologous recombination (HR) [7], [8], [9], [10]. Due to the inability of DNA polymerase to fully replicate the ends of linear DNA molecules, in the absence of compensatory mechanisms, telomeric DNA is lost at the rate of approximately 100 base pairs (bp) per telomere per cell division [11], [12], [13], [14], [15], [16], [17]. In normal somatic cells, this telomere erosion is used by the cell to monitor its division history, with moderate telomere shortening triggering either irreversible cell cycle arrest, termed replicative senescence, or apoptosis [18], [19], [20], [21]. This block to continued proliferation is thought to have evolved to prevent the development of cancer in long-lived organisms by restricting the uncontrolled outgrowth of transformed cell clones, and also by preventing further telomere erosion which would accompany such abnormal growth and eventually destabilize the telomeres leading to CIN [13], [22].
A current popular model for the involvement of telomere shortening in carcinogenesis posits that increasing numbers of cells experience telomere shortening as a person ages, which increases the pool size of cells that are in danger of experiencing eventual telomere dysfunction and prooncogenic CIN. In the vast majority of such cells, the senescence and apoptotic blocks are strictly enforced [23], [24], [25], [26], [27], [28]. However, this process eventually fails in rare cells which continue to replicate and eventually experience CIN due to critical telomere shortening [15], [29], [30], [31], [32], [33], [34], [35], [36], [37]. Notably, such cells may be more tolerant of rampant genomic instability due to their previous abrogation of the tumor suppressive telomere length checkpoints. However, if left unchecked, this instability will eventually reach lethal levels in the transforming cells, thereby presenting a second block to the development of cancer [37], [38], [39], [40]. This escalating telomere driven CIN creates a strong selective pressure for telomere maintenance in incipient cancer cell populations; a problem that is solved in one of two ways: activation of telomerase or alternative lengthening of telomeres (ALT). In the majority of human cancers, the telomere specific reverse transcriptase telomerase, which is stringently repressed in normal somatic cells, is activated, thereby restabilizing the telomeres, although cancer telomeres on average seem to remain very short [32], [41], [42], [43], [44], [45]. Whereas most cancers use telomerase to maintain telomere length, a significant minority of cancers (typically non-carcinomas) utilize ALT, a telomerase independent, homologous recombination based mechanism [46], [47], [48]. This mode of telomere maintenance results in extreme telomere length heterogeneity and, interestingly, better patient survival compared to their telomerase positive counterparts in several tumor types [49], [50], [51], [52]. These observations suggest that cancer cells utilizing ALT may have compromised their vitality in exchange for the unlimited replicative potential conferred by this telomere maintenance mechanism.
1.2. Centrosomes, the spindle assembly checkpoint, and tumorigenesis
The centrosome is the primary microtubule organizing center in dividing mammalian cells and is composed of a pair of centrioles surrounded by a cloud of proteins that promote microtubule nucleation [53], [54]. The centrosome is duplicated in a semiconservative fashion with one daughter centriole formed next to a preexisting mother centriole, and this process only occurs once in every cell cycle [53], [55]. Centrosome amplification, the presence of greater than two centrosomes during mitosis, is a common characteristic of most solid and hematological tumors that may induce multipolar mitoses, chromosome missegregation, and subsequent genetic imbalances that promote tumorigenesis [54], [56].
Centrosome amplification may be caused by diverse mechanisms, including centrosome overduplication [53], [57], [58], [59], de novo assembly, [60], [61], [62] and mitotic failure downstream from mono- [63] or multipolar division [64], [65], [66], [67], [68], [69]. The end result of these structural abnormalities is often cytokinesis failure, which can give rise to tetraploid binucleated cells and genome instability downstream. Over time, the net result is a small population of cells that harbor the ability to manage extra centrosomes, which could account for the accumulation of cancer cells with centrosome amplification and aneuploidy.
Catastrophic aneuploidy and nonviable daughter cells are a possible tumor suppressive consequence for centrosome abnormalities [70]. However, cancer cells have developed mechanisms that overcome this fate by clustering multiple centrosomes into a “pseudobipolar” state [59], [70], [71], [72]. Cancer cells may utilize this mechanism to dampen high level aneuploidy and extreme CIN, leading to better prognostic outcomes [73], [74]. Centrosome clustering in tumor cells is not completely understood, but it is likely to rely on microtubule associated proteins and motor proteins that bundle together microtubules and centrosomes [71]. Given that centrosome clustering may be advantageous for cancer cell survival, this process may be an attractive and specific therapeutic target [71], [75], [76]. In theory, the induction of multipolarity through declustering of supernumerary centrosomes will selectively target cancer cells without affecting healthy cells [71], [75], [76], [77].
Bipolar chromosome attachment during mitosis is ensured by a quality control mechanism known as the spindle assembly checkpoint. The assembly checkpoint senses tension across kinetochores as a measure of bipolar attachment of chromosomes, and prevents the onset of anaphase in the presence of unattached and/or misattached chromosomes [78]. Any failures to sense errors will compromise the checkpoint and, potentially, induce instability.
The assembly checkpoint relies upon kinase signaling to delay cell cycle progression and correct attachment errors. Aurora kinase B, for example, detects misattached chromosomes [79], [80] and overexpression of the kinase is sufficient to disrupt the checkpoint and promote tetraploidy [81]. Moreover, mutations or expression changes in other checkpoint gene products may compromise the checkpoint and favor tumorigenesis [82], [83], [84], [85]. Lastly, oncogenic cues, such as overexpression of Aurora kinase A, may override a functioning checkpoint and enable cells to enter anaphase despite misattached chromosomes [86]. Cancer cells may take advantage of the checkpoint for their own benefit. For example, checkpoint mediated delay provides time for centrosome clustering [71], which can be manipulated by disabling or restoring assembly checkpoint function [77]. The ability to manipulate or hijack the cell's innate quality control mechanism may act as a selection pressure, and cancer cells that possess this ability may have a growth advantage over others.
Correlation between aberrant centrosome numbers and aberrant chromosome numbers dates back over 100 years [87], yet there is still a debate whether supernumerary centrosomes are the cause or the result of genomic instability, or vice versa [54]. One interesting phenomenon that may shed light on this debate is the presence of a transient tetraploid state during tumorigenesis [88].
Tetraploidy arises after cytokinesis failure following prolonged activation of the assembly checkpoint, regardless of the reason for checkpoint activation [54]. Depending on the status of tumor protein 53 (TP53), a tumor suppressor, the aborted postmitotic cells will either undergo apoptosis after prolonged cell cycle arrest or continue to cycle [56], [89], [90], [91], [92]. In p53 null cells, a postmitotic checkpoint is compromised, which enables the cell to progress through a subsequent cell cycle with double the amount of centrosomes and genetic material [57], [89]. Consequently, each subsequent division for these tetraploid cells will be more error prone, generating more unstable and detrimental aneuploidy [88]. A TP53-dependent postmitotic checkpoint is frequently mutated during early stages of tumorigenesis [88], [90], [91], [92], which suggests that the tetraploid state serves as an intermediate for the aneuploid state observed in cancer cells [88]. In patients with Barrett's oesophagus, the presence of tetraploid cells is detected before aneuploid cells and correlates with early loss of TP53 [93]. Tetraploid cells were also isolated from p53-/− mouse mammary epithelial cells, and these cells formed tumors in nude mice and showed increased aberrant mitoses and genomic instability in culture [94]. Therefore, regardless of how centrosome amplification or genomic instability occurs in this “chicken or egg” argument, it is clear that either event is positively enhanced by the other in promoting tumorigenesis.
1.3. Epigenetic mechanisms contributing to genomic instability
A plethora of studies, including more recent genome-wide profiling, have demonstrated that epigenetic changes direct different cellular phenotypes in both normal and cancer cells [95], [96], [97]. Epigenetics refer to all heritable changes that may modify gene expression without changing the primary DNA sequence, such as DNA methylation and chromatin remodelling. Epigenetic modifications are established during differentiation and are stably inherited and maintained through multiple rounds of cell division. Epigenetic processes that lead to genomic instability and ultimately malignant transformation constitute heritable changes that modulate gene expression and can also affect DNA repair dynamics [95], [96], [97].
DNA methylation consists of the addition of a methyl group at the carbon 5 position of the cytosine pyrimidine ring or the number 6 nitrogen of the adenine purine ring [98], [99]. Most cytosine methylation occurs in the context of cytosine-phosphate-guanine (CpG) dinucleotides, and occurs via a group of DNA methyltransferase enzymes resulting in silencing of gene transcription [100], [101]. Aberrant changes in DNA methylation were among the first events to be recognized in cancer [102]. Global hypomethylation in repetitive sequences of the genome can occur early during tumorigenesis and may initially predispose premalignant cells to repetitive sequence genomic instability [103]. Furthermore, hypomethylation of the promoter of oncogenes can increase their expression [104] and lead to genomic instability [105], [106]. Similarly, aberrant sequence specific hypermethylation in cancer cells can lead to further genomic instability by the silencing of genes involved in cell cycle regulation and DNA repair [107]. A prominent example is the aberrant methylation of CpG islands in the promoter regions of DNA mismatch repair (MMR) genes that result in cancer cells with a “mutator phenotype” [108], [109].
In addition to DNA methylation, histone molecules that form the primary protein component of chromatin also regulate genome stability as well as gene transcription [110]. A number of posttranslational modifications such as acetylation, deacetylation, methylation, phosphorylation and ubiquitination have been identified that alter the function of histones [111]. Various combinations of these posttranslational histone modifications have been hypothesized to form a “histone code” that dictate distinct chromatin structures that can affect genome stability pathways and transcription [95], [97], [98], [112]. Acetylation of the lysine residues at the amino (N) terminus of histone proteins removes positive charges, thereby reducing the affinity between histones and DNA to facilitate access by ribonucleic acid (RNA) polymerase and transcription factors to gene promoter regions [112]. Therefore, in most cases, histone acetylation enhances transcription while histone deacetylation represses transcription. In addition, histone acetylation can affect DNA repair by promoting histone dynamics that stimulate a DNA damage response in response to ionizing radiation [113], [114], [115], [116], [117], [118]. Similarly, histone ubiquitination can also modify DNA repair capacity [119], [120], [121], [122], [123]. Briefly, ubiquitinated histones can lead to chromatin structures that are conducive to the assembly of nucleotide excision repair complexes on damaged DNA [124], as well as both types of DSB repair pathways and cell cycle checkpoint factors critical for the DNA damage response [119], [120], [121], [122], [123], [124], [125], [126]. Monoubiquitination of histones H2A and H2B prevents chromatin compaction and facilitates assembly of the repair machinery at the damaged sites [126]. Polyubiquitination of histone H2A and H2AX is important for the retention of repair proteins, such as TP53 binding protein 1 (53BP1) and breast cancer 1 (BRCA1), at damaged loci [120], [127]. Finally, histone phosphorylation is an early event following DNA damage and required for efficient DNA repair. Upon introduction of a DSB, hundreds of histone molecules become phosphorylated within minutes at the chromatin flanking the break site, thus providing a rapid and highly amplified detection system and a focus for the accumulation of many other proteins involved in DNA repair and chromatin remodelling [128]. These examples, and numerous other observations, suggest that a vast array of epigenetic mechanisms contribute to the genomic instability in cancer cells.
1.4. Mitochondrial DNA alteration in human cancers
Mitochondrial genetic reprogramming and energy balance within cancer cells play a pivotal role in tumorigenesis and are duly regarded as one of the hallmarks of human cancer [129]. In 1927, Otto Warburg [130], [131] identified mitochondrial dysfunction as a key component of tumorigenesis and numerous studies have since elaborated upon the role of mitochondrial DNA (mtDNA) alterations in different human cancers [132].
Mitochondria are the key component of the oxidative phosphorylation system to generate cellular adenosine triphosphate. They uniquely possess their own DNA and generate reactive oxygen species (ROS) [132]. Most human cells contain hundreds of nearly homoplasmic (identical) copies of mtDNA, which are maternally inherited [132]. Compared to the nuclear DNA, the mutation rate of mtDNA is nearly 10 times higher and alterations are much easier to detect due to their high copy number in cancer cells.
A substantial number of studies identified somatic mtDNA mutations involving coding and noncoding mtDNA regions in various cancers [132], [133], [134], [135], [136]. Among the noncoding mtDNA mutations, a poly C mononucleotide repeat (known as D310) was frequently altered in numerous cancers and appeared to be a mutational hot spot [132]. Notably, coding mtDNA mutations targeting respiratory complex I, III, IV or V were frequent in a variety of human cancers [132], [133], [134], [135], [136], [137], [138]. Moreover, alteration of mtDNA copy number could potentially be associated with mitochondrial dysfunction leading to disease progression [132], [133]. In recent studies, a correlation between mutations in mtDNA and epidermal growth factor receptor, or prostate-specific antigen expression, was established in lung and prostate cancer, respectively [134], [135]. These results suggest cross talk between mitochondria and nuclear genomes maintain tumor growth.
In order to understand their role, a number of studies introduced mtDNA mutations in cancer cells. Introduction of a mitochondrial mutant adenosine triphosphate 6 (ATP6) (complex V) or cyclooxygenase 1 (COXI) (complex IV) increased the growth of prostate cancer cells [136], [139] or induced cancer cell proliferation and altered reactive oxygen and nitrogen species [140], respectively. In a bladder cancer study, introduction of a mutant mitochondria encoded cytochrome B (CYTB) induced bladder cancer growth and invasion, accompanied with increased ROS, lactate production and oxygen consumption [141]. Moreover, the ROS-producing CYTB mutant tumor cells efficiently killed normal splenic immune effector cells, which may provide tumor cells with an immune evasion mechanism [141]. In addition, mutant CYTB overexpression in nontumorigenic bladder epithelial cells triggered an increased mitochondrial proliferation and inhibition of apoptosis [142]. As these mutations in mtDNA were detected in human patients, the preceding studies suggest a causative role for mtDNA alterations in tumorigenesis.
2. Repair pathways responsible for genetic fidelity and tumor suppression
DNA is replicated with extreme fidelity in normal cells with a mutation rate of 10−10 per base pair per cell division. DNA damage typically occurs through the following: (1) exposure to agents such as ultraviolet irradiation, genotoxic chemicals, and ionizing radiation; (2) spontaneous DNA damaging events, such as a basic site formation; and (3) failure in normal cellular DNA processing and replication events, such as stalled replication forks. These processes induce oxidation, alkylation, crosslinking, dimerization, and strand breaks in DNA, which must be resolved. As such, repair of this DNA damage is essential to preserving genome integrity and preventing cancer.
2.1. Excision repair pathways
Three excision repair pathways can repair single stranded DNA damage: nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR).
2.2. Nucleotide excision repair
Fidelity of genetic information transmission depends on NER, which serves to repair DNA damage caused by ultraviolet irradiation, alkylating and oxidizing agents, or chemotherapeutic drugs that form bulky, helix distorting adducts. Two sub-pathways have been identified. Global genome NER repairs damage in both strands of the DNA regardless of whether the gene is being actively transcribed [143], [144], [145]. Transcriptionally coupled NER, however, repairs transcriptionally active genes [143], [144], [145]. The two pathways are similar in that they use many of the same pathways, but global genome NER uses xeroderma pigmentosum complementation group C (XPC)-RAD23 homolog B (HR23B) and DNA damage binding protein 1 (DDB1)-DDB2/XPE proteins to recognize distortions in the double helix while transcriptionally coupled NER occurs at regions where RNA Polymerase II has stalled [146], [147], [148], [149], [150]. Genetic polymorphisms of NER gene products associate with human diseases, including xeroderma pigmentosum, which can lead to severe cases of skin cancer.
2.3. Base excision repair
The BER pathway fixes damaged DNA bases (reviewed in [151]). These lesions are recognized and removed by specific DNA glycosylases, which cleave the glycosidic bond between the damaged base and the sugar of the DNA backbone. In more complex lesions, proliferating cell nuclear antigen (PCNA), flap endonuclease 1 (FEN1), and DNA polymerase (POL) β, with or without POLδ/ɛ, act to repair the lesion. This complex set of events in BER is facilitated by poly (ADP-ribose) polymerase 1 (PARP1), which recruits proteins involved in the DNA repair response, such as X-ray repair cross-complementing protein (XRCC)1, DNA ligase, and DNA polymerase [152], [153].
Because cells are constantly subjected to DNA damaging conditions, the BER pathway is crucial to preserving genome integrity. This is exemplified by the embryonic lethality of mice that possess knockouts of key components of this pathway [154], [155], [156]. A biallelic germline defect within a DNA glycosylase, mutY Homology (MUTYH), was initially found in families that had excess colorectal tumors with somatic mutations in the adenomatous polyposis coli gene [157]. A subsequent larger study revealed that biallelic germline MUTYH defects conferred 93 fold excess risk of colon cancer with penetrance by age 60 [158], [159] and may also confer increased risk for endometrial cancer [160]. Mutations in another glycosylase, 8 Oxoguanine (OGG1), have been associated with laryngeal cancers [161] while gastric cancers harbor inactivating mutations in glycosylase nei endonuclease VIII-like 1 (NEIL1) [162]. Taken together, these studies confirm the importance of BER in the suppression of carcinogenesis.
2.4. DNA mismatch repair
Some evidence suggests that proofreading activity of replicative DNA polymerases and MMR machinery act in series in mammalian cells [163], [164], [165], [166]. MMR targets could generally be classified into base/base mispairs and large insertion–deletion loops. At the forefront of error recognition, MutS protein homolog (Msh) 2 pairs with Msh6 or Msh3, to form MutSα (Msh2/Msh6) and MutSβ (Msh2/Msh3). Whereas the former is mostly responsible for base/base mispairs, the latter targets large insertion–deletion loops. To initiate the repair process, MutL homolog 1(Mlh1)/Pms2 heterodimers (MutL homologues), in the presence of exonuclease1, interact with MutS complexes to create nicks in the 3′ and 5′ of the nascent strand containing the mismatch. Following nick creation, enzymes required to repair the damage site are recruited and resynthesis of the DNA is carried out by POLδ/ɛ. As a nexus for DNA damage sensing and cell death, MMR machinery play an important role in recognizing damaged DNA and relaying signals downstream to activate a G2/M cell cycle checkpoint.
2.5. Double strand break repair and cancer predisposition
The DSB is the most lethal form of DNA damage, as it can lead to significant DNA damage by multiple genomic changes, including translocation formation, deletions, and amplifications, resulting in heritable cellular genomic instability/damage that can lead to malignancy [167], [168], [169]. DSBs are repaired by both HR and NHEJ repair pathways. NHEJ repair occurs throughout the cell cycle, while HR prevails in S and G2 phase cells (reviewed, [167], [168], [169]). NHEJ repair joins broken DNA ends without identifying DNA sequence homology and is therefore highly error prone [169]. HR repair is dependent upon DNA sequence homology and therefore is relatively error free [168], [169], [170].
Errors in the NHEJ pathway may generate inappropriate dicentric chromosomes that are covalently joined (reviewed, [171], [172]). These dicentric chromosomes may break during anaphase, producing new dicentric chromosomes through further NHEJ [171], [172]. This process is known as the breakage fusion bridge (BFB) cycle, which is important in telomere related genome instability [172]. Damaged telomeres will be processed by NHEJ, unless the broken ends are healed by a new telomere, and continuation of the BFB cycle can result in complex chromosomal rearrangements that include gene loss, gene amplification and unbalanced translocations [171], [172]. BFB cycles are self-perpetuating and result in genetic heterogeneity in a variety of cancers [173].
HR repair involves multiple gene products some of which are involved in repair of stalled replication forks [170]. The DSB is recognized by a Mre11-Rad50-NBS1 complex which recruits many different proteins, including proteins with topoisomerase, endonuclease, and helicase activity. Eventually a “synaptic complex” is formed which allows homologous single strand DNA (ssDNA) to invade and anneal to complementary DNA. DNA polymerase then fills in the ssDNA gap and the synaptic complex is resolved. Both crossover and non-crossover products can be created by this process [167], [168], [169], [170]. Interestingly, loss or mutation of many of these gene products is associated with specific cancer prone diseases.
BRCA1 and BRCA2 are key players in the HR repair pathway and act as tumor suppressors by maintaining genome stability. Linkage studies in families with early onset breast cancer detected the presence of a breast cancer susceptibility gene, BRCA1 [174]. Subsequently, mutations in BRCA1 were confirmed in families with early onset breast and ovarian cancer [175], [176], [177], [178]. Later, a novel locus, encoding BRCA2, was discovered and linked to breast cancer susceptibility [179], [180], [181]. In one meta-analysis, cumulative breast and ovarian cancer risks for BRCA1 mutation carriers are 57 and 40%, respectively, while, for carriers of BRCA2 mutations, these risks are 49 and 18%, respectively [182]. Furthermore, hereditary BRCA mutations have been also linked to pancreatic, prostate, and colon cancers [183]. Of interest, germline mutations in BRCA1 versus BRCA2 associate with different subtypes of breast cancer. BRCA1 associated cancers are of the more aggressive triple negative subtype and appear at an earlier age than sporadic tumors. In contrast, BRCA2-associated tumors relate mostly with hormone receptor positive breast cancers.
Like BRCA1 and BRCA2, partner and localizer of BRCA1 (PALB2) promotes genome integrity through its role in DSB repair. It binds and colocalizes with BRCA2 in the nucleus to stabilize BRCA2 foci and facilitate the intra S phase checkpoint and HR repair [184]. Germline mutations in PALB2 confer a 2–5 fold increase in breast cancer risk [185], [186] and germline mutations have been recently found in African American breast cancer patients [187], [188]. PALB2 mutations have also been observed in 1% of Chinese women with early onset breast cancer [189]. Interestingly, exome sequencing identified PALB2 as a pancreatic cancer susceptibility gene [190] and PALB2 mutations have been found in patients with familial pancreatic cancers [191], [192].
Fanconi anemia (FA) is an autosomal recessive disorder characterized by congenital defects, CIN, hypersensitivity to DNA crosslinks, bone marrow failure, and predisposition to cancer [193], [194]. Fifteen FA or FA-like genes have been identified, all of which are involved in coordinating DNA repair through the FA/BRCA pathway. Interestingly, two of these genes are BRCA2 (FANCD1) and PALB2 (FANCN), thus revealing interplay between FA and HR [195], [196]. Patients with FA have increased susceptibility to breast, ovarian, and oral cancers. Additionally, heterozygote carriers of germline mutations in FA genes may also harbor an increased risk to develop cancer. Importantly, unlike the BRCA-associated cancers, tumors from FA patients, or with acquired FA defects, may be hypersensitive to crosslinking agents such as cisplatin and mitomycin C, and also are hypersensitive to radiation [197], [198], [199]. Finally, biallelic loss of ataxia telangiectasia mutated (ATM) results in ataxia telangiectasia, a disease characterized by a roughly 1000 fold increased lymphoma incidence [200], [201].
3. Therapeutic targeting of genomic instability
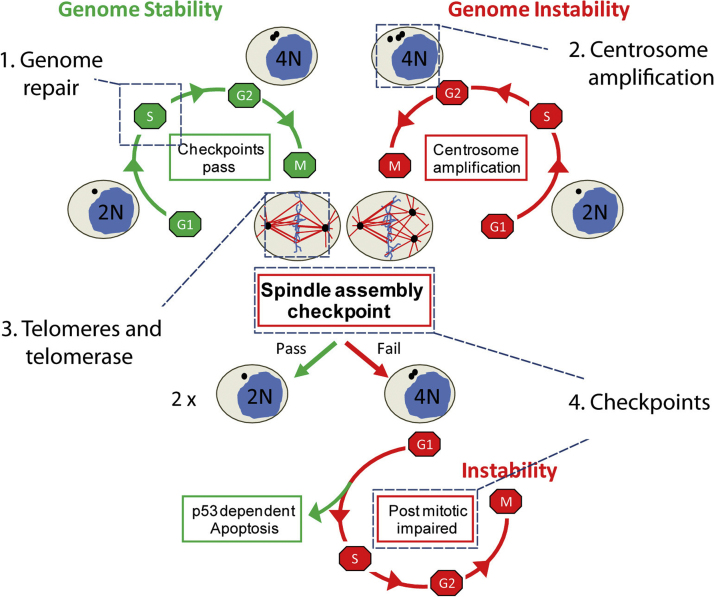
Current standard therapies for cancer often involve agents or strategies that damage the DNA, which can also damage noncancerous tissues. New treatments that target genomic instability (Fig. 1) may minimize these off target toxicities to normal tissues.
Fig. 1.
Genome stability is dependent on faithful DNA repair and chromosome segregation during cell division. During S phase, the centrosome and genomic material are replicated concurrently, and replication errors are repaired prior to mitotic entry (1). During mitosis, equal segregation of chromosomes requires a bipolar mitotic spindle, telomere preservation and the completion of the spindle assembly checkpoint. Ectopic amplification of centrosomes (2), telomerase dysfunction (3) and failure of the spindle assembly checkpoint (4) may result in aborted mitosis. Mitotic failure gives rise to a single tetraploid cell (4 N) instead of two diploid cells (2 N). This tetraploid cell can progress through the cell cycle should the TP53-dependent post mitotic checkpoint fail to induce apoptosis or senescence (4). Thus, genomic instability is propagated in subsequent cell cycles.
3.1. Targeting DNA repair pathways in cancer therapy
Drugs that target DNA repair proteins have shown preclinical and/or clinical efficacy in potentiating DNA damage (reviewed in [202]). Synthetic lethality, whereby deficiencies in parallel pathways are only cytotoxic when both pathways are defective, is a novel strategy that may selectively target cancerous cells defective in DNA repair [203]. Synthetic lethality was illustrated in BRCA deficient cells, which exhibited profound sensitivity to inhibition of PARP [204], [205]. This was due to conversion of unrepaired single strand breaks into DSBs during DNA replication due to the BRCA deficiency. Persistence of the unrepaired DSBs led to profound cellular cytotoxicity. The potential efficacy of PARP inhibitors in patients with BRCA-associated cancers has been reported in multiple clinical trials [206], [207], [208], [209], [210], [211]. Perhaps more important is the high therapeutic index of these compounds, since noncancerous cells in this patient population still maintain one wild type allele and thus remain HR proficient. Natural products have also been shown to act on PARP, such as isothiocyanates, which are found in Brassica vegetables. Isothiocyanates stimulate the proteolytic cleavage of PARP [212]. Recent work implicates vitamin D as a possible treatment mechanism to supplement PARP treatment [213]. BRCA1-deficient cells bypass growth arrest by activating cathepsin L (CTSL)-mediated degradation of 53BP1. Vitamin D depletes or inhibits CTSL leading to increased genomic instability and compromised cancer cell proliferation after irradiation and treatment with PARP inhibitors [213]. Selenocysteine can induce ROS formation, which can lead to DSBs in cancer cells but not in normal human fibroblasts [214]. Thus, DSB repair deficient cancers may be sensitive to treatment with selenium compounds.
In addition to defective DSB repair, inappropriate HR often results in a significant predisposition to cancer development [200], [201]. Many human malignancies with HR deficiency show increased sensitivity to chemotherapy agents that cause DSBs, such as ionizing radiation, bleomycin, and cisplatin. Additionally, agents which inhibit DNA replication, such as bifunctional alkylating agents and topoisomerase inhibitors, also preferentially inhibit the growth of HR deficient malignant cells [170], [215], [216]. There has been intense interest in identifying HR deficiencies in human tumors and targeting these tumors with DSB-inducing chemotherapeutic agents. Since ∼25% of human malignancies show HR defects [170], targeted treatment could eventually play a significant role in chemotherapy. As tumors often overexpress specific proteins involved in HR, this approach might also preferentially target tumor over normal tissue [170], [215], [216], [217]. A major challenge in this area is the efficient and accurate identification of HR deficiency in human malignancies.
Some of the proteins that have been proposed as useful targets to inhibit HR include ATM, checkpoint kinase (CHK)1&2, ataxia telangiectasia and Rad3 related (ATR), and the FA pathway proteins [170]. Many inhibitors that target these enzymes are either in preclinical development or in the early phases of clinical development [218], [219]. Resveratrol may activate sirtuin 1 (Sirt1) activity [220], which is a nicotinamide adenine dinucleotide-dependent deacetylase that is known to activate DNA repair. Studies in mice have shown that Sirt1+/−; p53+/− mice develop tumors in many different tissue types but mice treated with resveratrol display a reduced amount of tumorigenesis [221], indicating that resveratrol could act to prevent and/or treat cancers in patients that have reduced Sirt1 function. Although in early development, therapies that specifically alter HR are a promising area of research and may contribute to targeted chemotherapy regimens that are more personalized and effective.
3.2. Targeting microsatellite instability
MMR inactivation is associated with the lack of repair of replication errors leading to an increase in spontaneous mutation rate [222]. A marker of defective MMR is microsatellite instability (MSI), or numerous alterations in the lengths of microsatellites [223], [224]. Tumors displaying MSI are said to exhibit a “mutator phenotype”, with a dramatic predisposition to somatic mutations.
The critical role of MMR pathways in tumorigenesis is exemplified by the fact that germline mutations in the genes involved in MMR predispose to cancer development [225]. In the case of colorectal cancer (CRC), MMR deficiency is estimated to be present in 15 to 17% of all primary cancers, including both sporadic CRC and Lynch syndrome (then called hereditary nonpolyposis colorectal cancer), though through different mechanisms [223], [226], [227]. Lynch syndrome is characterized by inactivating germline mutations to MSH2, MSH6, PMS2, or MLH1, whereas MLH1 expression is silenced due to biallelic hypermethylation in sporadic CRC [228], [229], [230], [231], [232]. MLH1 methylation results from extensive aberrant promoter methylation [233], [234]. The 3′ end of the MLH1 promoter, proximal to the start codon, is most commonly methylated [227]. Methylation of the 5′ end of the MLH1 promoter can also occur, however, the methylation pattern must extends to the 3′ end to be deleterious [227]. Loss of MLH1 expression increases with age and protein expression is lost by ∼50% in patients who are 90 years or older [235]. The exact mechanism(s) behind MLH1 silencing remain unknown, but may result from abnormal methylation [236], structural chromatin changes that increase accessibility to promoter regions [237], or genomic damage [238]. Tumors with mutations in the MLH1 gene are rare, which suggests that hypermethylation of the MLH1 promoter is an important event in neoplastic transformation in sporadic CRC [234].
The extent of MSI in CRC has been classified as MSS (microsatellite–stable), MSI-H (with high level of instability), and MSI-L (with low level of instability) [239]. Classification between MSI-H and MSI-L depends on which MSI markers are present and their proportions [240]. These markers include mononucleotide repeats, such as BAT25, BAT26, and BAT40, and the dinucleotide repeats D5S346, D2S123, and D17S250 [241], [242], [243], [244]. Dinucleotide markers are present in both MSI-H and MSI-L cancers, whereas mononucleotide markers are specific for MSI-H cancers [241]. In MSI-H tumors, more than 30% of these markers are unstable, while in MSH-L tumors, 10–40% of these markers are unstable, and MSS tumors demonstrate no unstable markers [241], [245], [246], [247], [248]. MSI-L and MSS tumors are frequently grouped together due to similarities in their clinical features and gross abnormalities [248]. MSI-H is most prevalent in sporadic CRC, observed in 10–15% of all cases [240], [248].
The predictive value of MMR status as a marker of response to 5 fluorouracil, irinotecan and other drugs is still controversial [249]. Recently, two large retrospective analyses from several randomized trials confirmed the detrimental effect of a 5 fluorouracil-based adjuvant therapy in stage II colorectal patients [250], not applicable to stage III patients [251]. These latter authors, however, reported that MSI stage III tumors harboring genetic mutation in the MMR genes seem to benefit from the 5 fluorouracil adjuvant therapy. These data imply that molecular differences within the MSI subgroup influence the response to 5 fluorouracil.
The CRC MSI subgroup represents a cancer with a defined molecular etiology, a characterized mutational profile and an established genotype–phenotype relationship, which may enable synthetic lethal approaches that target MMR deficiency. High throughput experiments revealed synthetic lethal interactions between MSH2 and POLB, between MLH1 and Polymerase G gene [252], between RAD54B and FEN1 [253], [254], between MLH1/MSH2 and PTEN-induced putative kinase 1 gene [255], and the preferential effect of methotrexate in MMR deficient systems [256]. These synthetic interactions may induce or accumulate ROS [257], [258]. A phase II randomized clinical trial in MLH2-deficient metastatic CRC (NTC00952016) is currently underway [256]. Combination therapy with methotrexate and PARP inhibitors may be effective against tumors with MMR mutations. Methotrexate elevates ROS and DSBs and the combination of MMR mutation and PARP inhibition may attenuate repair and induce growth arrest or apoptosis [259], [260], [261].
3.3. Targeting gene expression of cell cycle and DNA repair components
RNA interference (RNAi) may enable personalized antitumor therapies. A number of RNAi-based studies have silenced genes responsible for tumor cell growth, metastasis, angiogenesis, and chemoresistance [262]. For example, siRNA targeting of Cyclin E suppressed tumor development [263]. Epigenetic regulation of gene expression is an alternative approach. Resveratrol, a phytoalexin produced by plants such as the Japanese knotweed, prevents hypermethylation of the BRCA1 promoter [264], and may be effective for triple negative or basal subtype breast cancers. Other natural compounds, like genistein and lycopene, can alter DNA methylation of the glutathione S transferase p1 (GSTP1) tumor suppressor gene [265].
3.4. Targeting centrosome abnormalities
Centrosome amplification is an important process during early stages of cancer development (see Section 1.2). Though the mechanism(s) behind centrosome amplification remain elusive, TP53 negatively regulates centrosome amplification through a TP53-p21-CDK2 signaling loop [266], [267]. Moreover, TP53 induces apoptosis through transactivation of proapoptotic genes and transrepression of antiapoptotic genes [268]. Thus, TP53 provides an interesting link between two major cancer processes, centrosome amplification and apoptosis dysregulation [268]. In one study, the loss of TP53, or treatment with 5 fluorouracil, promoted centrosome amplification in HCT116 cells and those cells with supernumerary centrosomes were more acutely sensitive to resveratrol [268]. However, TP53 defective cancer cells that resist 5 fluorouracil treatment are prone to centrosome amplification and downstream genome instability [269]. The presence of supernumerary centrosome can also be problematic for cancer cells. Clustering excess centrosomes may be a necessary prosurvival pathway for cancer cells and thus an attractive target [70]. Griseofulvin, an antifungal drug that suppresses proliferation in tumor cells without affecting non-transformed cells, declusters centrosome, although the precise mechanisms behind the drug's action remain unknown [71]. In a similar fashion, depletion of a kinesin-like motor protein can selectively kill tumor cells with supernumerary centrosomes [77]. Finally, the PARP inhibitor PJ34 also declusters supernumerary centrosomes without deleterious effects on spindle morphology, centrosome integrity, mitosis, or cell viability in normal cells [270].
4. Prevention of genomic instability and human cancer
There is no question that optimizing nutrient intake plays a significant role in stabilizing the genome. In recent years, an increasing number of biomarkers of genome integrity, including telomere length and mtDNA deletions, have been utilized in establishing recommended daily intakes for nutrients [271]. In several cases, such an approach has led to substantial changes in the levels of various nutrients that populations had been previously advised to consume. These findings highlight the need to better optimize an individual's diet to their personal genetic makeup, which in turn has prompted the emergence of nutrigenomics, a new field that aims to determine how a particular genotype or expression profile correlates to nutrient metabolism, absorption, etc. (reviewed in [272], [273]).
4.1. Vitamins–carotenoids
Since Peto et al. [274] concluded that the evidence pointed to a cancer preventive role for β carotene, many placebo-controlled carotenoid intervention trials have been carried out with disease and mortality as outcomes. Early findings were that, in subjects who were smokers and/or asbestos workers, there was a significant increase in lung cancer incidence [275]. A recent meta-analysis confirmed that a significant increase in mortality is associated with vitamin A, β carotene or vitamin E supplements [276]. When determining the effects that dietary supplements and other compounds have on cancer prevention, it is important to take into account the different types of data: conventional intervention studies, animal experiments, cell culture studies, or human intervention trials based on biomarkers. This is important because of the nature of each type of experiment and the information that can be obtained from them.
4.1.1. Human biomarker trials with molecular endpoints
Human trials with carotenoids [277], [278], [279], [280], [281], [282], [283], [284], [285], [286] were mostly cross sectional or case control studies, with a few intervention trials. In general, a negative correlation was seen between blood carotenoid levels and various biomarkers of DNA damage, and intervention trials tended to show a decrease in DNA damage or no effect.
4.1.2. Animal experiments
Most animal experiments [287], [288], [289], [290], [291], [292], [293], [294], [295], [296], [297], [298], [299], [300] involved treatment of rats or ferrets with genotoxic agents during or after carotenoid supplementation, and generally decreased levels of DNA damage were reported. Another study [301] looked at base oxidation in leukocytes and oxidation products in urine, reporting a carotenoid-induced decrease in the former (but no effect on urinary biomarkers). There was a decrease in endogenous DNA oxidation in liver of mice fed tomato paste (rich in lycopene) [302].
4.1.3. Cell culture experiments
We found eleven cell culture studies [287], [303], [304], [305], [306], [307], [308], [309], [310], [311], [312] with provitamin A carotenoids (α and β carotene, β cryptoxanthin, retinoic acid, retinal and retinol), and eight with nonvitamin A carotenoids (lycopene, lutein, astaxanthin or zeaxanthin) [294], [304], [313], [314], [315], [316], [317], [318]. Experiments in most cases involved cotreatment with DNA damaging agent and carotenoid. Concentrations of carotenoid varied widely, from less than 1–100 μM. Here we found a very clear pattern in the results, depending on the type of carotenoid and the concentration: while the non-vitamin A carotenoids invariably resulted in a decrease in DNA damage, the provitamin A carotenoids at low concentrations either had no effect or decreased DNA damage, while at concentrations above about 5 μM, increases in damage were the norm. Potential prooxidant effects of carotenoids can probably be ruled out as a cause of this DNA damage, since there is no obvious reason why provitamin A carotenoids should be more likely to act as prooxidants. Instead, we should perhaps be looking at downstream effects of vitamin A itself, on transcription, via retinoic acid and retinoic acid receptors binding to retinoic acid response elements present in the regulatory sequence of many genes.
In a review of effects of carotenoids on DNA repair [319], we found relatively few reports. Cells from the spleen of rats supplemented with carotenoids (plus nicotinamide and zinc) showed accelerated repair of DSBs induced by radiation, and lymphocytes from human subjects given the same supplement mix were faster at rejoining hydrogen peroxide induced breaks [184]. Mixed carotenes plus α tocopherol as a supplement in humans had no effect on DNA repair [320]. DSB rejoining was faster in Molt 17 cells in the presence of β carotene, lutein or β cryptoxanthin [321], and in HeLa and Caco2 cells with β cryptoxanthin [306]; but no effect was seen in lymphocytes incubated with β carotene or lycopene [322] or with vitamin A [311].
Lung cells from ferrets supplemented with β carotene were tested for BER capacity with an in vitro comet-based assay, and showed an increase in activity [323]. B cryptoxanthin enhanced BER of 8 oxoguanine in HeLa and Caco2 cells [306], but no effect of carotenoids was seen in Molt 17 cells [321] or lymphocytes [320].
The pattern that emerges from this survey of carotenoid effects on DNA damage is that, in cell culture at least, while nonvitamin A carotenoids tend to decrease damage (whether endogenous or induced), at whatever concentration, the provitamin A carotenoids show a clear tendency to cause or increase damage at high concentrations. Whether this can account for the apparent harmful effects of β carotene as seen in the human clinical trials is not possible to answer at present.
Glutathione is another important antioxidant that can improve outcomes for patients with cancer and can help reduce treatment toxicity. Some studies have shown that supplementation with glutathione can reduce the toxicity of chemotherapy agents such as cisplatin and cyclophosphamide during treatment [324], [325]. Interestingly, while the antioxidant properties of glutathione may reduce treatment toxicity, the same properties can make tumor cells resistant to chemotherapy when glutathione is present in high levels in the cells [326], [327]. A GSTP1 polymorphism (GSTP1 Ile105Val), which has a seven fold higher efficiency, has been linked to a reduced survival rate in cancer patients further emphasizing that while glutathione may be able to reduce treatment toxicity, it can potentially also confer an advantage to the tumor cells as well [328], [329].
An additional aspect that may lead to conflicting results regarding the efficacy of antioxidants in cancer treatment is the oxidative stress that tumor cells experience in their microenvironment. Tumor cells have been shown to undergo the Warburg effect, which is where they produce energy primarily through glycolysis rather than through aerobic respiration ([131]; reviewed in [330]). The production of lactate in tumor cells rather than pyruvate, an antioxidant, increases the load of ROS and increases oxidative stress in the cancer cells. Recent work has suggested that cancer cells might be targeted by using 3 bromopyruvate, an inhibitor of the glycolysis enzyme hexokinase II, to amplify the Warburg effect in cells [331].
4.2. Other vitamins
A range of B vitamins, including niacin (vitamin B3), folate (vitamin B9), and vitamin B12, significantly interact to maintain the stability of both nuclear and mitochondrial genomes. For example, a niacin deficiency, common in certain populations, impairs the function of the PARP family of enzymes, identified above as critical to DNA repair. A folate deficiency, especially in the presence of suboptimal levels of vitamin B6 and vitamin B12, may have significant effects on the expression of chromosomal fragile sites, leading to chromosome breaks, micronuclei and deletions of mtDNA. It may also lead to reduced telomere length. There are considerable interindividual differences in people's capacity to absorb and metabolize these vitamins, dependent upon genotype and epigenotype [332].
Vitamin C is considered an antioxidant, and is present at high concentrations (mM) in certain tissues such as the eye. Effects on various markers of genome stability were shown to depend on individual diet-derived vitamin C concentrations, and also on exposure to xenobiotics or oxidative stress [333]. Vitamin D is also critical in the maintenance of genome stability, possibly through protection against oxidative stress, chromosomal aberrations, telomere shortening and inhibition of telomerase activity [334].
4.3. Minerals
While a number of minerals are typically considered as toxicants, there are several that are essential micronutrients, albeit usually with a narrow window of efficacy as compared with toxicity. These include iron [335], selenium [336], and zinc [337].
Selenium provides a useful illustration of the complexities of reaching agreement on optimal population levels. The population as a whole shows a “U” shaped curve for functionality, where low and high selenium levels both increase genomic instability. Optimal levels of selenium may protect against DNA or chromosome breakage, chromosome gain or loss, damage to mtDNA, and detrimental effects on telomere length and function. One example of how selenium can function is by protecting genome stability through a BRCA1-dependent mechanism [338]. When cells are supplemented with selenium, there is reduced DNA breakage and the number of aneuploid cells is reduced when compared to control cells. Unfortunately, these optima differ among individuals and according to the form of selenium in the diet [336], [339]. Various genetic polymorphisms have been shown to affect the uptake and utilization of selenium among individuals.
4.4. Other dietary factors
Diets high in plant-based foods have been associated with decreased cancer risks [340]. Lim and Song [341] discuss how certain dietary components, common in plant foods, can alter DNA methylation levels, affecting genome stability and transcription of tumor suppressors and oncogenes. Much of the available data exist for folate, since this is a well-recognized nutritional factor in one-carbon metabolism, acting to supply the methyl units for DNA methylation. This has been shown to be especially important in the maternal diet as a lack of folate can lead to hypomethylation of some genes in their offspring. One well studied example of this is the Agouti mutation in mice, which affects coat color, as well as making the offspring more susceptible to cancer and obesity [342], [343]. The Agouti mutation has been shown to be a lack of methylation at the promoter of the Agouti gene [344]. Pregnant mice that were fed bisphenol A had offspring that exhibited hypomethylation of the Agouti gene but, by feeding them dietary supplements of folate, methylation status was rescued [345]. This data demonstrates the importance of the maternal diet during development to outcomes even later in life for their offspring. In other systems, folate supplements during pregnancy have also been shown to be protective against neuroectodermal brain tumors [346].
Alcohol, various polyphenols, phytoestrogens and lycopene also have demonstrable effects. Indeed, there is compelling evidence that a considerable range of plant polyphenols may stabilize genomic DNA, through various processes, including effects on DNA methylation [347]. Duthie [340] suggested that the evidence is particularly strong for berry phytochemicals, specifically anthocyanins (a class of flavonoids), which modulate various biomarkers of DNA damage and carcinogenesis, in both in vitro and in vivo animal studies. However, evidence for cancer preventive effects in human studies is currently weak.
Accumulating evidence shows that genome integrity is highly sensitive to nutrient status, and that optimal levels may differ among individuals. Many investigations to date are limited by considering only the effects of single nutrients, without looking at the potential interactions among these, and of nutrients with toxicants in the diet. Many currently available studies also suffer from a failure to consider the effects of genetic susceptibility. In subsequent work, it will be critical to consider modifying and interactive effects with deficiencies in nutrients required for effective DNA damage response, and DNA repair.
Hyperglycemia and a high fat diet have been shown to be positively correlated with an increased risk of cancers, such as breast and endometrial cancers. Hyperglycemic diets have been shown to increase the levels of many signaling molecules [348]. Rats that were fed high fat diets had an increased risk of breast cancer in their progeny [349]. These results were similar to those seen when mice are treated with estradiol. In addition to an increased risk of developing cancer, hyperglycemia, diabetes, and obesity have been linked to a worse prognosis. Advanced breast cancer patients with high blood glucose levels had a lower rate of survival than those with normal sugar levels [350] and obese adolescents with pediatric acute lymphoblastic leukemia had a higher likelihood of relapse than a normal cohort [351].
5. Complementary effects on the enabling characteristics of cancer while targeting genomic instability
Treatments that are less cytotoxic but can also act on multiple different cancers and pathways that contribute to cancer formation is an important goal (Table 1). Work focusing on the effects of vitamin D, vitamin B, selenium, carotenoids, PARP inhibitors, resveratrol, and isothiocyanates has shown promising results (Table 2).
Table 1.
Cross-validation for priority targets against genomic instability.
| Other cancer hallmarks | Priority targets for genomic instability |
||||
|---|---|---|---|---|---|
| Prevent DNA damage | Enhance DNA repair | Target deficient DNA repair | Block centrosome clustering | Inhibit telomerase | |
| Sustained proliferative signaling | 0 | 0 | 0 | 0 | + [432], [433] |
| Tumor-promoting inflammation | − [434], [435], [436] |
− [434], [435], [436] |
− [434], [435], [436] |
+ [437], [438] |
+ [439], [440] |
| Evasion of anti-growth signaling | + [441], [442] |
+ [441], [442], [443] |
+ [444] |
+ [445], [446] |
+ [447] |
| Resistance to apoptosis | ± [448] |
± [448] |
± [448] |
0 | + [449] |
| Replicative immortality | + [450], [451] |
0 | 0 | + [452] |
n/a – same target |
| Dysregulated metabolism | ± [453] |
± [453] |
± [453] |
+ [454] |
+ [455], [456] |
| Immune system invasion | + [352] |
+ [352] |
+ [352] |
0 | 0 |
| Angiogenesis | − [457], [458], [459], [460], [461] |
− [457], [458], [459], [460], [461] |
0 | + [462] |
+ [463], [464], [465] |
| Tissue invasion and metastasis | + [466], [467] |
+ [466], [467] |
+ [466], [467] |
+ [468], [469] |
+ [470], [471], [472], [473], [474] |
| Tumor microenvironment | + | + [475] |
+ [476] |
+ [437] |
+ [477] |
Priority targets that were not only relevant for genomic instability, but also relevant for other aspects of cancer's biology (i.e., anticarcinogenic) were noted as having complementary effects (+). Those targets that were found to have procarcinogenic actions were noted as having contrary effects (−). In instances where reports on relevant actions in other aspects of cancer biology were mixed (i.e., reports showing both procarcinogenic potential and anticarcinogenic potential), the designation (±) was used. Finally, we indicate (0) in instances where no literature support was found to document the relevance of a target in a particular aspect of cancer's biology.
Table 2.
Cross-validation for priority approaches against genomic instability.
| Other cancer hallmarks | Priority approaches for genomic instability |
||||||
|---|---|---|---|---|---|---|---|
| Vitamin D | Vitamin B | Selenium | Carotenoids | PARP inhibitor | Resveratrol | Isothiocyanates | |
| Sustained proliferative signaling | + [478] |
± [259], [426] |
± [357], [479] |
+ [480], [481], [482] |
+ [358], [483], [484] |
+ [354], [355] |
+ [485], [486] |
| Tumor-promoting inflammation | + [487], [488] |
+ [393], [394] |
− [395], [396], [397] |
+ [398], [399] |
+ [400], [401] |
+ [402], [489] |
+ [403], [404] |
| Evasion of anti-growth signaling | + [490], [491], [492] |
0 |
+ [493], [494] |
± [366], [495] |
+ [358] |
+ [496], [497] |
+ [360], [361], [364] |
| Resistance to apoptosis | + [380] |
0 | + [378] |
+ [377] |
+ [498] |
+ [382] |
+ [379] |
| Replicative immortality | + [369], [370], [374] |
0 | + [182] |
+ [369], [372], [373] |
+ [368], [499] |
+ [375], [376] |
+ [364], [371] |
| Dysregulated metabolism | 0 | 0 | 0 | + [500] |
0 | + [268], [501], [502], [503], [504], [505], [506], [507], [508] |
+ [364], [383], [509] |
| Immune system invasion | + [406] |
0 | 0 | + [405] |
0 | + [407], [510], [511], [512], [513] |
0 |
| Angiogenesis | + [514] |
± [408], [409], [410], [412], [414] |
± [411], [413], [415], [417] |
+ [416] |
+ [418] |
+ [515], [516] |
+ [419] |
| Tissue invasion and metastasis | + [517] |
+ [425], [426] |
+ [351], [427], [428] |
+ [429], [431], [518] |
+ [484] |
+ [519] |
+ [363], [420], [430] |
| Tumor microenvironment | + [520] |
0 |
+ [395] |
+ [521] |
+ [476] |
+ [522] |
+ [523] |
Approaches that are not only relevant for genomic instability, but also relevant for other aspects of cancer's biology were noted as having complementary effects (+). Those approaches that were found to have procarcinogenic actions were noted as having contrary effects (−). In instances where reports on relevant actions in other aspects of cancer biology were mixed, the designation (±) was used. Finally, we indicate (0) in instances where no literature support was found to document the relevance of an approach in a particular aspect of cancer's biology.
During cancer formation, genetic instability interacts with many other pathways that are integral to the survival and proliferation of the cancer cells, such as inflammation, immune system evasion, or apoptosis resistance. Preventing and/or treating genomic instability can cause tumor cells to lose: (1) their replicative immortality; (2) their ability to evade the immune system; and/or, (3) their ability to evade antigrowth signaling (Table 1). For example, in MSI-H CRCs, the immune response can be evaded by mutations in the neoantigens caused by defects in DNA repair machinery [352]. In this case, by preventing genomic instability, it could be possible to minimize the mutations that lead to immune system evasion.
Sustained proliferative signaling is required for cancer cell growth and vitamin D and resveratrol are able to inhibit this signaling [353], [354], [355]. There are no known interactions for the other compounds, except selenium, which inhibits growth in some cases while inducing it in others [356], [357]. In a related characteristic of cancer cells, evasion of antigrowth signaling, all compounds are able to inhibit growth except vitamin B, which shows no relationship, and carotenoids, which have mixed results [255], [358], [359], [360], [361], [362], [363], [364], [365], [366]. Similarly, all of the compounds are able to prevent replicative immortality by impairing telomerase activity or inducing senescence [364], [367], [368], [369], [370], [371], [372], [373], [374], [375], [376] and increasing cell death except for vitamin B, which has no known relationship to apoptosis [377], [378], [379], [380], [381], [382]. Dysregulated metabolism also contributes to cancer cell growth and all of the compounds had complementary effects on metabolic pathways except vitamin D and PARP inhibitors, which have no known link [268], [338], [364], [383], [384], [385], [386].
Cancer cells use inflammatory agents in the microenvironment to promote their proliferation and survival. One important inflammatory signaling molecule is nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB), a transcription factor whose aberrant regulation has been linked to cancer [387], [388]. Inflammatory signaling, including that of NFκB, can be affected by the diet. It has been shown that compounds found in cruciferous vegetables can reduce NFκB signaling in pancreatic cancer cells [389]. Polyphenols have also been shown to suppress transcription factors upstream of NFκB [390]. Inflammation, in general, is also inhibited by all of the compounds, except vitamin B [354], [391], [392], [393], [394], [395], [396], [397], [398], [399], [400], [401], [402], [403], [404], while only vitamin D, carotenoids, and resveratrol prevent the tumor cells from evading the immune system [405], [406], [407].
Tumors need specialized environments to grow and thrive in. As the tumor grows, new blood vessels need to form to provide the cells with oxygen and all of the treatment options selected are able to inhibit angiogenesis, except vitamin B and selenium, both of which show mixed results [408], [409], [410], [411], [412], [413], [414], [415], [416], [417], [418], [419]. Interestingly though, in regards to other factors that contribute to the tumor microenvironment, all compounds are able to provide therapeutic value [395], [420], [421], [422], [423], [424].
Effective treatments to prevent tissue invasion and metastasis are important as these stages of cancer are associated with poor outcomes. It has been found that all of the targeted treatments are able to inhibit/prevent these pathways except for resveratrol and PARP inhibitors, which have no known relationship [363], [420], [421], [425], [426], [427], [428], [429], [430], [431]. Further work and clinical trials will have to be performed to understand the full benefit of these compounds in regards to cancer treatment.
6. Conclusion
Genomic instability plays a critical role in cancer initiation and progression. The fidelity of the genome is protected at every stage of the cell cycle. In cancer, the presence of aneuploid or tetraploid cells indicates the failure of one or many of these safety nets. The resultant genomic heterogeneity may offer the cancer “tissue” a selection advantage against standard of care and emerging therapies. Understanding these safety nets, and how they are bypassed in cancer cells, may highlight new and more specific mechanisms for cancer prevention or therapeutic attack.
The therapeutic targeting of genomic instability may dampen other enabling characteristic of tumors cells, such as replicative immortality, evasion of antigrowth signaling, and tumor promoting inflammation. To this end, vitamins, minerals, and antioxidants, such as vitamin D, vitamin B, selenium, and carotenoids, as well as nutraceuticals, such as resveratrol, have shown remarkable plasticity in elucidating antitumor responses. In addition to alleviating genomic instability, these compounds are known to inhibit proliferative signaling [353], [354], [355], attenuate oncogenic metabolism [268], [338], [364], [383], [384], [385], [386], and block inflammation [354], [391], [392], [393], [394], [395], [396], [397], [398], [399], [400], [401], [402], [403], [404]. However, caution must be applied as certain antioxidants, such β carotene, may promote carcinogenic processes in a dose- and context-dependent manner.
While mortality rates associated with heart disease and stroke have been reduced ∼70% in the last 50 years, mortality rates associated with cancer remain largely unchanged. This is likely due to our ability to manage the risk factors for heart disease and stroke and our inability to detect and prevent genomic instability and cancer. However, diet and lifestyle are two of our great hopes in this area. In particular, antioxidants are critical for the prevention of DNA damage that enables cancer initiation and growth. Growing evidence shows that vitamins, minerals, and other dietary factors have profound and protective effects against cancer cells, whether they are grown in the lab, in animals, or studied in human populations. A better understanding of the effects and synergy of these dietary factors in the prevention and treatment of genomic instability is critical to the future reduction of mortality associated with cancer.
Author's contributions
The following authors composed the indicated sections: HC, CAM, AKM (Mechanisms underlying genomic instability), PTT (epigenetic mechanisms), SD and DS (Mitochondrial genetics), GD, ESY, SR (DNA repair pathways), ARC and LRF (cancer prevention), SP and MM (targeting with RNAi). AA, AA, SSA, KA, ASA, DB, AB, CSB, SC, MC, MRC, HJ, GG, DH, BH, WNK, SM, EN, XY and KH (cross validation in Table 1, Table 2). LRF, MC, and CAM integrated and edited the sections.
Conflict of interest statement
The following authors declare that there are no conflicts of interest: LRF, HC, ARC, MC, GD, SD, MM, AKM, AA, AA, SSA, KA, ASA, DB, AB, CSB, SC, MRC, HF, GG, DH, WGH, WNK, SIM, EN, XY, KH, VRP, PR, SR, RS, DS, PTT, and CAM. ESY has material transfer agreements with AbbVie, Eli Lilly, Bristol Myers Squibb, and Cerion NRx.
Acknowledgments
The following funding agencies supported the research: Italian Ministry of University and Research and the University of Florence (A. Amedia); Terry Fox Foundation, UAEU Program for Advanced Research, Al-Jalila Foundation and Zayed Center for Health Sciences (A. Amin); Italia Ministry of Education, University and Research- Miur, PRIN 20125S38FA_002 (K. Aquilano); University of Glasgow, Beatson Oncology Centre Fund, CRUK grant C301/A14762 (A. Bilsland and W.N. Keith); Child and Family Research Institute PhD studentship (H. Chen); Michael Cuccione childhood cancer research foundation fellowship (M. Connell); Ovarian Prostate Cancer Research Trust Laboratory (S. Chen); University of Oslo (A.R. Collins); Italian Association for Cancer Research AIRC IG 10636 (M.R. Ciriolo); Elsa U Pardee Foundation (S. Dasgupta); Auckland Cancer Society Research Centre, University of Auckland (L. Ferguson and V. Parslow); National Institutes of Health (H.D. Halicka); Ministry of Education, Culture, Sports, Science and Technology, Japan, No. 24590493 (K. Honoki); CIHR, in partnership with the Avon Foundation for Women, OBC 134038 (C.A. Maxwell); NIH/NCI RO1CA172380, DOD W81WH1210545, DOD W81XWH12PCRPTIA, NIH/NCI P30CA006973, NIH/NCI RCI RFA0D09009, NIH/NCI 5R01 CA132996-03 (A. Meeker); Italian Ministry of University and University of Italy (E. Niccolai); CIHR MOP 64308 (S. Prakash); Department of Biology, University of Rochester (S. Rezazadeh); Patrick C. Walsh Prostate Cancer Research Fund, the Department of Defense (W81XWH-11-1-0272 and W81XWH-13-1-0182), Commonwealth Foundation, Uniting Against Lung Cancer, a Sidney Kimmel Translational Scholar award (SKF-13-021), an ACS Scholar award (122688-RSG-12-196-01-TBG) and the NIH (1R01CA166348) (P.T. Tran); Susan G. Komen Foundation, AACR, Gabrielle's Angel Foundation, Eli Lilly, and Bristol Myers Squibb (E.S. Yang).
Contributor Information
Lynnette R. Ferguson, Email: l.ferguson@auckland.ac.nz.
Christopher A. Maxwell, Email: cmaxwell@cfri.ca.
References
- 1.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller H.J. The remaking of chromosomes. Collecting Net. 1938;13:181–198. [Google Scholar]
- 3.Wong K.K., DePinho R.A. Walking the telomere plank into cancer. J Natl Cancer Inst. 2003;95:1184–1186. doi: 10.1093/jnci/djg028. [DOI] [PubMed] [Google Scholar]
- 4.Wright W.E., Shay J.W. Telomere biology in aging and cancer. J Am Geriatr Soc. 2005;53:S292–S294. doi: 10.1111/j.1532-5415.2005.53492.x. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E.H. Telomeres and telomerase. Keio J Med. 2000;49:59–65. doi: 10.2302/kjm.49.59. [DOI] [PubMed] [Google Scholar]
- 6.Greider C.W. Telomeres. Curr Opin Cell Biol. 1991;3:444–451. doi: 10.1016/0955-0674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- 7.Konishi A., de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlseder J., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hockemeyer D., Sfeir A.J., Shay J.W., Wright W.E., de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol. 2010;75:167–177. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- 11.Watson J.D. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 12.Olovnikov A.M. Principle of marginotomy in template synthesis of polynucleotides. Dokl Akad Nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- 13.Harley C.B. Telomere loss: mitotic clock or genetic time bomb. Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 14.Levy M.Z., Allsopp R.C., Futcher A.B., Greider C.W., Harley C.B. Telomere end-replication problem and cell aging. J Mol Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 15.Hastie N.D., Dempster M., Dunlop M.G., Thompson A.M., Green D.K., Allshire R.C. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 16.Aubert G., Lansdorp P.M. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 17.Baerlocher G.M., Lansdorp P.M. Telomere length measurements in leukocyte subsets by automated multicolor flow-FISH. Cytom A: J Int Soc Anal Cytol. 2003;55:1–6. doi: 10.1002/cyto.a.10064. [DOI] [PubMed] [Google Scholar]
- 18.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 19.Hayflick L., Moorhead P.S. The serial cultivation of human diploid strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 20.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 21.Campisi J. Cancer aging and cellular senescence. In Vivo. 2000;14:183–188. [PubMed] [Google Scholar]
- 22.Harley C.B., Sherwood S.W. Telomerase, checkpoints and cancer. Cancer Surv. 1997;29:263–284. [PubMed] [Google Scholar]
- 23.Meeker A.K., Hicks J.L., Platz E.A., March G.E., Bennett C.J., Delannoy M.J. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–6409. [PubMed] [Google Scholar]
- 24.Meeker A.K., Argani P. Telomere shortening occurs early during breast tumorigenesis: a cause of chromosome destabilization underlying malignant transformation. J Mammary Gland Biol Neoplasia. 2004;9:285–296. doi: 10.1023/B:JOMG.0000048775.04140.92. [DOI] [PubMed] [Google Scholar]
- 25.Meeker A.K., Hicks J.L., Iacobuzio-Donahue C.A., Montgomery E.A., Westra W.H., Chan T.Y. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin Cancer Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 26.van Heek N.T., Meeker A.K., Kern S.E., Yeo C.J., Lillemoe K.D., Cameron J.L. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 28.Raynaud C.M., Jang S.J., Nuciforo P., Lantuejoul S., Brambilla E., Mounier N. Telomere shortening is correlated with the DNA damage response and telomeric protein down-regulation in colorectal preneoplastic lesions. Ann Oncol. 2008;19:1875–1881. doi: 10.1093/annonc/mdn405. [DOI] [PubMed] [Google Scholar]
- 29.Smith J.K., Yeh G. Telomere reduction in endometrial adenocarcinoma. Am J Obstet Gynecol. 1992;167:1883–1887. doi: 10.1016/0002-9378(92)91791-8. [DOI] [PubMed] [Google Scholar]
- 30.Hiyama E., Hiyama K., Yokoyama T., Ichikawa T., Matsuura Y. Length of telomeric repeats in neuroblastoma: correlation with prognosis and other biological characteristics. Jpn J Cancer Res. 1992;83:159–164. doi: 10.1111/j.1349-7006.1992.tb00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehle C., Ljungberg B., Roos G. Telomere shortening in renal cell carcinoma. Cancer Res. 1994;54:236–241. [PubMed] [Google Scholar]
- 32.Counter C.M., Hirte H.W., Bacchetti S., Harley C.B. Telomerase activity in human ovarian carcinoma. Proc Natl Acad Sci U S A. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Artandi S.E., Chang S., Lee S.L., Alson S., Gottlieb G.J., Chin L. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 34.O’Hagan R.C., Chang S., Maser R.S., Mohan R., Artandi S.E., Chin L. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 35.O'Sullivan J.N., Bronner M.P., Brentnall T.A., Finley J.C., Shen W.T., Emerson S. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32:280–284. doi: 10.1038/ng989. [DOI] [PubMed] [Google Scholar]
- 36.Davoli T., Denchi E.L., de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudolph K.L., Millard M., Bosenberg M.W., DePinho R.A. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 38.Wong K.K., Maser R.S., Bachoo R.M., Menon J., Carrasco D.R., Gu Y. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- 39.Farazi P.A., Glickman J., Jiang S., Yu A., Rudolph K.L., DePinho R.A. Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res. 2003;63:5021–5027. [PubMed] [Google Scholar]
- 40.Artandi S.E., DePinho R.A. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr Opin Genet Dev. 2000;10:39–46. doi: 10.1016/s0959-437x(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 41.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 42.Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–336. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 43.Morin G.B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 44.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 45.Avilion A.A., Piatyszek M.A., Gupta J., Shay J.W., Bacchetti S., Greider C.W. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645–650. [PubMed] [Google Scholar]
- 46.Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 47.Shay J.W., Reddel R.R., Wright W.E. Cancer. Cancer and telomeres – an ALTernative to telomerase. Science. 2012;336:1388–1390. doi: 10.1126/science.1222394. [DOI] [PubMed] [Google Scholar]
- 48.Heaphy C.M., Subhawong A.P., Hong S.M., Goggins M.G., Montgomery E.A., Gabrielson E. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulaner G.A., Huang H.Y., Otero J., Zhao Z., Ben-Porat L., Satagopan J.M. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. 2003;63:1759–1763. [PubMed] [Google Scholar]
- 50.Costa A., Daidone M.G., Daprai L., Villa R., Cantu S., Pilotti S. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. 2006;66:8918–8924. doi: 10.1158/0008-5472.CAN-06-0273. [DOI] [PubMed] [Google Scholar]
- 51.Venturini L., Daidone M.G., Motta R., Cimino-Reale G., Hoare S.F., Gronchi A. Telomere maintenance mechanisms in malignant peripheral nerve sheath tumors: expression and prognostic relevance. Neuro Oncol. 2012;14:736–744. doi: 10.1093/neuonc/nos083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald K.L., McDonnell J., Muntoni A., Henson J.D., Hegi M.E., von Deimling A. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J Neuropathol Exp Neurol. 2010;69:729–736. doi: 10.1097/NEN.0b013e3181e576cf. [DOI] [PubMed] [Google Scholar]
- 53.Doxsey S. Re-evaluating centrosome function. Nat Rev Mol Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- 54.Nigg E.A. Centrosome aberrations: cause or consequence of cancer progression. Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 55.Nigg E.A., Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarapore P., Fukasawa K. Loss of p53 and centrosome hyperamplification. Oncogene. 2002;21:6234–6240. doi: 10.1038/sj.onc.1205707. [DOI] [PubMed] [Google Scholar]
- 57.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 58.Ko M.A., Rosario C.O., Hudson J.W., Kulkarni S., Pollett A., Dennis J.W. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 59.Pellegrino R., Calvisi D.F., Ladu S., Ehemann V., Staniscia T., Evert M. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51:857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
- 60.Khodjakov A. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.La Terra S., English C.N., Hergert P., McEwen B.F., Sluder G., Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peel N., Stevens N., Basto R., Raff J. Overexpression centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glover D.M., Leibowitz M.H., McLean D.A., Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;891:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 64.Haren L., Gnadt N., Wright M., Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res Notes. 2009;2:64. doi: 10.1186/1756-0500-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luca M.D., Lavia P., Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at the spindle poles. Cell Cycle. 2006;5 doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- 66.Maxwell C.A., Keats J.J., Belch A.R., Pilarski L.M., Reiman T. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myelomana nd maintains mitotic integrity. Cancer Res. 2005;56:850–860. [PubMed] [Google Scholar]
- 67.Maxwell C.A., Keats J.J., Crainie M., Sun X., Yen T., Shibuya E. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol Biol Cell. 2003;14:2262–2276. doi: 10.1091/mbc.E02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merdes A., Ramyar K., Vechio D., Cleveland D.W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 69.Silk A.D., Holland A.J., Cleveland D.W. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J Cell Biol. 2009;184:677–690. doi: 10.1083/jcb.200810091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kramer A., Maier B., Bartek J. Centrosome clustering and chromosomal (in)stability: a matter of life and death. Mol Oncol. 2011;5:324–335. doi: 10.1016/j.molonc.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogden A., Rida P.C., Aneja R. Let's huddle to prevent a muddle: centrosome declustering as an attractive anticancer strategy. Cell Death Differ. 2012;19:1255–1267. doi: 10.1038/cdd.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quintyne N.J., Reing J.E., Hoffelder D.R., Gollin S.M., Saunders W.S. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 73.Birkbak N.J., Eklund A.C., Li Q., McClelland S.E., Endesfelder D., Tan P. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 75.Gergely F., Basto R. Multiple centrosomes: together they stand, divided they fall. Genes Dev. 2008;22:2291–2296. doi: 10.1101/gad.1715208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marthien V., Piel M., Basto R. Never tear us apart – the importance of centrosome clustering. J Cell Sci. 2012;125:3281–3292. doi: 10.1242/jcs.094797. [DOI] [PubMed] [Google Scholar]
- 77.Kwon M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 79.Foley E.A., Kapoor T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lens S.M., Voest E.E., Medema R.H. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 81.Nguyen H.G., Makitalo M., Yang D., Chinnappan D., St Hilaire C., Ravid K. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. FASEB J. 2009;23:2741–2748. doi: 10.1096/fj.09-130963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan L.Y., Amon A. The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev. 2009;23:1639–1649. doi: 10.1101/gad.1804609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang X., Zhang R. Aneuploidy and tumourigenesis. Semin Cell Dev Biol. 2011;22:595–601. doi: 10.1016/j.semcdb.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grabsch H., Takeno S., Parsons W.J., Pomjanski N., Boecking A., Gabbert H.E. Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer – association with tumour cell proliferation. J Pathol. 2003;200:16–22. doi: 10.1002/path.1324. [DOI] [PubMed] [Google Scholar]
- 85.Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 86.Anand P., Lowe V. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 87.Hardy P.A., Zacharias H. Reappraisal of the Hansemann–Boveri hypothesis on the origin of tumors. Cell Biol Int. 2005;29:983–992. doi: 10.1016/j.cellbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 88.Shackney S.E., Smith C.A., Miller B.W., Burholt D.R., Murtha K., Giles H.R. Model for the genetic evolution of human solid tumors. Cancer Res. 1989;49:3344–3354. [PubMed] [Google Scholar]
- 89.Andreassen P.R., Lohez O.D., Lacroix F.B., Margolis R.L. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borel F., Lohez O.D., Lacroix F.B., Margolis R.L. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci U S A. 2002;99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lanni J.S., Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18 doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Livingstone L.R., White A., Sprouse J., Livanos E., Jacks T., Tlsty T.D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 93.Galipeau P.C., Cowan D.S., Sanchez C.A., Barretr M.T., Emondt M.J., Levinet D.S. Progression to aneuploidy in Barrett’ s esophagus. Cell. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujiwara T., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 95.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berger S.L., Kouzarides T., Shiekhattar R., Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nowsheen S., Aziz K., Tran P.T., Gorgoulis V.G., Yang E.S., Georgakilas A.G. Epigenetic inactivation of DNA repair in breast cancer. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 98.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 99.Robertson K.D. DNA methylation and chromatin – unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 100.Das P.M., Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 101.Baylin S.B., Esteller M., Rountree M.R., Bachman K.E., Schuebel K., Herman J.G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 102.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 103.Robertson K.D. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 104.Baylin S.B., Ohm J.E. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 105.Felsher D.W., Bishop J.M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Denko N.C., Giaccia A.J., Stringer J.R., Stambrook P.J. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci. 1994;91:5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin B., Robertson K.D. DNA methyltransferases, DNA damage repair and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hitchins M.P. Inheritance of epigenetic aberrations (constitutional epimutations) in cancer susceptibility. Adv Genet. 2010;70:201–243. doi: 10.1016/B978-0-12-380866-0.60008-3. [DOI] [PubMed] [Google Scholar]
- 109.Goel A., Nagasaka T., Arnold C.N., Inoue T., Hamilton C., Niedzwiecki D. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 110.Sproul D., Gilbert N., Bickmore W.A. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- 111.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 112.Turner B.M. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 113.Gorrini C., Squatrito M., Luise C., Syed N., Perna D., Wark L. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448 doi: 10.1038/nature06055. 1063-U12. [DOI] [PubMed] [Google Scholar]
- 114.Tang Y., Luo J.Y., Zhang W.Z., Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 115.Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugarna R. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun Y.L., Jiang X.F., Chen S.J., Fernandes N., Price B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xie W., Song C.Y., Young N.L., Sperling A.S., Xu F., Sridharan R. Histone H3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Das C., Lucia M.S., Hansen K.C., Tyler J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459 doi: 10.1038/nature07861. 113-U23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Osley M.A. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomic Proteomic. 2006;5:179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- 120.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 121.Huen M.S.Y., Grant R., Manke I., Minn K., Yu X.C., Yaffe M.B. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kao C.F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M.A. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turner S.D., Ricci A.R., Petropoulos H., Genereaux J., Skerjanc I.S., Brandl C.J. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol Cell Biol. 2002;22:4011–4019. doi: 10.1128/MCB.22.12.4011-4019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luijsterburg M.S., Goedhart J., Moser J., Kool H., Geverts B., Houtsmuller A.B. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J Cell Sci. 2007;120:2706–2716. doi: 10.1242/jcs.008367. [DOI] [PubMed] [Google Scholar]
- 125.Nicassio F., Corrado N., Vissers J.H.A., Areces L.B., Bergink S., Marteijn J.A. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 126.Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.Y. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 128.Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S. OPINION gamma H2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 130.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 132.Chatterjee A., Dasgupta S., Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila) 2011;4:638–654. doi: 10.1158/1940-6207.CAPR-10-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dasgupta S., Shao C., Keane T.E., Duberow D.P., Mathies R.A., Fisher P.B. Detection of mitochondrial deoxyribonucleic acid alterations in urine from urothelial cell carcinoma patients. Int J Cancer. 2012;131:158–164. doi: 10.1002/ijc.26357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dasgupta S., Soudry E., Mukhopadhyay N., Shao C., Yee J., Lam S. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol. 2012;227:2451–2460. doi: 10.1002/jcp.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kloss-Brandstatter A., Schafer G., Erhart G., Huttenhofer A., Coassin S., Seifarth C. Somatic mutations throughout the entire mitochondrial genome are associated with elevated PSA levels in prostate cancer patients. Am J Hum Genet. 2010;87:802–812. doi: 10.1016/j.ajhg.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Petros J.A., Baumann A.K., Ruiz-Pesini E., Amin M.B., Sun C.Q., Hall J. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dasgupta S., Yung R.C., Westra W.H., Rini D.A., Brandes J., Sidransky D. Following mitochondrial footprints through a long mucosal path to lung cancer. PLoS ONE. 2009;4:e6533. doi: 10.1371/journal.pone.0006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dasgupta S., Koch R., Westra W.H., Califano J.A., Ha P.K., Sidransky D. Mitochondrial DNA mutation in normal margins and tumors of recurrent head and neck squamous cell carcinoma patients. Cancer Prev Res (Phila) 2010;3:1205–1211. doi: 10.1158/1940-6207.CAPR-10-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cook C.C., Kim A., Terao S., Gotoh A., Higuchi M. Consumption of oxygen: a mitochondrial-generated progression signal of advanced cancer. Cell Death Dis. 2012;3:e258. doi: 10.1038/cddis.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arnold R.S., Sun C.Q., Richards J.C., Grigoriev G., Coleman I.M., Nelson P.S. Mitochondrial DNA mutation stimulates prostate cancer growth in bone stromal environment. Prostate. 2009;69:1–11. doi: 10.1002/pros.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Arnold R.S., Sun Q., Sun C.Q., Richards J.C., O’Hearn S., Osunkoya A.O. An inherited heteroplasmic mutation in mitochondrial gene COI in a patient with prostate cancer alters reactive oxygen, reactive nitrogen and proliferation. Biomed Res Int. 2013;2013:239257. doi: 10.1155/2013/239257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dasgupta S., Hoque M.O., Upadhyay S., Sidransky D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008;68:700–706. doi: 10.1158/0008-5472.CAN-07-5532. [DOI] [PubMed] [Google Scholar]
- 143.Aguilera A., Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 144.Houtgraaf J.H., Versmissen J., van der Giessen W.J. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7:165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 145.Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 146.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 147.Wei K., Kucherlapati R., Edelmann W. Mouse models for human DNA mismatch-repair gene defects. Trends Mol Med. 2002;8:346–353. doi: 10.1016/s1471-4914(02)02359-6. [DOI] [PubMed] [Google Scholar]
- 148.Taketo M.M., Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136:780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 149.Hollander M.C., Philburn R.T., Patterson A.D., Velasco-Miguel S., Friedberg E.C., Linnoila R.I. Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13200–13205. doi: 10.1073/pnas.0503133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thoms K.M., Kuschal C., Emmert S. Lessons learned from DNA repair defective syndromes. Exp Dermatol. 2007;16:532–544. doi: 10.1111/j.1600-0625.2007.00559.x. [DOI] [PubMed] [Google Scholar]
- 151.Maynard S., Schurman S.H., Harboe C., de Souza-Pinto N.C., Bohr V.A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rouleau M., Patel A., Hendzel M.J., Kaufmann S.H., Poirier G.G. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hassa P.O., Hottiger M.O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 154.Larsen E., Gran C., Saether B.E., Seeberg E., Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sobol R.W., Horton J.K., Kuhn R., Gu H., Singhal R.K., Prasad R. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 156.Tebbs R.S., Flannery M.L., Meneses J.J., Hartmann A., Tucker J.D., Thompson L.H. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 157.Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T. Inherited variants of MYH associated with somatic G:C → T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 158.Farrington S.M., Tenesa A., Barnetson R., Wiltshire A., Prendergast J., Porteous M. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am J Hum Genet. 2005;77:112–119. doi: 10.1086/431213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tenesa A., Farrington S.M., Dunlop M.G. Re: association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst. 2005;97:320–321. doi: 10.1093/jnci/dji051. author reply 1–2. [DOI] [PubMed] [Google Scholar]
- 160.Barnetson R.A., Devlin L., Miller J., Farrington S.M., Slater S., Drake A.C. Germline mutation prevalence in the base excision repair gene, MYH, in patients with endometrial cancer. Clin Genet. 2007;72:551–555. doi: 10.1111/j.1399-0004.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 161.Mahjabeen I., Masood N., Baig R.M., Sabir M., Inayat U., Malik F.A. Novel mutations of OGG1 base excision repair pathway gene in laryngeal cancer patients. Fam Cancer. 2012;11:587–593. doi: 10.1007/s10689-012-9554-2. [DOI] [PubMed] [Google Scholar]
- 162.Shinmura K., Tao H., Goto M., Igarashi H., Taniguchi T., Maekawa M. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25:2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 163.Kunkel T.A. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Morrison A., Johnson A.L., Johnston L.H., Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Albertson T.M., Ogawa M., Bugni J.M., Hays L.E., Chen Y., Wang Y. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kolodner R.D., Marsischky G.T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 167.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Helleday T., Lo J., van Gent D.C., Engelward B.P. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Rep. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 169.Moynahan M.E., Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Willers H., Pfäffle H.N., Zou L. In: Targeting homologous recombination repair in cancer: molecular targets and clinical applications. Kelley M.R., editor. Academic Press, Elsevier; 2011. pp. 119–160. [Google Scholar]
- 171.Riha K., Heacock M.L., Shippen D.E. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 172.De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 173.McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci U S A. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hall J.M., Lee M.K., Newman B., Morrow J.E., Anderson L.A., Huey B. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 175.Friedman L.S., Ostermeyer E.A., Szabo C.I., Dowd P., Lynch E.D., Rowell S.E. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 176.Castilla L.H., Couch F.J., Erdos M.R., Hoskins K.F., Calzone K., Garber J.E. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nat Genet. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 177.Futreal P.A., Liu Q., Shattuck-Eidens D., Cochran C., Harshman K., Tavtigian S. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 178.Ford D., Easton D.F., Bishop D.T., Narod S.A., Goldgar D.E. Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet. 1994;343:692–695. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- 179.Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 180.Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 181.Collins N., McManus R., Wooster R., Mangion J., Seal S., Lakhani S.R. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- 182.Chen S., Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Friedenson B. BRCA1 and BRCA2 pathways and the risk of cancers other than breast or ovarian. MedGenMed. 2005;7:60. [PMC free article] [PubMed] [Google Scholar]
- 184.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 185.Tischkowitz M., Capanu M., Sabbaghian N., Li L., Liang X., Vallee M.P. Rare germline mutations in PALB2 and breast cancer risk: a population-based study. Hum Mutat. 2012;33:674–680. doi: 10.1002/humu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rahman N., Seal S., Thompson D., Kelly P., Renwick A., Elliott A. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zheng Y., Zhang J., Niu Q., Huo D., Olopade O.I. Novel germline PALB2 truncating mutations in African American breast cancer patients. Cancer. 2012;118:1362–1370. doi: 10.1002/cncr.26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ding Y.C., Steele L., Chu L.H., Kelley K., Davis H., John E.M. Germline mutations in PALB2 in African-American breast cancer cases. Breast Cancer Res Treat. 2011;126:227–230. doi: 10.1007/s10549-010-1271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cao A.Y., Huang J., Hu Z., Li W.F., Ma Z.L., Tang L.L. The prevalence of PALB2 germline mutations in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res Treat. 2009;114:457–462. doi: 10.1007/s10549-008-0036-z. [DOI] [PubMed] [Google Scholar]
- 190.Jones S., Hruban R.H., Kamiyama M., Borges M., Zhang X., Parsons D.W. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tischkowitz M.D., Sabbaghian N., Hamel N., Borgida A., Rosner C., Taherian N. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Slater E.P., Langer P., Niemczyk E., Strauch K., Butler J., Habbe N. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 193.Kee Y., D’Andrea A.D. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.D’Andrea A.D. Susceptibility pathways in Fanconi's anemia and breast cancer. New Engl J Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tischkowitz M., Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70:7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Reid S., Schindler D., Hanenberg H., Barker K., Hanks S., Kalb R. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 197.Chirnomas D., Taniguchi T., de la Vega M., Vaidya A.P., Vasserman M., Hartman A.R. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol Cancer Ther. 2006;5:952–961. doi: 10.1158/1535-7163.MCT-05-0493. [DOI] [PubMed] [Google Scholar]
- 198.Alter B.P. Radiosensitivity in Fanconi's anemia patients. Radiother Oncol. 2002;62:345–347. doi: 10.1016/s0167-8140(01)00474-1. [DOI] [PubMed] [Google Scholar]
- 199.Otsuki T., Wang J., Demuth I., Digweed M., Liu J.M. Assessment of mitomycin C sensitivity in Fanconi anemia complementation group C gene (Fac) knock-out mouse cells. Int J Hematol. 1998;67:243–248. doi: 10.1016/s0925-5710(98)00012-7. [DOI] [PubMed] [Google Scholar]
- 200.Stracker T.H., Roig I., Knobel P.A., Marjanovic M. The ATM signaling network in development and disease. Front Genet. 2013;4:37. doi: 10.3389/fgene.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kobayashi H., Ohno S., Sasaki Y., Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (review) Oncol Rep. 2013 doi: 10.3892/or.2013.2541. [DOI] [PubMed] [Google Scholar]
- 202.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 203.Sajesh B.V., Guppy B.J., McManus K.J. Synthetic genetic targeting of genome instability in cancer. Cancers. 2013;5:739–761. doi: 10.3390/cancers5030739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 205.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 206.Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. New Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 207.Fong P.C., Yap T.A., Boss D.S., Carden C.P., Mergui-Roelvink M., Gourley C. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 208.Irshad S., Ashworth A., Tutt A. Therapeutic potential of PARP inhibitors for metastatic breast cancer. Expert Rev Anticancer Ther. 2011;11:1243–1251. doi: 10.1586/era.11.52. [DOI] [PubMed] [Google Scholar]
- 209.Sessa C. Update on PARP1 inhibitors in ovarian cancer. Ann Oncol. 2011;22(Suppl. 8):viii72–viii76. doi: 10.1093/annonc/mdr528. [DOI] [PubMed] [Google Scholar]
- 210.Telli M.L., Ford J.M. PARP inhibitors in breast cancer. Clin Adv Hematol Oncol. 2010;8:629–635. [PubMed] [Google Scholar]
- 211.Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 212.Fowke J.H., Gao Y.T., Chow W.H., Cai Q., Shu X.O., Li H.L. Urinary isothiocyanate levels and lung cancer risk among non-smoking women: a prospective investigation. Lung Cancer. 2011;73:18–24. doi: 10.1016/j.lungcan.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Grotsky D.A., Gonzalez-Suarez I., Novell A., Neumann M.A., Yaddanapudi S.C., Croke M. BRCA1 loss activates cathepsin l-mediated degradation of 53BP1 in breast cancer cells. J Cell Biol. 2013;200:187–202. doi: 10.1083/jcb.201204053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen T., Wong Y.S. Selenocystine induces caspase-independent apoptosis in MCF-7 human breast carcinoma cells with involvement of p53 phosphorylation and reactive oxygen species generation. Int J Biochem Cell Biol. 2009;41:666–676. doi: 10.1016/j.biocel.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 215.Taniguchi T., Tischkowitz M., Ameziane N., Hodgson S.V., Mathew C.G., Joenje H. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 216.Swisher E.M., Sakai W., Karlan B.Y., Wurz K., Urban N., Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tennstedt P., Fresow R., Simon R., Marx A., Terracciano L., Petersen C. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int J Cancer. 2013;132:2118–2126. doi: 10.1002/ijc.27907. [DOI] [PubMed] [Google Scholar]
- 218.Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 219.Dent P., Tang Y., Yacoub A., Dai Y., Fisher P.B., Grant S. CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Mol Interv. 2011;11:133–140. doi: 10.1124/mi.11.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Poulsen M.M., Jorgensen J.O., Jessen N., Richelsen B., Pedersen S.B. Resveratrol in metabolic health: an overview of the current evidence and perspectives. Ann N Y Acad Sci. 2013;1290:74–82. doi: 10.1111/nyas.12141. [DOI] [PubMed] [Google Scholar]
- 221.Wang R.H., Zheng Y., Kim H.S., Xu X., Cao L., Luhasen T. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hoeijmakers J.H. DNA damage, aging, and cancer. New Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 223.Imai K., Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 224.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 225.Vilar E., Gruber S.B. Microsatellite instability in colorectal cancer – the stable evidence. Nat Rev Clin Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Popat S., Hubner R., Houlston R.S. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 227.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Papadopoulos N., Nicolaides N.C., Wei Y.F., Ruben S.M., Carter K.C., Rosen C.A. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 229.Nicolaides N.C., Papadopoulos N., Liu B., Wei Y.F., Carter K.C., Ruben S.M. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 230.Bronner C.E., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 231.Leach F.S., Nicolaides N.C., Papadopoulos N., Liu B., Jen J., Parsons R. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 232.Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1994;77 1 p. following 166. [PubMed] [Google Scholar]
- 233.Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cunningham J.M., Christensen E.R., Tester D.J., Kim C.Y., Roche P.C., Burgart L.J. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 235.Kakar S., Burgart L.J., Thibodeau S.N., Rabe K.G., Petersen G.M., Goldberg R.M. Frequency of loss of hMLH1 expression in colorectal carcinoma increases with advancing age. Cancer. 2003;97:1421–1427. doi: 10.1002/cncr.11206. [DOI] [PubMed] [Google Scholar]
- 236.Baylin S.B., Herman J.G., Graff J.R., Vertino P.M., Issa J.P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 237.Pfeifer G.P., Steigerwald S.D., Hansen R.S., Gartler S.M., Riggs A.D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wachsman J.T. DNA methylation and the association between genetic and epigenetic changes: relation to carcinogenesis. Mutat Res. 1997;375:1–8. doi: 10.1016/s0027-5107(97)00003-1. [DOI] [PubMed] [Google Scholar]
- 239.Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Ruschoff J. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pawlik T.M., Raut C.P., Rodriguez-Bigas M.A. Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers. 2004;20:199–206. doi: 10.1155/2004/368680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Dietmaier W., Wallinger S., Bocker T., Kullmann F., Fishel R., Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 242.Stone J.G., Tomlinson I.P., Houlston R.S. Optimising methods for determining RER status in colorectal cancers. Cancer Lett. 2000;149:15–20. doi: 10.1016/s0304-3835(99)00324-9. [DOI] [PubMed] [Google Scholar]
- 243.Zhou X.P., Hoang J.M., Cottu P., Thomas G., Hamelin R. Allelic profiles of mononucleotide repeat microsatellites in control individuals and in colorectal tumors with and without replication errors. Oncogene. 1997;15:1713–1718. doi: 10.1038/sj.onc.1201337. [DOI] [PubMed] [Google Scholar]
- 244.Hoang J.M., Cottu P.H., Thuille B., Salmon R.J., Thomas G., Hamelin R. BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines. Cancer Res. 1997;57:300–303. [PubMed] [Google Scholar]
- 245.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 246.Gryfe R., Kim H., Hsieh E.T., Aronson M.D., Holowaty E.J., Bull S.B. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. New Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 247.Tomlinson I., Halford S., Aaltonen L., Hawkins N., Ward R. Does MSI-low exist? J Pathol. 2002;197:6–13. doi: 10.1002/path.1071. [DOI] [PubMed] [Google Scholar]
- 248.Watson M.M., Berg M., Soreide K. Prevalence and implications of elevated microsatellite alterations at selected tetranucleotides in cancer. Br J Cancer. 2014 doi: 10.1038/bjc.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Vilar E., Tabernero J. Molecular dissection of microsatellite instable colorectal cancer. Cancer Discov. 2013;3:502–511. doi: 10.1158/2159-8290.CD-12-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sargent D.J., Marsoni S., Monges G., Thibodeau S.N., Labianca R., Hamilton S.R. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sinicrope F.A., Foster N.R., Thibodeau S.N., Marsoni S., Monges G., Labianca R. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Martin S.A., McCabe N., Mullarkey M., Cummins R., Burgess D.J., Nakabeppu Y. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.van Pel D.M., Barrett I.J., Shimizu Y., Sajesh B.V., Guppy B.J., Pfeifer T. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLOS Genet. 2013;9:e1003254. doi: 10.1371/journal.pgen.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.McManus K.J., Barrett I.J., Nouhi Y., Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc Natl Acad Sci U S A. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Martin S.A., Hewish M., Sims D., Lord C.J., Ashworth A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res. 2011;71:1836–1848. doi: 10.1158/0008-5472.CAN-10-2836. [DOI] [PubMed] [Google Scholar]
- 256.Martin S.A., McCarthy A., Barber L.J., Burgess D.J., Parry S., Lord C.J. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO Mol Med. 2009;1:323–337. doi: 10.1002/emmm.200900040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Martin S.A., Lord C.J., Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res. 2010;16:5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- 258.Sajesh B.V., Bailey M., Lichtensztejn Z., Hieter P., McManus K.J. Synthetic lethal targeting of superoxide dismutase 1 selectively kills RAD54B-deficient colorectal cancer cells. Genetics. 2013;195:757–767. doi: 10.1534/genetics.113.156836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.McCabe N., Turner N.C., Lord C.J., Kluzek K., Bialkowska A., Swift S. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 260.Vilar E., Bartnik C.M., Stenzel S.L., Raskin L., Ahn J., Moreno V. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011;71:2632–2642. doi: 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Miquel C., Jacob S., Grandjouan S., Aime A., Viguier J., Sabourin J.C. Frequent alteration of DNA damage signalling and repair pathways in human colorectal cancers with microsatellite instability. Oncogene. 2007;26:5919–5926. doi: 10.1038/sj.onc.1210419. [DOI] [PubMed] [Google Scholar]
- 262.Phalon C., Rao D.D., Nemunaitis J. Potential use of RNA interference in cancer therapy. Expert Rev Mol Med. 2010:2010. doi: 10.1017/S1462399410001584. [DOI] [PubMed] [Google Scholar]
- 263.Liang Y., Gao H., Lin S.Y., Goss J.A., Brunicardi F.C., Li K. siRNA-based targeting of cyclin E overexpression inhibits breast cancer cell growth and suppresses tumor development in breast cancer mouse model. PLoS ONE. 2010;5:e12860. doi: 10.1371/journal.pone.0012860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Papoutsis A.J., Borg J.L., Selmin O.I., Romagnolo D.F. BRCA-1 promoter hypermethylation and silencing induced by the aromatic hydrocarbon receptor-ligand TCDD are prevented by resveratrol in MCF-7 cells. J Nutr Biochem. 2012;23:1324–1332. doi: 10.1016/j.jnutbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 265.King-Batoon A., Leszczynska J.M., Klein C.B. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 266.Fukasawa K. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta. 2008;1786:15–23. doi: 10.1016/j.bbcan.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lacey K.R., Jackson P.K., Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci U S A. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lee S.C., Chan J.Y., Pervaiz S. Spontaneous and 5-fluorouracil-induced centrosome amplification lowers the threshold to resveratrol-evoked apoptosis in colon cancer cells. Cancer Lett. 2010;288:36–41. doi: 10.1016/j.canlet.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 269.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 270.Castiel A., Visochek L., Mittelman L., Dantzer F., Izraeli S., Cohen-Armon M. A phenanthrene derived PARP inhibitor is an extra-centrosomes de-clustering agent exclusively eradicating human cancer cells. BMC Cancer. 2011;11:412. doi: 10.1186/1471-2407-11-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Otten J.J., Hellwig J.P., Meyers L.D. National Academies Press; Washington, DC: 2006. DRI, dietary reference intakes: the essential guide to nutrient requirements. [Google Scholar]
- 272.Neeha V.S., Kinth P. Nutrigenomics research: a review. J Food Sci Technol. 2013;50:415–428. doi: 10.1007/s13197-012-0775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.German J.B., Zivkovic A.M., Dallas D.C., Smilowitz J.T. Nutrigenomics and personalized diets: what will they mean for food? Ann Rev Food Sci Technol. 2011;2:97–123. doi: 10.1146/annurev.food.102308.124147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Peto R., Doll R., Buckley J.D., Sporn M. Can dietary beta-carotene materially reduce human cancer rates. Nature. 1981;290:201–208. doi: 10.1038/290201a0. [DOI] [PubMed] [Google Scholar]
- 275.Omenn G.S., Goodman G.E., Thornquist M.D., Balmes J., Cullen M.R., Glass A. Risk factors for lung cancer and for intervention effects in CARET, the Beta-Carotene and Retinol Efficacy Trial. J Natl Cancer Inst. 1996;88:1550–1559. doi: 10.1093/jnci/88.21.1550. [DOI] [PubMed] [Google Scholar]
- 276.Bjelakovic G., Nikolova D., Gluud L.L., Simonetti R.G., Gluud C. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: systematic review and meta-analysis. JAMA. 2007;297:842–857. doi: 10.1001/jama.297.8.842. [DOI] [PubMed] [Google Scholar]
- 277.Foksinski M., Gackowski D., Rozalski R., Siomek A., Guz J., Szpila A. Effects of basal level of antioxidants on oxidative DNA damage in humans. Eur J Nutr. 2007;46:174–180. doi: 10.1007/s00394-006-0642-7. [DOI] [PubMed] [Google Scholar]
- 278.Çalışkan-Can E., Fırat H., Ardıç S., Şimşek B., Torun M., Yardim-Akaydin S. Increased levels of 8-hydroxydeoxyguanosine and its relationship with lipid peroxidation and antioxidant vitamins in lung cancer. Clin Chem Lab Med. 2008;46:107–112. doi: 10.1515/CCLM.2008.010. [DOI] [PubMed] [Google Scholar]
- 279.Sram R., Farmer P., Singh R., Garte S., Kalina I., Popov T. Effect of vitamin levels on biomarkers of exposure and oxidative damage – the EXPAH study. Mutat Res. 2009;672:129–134. doi: 10.1016/j.mrgentox.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 280.Kelvin E.A., Edwards S., Jedrychowski W., Schleicher R.L., Camann D., Tang D. Modulation of the effect of prenatal PAH exposure on PAH-DNA adducts in cord blood by plasma antioxidants. Cancer Epidemiol Biomark. 2009;18:2262–2268. doi: 10.1158/1055-9965.EPI-09-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Park E., Park Y.K., Kim S.-M., Lee H.-J., Kang M.-H. Susceptibility to oxidative stress is greater in Korean Patients with Coronary Artery Disease than Healthy Subjects. J Clin Biochem Nutr. 2009;45:341. doi: 10.3164/jcbn.09-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lagadu S., Lechevrel M., Sichel F., Breton J., Pottier D., Couderc R. 8-Oxo-7, 8-dihydro-2’-deoxyguanosine as a biomarker of oxidative damage in oesophageal cancer patients: lack of association with antioxidant vitamins and polymorphism of hOGG1 and GST. J Exp Clin Cancer Res. 2010;29:157. doi: 10.1186/1756-9966-29-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Briviba K., Bub A., Möseneder J., Schwerdtle T., Hartwig A., Kulling S. No differences in DNA damage and antioxidant capacity between intervention groups of healthy, nonsmoking men receiving 2, 5, or 8 servings/day of vegetables and fruit. Nutr Cancer. 2008;60:164–170. doi: 10.1080/01635580701621346. [DOI] [PubMed] [Google Scholar]
- 284.Cho S., Lee D.H., Won C.-H., Kim S.M., Lee S., Lee M.-J. Differential effects of low-dose and high-dose beta-carotene supplementation on the signs of photoaging and type I procollagen gene expression in human skin in vivo. Dermatology. 2010;221:160–171. doi: 10.1159/000305548. [DOI] [PubMed] [Google Scholar]
- 285.Devaraj S., Mathur S., Basu A., Aung H.H., Vasu V.T., Meyers S. A dose-response study on the effects of purified lycopene supplementation on biomarkers of oxidative stress. J Am Coll Nutr. 2008;27:267–273. doi: 10.1080/07315724.2008.10719699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Park J.S., Chyun J.H., Kim Y.K., Line L.L., Chew B.P. Astaxanthin decreased oxidative stress and inflammation and enhanced immune response in humans. Nutr Metab. 2010;7 doi: 10.1186/1743-7075-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.van Helden Y.G., Keijer J., Heil S.G., Picó C., Palou A., Oliver P. Beta-carotene affects oxidative stress-related DNA damage in lung epithelial cells and in ferret lung. Carcinogenesis. 2009;30:2070–2076. doi: 10.1093/carcin/bgp186. [DOI] [PubMed] [Google Scholar]
- 288.Velanganni A., Dharaneedharan S., Geraldine P., Balasundram C. Dietary supplementation of vitamin A, C and E prevents p-dimethylamino-azobenzene induced hepatic DNA damage in rats. Indian J Biochem Biophys. 2007;44:157. [PubMed] [Google Scholar]
- 289.Liu C., Bronson R.T., Russell R.M., Wang X.-D. β-Cryptoxanthin supplementation prevents cigarette smoke-induced lung inflammation, oxidative damage, and squamous metaplasia in ferrets. Cancer Prev Res. 2011;4:1255–1266. doi: 10.1158/1940-6207.CAPR-10-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Moreno F.S., Toledo L.P., de Conti A., Heidor R., Jordão A., Jr., Vannucchi H. Lutein presents suppressing but not blocking chemopreventive activity during diethylnitrosamine-induced hepatocarcinogenesis and this involves inhibition of DNA damage. Chem Biol Interact. 2007;168:221–228. doi: 10.1016/j.cbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 291.Ferreira A.L.A., Salvadori D.M.F., Nascimento M.C.M.O., Rocha N.S., Correa C.R., Pereira E.J. Tomato-oleoresin supplement prevents doxorubicin-induced cardiac myocyte oxidative DNA damage in rats. Mutat Res. 2007;631:26–35. doi: 10.1016/j.mrgentox.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 292.Tripathi D., Jena G. Astaxanthin inhibits cytotoxic and genotoxic effects of cyclophosphamide in mice germ cells. Toxicology. 2008;248:96–103. doi: 10.1016/j.tox.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 293.Nakajima Y., Inokuchi Y., Shimazawa M., Otsubo K., Ishibashi T., Hara H. Astaxanthin, a dietary carotenoid, protects retinal cells against oxidative stress in-vitro and in mice in-vivo. J Pharm Pharmacol. 2008;60:1365–1374. doi: 10.1211/jpp/60.10.0013. [DOI] [PubMed] [Google Scholar]
- 294.Scolastici C., Lopes G.A., Barbisan L.F., Salvadori D.M. Tomato oleoresin inhibits DNA damage but not diethylnitrosamine-induced rat hepatocarcinogenesis. Exp Toxicol Pathol. 2008;60:59–68. doi: 10.1016/j.etp.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 295.Tripathi D., Jena G. Intervention of astaxanthin against cyclophosphamide-induced oxidative stress and DNA damage: a study in mice. Chem Biol Interact. 2009;180:398–406. doi: 10.1016/j.cbi.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 296.Serpeloni J.M., Grotto D., Mercadante A.Z., de Lourdes Pires Bianchi M., Antunes L.M.G. Lutein improves antioxidant defense in vivo and protects against DNA damage and chromosome instability induced by cisplatin. Arch Toxicol. 2010;84:811–822. doi: 10.1007/s00204-010-0576-y. [DOI] [PubMed] [Google Scholar]
- 297.Sasaki M., Yuki K., Kurihara T., Miyake S., Noda K., Kobayashi S. Biological role of lutein in the light-induced retinal degeneration. J Nutr Biochem. 2012;23:423–429. doi: 10.1016/j.jnutbio.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 298.Chew B.P., Mathison B.D., Hayek M.G., Massimino S., Reinhart G.A., Park J.S. Dietary astaxanthin enhances immune response in dogs. Vet Immunol Immunopathol. 2011;140:199–206. doi: 10.1016/j.vetimm.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 299.Tripathi D., Jena G. Astaxanthin intervention ameliorates cyclophosphamide-induced oxidative stress, DNA damage and early hepatocarcinogenesis in rat: role of Nrf2, p53, p38 and phase-II enzymes. Mutat Res. 2010;696:69–80. doi: 10.1016/j.mrgentox.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 300.Huang C.-S., Hu M.-L. Lycopene inhibits DNA damage and reduces hMTH1 mRNA expression in the liver of Mongolian gerbils treated with ferric nitrilotriacetate. Food Chem Toxicol. 2011;49:1381–1386. doi: 10.1016/j.fct.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 301.Morin B., Narbonne J.-F., Ribera D., Badouard C., Ravanat J.-L. Effect of dietary fat-soluble vitamins A and E and proanthocyanidin-rich extract from grape seeds on oxidative DNA damage in rats. Food Chem Toxicol. 2008;46:787–796. doi: 10.1016/j.fct.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 302.Konijeti R., Henning S., Moro A., Sheikh A., Elashoff D., Shapiro A. Chemoprevention of prostate cancer with lycopene in the TRAMP model. Prostate. 2010;70:1547–1554. doi: 10.1002/pros.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Zhou G.-D., Richardson M., Fazili I.S., Wang J., Donnelly K.C., Wang F. Role of retinoic acid in the modulation of benzo (a) pyrene-DNA adducts in human hepatoma cells: implications for cancer prevention. Toxicol Appl Pharm. 2010;249:224–230. doi: 10.1016/j.taap.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Reddy L., Odhav B., Bhoola K. Aflatoxin B1-induced toxicity in HepG2 cells inhibited by carotenoids: morphology, apoptosis and DNA damage. Biol Chem. 2006;387:87–93. doi: 10.1515/BC.2006.012. [DOI] [PubMed] [Google Scholar]
- 305.Bergström T., Bergman J., Möller L. Vitamin A and C compounds permitted in supplements differ in their abilities to affect cell viability, DNA and the DNA nucleoside deoxyguanosine. Mutagenesis. 2011;26:735–744. doi: 10.1093/mutage/ger041. [DOI] [PubMed] [Google Scholar]
- 306.Lorenzo Y., Azqueta A., Luna L., Bonilla F., Domínguez G., Collins A.R. The carotenoid β-cryptoxanthin stimulates the repair of DNA oxidation damage in addition to acting as an antioxidant in human cells. Carcinogenesis. 2009;30:308–314. doi: 10.1093/carcin/bgn270. [DOI] [PubMed] [Google Scholar]
- 307.Rosignoli P., Fabiani R., De Bartolomeo A., Fuccelli R., Pelli M.A., Morozzi G. Genotoxic effect of bile acids on human normal and tumour colon cells and protection by dietary antioxidants and butyrate. Eur J Nutr. 2008;47:301–309. doi: 10.1007/s00394-008-0725-8. [DOI] [PubMed] [Google Scholar]
- 308.Yurtcu E., Iseri O.D., Sahin F.I. Effects of ascorbic acid and β-carotene on HepG2 human hepatocellular carcinoma cell line. Mol Biol Rep. 2011;38:4265–4272. doi: 10.1007/s11033-010-0549-5. [DOI] [PubMed] [Google Scholar]
- 309.van Helden Y.G., Keijer J., Knaapen A.M., Heil S.G., Briedé J.J., Van Schooten F.J. β-Carotene metabolites enhance inflammation-induced oxidative DNA damage in lung epithelial cells. Free Rad Biol Med. 2009;46:299–304. doi: 10.1016/j.freeradbiomed.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 310.Shiau R.-J., Chen K.-Y., Wen Y.-D., Chuang C.-H., Yeh S.-L. Genistein and β-carotene enhance the growth-inhibitory effect of trichostatin A in A549 cells. Eur J Nutr. 2010;49:19–25. doi: 10.1007/s00394-009-0044-8. [DOI] [PubMed] [Google Scholar]
- 311.Kontek R., Drozda R., Śliwiński M., Grzegorczyk K. Genotoxicity of irinotecan and its modulation by vitamins A, C and E in human lymphocytes from healthy individuals and cancer patients. Toxicol In Vitro. 2010;24:417–424. doi: 10.1016/j.tiv.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 312.Jang S.H., Lim J.W., Kim H. Mechanism of β-carotene-induced apoptosis of gastric cancer cells: involvement of ataxia-telangiectasia-mutated. Ann N Y Acad Sci. 2009;1171:156–162. doi: 10.1111/j.1749-6632.2009.04711.x. [DOI] [PubMed] [Google Scholar]
- 313.Santocono M., Zurria M., Berrettini M., Fedeli D., Falcioni G. Lutein, zeaxanthin and astaxanthin protect against DNA damage in SK-N-SH human neuroblastoma cells induced by reactive nitrogen species. J Photochem Photobiol B: Biol. 2007;88:1–10. doi: 10.1016/j.jphotobiol.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 314.Scolastici C., de Lima R., Barbisan L., Ferreira A., Ribeiro D., Salvadori D. Lycopene activity against chemically induced DNA damage in Chinese hamster ovary cells. Toxicol In Vitro. 2007;21:840–845. doi: 10.1016/j.tiv.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 315.Srinivasan M., Sudheer A.R., Pillai K.R., Kumar P.R., Sudhakaran P., Menon V. Lycopene as a natural protector against γ-radiation induced DNA damage, lipid peroxidation and antioxidant status in primary culture of isolated rat hepatocytes in vitro. Biochim Biophys Acta. 2007;1770:659–665. doi: 10.1016/j.bbagen.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 316.Srinivasan M., Devipriya N., Kalpana K., Menon V.P. Lycopene: an antioxidant and radioprotector against γ-radiation-induced cellular damages in cultured human lymphocytes. Toxicology. 2009;262:43–49. doi: 10.1016/j.tox.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 317.Kowalczyk M.C., Walaszek Z., Kowalczyk P., Kinjo T., Hanausek M., Slaga T.J. Differential effects of several phytochemicals and their derivatives on murine keratinocytes in vitro and in vivo: implications for skin cancer prevention. Carcinogenesis. 2009;30:1008–1015. doi: 10.1093/carcin/bgp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zini A., San Gabriel M., Libman J. Lycopene supplementation in vitro can protect human sperm deoxyribonucleic acid from oxidative damage. Fertil Steril. 2010;94:1033–1036. doi: 10.1016/j.fertnstert.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 319.Collins A.R., Azqueta A., Langie S.A. Effects of micronutrients on DNA repair. Eur J Nutr. 2012;51:261–279. doi: 10.1007/s00394-012-0318-4. [DOI] [PubMed] [Google Scholar]
- 320.Astley S.B., Elliott R.M., Archer D.B., Susan Southon S. Evidence that dietary supplementation with carotenoids and carotenoid-rich foods modulates the DNA damage:repair balance in human lymphocytes. Br J Nutr. 2004;91:63–72. doi: 10.1079/bjn20031001. [DOI] [PubMed] [Google Scholar]
- 321.Astley S.B., Elliott R.M., Archer D.B., Southon S. Increased cellular carotenoid levels reduce the persistence of DNA single-strand breaks after oxidative challenge. Nutr Cancer. 2002;43:202–213. doi: 10.1207/S15327914NC432_11. [DOI] [PubMed] [Google Scholar]
- 322.Glei M., Liegibel U.M., Ebert M.N., Böhm V., Pool-Zobel B.L. β-Carotene reduces bleomycin-induced genetic damage in human lymphocytes. Toxicol Appl Pharm. 2002;179:65–73. doi: 10.1006/taap.2001.9329. [DOI] [PubMed] [Google Scholar]
- 323.van Helden Y.G., Keijer J., Heil S.G., Pico C., Palou A., Oliver P. Beta-carotene affects oxidative stress-related DNA damage in lung epithelial cells and in ferret lung. Carcinogenesis. 2009;30:2070–2076. doi: 10.1093/carcin/bgp186. [DOI] [PubMed] [Google Scholar]
- 324.Di Re F., Bohm S., Oriana S., Spatti G.B., Zunino F. Efficacy and safety of high-dose cisplatin and cyclophosphamide with glutathione protection in the treatment of bulky advanced epithelial ovarian cancer. Cancer Chemother Pharmacol. 1990;25:355–360. doi: 10.1007/BF00686237. [DOI] [PubMed] [Google Scholar]
- 325.Smyth J.F., Bowman A., Perren T., Wilkinson P., Prescott R.J., Quinn K.J. Glutathione reduces the toxicity and improves quality of life of women diagnosed with ovarian cancer treated with cisplatin: results of a double-blind, randomised trial. Ann Oncol. 1997;8:569–573. doi: 10.1023/a:1008211226339. [DOI] [PubMed] [Google Scholar]
- 326.Calvert P., Yao K.S., Hamilton T.C., O’Dwyer P.J. Clinical studies of reversal of drug resistance based on glutathione. Chem Biol Interact. 1998;111–112:213–224. doi: 10.1016/s0009-2797(98)00008-8. [DOI] [PubMed] [Google Scholar]
- 327.Estrela J.M., Ortega A., Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Labor Sci. 2006;43:143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 328.Ali-Osman F., Akande O., Antoun G., Mao J.X., Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 329.Stoehlmacher J., Park D.J., Zhang W., Groshen S., Tsao-Wei D.D., Yu M.C. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94:936–942. doi: 10.1093/jnci/94.12.936. [DOI] [PubMed] [Google Scholar]
- 330.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.El Sayed S.M., Mahmoud A.A., El Sawy S.A., Abdelaal E.A., Fouad A.M., Yousif R.S. Warburg effect increases steady-state ROS condition in cancer cells through decreasing their antioxidant capacities (anticancer effects of 3-bromopyruvate through antagonizing Warburg effect) Med Hypotheses. 2013;81:866–870. doi: 10.1016/j.mehy.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 332.van Ommen B., El-Sohemy A., Hesketh J., Kaput J., Fenech M., Evelo C.T. The Micronutrient Genomics Project: a community-driven knowledge base for micronutrient research. Genes Nutr. 2010;5:285–296. doi: 10.1007/s12263-010-0192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Sram R.J., Binkova B., Rossner P., Jr. Vitamin C for DNA damage prevention. Mutat Res. 2012;733:39–49. doi: 10.1016/j.mrfmmm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 334.Nair-Shalliker V., Armstrong B.K., Fenech M. Does vitamin D protect against DNA damage? Mutat Res. 2012;733:50–57. doi: 10.1016/j.mrfmmm.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 335.Prá D., Franke S.I.R., Henriques J.A.P., Fenech M. Iron and genome stability: an update. Mutat Res. 2012;733:92–99. doi: 10.1016/j.mrfmmm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 336.Ferguson L.R., Karunasinghe N., Zhu S., Wang A.H. Selenium and its’ role in the maintenance of genomic stability. Mutat Res. 2012;733:100–110. doi: 10.1016/j.mrfmmm.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 337.Sharif R., Thomas P., Zalewski P., Fenech M. The role of zinc in genomic stability. Mutat Res. 2012;733:111–121. doi: 10.1016/j.mrfmmm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 338.Ferguson L., Schlothauer R. The potential role of nutritional genomics tools in validating high health foods for cancer control: broccoli as example. Mol Nutr Food Res. 2012;56:126–146. doi: 10.1002/mnfr.201100507. [DOI] [PubMed] [Google Scholar]
- 339.Karunasinghe N., Han D.Y., Zhu S., Yu J., Lange K., Duan H. Serum selenium and single-nucleotide polymorphisms in genes for selenoproteins: relationship to markers of oxidative stress in men from Auckland, New Zealand. Genes Nutr. 2012;7:179–190. doi: 10.1007/s12263-011-0259-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Duthie S.J. Berry phytochemicals, genomic stability and cancer: evidence for chemoprotection at several stages in the carcinogenic process. Mol Nutr Food Res. 2007;51:665–674. doi: 10.1002/mnfr.200600257. [DOI] [PubMed] [Google Scholar]
- 341.Lim U., Song M.-A. Springer; 2012. Dietary and lifestyle factors of DNA methylation. Cancer epigenetics; pp. 359–376. [DOI] [PubMed] [Google Scholar]
- 342.Hansen L.A., Malarkey D.E., Wilkinson J.E., Rosenberg M., Woychik R.E., Tennant R.W. Effect of the viable-yellow (A(vy)) agouti allele on skin tumorigenesis and humoral hypercalcemia in v-Ha-ras transgenic TGxAC mice. Carcinogenesis. 1998;19:1837–1845. doi: 10.1093/carcin/19.10.1837. [DOI] [PubMed] [Google Scholar]
- 343.Kuklin A.I., Mynatt R.L., Klebig M.L., Kiefer L.L., Wilkison W.O., Woychik R.P. Liver-specific expression of the agouti gene in transgenic mice promotes liver carcinogenesis in the absence of obesity and diabetes. Mol Cancer. 2004;3:17. doi: 10.1186/1476-4598-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Morgan H.D., Sutherland H.G., Martin D.I., Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 345.Dolinoy D.C., Huang D., Jirtle R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Bunin G.R., Kuijten R.R., Buckley J.D., Rorke L.B., Meadows A.T. Relation between maternal diet and subsequent primitive neuroectodermal brain tumors in young children. New Engl J Med. 1993;329:536–541. doi: 10.1056/NEJM199308193290804. [DOI] [PubMed] [Google Scholar]
- 347.Ferguson L.R. Role of plant polyphenols in genomic stability. Mutat Res. 2001;475:89–111. doi: 10.1016/s0027-5107(01)00073-2. [DOI] [PubMed] [Google Scholar]
- 348.Lopez R., Arumugam A., Joseph R., Monga K., Boopalan T., Agullo P. Hyperglycemia enhances the proliferation of non-tumorigenic and malignant mammary epithelial cells through increased leptin/IGF1R signaling and activation of AKT/mTOR. PLOS ONE. 2013;8:e79708. doi: 10.1371/journal.pone.0079708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hilakivi-Clarke L., Clarke R., Onojafe I., Raygada M., Cho E., Lippman M. A maternal diet high in n-6 polyunsaturated fats alters mammary gland development, puberty onset, and breast cancer risk among female rat offspring. Proc Natl Acad Sci U S A. 1997;94:9372–9377. doi: 10.1073/pnas.94.17.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Villarreal-Garza C., Shaw-Dulin R., Lara-Medina F., Bacon L., Rivera D., Urzua L. Impact of diabetes and hyperglycemia on survival in advanced breast cancer patients. Exp Diabetes Res. 2012;2012:732027. doi: 10.1155/2012/732027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Butturini A.M., Dorey F.J., Lange B.J., Henry D.W., Gaynon P.S., Fu C. Obesity and outcome in pediatric acute lymphoblastic leukemia. J Clin Oncol. 2007;25:2063–2069. doi: 10.1200/JCO.2006.07.7792. [DOI] [PubMed] [Google Scholar]
- 352.Kloor M., Michel S., von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer. 2010;127:1001–1010. doi: 10.1002/ijc.25283. [DOI] [PubMed] [Google Scholar]
- 353.Liu W., Asa S.L., Fantus I.G., Walfish P.G., Ezzat S. Vitamin D arrests thyroid carcinoma cell growth and induces p27 dephosphorylation and accumulation through PTEN/akt-dependent and -independent pathways. Am J Pathol. 2002;160:511–519. doi: 10.1016/S0002-9440(10)64870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Zhang M., Zhou X., Zhou K. Resveratrol inhibits human nasopharyngeal carcinoma cell growth via blocking pAkt/p70S6K signaling pathways. Int J Mol Med. 2013;31:621–627. doi: 10.3892/ijmm.2013.1237. [DOI] [PubMed] [Google Scholar]
- 355.Amiri F., Zarnani A.H., Zand H., Koohdani F., Jeddi-Tehrani M., Vafa M. Synergistic anti-proliferative effect of resveratrol and etoposide on human hepatocellular and colon cancer cell lines. Eur J Pharmacol. 2013;718:34–40. doi: 10.1016/j.ejphar.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 356.Zhang S., Li F., Younes M., Liu H., Chen C., Yao Q. Reduced selenium-binding protein 1 in breast cancer correlates with poor survival and resistance to the anti-proliferative effects of selenium. PLOS ONE. 2013;8:e63702. doi: 10.1371/journal.pone.0063702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Lee Y.C., Tang Y.C., Chen Y.H., Wong C.M., Tsou A.P. Selenite-induced survival of HuH7 hepatoma cells involves activation of focal adhesion kinase-phosphatidylinositol 3-kinase-Akt pathway and Rac1. J Biol Chem. 2003;278:39615–39624. doi: 10.1074/jbc.M304095200. [DOI] [PubMed] [Google Scholar]
- 358.Chuang H.-C., Kapuriya N., Kulp S., Chen C.-S., Shapiro C. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Br Cancer Res Treat. 2012;134:649–659. doi: 10.1007/s10549-012-2106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Chun J., Nadiminty N., Lee S., Onate S., Lou W., Gao A. Mechanisms of selenium down-regulation of androgen receptor signaling in prostate cancer. Mol Cancer Ther. 2006;5:913–918. doi: 10.1158/1535-7163.MCT-05-0389. [DOI] [PubMed] [Google Scholar]
- 360.Jakubikova J., Cervi D., Ooi M., Kim K., Nahar S., Klippel S. Anti-tumor activity and signaling events triggered by the isothiocyanates, sulforaphane and phenethyl isothiocyanate, in multiple myeloma. Haematologica. 2011;96:1170–1179. doi: 10.3324/haematol.2010.029363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kang L., Wang Z.-Y. Breast cancer cell growth inhibition by phenethyl isothiocyanate is associated with down-regulation of oestrogen receptor-alpha36. J Cell Mol Med. 2010;14:1485–1493. doi: 10.1111/j.1582-4934.2009.00877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Keith M., Laporta E., Welsh J. Stable expression of human VDR in murine VDR-null cells recapitulates vitamin D mediated anti-cancer signaling. Mol Carcinog. 2013 doi: 10.1002/mc.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Kim S.-H., Nagalingam A., Saxena N., Singh S., Sharma D. Benzyl isothiocyanate inhibits oncogenic actions of leptin in human breast cancer cells by suppressing activation of signal transducer and activator of transcription 3. Carcinogenesis. 2011;32:359–367. doi: 10.1093/carcin/bgq267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Lamy E., Herz C., Lutz-Bonengel S., Hertrampf A., Márton M.-R., Mersch-Sundermann V. The MAPK pathway signals telomerase modulation in response to isothiocyanate-induced DNA damage of human liver cancer cells. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0053240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Lowe L., Hansen C., Senaratne S., Colston K. Mechanisms implicated in the growth regulatory effects of vitamin D compounds in breast cancer cells. Recent Results Cancer Res. 2003;164:99–110. doi: 10.1007/978-3-642-55580-0_6. [DOI] [PubMed] [Google Scholar]
- 366.Palozza P., Serini S., Di Nicuolo F., Calviello G. Mitogenic and apoptotic signaling by carotenoids: involvement of a redox mechanism. IUBMB Life. 2001;52:77–81. doi: 10.1080/15216540252774810. [DOI] [PubMed] [Google Scholar]
- 367.Błasiak J., Kadłubek M., Kowalik J., Romanowicz-Makowska H., Pertyński T. Inhibition of telomerase activity in endometrial cancer cells by selenium-cisplatin conjugate despite suppression of its DNA-damaging activity by sodium ascorbate. Teratog Carcinog Mutagen. 2002;22:73–82. doi: 10.1002/tcm.1040. [DOI] [PubMed] [Google Scholar]
- 368.Efimova E., Mauceri H., Golden D., Labay E., Bindokas V., Darga T. Poly(ADP-ribose) polymerase inhibitor induces accelerated senescence in irradiated breast cancer cells and tumors. Cancer Res. 2010;70:6277–6282. doi: 10.1158/0008-5472.CAN-09-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ikeda N., Uemura H., Ishiguro H., Hori M., Hosaka M., Kyo S. Combination treatment with 1alpha,25-dihydroxyvitamin D3 and 9-cis-retinoic acid directly inhibits human telomerase reverse transcriptase transcription in prostate cancer cells. Mol Cancer Ther. 2003;2:739–746. [PubMed] [Google Scholar]
- 370.Kasiappan R., Shen Z., Tse A., Jinwal U., Tang J., Lungchukiet P. 1,25-Dihydroxyvitamin D3 suppresses telomerase expression and human cancer growth through microRNA-498. J Biol Chem. 2012;287:41297–41309. doi: 10.1074/jbc.M112.407189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Lamy E., Oey D., Eißmann F., Herz C., Münstedt K., Tinneberg H.-R. Erucin and benzyl isothiocyanate suppress growth of late stage primary human ovarian carcinoma cells and telomerase activity in vitro. Phytother Res. 2013;27:1036–1041. doi: 10.1002/ptr.4798. [DOI] [PubMed] [Google Scholar]
- 372.Liu L., Berletch J.B., Green J.G., Pate M.S., Andrews L.G., Tollefsbol T.O. Telomerase inhibition by retinoids precedes cytodifferentiation of leukemia cells and may contribute to terminal differentiation. Mol Cancer Ther. 2004;3:1003–1009. [PubMed] [Google Scholar]
- 373.Love W., Berletch J., Andrews L., Tollefsbol T. Epigenetic regulation of telomerase in retinoid-induced differentiation of human leukemia cells. Int J Oncol. 2008;32:625–631. [PMC free article] [PubMed] [Google Scholar]
- 374.Zhang X., Nicosia S.V., Bai W. Vitamin D receptor is a novel drug target for ovarian cancer treatment. Curr Cancer Drug Targets. 2006;6:229–244. doi: 10.2174/156800906776842939. [DOI] [PubMed] [Google Scholar]
- 375.Fuggetta M.P., Lanzilli G., Tricarico M., Cottarelli A., Falchetti R., Ravagnan G. Effect of resveratrol on proliferation and telomerase activity of human colon cancer cells in vitro. J Exp Clin Cancer Res. 2006;25:189–193. [PubMed] [Google Scholar]
- 376.Lanzilli G., Fuggetta M.P., Tricarico M., Cottarelli A., Serafino A., Falchetti R. Resveratrol down-regulates the growth and telomerase activity of breast cancer cells in vitro. Int J Oncol. 2006;28:641–648. [PubMed] [Google Scholar]
- 377.Rengarajan T., Rajendran P., Nandakumar N., Balasubramanian M., Nishigaki I. Cancer preventive efficacy of marine carotenoid fucoxanthin: cell cycle arrest and apoptosis. Nutrients. 2013;5:4978–4989. doi: 10.3390/nu5124978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Sanmartín C., Plano D., Sharma A., Palop J. Selenium compounds, apoptosis and other types of cell death: an overview for cancer therapy. Int J Mol Sci. 2012;13:9649–9672. doi: 10.3390/ijms13089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Thomson S., Brown K., Pullar J., Hampton M. Phenethyl isothiocyanate triggers apoptosis in Jurkat cells made resistant by the overexpression of Bcl-2. Cancer Res. 2006;66:6772–6777. doi: 10.1158/0008-5472.CAN-05-3809. [DOI] [PubMed] [Google Scholar]
- 380.Ting H.-J., Hsu J., Bao B.-Y., Lee Y.-F. Docetaxel-induced growth inhibition and apoptosis in androgen independent prostate cancer cells are enhanced by 1alpha,25-dihydroxyvitamin D3. Cancer Lett. 2007;247:122–129. doi: 10.1016/j.canlet.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 381.Zhang J., Kan Y., Tian Y., Wang Z., Zhang J. Effects of poly (ADP-ribosyl) polymerase (PARP) inhibitor on cisplatin resistance & proliferation of the ovarian cancer C13* cells. Indian J Med Res. 2013;137:527–532. [PMC free article] [PubMed] [Google Scholar]
- 382.Wesolowska O., Wisniewski J., Bielawska-Pohl A., Paprocka M., Duarte N., Ferreira M.J. Stilbenes as multidrug resistance modulators and apoptosis inducers in human adenocarcinoma cells. Anticancer Res. 2010;30:4587–4593. [PubMed] [Google Scholar]
- 383.Chen H., Landen C., Li Y., Alvarez R., Tollefsbol T. Epigallocatechin gallate and sulforaphane combination treatment induce apoptosis in paclitaxel-resistant ovarian cancer cells through hTERT and Bcl-2 down-regulation. Exp Cell Res. 2013;319:697–706. doi: 10.1016/j.yexcr.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.El-Bayoumy K. The protective role of selenium on genetic damage and on cancer. Mutat Res. 2001;475:123–139. doi: 10.1016/s0027-5107(01)00075-6. [DOI] [PubMed] [Google Scholar]
- 385.Moon D.-O., Kang S.-H., Kim K.-C., Kim M.-O., Choi Y., Kim G.-Y. Sulforaphane decreases viability and telomerase activity in hepatocellular carcinoma Hep3B cells through the reactive oxygen species-dependent pathway. Cancer Lett. 2010;295:260–266. doi: 10.1016/j.canlet.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 386.Donkena K.V., Yuan H., Young C.Y. Vitamin Bs, one carbon metabolism and prostate cancer. Mini Rev Med Chem. 2010;10:1385–1392. doi: 10.2174/138955710793564106. [DOI] [PubMed] [Google Scholar]
- 387.Rayet B., Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 388.Lerebours F., Vacher S., Andrieu C., Espie M., Marty M., Lidereau R. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Batra S., Sahu R.P., Kandala P.K., Srivastava S.K. Benzyl isothiocyanate-mediated inhibition of histone deacetylase leads to NF-kappaB turnoff in human pancreatic carcinoma cells. Mol Cancer Ther. 2010;9:1596–1608. doi: 10.1158/1535-7163.MCT-09-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Choi K.C., Jung M.G., Lee Y.H., Yoon J.C., Kwon S.H., Kang H.B. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69:583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 391.Hopkins M.H., Owen J., Ahearn T., Fedirko V., Flanders W.D., Jones D.P. Effects of supplemental vitamin D and calcium on biomarkers of inflammation in colorectal adenoma patients: a randomized, controlled clinical trial. Cancer Prev Res. 2011;4:1645–1654. doi: 10.1158/1940-6207.CAPR-11-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Krishnan A.V., Moreno J., Nonn L., Swami S., Peehl D.M., Feldman D. Calcitriol as a chemopreventive and therapeutic agent in prostate cancer: role of anti-inflammatory activity. J Bone Miner Res. 2007;22(Suppl 2):V74–V80. doi: 10.1359/jbmr.07s213. [DOI] [PubMed] [Google Scholar]
- 393.Zhang X.-H., Ma J., Smith-Warner S., Lee J., Giovannucci E. Vitamin B6 and colorectal cancer: current evidence and future directions. World J Gastroenterol. 2013;19:1005–1010. doi: 10.3748/wjg.v19.i7.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sharp L., Carsin A.E., Cantwell M.M., Anderson L.A., Murray L.J., Group F.S. Intakes of dietary folate and other B vitamins are associated with risks of esophageal adenocarcinoma, Barrett's esophagus, and reflux esophagitis. J Nutr. 2013;143:1966–1973. doi: 10.3945/jn.113.174664. [DOI] [PubMed] [Google Scholar]
- 395.Barrett C., Singh K., Motley A., Lintel M., Matafonova E., Bradley A. Dietary selenium deficiency exacerbates DSS-induced epithelial injury and AOM/DSS-induced tumorigenesis. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0067845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Kristal A., Arnold K., Neuhouser M., Goodman P., Platz E., Albanes D. Diet, supplement use, and prostate cancer risk: results from the prostate cancer prevention trial. Am J Epidemiol. 2010;172:566–577. doi: 10.1093/aje/kwq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Ozten N., Horton L., Lasano S., Bosland M. Selenomethionine and alpha-tocopherol do not inhibit prostate carcinogenesis in the testosterone plus estradiol-treated NBL rat model. Cancer Prev Res (Phila) 2010;3:371–380. doi: 10.1158/1940-6207.CAPR-09-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Trejo-Solís C., Pedraza-Chaverrí J., Torres-Ramos M., Jiménez-Farfán D., Cruz Salgado A., Serrano-García N. Multiple molecular and cellular mechanisms of action of lycopene in cancer inhibition. Evid Based Complement Alternat Med. 2013;2013:705121. doi: 10.1155/2013/705121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Wertz K. Lycopene effects contributing to prostate health. Nutr Cancer. 2009;61:775–783. doi: 10.1080/01635580903285023. [DOI] [PubMed] [Google Scholar]
- 400.Swindall A., Stanley J., Yang E. PARP-1: friend or foe of DNA damage and repair in tumorigenesis. Cancers. 2013;5:943–958. doi: 10.3390/cancers5030943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Masutani M., Fujimori H. Poly(ADP-ribosyl)ation in carcinogenesis. Mol Asp Med. 2013;34:1202–1216. doi: 10.1016/j.mam.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 402.Chen M.C., Chang W.W., Kuan Y.D., Lin S.T., Hsu H.C., Lee C.H. Resveratrol inhibits LPS-induced epithelial–mesenchymal transition in mouse melanoma model. Innate Immun. 2012;18:685–693. doi: 10.1177/1753425912436589. [DOI] [PubMed] [Google Scholar]
- 403.Fahey J., Wehage S., Holtzclaw W., Kensler T., Egner P., Shapiro T. Protection of humans by plant glucosinolates: efficiency of conversion of glucosinolates to isothiocyanates by the gastrointestinal microflora. Cancer Prev Res (Phila) 2012;5:603–611. doi: 10.1158/1940-6207.CAPR-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Li R., Li C., Wang T. Transcriptomic alterations in human prostate cancer cell LNCaP tumor xenograft modulated by dietary phenethyl isothiocyanate. Mol Carcinog. 2013;52:426–437. doi: 10.1002/mc.21873. [DOI] [PubMed] [Google Scholar]
- 405.Kusmartsev S., Cheng F., Yu B., Nefedova Y., Sotomayor E., Lush R. All-trans-retinoic acid eliminates immature myeloid cells from tumor-bearing mice and improves the effect of vaccination. Cancer Res. 2003;63:4441–4449. [PubMed] [Google Scholar]
- 406.Najjar Y., Finke J. Clinical perspectives on targeting of myeloid derived suppressor cells in the treatment of cancer. Front Oncol. 2013;3:49. doi: 10.3389/fonc.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Sayeed A., Luciani-Torres G., Meng Z., Bennington J.L., Moore D.H., Dairkee S.H. Aberrant regulation of the BST2 (Tetherin) promoter enhances cell proliferation and apoptosis evasion in high grade breast cancer cells. PLOS ONE. 2013;8:e67191. doi: 10.1371/journal.pone.0067191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Aroni K., Anagnostopoulou K., Tsagroni E., Ioannidis E. Skin hyperpigmentation and increased angiogenesis secondary to vitamin B12 deficiency in a young vegetarian woman. Acta Derm Venereol. 2008;88:191–192. doi: 10.2340/00015555-0377. [DOI] [PubMed] [Google Scholar]
- 409.Gadau S., Emanueli C., Van Linthout S., Graiani G., Todaro M., Meloni M. Benfotiamine accelerates the healing of ischaemic diabetic limbs in mice through protein kinase B/Akt-mediated potentiation of angiogenesis and inhibition of apoptosis. Diabetologia. 2006;49:405–420. doi: 10.1007/s00125-005-0103-5. [DOI] [PubMed] [Google Scholar]
- 410.Huang P.H., Lin C.P., Wang C.H., Chiang C.H., Tsai H.Y., Chen J.S. Niacin improves ischemia-induced neovascularization in diabetic mice by enhancement of endothelial progenitor cell functions independent of changes in plasma lipids. Angiogenesis. 2012;15:377–389. doi: 10.1007/s10456-012-9267-z. [DOI] [PubMed] [Google Scholar]
- 411.Jiang C., Jiang W., Ip C., Ganther H., Lu J. Selenium-induced inhibition of angiogenesis in mammary cancer at chemopreventive levels of intake. Mol Carcinog. 1999;26:213–225. doi: 10.1002/(sici)1098-2744(199912)26:4<213::aid-mc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 412.Lin S.Y., Lee W.R., Su Y.F., Hsu S.P., Lin H.C., Ho P.Y. Folic acid inhibits endothelial cell proliferation through activating the cSrc/ERK 2/NF-kappaB/p53 pathway mediated by folic acid receptor. Angiogenesis. 2012;15:671–683. doi: 10.1007/s10456-012-9289-6. [DOI] [PubMed] [Google Scholar]
- 413.McAuslan B.R., Reilly W. Selenium-induced cell migration and proliferation: relevance to angiogenesis and microangiopathy. Microvasc Res. 1986;32:112–120. doi: 10.1016/0026-2862(86)90047-6. [DOI] [PubMed] [Google Scholar]
- 414.Premkumar V.G., Yuvaraj S., Sathish S., Shanthi P., Sachdanandam P. Anti-angiogenic potential of CoenzymeQ10, riboflavin and niacin in breast cancer patients undergoing tamoxifen therapy. Vasc Pharmacol. 2008;48:191–201. doi: 10.1016/j.vph.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 415.Sun D., Liu Y., Yu Q., Zhou Y., Zhang R., Chen X. The effects of luminescent ruthenium(II) polypyridyl functionalized selenium nanoparticles on bFGF-induced angiogenesis and AKT/ERK signaling. Biomaterials. 2013;34:171–180. doi: 10.1016/j.biomaterials.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 416.Arensman R.M., Stolar C.J. Vitamin A effect on tumor angiogenesis. J Pediatr Surg. 1979;14:809–813. doi: 10.1016/s0022-3468(79)80271-7. [DOI] [PubMed] [Google Scholar]
- 417.Bajpai S., Mishra M., Kumar H., Tripathi K., Singh S., Pandey H. Effect of selenium on connexin expression, angiogenesis, and antioxidant status in diabetic wound healing. Biol Trace Elem Res. 2011;144:327–338. doi: 10.1007/s12011-011-9097-7. [DOI] [PubMed] [Google Scholar]
- 418.Rajesh M., Mukhopadhyay P., Bátkai S., Godlewski G., Haskó G., Liaudet L. Pharmacological inhibition of poly(ADP-ribose) polymerase inhibits angiogenesis. Biochem Biophys Res Commun. 2006;350:352–357. doi: 10.1016/j.bbrc.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Thejass P., Kuttan G. Allyl isothiocyanate (AITC) and phenyl isothiocyanate (PITC) inhibit tumour-specific angiogenesis by downregulating nitric oxide (NO) and tumour necrosis factor-alpha (TNF-alpha) production. Nitric Oxide. 2007;16:247–257. doi: 10.1016/j.niox.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 420.Boreddy S., Sahu R., Srivastava S. Benzyl isothiocyanate suppresses pancreatic tumor angiogenesis and invasion by inhibiting HIF-α/VEGF/Rho-GTPases: pivotal role of STAT-3. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0025799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Krishnan A., Swami S., Feldman D. Equivalent anticancer activities of dietary vitamin D and calcitriol in an animal model of breast cancer: importance of mammary CYP27B1 for treatment and prevention. J Steroid Biochem Mol Biol. 2013;136:289–295. doi: 10.1016/j.jsbmb.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Puduvalli V., Sampath D., Bruner J., Nangia J., Xu R., Kyritsis A. TRAIL-induced apoptosis in gliomas is enhanced by Akt-inhibition and is independent of JNK activation. Apoptosis. 2005;10:233–243. doi: 10.1007/s10495-005-6078-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Soares N., Teodoro A., Oliveira F., Santos C., Takiya C., Junior O. Influence of lycopene on cell viability, cell cycle, and apoptosis of human prostate cancer and benign hyperplastic cells. Nutr Cancer. 2013;65:1076–1085. doi: 10.1080/01635581.2013.812225. [DOI] [PubMed] [Google Scholar]
- 424.Zastre J., Sweet R., Hanberry B., Ye S. Linking vitamin B1 with cancer cell metabolism. Cancer Metab. 2013;1:16. doi: 10.1186/2049-3002-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Park E., Wilder E., Lane M. Retinol inhibits the invasion of retinoic acid-resistant colon cancer cells in vitro and decreases matrix metalloproteinase mRNA, protein, and activity levels. Nutr Cancer. 2007;57:66–77. doi: 10.1080/01635580701268238. [DOI] [PubMed] [Google Scholar]
- 426.Park E.Y., Pinali D., Lindley K., Lane M.A. Hepatic vitamin A preloading reduces colorectal cancer metastatic multiplicity in a mouse xenograft model. Nutr Cancer. 2012;64:732–740. doi: 10.1080/01635581.2012.687425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Raffoul J., Heydari A., Hillman G., DNA repair and cancer therapy Targeting APE1/Ref-1 using dietary agents. J Oncol. 2012;2012:370481. doi: 10.1155/2012/370481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Rao L., Puschner B., Prolla T.A. Gene expression profiling of low selenium status in the mouse intestine: transcriptional activation of genes linked to DNA damage, cell cycle control and oxidative stress. J Nutr. 2001;131:3175–3181. doi: 10.1093/jn/131.12.3175. [DOI] [PubMed] [Google Scholar]
- 429.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 430.Yang M.-D., Lai K.-C., Lai T.-Y., Hsu S.-C., Kuo C.-L., Yu C.-S. Phenethyl isothiocyanate inhibits migration and invasion of human gastric cancer AGS cells through suppressing MAPK and NF-kappaB signal pathways. Anticancer Res. 2010;30:2135–2143. [PubMed] [Google Scholar]
- 431.Nishino H., Murakosh M., Ii T., Takemura M., Kuchide M., Kanazawa M. Carotenoids in cancer chemoprevention. Cancer Metastasis Rev. 2002;21:257–264. doi: 10.1023/a:1021206826750. [DOI] [PubMed] [Google Scholar]
- 432.Damm K., Hemmann U., Garin-Chesa P., Hauel N., Kauffmann I., Priepke H. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Deeb D., Gao X., Liu Y., Kim S.H., Pindolia K.R., Arbab A.S. Inhibition of cell proliferation and induction of apoptosis by oleanane triterpenoid (CDDO-Me) in pancreatic cancer cells is associated with the suppression of hTERT gene expression and its telomerase activity. Biochem Biophys Res Commun. 2012;422:561–567. doi: 10.1016/j.bbrc.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Ermolaeva M.A., Schumacher B. Systemic DNA damage responses: organismal adaptations to genome instability. Trends Genet. 2014;30:95–102. doi: 10.1016/j.tig.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Etchegaray J.P., Zhong L., Mostoslavsky R. The histone deacetylase SIRT6: at the crossroads between epigenetics, metabolism and disease. Curr Top Med Chem. 2013;13:2991–3000. doi: 10.2174/15680266113136660213. [DOI] [PubMed] [Google Scholar]
- 436.Szabo G., Lippai D. Molecular hepatic carcinogenesis: impact of inflammation. Dig Dis. 2012;30:243–248. doi: 10.1159/000336913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Xia X., Liu S., Xiao Z., Zhu F., Song N.Y., Zhou M. An IKKalpha-nucleophosmin axis utilizes inflammatory signaling to promote genome integrity. Cell Rep. 2013;5:1243–1255. doi: 10.1016/j.celrep.2013.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Mishra A., Liu S., Sams G.H., Curphey D.P., Santhanam R., Rush L.J. Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell. 2012;22:645–655. doi: 10.1016/j.ccr.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Ghosh A., Saginc G., Leow S.C., Khattar E., Shin E.M., Yan T.D. Telomerase directly regulates NF-kappaB-dependent transcription. Nat Cell Biol. 2012;14:1270–1281. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 440.Nassani N., Khayat G., Raad I., Jiang Y., Alaaeddine N., Hilal G. Telomerase as a potential marker for inflammation and cancer detection in bronchial washing: a prospective study. Clin Biochem. 2013;46:1701–1704. doi: 10.1016/j.clinbiochem.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 441.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability – an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 442.Yun U.J., Park S.E., Jo Y.S., Kim J., Shin D.Y. DNA damage induces the IL-6/STAT3 signaling pathway, which has anti-senescence and growth-promoting functions in human tumors. Cancer Lett. 2012;323:155–160. doi: 10.1016/j.canlet.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 443.Nishikawa K., Rosenblum M.G., Newman R.A., Pandita T.K., Hittelman W.N., Donato N.J. Resistance of human cervical carcinoma cells to tumor necrosis factor correlates with their increased sensitivity to cisplatin: evidence of a role for DNA repair and epidermal growth factor receptor. Cancer Res. 1992;52:4758–4765. [PubMed] [Google Scholar]
- 444.Shen S., Mao C.Q., Yang X.Z., Du X.J., Liu Y., Zhu Y.H. Cationic lipid-assisted polymeric nanoparticle mediated GATA2 siRNA delivery for synthetic lethal therapy of KRAS mutant non-small-cell lung carcinoma. Mol Pharm. 2014 doi: 10.1021/mp400714z. [DOI] [PubMed] [Google Scholar]
- 445.Nam H.J., Chae S., Jang S.H., Cho H., Lee J.H. The PI3K-Akt mediates oncogenic Met-induced centrosome amplification and chromosome instability. Carcinogenesis. 2010;31:1531–1540. doi: 10.1093/carcin/bgq133. [DOI] [PubMed] [Google Scholar]
- 446.Xu X., Weaver Z., Linke S.P., Li C., Gotay J., Wang X.W. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 447.Shay J.W. Telomerase therapeutics: telomeres recognized as a DNA damage signal: commentary re: K. Kraemer et al., antisense-mediated hTERT inhibition specifically reduces the growth of human bladder cancer cells. Clin. Cancer Res., 9: 3794–3800, 2003. Clin Cancer Res. 2003;9:3521–3525. [PubMed] [Google Scholar]
- 448.Ghosal G., Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Poynter K.R., Sachs P.C., Bright A.T., Breed M.S., Nguyen B.N., Elmore L.W. Genetic inhibition of telomerase results in sensitization and recovery of breast tumor cells. Mol Cancer Ther. 2009;8:1319–1327. doi: 10.1158/1535-7163.MCT-08-0849. [DOI] [PubMed] [Google Scholar]
- 450.Aguilera A., Garcia-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 451.Cha H.J., Yim H. The accumulation of DNA repair defects is the molecular origin of carcinogenesis. Tumour Biol. 2013;34:3293–3302. doi: 10.1007/s13277-013-1038-y. [DOI] [PubMed] [Google Scholar]
- 452.Ohshima S. Centrosome aberrations associated with cellular senescence and p53 localization at supernumerary centrosomes. Oxid Med Cell Longev. 2012;2012:217594. doi: 10.1155/2012/217594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Tamae D., Lim P., Wuenschell G.E., Termini J. Mutagenesis and repair induced by the DNA advanced glycation end product N2-1-(carboxyethyl)-2’-deoxyguanosine in human cells. Biochemistry. 2011;50:2321–2329. doi: 10.1021/bi101933p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Dai C., Sun F., Zhu C., Hu X. Tumor environmental factors glucose deprivation and lactic acidosis induce mitotic chromosomal instability – an implication in aneuploid human tumors. PLOS ONE. 2013;8:e63054. doi: 10.1371/journal.pone.0063054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Li Y., Liu L., Tollefsbol T.O. Glucose restriction can extend normal cell lifespan and impair precancerous cell growth through epigenetic control of hTERT and p16 expression. FASEB J. 2010;24:1442–1453. doi: 10.1096/fj.09-149328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Mai W., Kawakami K., Shakoori A., Kyo S., Miyashita K., Yokoi K. Deregulated GSK3{beta} sustains gastrointestinal cancer cells survival by modulating human telomerase reverse transcriptase and telomerase. Clin Cancer Res. 2009;15:6810–6819. doi: 10.1158/1078-0432.CCR-09-0973. [DOI] [PubMed] [Google Scholar]
- 457.Chavakis T., Orlova V.V., Langer H.F. A possible crosstalk between DNA repair pathways and angiogenesis. Cell Cycle. 2009;8:3438–3439. doi: 10.4161/cc.8.21.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Rankin E.B., Giaccia A.J., Hammond E.M. Bringing H2AX into the angiogenesis family. Cancer Cell. 2009;15:459–461. doi: 10.1016/j.ccr.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Economopoulou M., Langer H.F., Celeste A., Orlova V.V., Choi E.Y., Ma M. Histone H2AX is integral to hypoxia-driven neovascularization. Nat Med. 2009;15:553–558. doi: 10.1038/nm.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Coleman M.L., Ratcliffe P.J. Angiogenesis: escape from hypoxia. Nat Med. 2009;15:491–493. doi: 10.1038/nm0509-491. [DOI] [PubMed] [Google Scholar]
- 462.Taylor S.M., Nevis K.R., Park H.L., Rogers G.C., Rogers S.L., Cook J.G. Angiogenic factor signaling regulates centrosome duplication in endothelial cells of developing blood vessels. Blood. 2010;116:3108–3117. doi: 10.1182/blood-2010-01-266197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Kim Y.M., Yang S., Xu W., Li S., Yang X. Continuous in vitro exposure to low-dose genistein induces genomic instability in breast epithelial cells. Cancer Genet Cytogen. 2008;186:78–84. doi: 10.1016/j.cancergencyto.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Li Y., Liu L., Andrews L.G., Tollefsbol T.O. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009;125:286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Holysz H., Lipinska N., Paszel-Jaworska A., Rubis B. Telomerase as a useful target in cancer fighting-the breast cancer case. Tumour Biol. 2013;34:1371–1380. doi: 10.1007/s13277-013-0757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 467.Olaussen K.A., Dunant A., Fouret P., Brambilla E., Andre F., Haddad V. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 468.Ghadimi B.M., Sackett D.L., Difilippantonio M.J., Schrock E., Neumann T., Jauho A. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosom Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 469.Lingle W.L., Barrett S.L., Negron V.C., D’Assoro A.B., Boeneman K., Liu W. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Chen L., Lu M.H., Zhang D., Hao N.B., Fan Y.H., Wu Y.Y. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5:e1034. doi: 10.1038/cddis.2013.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Chen P.C., Peng J.R., Huang L., Li W.X., Wang W.Z., Cui Z.Q. Overexpression of human telomerase reverse transcriptase promotes the motility and invasiveness of HepG2 cells in vitro. Oncol Rep. 2013;30:1157–1164. doi: 10.3892/or.2013.2563. [DOI] [PubMed] [Google Scholar]
- 472.Dikmen Z.G., Ozgurtas T., Gryaznov S.M., Herbert B.S. Targeting critical steps of cancer metastasis and recurrence using telomerase template antagonists. Biochim Biophys Acta. 2009;1792:240–247. doi: 10.1016/j.bbadis.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 473.Griffith J.K., Bryant J.E., Fordyce C.A., Gilliland F.D., Joste N.E., Moyzis R.K. Reduced telomere DNA content is correlated with genomic instability and metastasis in invasive human breast carcinoma. Breast Cancer Res Treat. 1999;54:59–64. doi: 10.1023/a:1006128228761. [DOI] [PubMed] [Google Scholar]
- 474.Heaphy C.M., Baumgartner K.B., Bisoffi M., Baumgartner R.N., Griffith J.K. Telomere DNA content predicts breast cancer-free survival interval. Clin Cancer Res. 2007;13:7037–7043. doi: 10.1158/1078-0432.CCR-07-0432. [DOI] [PubMed] [Google Scholar]
- 475.Alderton G.K. Microenvironment secreted DNA damage? Nat Rev Cancer. 2013;13:77. doi: 10.1038/nrc3455. [DOI] [PubMed] [Google Scholar]
- 476.Chan N., Pires I.M., Bencokova Z., Coackley C., Luoto K.R., Bhogal N. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res. 2010;70:8045–8054. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Falchetti M.L., Mongiardi M.P., Fiorenzo P., Petrucci G., Pierconti F., D’Agnano I. Inhibition of telomerase in the endothelial cells disrupts tumor angiogenesis in glioblastoma xenografts. Int J Cancer. 2008;122:1236–1242. doi: 10.1002/ijc.23193. [DOI] [PubMed] [Google Scholar]
- 478.Liu W., Asa S., Fantus I., Walfish P., Ezzat S. Vitamin D arrests thyroid carcinoma cell growth and induces p27 dephosphorylation and accumulation through PTEN/akt-dependent and-independent pathways. Am J Pathol. 2002 doi: 10.1016/S0002-9440(10)64870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Zhang S., Li F., Younes M., Liu H., Chen C., Yao Q. Reduced selenium-binding protein 1 in breast cancer correlates with poor survival and resistance to the anti-proliferative effects of selenium. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0063702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Haddad N.F., Teodoro A.J., Leite de Oliveira F., Soares N., de Mattos R.M., Hecht F. Lycopene and beta-carotene induce growth inhibition and proapoptotic effects on ACTH-secreting pituitary adenoma cells. PLOS ONE. 2013;8:e62773. doi: 10.1371/journal.pone.0062773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Yang E.S., Nowsheen S., Rahman M.A., Cook R.S., Xia F. Targeting BRCA1 localization to augment breast tumor sensitivity to poly(ADP-Ribose) polymerase inhibition. Cancer Res. 2012;72:5547–5555. doi: 10.1158/0008-5472.CAN-12-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Yang C.M., Lu Y.L., Chen H.Y., Hu M.L. Lycopene and the LXRalpha agonist T0901317 synergistically inhibit the proliferation of androgen-independent prostate cancer cells via the PPARgamma-LXRalpha-ABCA1 pathway. J Nutr Biochem. 2012;23:1155–1162. doi: 10.1016/j.jnutbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 483.Cardnell R.J., Feng Y., Diao L., Fan Y.H., Masrorpour F., Wang J. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res. 2013;19:6322–6328. doi: 10.1158/1078-0432.CCR-13-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Wang Z., Li Y., Lv S., Tian Y. Inhibition of proliferation and invasiveness of ovarian cancer C13* cells by a poly(ADP-ribose) polymerase inhibitor and the role of nuclear factor-kappaB. J Int Med Res. 2013;41:1577–1585. doi: 10.1177/0300060513480913. [DOI] [PubMed] [Google Scholar]
- 485.Melchini A., Traka M.H., Catania S., Miceli N., Taviano M.F., Maimone P. Antiproliferative activity of the dietary isothiocyanate erucin, a bioactive compound from cruciferous vegetables, on human prostate cancer cells. Nutr Cancer. 2013;65:132–138. doi: 10.1080/01635581.2013.741747. [DOI] [PubMed] [Google Scholar]
- 486.Stan S.D., Singh S.V., Whitcomb D.C., Brand R.E. Phenethyl isothiocyanate inhibits proliferation and induces apoptosis in pancreatic cancer cells in vitro and in a MIAPaca2 xenograft animal model. Nutr Cancer. 2013 doi: 10.1080/01635581.2013.795979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Hopkins M., Owen J., Ahearn T., Fedirko V., Flanders W., Jones D. Effects of supplemental vitamin D and calcium on biomarkers of inflammation in colorectal adenoma patients: a randomized, controlled clinical trial. Cancer Prev Res (Phila) 2011;4:1645–1654. doi: 10.1158/1940-6207.CAPR-11-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Krishnan A., Moreno J., Nonn L., Swami S., Peehl D., Feldman D. Calcitriol as a chemopreventive and therapeutic agent in prostate cancer: role of anti-inflammatory activity. J Bone Min Res. 2007;22(Suppl. 2):80. doi: 10.1359/jbmr.07s213. [DOI] [PubMed] [Google Scholar]
- 489.Gatz S.A., Wiesmuller L. Take a break – resveratrol in action on DNA. Carcinogenesis. 2008;29:321–332. doi: 10.1093/carcin/bgm276. [DOI] [PubMed] [Google Scholar]
- 490.Hager G., Formanek M., Gedlicka C., Thurnher D., Knerer B., Kornfehl J. 1,25(OH)2 vitamin D3 induces elevated expression of the cell cycle-regulating genes P21 and P27 in squamous carcinoma cell lines of the head and neck. Acta Otolaryngol. 2001;121:103–109. doi: 10.1080/000164801300006353. [DOI] [PubMed] [Google Scholar]
- 491.Hager G., Kornfehl J., Knerer B., Weigel G., Formanek M. Molecular analysis of p21 promoter activity isolated from squamous carcinoma cell lines of the head and neck under the influence of 1,25(OH)2 vitamin D3 and its analogs. Acta Otolaryngol. 2004;124:90–96. doi: 10.1080/00016480310015353. [DOI] [PubMed] [Google Scholar]
- 492.Wong G., Gupta R., Dixon K.M., Deo S.S., Choong S.M., Halliday G.M. 1,25-Dihydroxyvitamin D and three low-calcemic analogs decrease UV-induced DNA damage via the rapid response pathway. J Steroid Biochem Mol Biol. 2004;89-90:567–570. doi: 10.1016/j.jsbmb.2004.03.072. [DOI] [PubMed] [Google Scholar]
- 493.Bera S., De Rosa V., Rachidi W., Diamond A.M. Does a role for selenium in DNA damage repair explain apparent controversies in its use in chemoprevention. Mutagenesis. 2013;28:127–134. doi: 10.1093/mutage/ges064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Hamilton E.E., Fanwick P.E., Wilker J.J. Alkylation of inorganic oxo compounds and insights on preventing DNA damage. J Am Chem Soc. 2006;128:3388–3395. doi: 10.1021/ja056568v. [DOI] [PubMed] [Google Scholar]
- 495.Collins A.R., Harrington V., Drew J., Melvin R. Nutritional modulation of DNA repair in a human intervention study. Carcinogenesis. 2003;24:511–515. doi: 10.1093/carcin/24.3.511. [DOI] [PubMed] [Google Scholar]
- 496.He X., Wang Y., Zhu J., Orloff M., Eng C. Resveratrol enhances the anti-tumor activity of the mTOR inhibitor rapamycin in multiple breast cancer cell lines mainly by suppressing rapamycin-induced AKT signaling. Cancer Lett. 2011;301:168–176. doi: 10.1016/j.canlet.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 497.Kim K.H., Back J.H., Zhu Y., Arbesman J., Athar M., Kopelovich L. Resveratrol targets transforming growth factor-beta2 signaling to block UV-induced tumor progression. J Investig Dermatol. 2011;131:195–202. doi: 10.1038/jid.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Zhang J., Kan Y., Tian Y., Wang Z., Zhang J. Effects of poly (ADP-ribosyl) polymerase (PARP) inhibitor on cisplatin resistance & proliferation of the ovarian cancer C13* cells. Indian J Med Res. 2013;137:527–532. [PMC free article] [PubMed] [Google Scholar]
- 499.Yao H., Sundar I., Gorbunova V., Rahman I. P21-PARP-1 pathway is involved in cigarette smoke-induced lung DNA damage and cellular senescence. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0080007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Schabath M.B., Grossman H.B., Delclos G.L., Hernandez L.M., Day R.S., Davis B.R. Dietary carotenoids and genetic instability modify bladder cancer risk. J Nutr. 2004;134:3362–3369. doi: 10.1093/jn/134.12.3362. [DOI] [PubMed] [Google Scholar]
- 501.Ren Z., Wang L., Cui J., Huoc Z., Xue J., Cui H. Resveratrol inhibits NF-kB signaling through suppression of p65 and IkappaB kinase activities. Pharmazie. 2013;68:689–694. [PubMed] [Google Scholar]
- 502.Khuda-Bukhsh A.R., Das S., Saha S.K. Molecular approaches toward targeted cancer prevention with some food plants and their products: inflammatory and other signal pathways. Nutr Cancer. 2014;66:194–205. doi: 10.1080/01635581.2014.864420. [DOI] [PubMed] [Google Scholar]
- 503.Rayalam S., Della-Fera M.A., Yang J.Y., Park H.J., Ambati S., Baile C.A. Resveratrol potentiates genistein's antiadipogenic and proapoptotic effects in 3T3-L1 adipocytes. J Nutr. 2007;137:2668–2673. doi: 10.1093/jn/137.12.2668. [DOI] [PubMed] [Google Scholar]
- 504.Gomez L.S., Zancan P., Marcondes M.C., Ramos-Santos L., Meyer-Fernandes J.R., Sola-Penna M. Resveratrol decreases breast cancer cell viability and glucose metabolism by inhibiting 6-phosphofructo-1-kinase. Biochimie. 2013;95:1336–1343. doi: 10.1016/j.biochi.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 505.Fouad M.A., Agha A.M., Merzabani M.M., Shouman S.A. Resveratrol inhibits proliferation, angiogenesis and induces apoptosis in colon cancer cells: calorie restriction is the force to the cytotoxicity. Hum Exp Toxicol. 2013;32:1067–1080. doi: 10.1177/0960327113475679. [DOI] [PubMed] [Google Scholar]
- 506.Filomeni G., Graziani I., Rotilio G., Ciriolo M.R. trans-Resveratrol induces apoptosis in human breast cancer cells MCF-7 by the activation of MAP kinases pathways. Genes Nutr. 2007;2:295–305. doi: 10.1007/s12263-007-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Kang N.H., Hwang K.A., Lee H.R., Choi D.W., Choi K.C. Resveratrol regulates the cell viability promoted by 17beta-estradiol or bisphenol A via down-regulation of the cross-talk between estrogen receptor alpha and insulin growth factor-1 receptor in BG-1 ovarian cancer cells. Food Chem Toxicol. 2013;59:373–379. doi: 10.1016/j.fct.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 508.Vanamala J., Reddivari L., Radhakrishnan S., Tarver C. Resveratrol suppresses IGF-1 induced human colon cancer cell proliferation and elevates apoptosis via suppression of IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer. 2010;10:238. doi: 10.1186/1471-2407-10-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Moon A.F., Garcia-Diaz M., Batra V.K., Beard W.A., Bebenek K., Kunkel T.A. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Lee-Chang C., Bodogai M., Martin-Montalvo A., Wejksza K., Sanghvi M., Moaddel R. Inhibition of breast cancer metastasis by resveratrol-mediated inactivation of tumor-evoked regulatory B cells. J Immunol. 2013;191:4141–4151. doi: 10.4049/jimmunol.1300606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Iwasaki K., Ray P.D., Huang B.W., Sakamoto K., Kobayashi T., Tsuji Y. Role of AMP-activated protein kinase in ferritin H gene expression by resveratrol in human T cells. Biochemistry. 2013;52:5075–5083. doi: 10.1021/bi400399f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Noh K.T., Chae S.H., Chun S.H., Jung I.D., Kang H.K., Park Y.M. Resveratrol suppresses tumor progression via the regulation of indoleamine 2,3-dioxygenase. Biochem Biophys Res Commun. 2013;431:348–353. doi: 10.1016/j.bbrc.2012.12.093. [DOI] [PubMed] [Google Scholar]
- 513.Buttari B., Profumo E., Facchiano F., Ozturk E.I., Segoni L., Saso L. Resveratrol prevents dendritic cell maturation in response to advanced glycation end products. Oxid Med Cell Longev. 2013;2013:574029. doi: 10.1155/2013/574029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Oikawa T., Hirotani K., Ogasawara H., Katayama T., Nakamura O., Iwaguchi T. Inhibition of angiogenesis by vitamin D3 analogues. Eur J Pharmacol. 1990;178:247–250. doi: 10.1016/0014-2999(90)90483-m. [DOI] [PubMed] [Google Scholar]
- 515.Chen Y., Tseng S.H. Review. Pro- and anti-angiogenesis effects of resveratrol. In Vivo. 2007;21:365–370. [PubMed] [Google Scholar]
- 516.Martin D.N., Boersma B.J., Yi M., Reimers M., Howe T.M., Yfantis H.G. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS ONE. 2009;4:e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Bao B.-Y., Yeh S.-D., Lee Y.-F. 1α,25-Dihydroxyvitamin D3 inhibits prostate cancer cell invasion via modulation of selective proteases. Carcinogenesis. 2006;27:32–42. doi: 10.1093/carcin/bgi170. [DOI] [PubMed] [Google Scholar]
- 518.Zhu W.Y., Zheng J., Fang W.G. The inhibition effect of retinoic acid on the invasion and metastasis abilities of metastatic human lung cancer cell subline. Zhonghua Bing Li Xue Za Zhi. 1994;23:347–350. [PubMed] [Google Scholar]
- 519.Ji Q., Liu X., Fu X., Zhang L., Sui H., Zhou L. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLOS ONE. 2013;8:e78700. doi: 10.1371/journal.pone.0078700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 520.Ooi L.L., Zhou H., Kalak R., Zheng Y., Conigrave A.D., Seibel M.J. Vitamin D deficiency promotes human breast cancer growth in a murine model of bone metastasis. Cancer Res. 2010;70:1835–1844. doi: 10.1158/0008-5472.CAN-09-3194. [DOI] [PubMed] [Google Scholar]
- 521.Simone R.E., Russo M., Catalano A., Monego G., Froehlich K., Boehm V. Lycopene inhibits NF-kB-mediated IL-8 expression and changes redox and PPARgamma signalling in cigarette smoke-stimulated macrophages. PLoS ONE. 2011;6:e19652. doi: 10.1371/journal.pone.0019652. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 522.Shamim U., Hanif S., Albanyan A., Beck F.W., Bao B., Wang Z. Resveratrol-induced apoptosis is enhanced in low pH environments associated with cancer. J Cell Physiol. 2012;227:1493–1500. doi: 10.1002/jcp.22865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Gupta P., Adkins C., Lockman P., Srivastava S.K. Metastasis of breast tumor cells to brain is suppressed by phenethyl isothiocyanate in a novel metastasis model. PLOS ONE. 2013;8:e67278. doi: 10.1371/journal.pone.0067278. [DOI] [PMC free article] [PubMed] [Google Scholar]