To the Editor
Recent translational studies of airway inflammation have shown that nasal epithelial cells are a good surrogate for bronchial epithelial cells1, 2 in asthma3, 4. The standard method of nasal sampling, however, requires use of a nasal speculum and specialized training. In pediatric studies requiring longitudinal specimen collection, sampling by this method may be limited by subject refusal and technical challenges.
Alternate methods of nasal sampling have been proposed. Different instruments for collection have been used, ranging from polyester tipped swabs, plastic curettes, to cytology brushes. Different sampling locations have been proposed, such as beneath the inferior turbinate or the anterior nares, where respiratory epithelial cells are also located5. Nasal epithelial cells obtained with use of a cytology brush beneath the inferior turbinate has been the most commonly used method; it has been validated as a surrogate for bronchial epithelial cells2, and has been shown to be clinically important in translational asthma studies4. This method has also been shown in preliminary studies to be more difficult to tolerate6. Whether a more comfortable method of sampling exists, and whether this method can provide equivalent cytologic, gene expression, and epigenetic results, is undetermined.
Here, we compared nasal epithelial cells sampled from the anterior nares using either a polyester swab or a cytology brush with the standard collection method - cytology brush sampling from beneath the inferior turbinate. The benefit of the former method is that it does not require the use of a speculum to visualize nasal anatomy, is technically easy to perform, and with the swab method, is already widely used in clinical practice for obtaining microbiologic samples.
Informed consent was obtained from 12 healthy adults. Four samples were collected from each subject; for each nare, paired inferior turbinate and anterior nare samples were collected. Inferior turbinate samples were collected with cytology brushes using nasal speculums for direct visualization. Anterior nare samples were obtained by inserting either a brush or swab inferior to the nasal bone, and vigorously rubbing along the nares (detailed methods in Online Repository). Each sample was immediately aliquoted for cytology, RNA, and DNA extraction. Paired samples with sufficient nucleic acid yield for downstream microarray analysis were analyzed for whole genome expression (Illumina HumanHT-12 v4 Expression BeadChip) and methylation (Illumina Beadchip Infinium HD array) levels. Subjects were asked to rate their discomfort level immediately after each collection using a numerical 0 to 10 point rating scale.
The largest observed difference between sample collection methods for measured parameters was between the location of sampling (inferior turbinate vs. anterior nares) rather than the use of a cytology brush vs. swab (Table 1). Average discomfort levels were significantly higher for inferior turbinate compared to anterior nares sampling (median 3.5 [IQR 2.9-7.0] vs. 1 [0.4-2.0], p < 0.001). Inferior turbinate samples had significantly more respiratory epithelial cells than anterior nares samples (median 99.2 [IQR 92.2 - 100.0] % vs 65.4 [46.8- 84.7] %, p < 0.001, Supplemental Figure E1); proportions for squamous epithelial cells were reversed. Regardless of sampling method, there were very few inflammatory cells in all samples (median 0, IQR 0–1.2%).
Table 1.
All measured parameters comparing inferior turbinate to anterior nares sampling locations. Parameters expressed as median (interquartile range). 48 samples were collected from 12 subjects. 24 samples from 8 subjects underwent downstream whole genome expression and methylation analysis.
| Parameter | Inferior turbinate | Anterior nares | p-valuea |
|---|---|---|---|
| Discomfort b | 3.5 [2.9 – 7.0] | 1.0 [0.4 – 2.0] | <0.001 |
| Proportion respiratory epithelial cells (%) | 99.2 [92.2 – 100.0] % | 65.4 [46.8 – 84.7] % c | <0.001 |
| RNA yield (ng) | 4620 [2364 – 6,993] ng | 156 [66 – 606] ng d | <0.001 |
| RNA integrity number (RIN) | 8.9 [8.6 – 9.5] | 2.2 [1.0 – 3.9] e | <0.001 |
| DNA yield (ng) | 4,288 [2,081 – 7,600] ng | 875 [181 – 2375] ng f | <0.001 |
| Gene expression (correlation) g | 0.94 [0.93 – 0.96] | 0.91 [0.78 – 0.94] | - |
| Gene expression (average relative error) g | 6.1 [3.0 – 6.0] % | 9.1 [3.7 – 15.3] % | - |
| Methylation, all genes (correlation) g | 0.97 [0.96 – 0.98] | 0.93 [0.81 – 0.98] | - |
| Methylation, all genes (average relative error) g | 6.1 [5.2 – 7.4] % | 7.0 [4.7 – 8.8] % | - |
| Methylation, asthma genes (correlation) g | 0.98 [0.97 – 0.99] | 0.95 [0.88 – 0.93] | - |
| Methylation, asthma genes (average relative error) g | 5.8 [5.0 – 6.1] % | 7.6 [3.1 – 10.4] % | - |
p-value comparing parameters from inferior turbinate sampling vs. anterior nares sampling
Subjects asked to rate discomfort level on a standardized 0–10 numerical rating scale, where 0 indicates no discomfort and 10 indicates maximum discomfort. For anterior nares location, no significant differences between brush (1.0 [0.63 – 2.8]) and swab (1.3 [0.3 – 2.0]) method, p=0.51.
For anterior nares location, no statistically significant differences between brush (82.4 [59.6 – 84.9]) and swab (50.3 [42.3 – 71.5]) method, p=0.28. For anterior nares location, brush (11.0 [5.8 – 39.3]) had lower proportion of squamous epithelial cells compared to swab (49.7 [24.4 – 50.9]) method (p < 0.001).
For anterior nares location, no statistically significant differences between brush (192 [81 – 615]) and swab (156 [60 – 588]) method, p=0.58.
For anterior nares location, brush (3.9 [2.7 – 5.3]) with higher RIN compared to swab (1.1 [1 – 1.8]) method, p=0.047.
For anterior nares location, no statistically significant differences between brush (850.0 [175.0 – 2343.8]) and swab (875.0 [212.5 – 2281.3]) method, p=0.87.
Correlation and average relative error calculated between left and right inferior turbinate samples from same subject, and between inferior turbinate and anterior nares samples from same subject.
Inferior turbinate samples had higher RNA yields (4,620 vs. 156 ng, p < 0.001) and higher RNA integrity numbers (8.9 vs 2.2, p < 0.001) than anterior nares samples (Table 1). The same was true for DNA yields (4,288 vs. 875 ng, p < 0.001). Importantly, 91.7% of inferior turbinate samples but only 33.3% of anterior nares samples yielded sufficient RNA, and 100% of inferior turbinate samples but only 50% of anterior nares samples yielded sufficient DNA, for downstream microarray analysis.
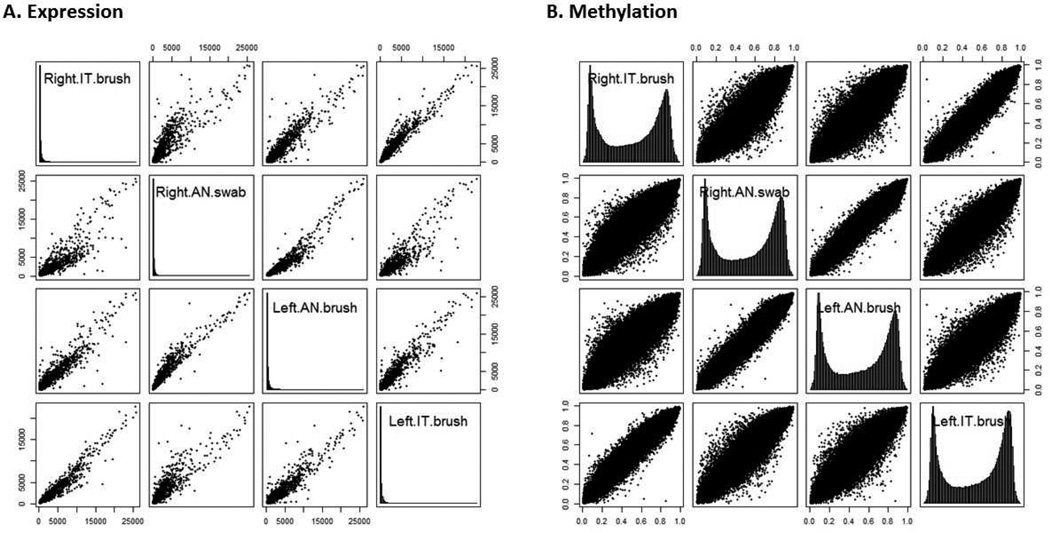
24 samples were selected for paired whole genome expression and methylation analysis. 7 failed to hybridize to the expression microarray; of these, all were anterior nares samples with low RNA yields. Despite the low RNA integrity number in remaining anterior nares samples, average expression intensity was highly correlated in expressed genes when comparing inferior turbinate with anterior nares samples from the same individual (correlation 0.91 [0.78-0.94], Figure 1A). To determine the accuracy of the two sampling techniques, we calculated the relative error averaged over all genes. Average relative error comparing anterior nares to inferior turbinate samples was not significantly different from comparing left and right inferior turbinate samples from the same individual (9.1 [3.7-15.3] % vs. 6.1 [3.0-6.0] %, p = 0.19).
Figure 1. Correlation of expression and methylation patterns from different sampling locations.
Figure depicts samples obtained from subject 10 as a representative figure. 2A. Scatterplot of gene-level expression intensities from Illumina HumanHT-12 v4 Expression BeadChip array. Nasal samples from inferior turbinate (IT) and anterior nares (AN) locations collected using cytology brushes or polyester swabs. Histogram of expression intensities for each sample plotted on the diagonal. 2B. Scatterplot of all varying methylation sites from Illumina Beadchip Infinium HD array. Note that anterior nares and inferior turbinate samples from the same individual are highly correlated for both gene expression and methylation.
All 24 DNA samples were successfully hybridized to the methylation array. Among all variable methylation sites, methylation was again highly correlated between inferior turbinate and anterior nares samples from the same individual (correlation 0.93 [0.81-0.98], Figure 1B). Average relative error was also not significantly different (6.1 [5.2-7.4] % vs. 7.0 [4.7-8.8] %, p=0.67). When we looked only at genes previously reported to be associated with asthma (Supplemental Table E1), methylation was highly correlated between inferior turbinate and anterior nares samples (Supplemental Figure E2, correlation 0.95 [0.88-0.93]).
High dimensional genomics studies have recently focused on the use of nasal epithelial cells as a noninvasive surrogate for bronchial epithelial cells in asthma, with the hope that prognostic biomarkers discovered can ultimately be translated to the bedside. While nasal epithelial cell collection from beneath the inferior turbinate has been the standard collection method, it requires specialized training and equipment, can be uncomfortable, and is unlikely to be broadly implemented given these challenges. Nasal sampling using polyester swabs, however, is already widely used by clinicians for microbiologic testing. This is the first study to comprehensively evaluate cytologic, expression, and methylation patterns in epithelial cells obtained from inferior turbinate vs anterior nares sampling. Although one limitation of our study is that it was performed in healthy subjects, we identified anterior nares sampling as a promising alternative way to obtain nasal epithelial cells. Both expression and methylation markers are highly correlated between anterior nares and inferior turbinate samples with comparable relative error. RNA quantity and degradation in anterior nares samples likely limits the use of this method for expression studies. In methylation studies involving children, or in biomarker discovery studies targeted towards clinical practice, anterior nares sampling represents a promising alternative and should be explored in future asthma trials.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Casey J Brennan and Kaylene Lin for their help in sample processing and nucleic acid extraction.
Support: This work was supported in part by NIH grants R01 AI 073964, U01 AI 110397, K24 AI 106822, U10HL098102 (PI: Phipatanakul), NIH-NIEHS P01ES018181, K23ES023700, EPA RD83451501, AAAAI/ALA Respiratory Faculty Award, and a Pilot Project grant from the HSPH-NIEHS Center for Environmental Health (P30ES000002). This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no conflicts of interest to disclose.
References
- 1.McDougall CM, Blaylock MG, Douglas JG, Brooker RJ, Helms PJ, Walsh GM. Nasal epithelial cells as surrogates for bronchial epithelial cells in airway inflammation studies. Am J Respir Cell Mol Biol. 2008;39:560–568. doi: 10.1165/rcmb.2007-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Sebastiani P, Liu G, Schembri F, Zhang X, Dumas YM, et al. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 2010;41:1–8. doi: 10.1152/physiolgenomics.00167.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guajardo JR, Schleifer KW, Daines MO, Ruddy RM, Aronow BJ, Wills-Karp M, et al. Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. J Allergy Clin Immunol. 2005;115:243–251. doi: 10.1016/j.jaci.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O'Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133:670–678. doi: 10.1016/j.jaci.2013.11.025. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pipkorn U, Karlsson G. Methods for obtaining specimens from the nasal mucosa for morphological and biochemical analysis. Eur Respir J. 1988;1:856–862. [PubMed] [Google Scholar]
- 6.Stokes AB, Kieninger E, Schogler A, Kopf BS, Casaulta C, Geiser T, et al. Comparison of three different brushing techniques to isolate and culture primary nasal epithelial cells from human subjects. Exp Lung Res. 2014;40:327–332. doi: 10.3109/01902148.2014.925987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.