Abstract
AIM: To investigate the expression characteristics of peroxiredoxin 1 (PRDX1) mRNA and protein in liver cancer cell lines and tissues.
METHODS: The RNA sequencing data from 374 patients with liver cancer were obtained from The Cancer Genome Atlas. The expression and clinical characteristics of PRDX1 mRNA were analyzed in this dataset. The Kaplan-Meier and Cox regression survival analysis was performed to determine the relationship between PRDX1 levels and patient survival. Subcellular fractionation and Western blotting were used to demonstrate the expression of PRDX1 protein in six liver cancer cell lines and 29 paired fresh tissue specimens. After bioinformatics prediction, a putative post-translational modification form of PRDX1 was observed using immunofluorescence under confocal microscopy and immunoprecipitation analysis in liver cancer cells.
RESULTS: The mRNA of PRDX1 gene was upregulated about 1.3-fold in tumor tissue compared with the adjacent non-tumor control (P = 0.005). Its abundance was significantly higher in men than women (P < 0.001). High levels of PRDX1 mRNA were associated with a shorter overall survival time (P = 0.04) but not with recurrence-free survival. The Cox regression analysis demonstrated that patients with high PRDX1 mRNA showed about 1.9-fold increase of risk for death (P = 0.03). In liver cancer cells, PRDX1 protein was strongly expressed with multiple different bands. PRDX1 in the cytosol fraction existed near the theoretical molecular weight, whereas two higher molecular weight bands were present in the membrane/organelle and nuclear fractions. Importantly, the theoretical PRDX1 band was increased, whereas the high molecular weight form was decreased in tumor tissues. Subsequent experiments revealed that the high molecular weight bands of PRDX1 might result from the post-translational modification by small ubiquitin-like modifier-1 (SUMO1).
CONCLUSION: PRDX1 was overexpressed in the tumor tissues of liver cancer and served as an independent poor prognostic factor for overall survival. PRDX1 can be modified by SUMO to play specific roles in hepatocarcinogenesis.
Keywords: Peroxiredoxin 1, Liver cancer, Prognostic factor, Post-translational modification, SUMOylation
Core tip: Peroxiredoxin 1 (PRDX1) is an antioxidant enzyme, and, therefore, it is considered a tumor suppressor gene. However, only recently has various data revealed that PRDX1 not only functions in peroxide detoxification but also in tumorigenesis. Here, we found that PRDX1 was overexpressed in liver cancer at the transcriptional level, and it was an independent unfavorable prognostic factor for overall survival. In liver cancer cells, PRDX1 is post-translationally modified by small ubiquitin-like modifier. The downregulation of sumoylated PRDX1 in tumors might participate in hepatocarcinogenesis. PRDX1 represents both a prognostic biomarker and therapeutic target for liver cancer.
INTRODUCTION
Liver cancer is the second leading cause of cancer death worldwide, accounting for about 9.1% of total cancer deaths. It was estimated that 782500 new cases and 745500 deaths occurred globally in 2012, and China alone accounted for about 50% of the total number of cases and deaths[1]. Overwhelmingly, chronic infection with hepatitis B or C virus, alcoholic liver disease, and nonalcoholic fatty liver disease are the major risk factors of liver cancer[2]. The exposures of these factors generally contribute to the multistep development of liver cancer by promoting extensive oxidative stress, liver inflammation, and immune response[3]. Among them, reactive oxygen species (ROS) can promote many aspects of tumor development and progression via oxidative DNA and protein damage, lipid peroxidation, damage to tumor suppressor genes, and enhanced expression of proto-oncogenes etc.[4].
Under normal physiological conditions, the intracellular ROS are detoxified by non-enzymatic molecules (i.e., glutathione, flavonoids, and vitamins A, C, and E) or antioxidant enzymes. There are at least five families of antioxidant enzymes with specifically scavenging capacity, including superoxide dismutases, catalases, peroxiredoxins (PRDXs), thioredoxins, and glutathione system etc.[4]. Among them, PRDXs use thioredoxin as the electron donor to catalyze the reduction of hydrogen peroxide, organic hydroperoxides, and peroxynitrite. Mammalian cells express six isoforms of PRDXs, which are classified into three subfamilies based on the location or absence of the essential catalytic cysteine (Cys) residue, 2-Cys (PRDX1, 2, 3, 4), atypical 2-Cys (PRDX5), and 1-Cys (PRDX6). PRDX1 is the most abundant and ubiquitously distributed isoform.
Notably, recent evidence suggests that hydrogen peroxide may serve as an intracellular signaling messenger molecule in response to stimulation in many mammalian cell types[4-7]. It oxidizes the critical residues of its effectors, as exemplified by the inhibition of protein-tyrosine phosphatases and the tumor suppressor phosphatase and tensin homolog (PTEN)[8,9]. Actually, PRDX1 has been reported to act as an intermediate in cell signaling via oxidizing several signaling proteins[10-12] to regulate cell proliferation, differentiation, apoptosis, migration, angiogenesis, and senescence[12-15]. Therefore, PRDX1 has a dual function in tumorigenesis. On the one hand, it functions as a tumor suppressor gene. Prdx1-/- mice have a shortened lifespan due to severe hemolytic anemia and several malignant cancers, including liver cancer[16]. In Prdx1-/- fibroblasts and mammary epithelial cells, it was shown to act as a safeguard for the lipid phosphatase activity of PTEN to suppress H-Ras and ErbB-2-induced cell transformation[17]. On the other hand, PRDX1 can act in a manner independent of its peroxide detoxifying function. The high level of PRDX1 was associated with a high potential for recurrence in squamous cell carcinoma of the tongue[18] and diminished overall survival and disease-free survival in gallbladder cancer, ovarian serous carcinomas, lung cancer, and pancreatic cancer[19-22]. In addition, inhibition of PRDX1 increases radio- and chemosensitivity in glioma and lung cancer[23-26]. In prostate cancer, it enhances the transactivation of androgen receptor[27].
In liver cancer, the overexpression of PRDX1 mRNA and protein has been observed in limited clinical specimens. Increased PRDX1 expression was associated with tumor angiogenesis, progression, and tumor necrosis factor alpha related apoptosis inducing ligand resistance and served as an independent poor prognosis factor[28,29]. Silencing PRDX1 in HepG2 cells partially reversed the tumor phenotype via the downregulation of proteins involved in cell proliferation and differentiation[30]. In this study, we investigated the expression and clinical significance of PRDX1 mRNA in liver cancer using an RNA sequencing dataset from The Cancer Genome Atlas (TCGA) (n = 374). Meanwhile, according to the protein expression and subcellular localization of PRDX1, a novel post-translational modification form of PRDX1 was explored in liver cancer cells.
MATERIALS AND METHODS
Cell lines and cell cultures
The human liver cancer cell lines HepG2, Hep3B, and SK-HEP-1 were obtained from the American Type Culture Collection (Rockville, MD, United States). Bel-7402, Bel-7404, and SMMC-7721 liver cancer cells were purchased form Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China); HLE cell was purchased from the Human Science Research Resources Bank (Osaka, Japan). They were maintained in recommended media at 37 °C with 5% CO2.
Protein extraction from cells
For total proteins extraction, cells in the exponential phase of growth were harvested using a protein lysis buffer (pH 7.4) containing 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS), 10 mmol/L N-methylmaleylimide (Sigma-Aldrich, St. Louis, MO, United States) and protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). The protein content was determined by Coomassie Plus Protein Assay (Pierce, Rockford, IL, United States). In addition, subcellular protein extraction was performed using ProteoExtractTM Subcellular Proteome Extraction Kit (Calbiochem, Billerica, MA, United States) according to the manufacturer’s guidelines.
Clinical specimen collection and preparation
Surgical tissues from liver cancer patients were collected after obtaining informed consent and approval from the Institutional Review Board of the Cancer Institute and Hospital of Chinese Academy of Medical Sciences (Beijing, China). All patients were diagnosed by two senior pathologists without chemo/radiotherapy before surgical operation. A total of 29 fresh tumor and paired adjacent non-tumor liver tissue samples were collected from patients (26 male, three female; median age, 54 ± 12; range 32-78 years) undergoing resection during the period from May 2006 to November 2007. Among them, 17 cases were α-fetoprotein (AFP)-normal, while 12 were AFP-positive. The tissue samples were collected and washed right after surgical resection. They were then snap-frozen in liquid nitrogen immediately and stored at -80 °C. Fresh tissue samples were homogenized and the proteins were extracted using the protein lysis buffer described above.
Western blot analysis
Approximately 15 μg of total proteins or subcellular proteins were diluted in Laemmli buffer containing 10% β-mercaptoethanol and boiled at 95 °C for 10 min. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes. After blocking, the membranes were incubated with anti-PRDX1 (ab15571, Abcam, Cambridge, United Kingdom) and anti-β-actin (Sigma-Aldrich) antibodies. Following intensive washing, the membranes were developed with horseradish peroxidase conjugated second antibodies (Jackson Immunoresearch Lab., West Grove, PA, United States) and visualized using an enhanced chemiluminescence system (Santa Cruz Biotech., Dallas, TX, United States). The upregulation or downregulation of PRDX1 was defined as higher or lower relative band intensity in tumors compared with their paired adjacent normal liver tissues.
Immunofluorescence under confocal microscopy
HepG2, Hep3B, and SK-HEP-1 cells were grown in 0.01% poly-l-Lysine coated slides for 24 h. After fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with PBS (pH 7.4), the cells were blocked with 1% bovine serum albumin (BSA) and 0.1% Triton X-100 for 30 min at room temperature. Washed cells were incubated for 30 min with rabbit anti-PRDX1 and mouse anti-SUMO1 (Zymed Lab., San Francisco, CA, United States) antibodies. Then, the cells were incubated in the dark for 60 min with Alexa Fluor 488-conjugated goat anti-rabbit and Alexa Fluor 594-conjugated goat anti-mouse (Life Technologies, Carlsbad, CA, United States) secondary IgG. The fluorescence signals were captured under a TCS SP2 laser confocal microscope (Leica Microsystems, Wetzlar, Germany).
Immunoprecipitation
For co-immunoprecipitation analysis, 1.5 mg of whole cell lysate of HepG2 was precleared by incubating with protein G-agarose beads (Roche Diagnostics, Basel, Switzerland) at 4 °C for 1 h. The collected supernatant was incubated at 4 °C with 4 μg of rabbit anti-PRDX1 antibody, mouse anti-SUMO1 antibody, or nonimmune rabbit/mouse IgG (Zhongshan Biotechnology, Beijing, China) overnight with rotation. The immune complex was precipitated by incubation with 50 μL of protein G-agarose for 3 h at 4 °C. The agarose beads were pelleted by centrifugation and washed three times with lysis buffer. The beads were suspended in 2 × Laemmli sample buffer and boiled for 5 min. Protein G-agarose beads were removed from the complex by centrifugation at 10000 g for 5 min. The supernatant was loaded onto 10% SDS-PAGE for Western blot analysis with respective antibodies to PRDX1 and SUMO1.
TCGA RNA sequencing data mining and statistical analysis
The liver cancer transcriptome dataset was obtained from TCGA. The RNA sequencing data from 49 non-tumor liver tissues and 374 tumor tissues were available. The expression of PRDX1 mRNA and its clinical significance was analyzed. Mann-Whitney U test was used to compare the Read per Million (RPM) between two groups. The Kaplan-Meier method was used to determine the relationship between the RPM of PRDX1 and patient survival, and log-rank analysis was performed to compare survival curves. Univariate and multivariate analyses were performed using the Cox regression model. In addition, the bioinformatic tool SUMOplot (http://www.abgent.com/sumoplot) was used to predict the putative SUMOylation sites of PRDX1. P values < 0.05 were considered significant. All analyses were performed using Graphpad prism 6.0 (GraphPad Software Inc., La Jolla, CA, United States).
RESULTS
Upregulation of PRDX1 mRNA in human liver cancer tissues
According to the RNA sequencing data from TCGA, PRDX1 mRNA was upregulated approximately 1.3-fold in tumor tissues (n = 374) compared with adjacent non-tumor livers (n = 49) (Figure 1A, P = 0.005). Based on the median RPM value of PRDX1 in tumor tissues, all 374 cases were divided into two groups, high level group and low level group. Kaplan-Meier survival analysis with a log-rank test showed a significant correlation between high PRDX1 mRNA expression and shorter overall survival time (P = 0.04) in liver cancer patients (Figure 1B, left panel). The median survival times of high and low expression groups were 635 and 498 d, respectively. However, the expression of PRDX1 mRNA was not associated with recurrence of the patients with liver cancer (Figure 1B, right panel). Even for patients who had received a curative resection (R0 resection, n = 310), high levels of PRDX1 mRNA also was correlated with shorter overall survival (P = 0.05) but not with recurrence (Supplementary file).
Figure 1.
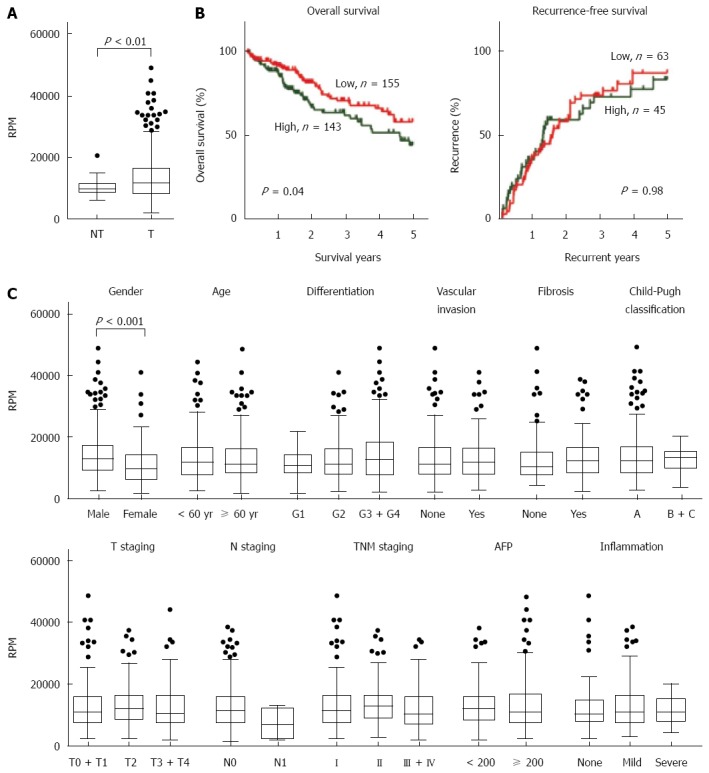
Expression and clinicopathological characteristics of peroxiredoxin 1 mRNA presented in The Cancer Genome Atlas liver cancer RNA sequencing dataset. A: Peroxiredoxin 1 (PRDX1) mRNA was significantly up-regulated in tumor tissues (n = 374) compared with the adjacent non-tumor tissues (n = 50); B: Kaplan-Meier curves of overall survival (left panel) and recurrence (right panel) according to the PRDX1 levels in tumor samples. Log-rank test was performed; C: The clinicopathological characteristics analysis of PRDX1 expression in 374 liver cancer cases. RPM: Read per Million; RPM: Read per Million; AFP: α-fetoprotein.
These findings were further confirmed by both univariate and multivariate Cox regression analysis (Tables 1 and 2). In the univariate analysis, compared with the low PRDX1 mRNA expression group, patients with the high PRDX1 mRNA exhibited a 1.55-fold increase of relative risk (RR) for overall survival (P = 0.04). Other significant risk factors included Child-Pugh classification (RR = 2.49, P = 0.01) and tumor, node, metastases (TNM) staging (RR = 2.39, P < 0.001). As concluded by the multivariate analysis, TNM staging (RR = 2.36, P = 0.007), Child-Pugh classification (RR = 2.30, P = 0.03), and PRDX1 mRNA expression (RR = 1.89, P = 0.03) were the independent prognostic factors for death. For recurrence-free survival, in the univariate analysis, residual tumor (RR = 2.25, P = 0.05), vascular invasion (RR = 1.65, P = 0.05), and TNM staging (RR = 4.32, P < 0.001) were associated with increased risk of recurrence. Only TNM staging (RR = 5.04, P < 0.001) was considered as an independent recurrent factor.
Table 1.
Univariate and multivariate survival analysis for overall survival and recurrence-free survival in The Cancer Genome Atlas patients with hepatocellular carcinoma
| Variables |
Overall survival |
Recurrence-free survival |
||
| Relative risk (95%CI) | P value | Relative risk (95%CI) | P value | |
| Univariate | ||||
| Age (> 60 vs ≤ 60 yr) | 1.17 (0.76-1.80) | 0.48 | 1.33 (0.83-2.14) | 0.24 |
| Gender (female vs male) | 1.21 (0.78-1.86) | 0.39 | 0.91 (0.72-1.15) | 0.44 |
| Differentiation (Poorly vs well and moderately) | 1.27 (0.83-1.95) | 0.28 | 0.76 (0.48-1.21) | 0.25 |
| Residual tumor (R1 + R2 vs R0) | 1.95 (0.94-4.06) | 0.07 | 2.25 (1.02-4.98) | 0.05 |
| Child-Pugh classification (grade B vs A) | 2.49 (1.21-5.12) | 0.01 | 2.22 (0.95-5.19) | 0.07 |
| Vascular invasion (macro + micro vs none) | 1.52 (0.94-2.46) | 0.08 | 1.65 (1.00-2.71) | 0.05 |
| TNM staging (III + IV vs I + II) | 2.39 (1.51-3.79) | < 0.001 | 4.32 (2.50-7.47) | < 0.001 |
| Fibrosis (fibrosis + cirrhosis vs none) | 0.98 (0.88-1.08) | 0.63 | 0.81 (0.46-1.41) | 0.45 |
| AFP (≥ 200 ng/mL vs < 200 ng/mL) | 1.15 (0.68-1.92) | 0.61 | 0.85 (0.49-1.49) | 0.58 |
| PRDX1 expression (high vs low) | 1.55 (1.01-2.36) | 0.04 | 1.01 (0.63-1.60) | 0.98 |
| Inflammation in adjacent liver (severe + mild vs none) | 1.20 (0.79-1.80) | 0.39 | 1.30 (0.82-2.07) | 0.26 |
| Multivariate | ||||
| Residual tumor (R1 + R2 vs R0) | - | - | 0. 89 (0.28-2.89) | 0.77 |
| Child-Pugh classification (grade B vs A) | 2.30 (1.10-4.78) | 0.03 | - | - |
| Vascular invasion (macro + micro vs none) | - | - | 1.42 (0.79-2.55) | 0.23 |
| TNM staging (III + IV vs I + II) | 2.36 (1.26-4.41) | 0.007 | 5.04 (2.76-9.22) | < 0.001 |
| PRDX1 expression (high vs low) | 1.89 (1.07-3.36) | 0.03 | - | - |
AFP: α-fetoprotein; PRDX1: Peroxiredoxin 1.
Table 2.
Expression of peroxiredoxin 1 protein and its clinical significance
|
22 kDa band |
P value |
50 kDa band |
P value | |||
| n | Up-regulated (%) | n | Down-regulated (%) | |||
| Gender | ||||||
| Male | 26 | 45.2 (12/26) | 1.00 | 26 | 69.2 (18/26) | 1.00 |
| Female | 3 | 33.3 (1/3) | 3 | 66.7 (2/3) | ||
| Age | ||||||
| ≥ 60 | 10 | 30.0 (3/10) | 0.43 | 10 | 40.0 (4/10) | 0.03 |
| < 60 | 19 | 52.6 (10/19) | 19 | 65.5 (16/19) | ||
| Tumor size | ||||||
| ≤ 3 cm | 7 | 42.9 (3/7) | 1.00 | 7 | 42.9 (3/7) | 0.16 |
| > 3 cm | 22 | 45.5 (10/22) | 22 | 77.3 (17/22) | ||
| Differentiation | ||||||
| Well | 7 | 28.6 (2/7) | 0.41 | 7 | 71.45 (5/7) | 1.00 |
| Moderately | 18 | 55.6 (10/18) | 18 | 66.7 (12/18) | ||
| Poorly | 4 | 25.0 (1/4) | 4 | 75.0 (3/4) | ||
| TNM staging | ||||||
| I-II | 20 | 35.0 (7/20) | 0.23 | 20 | 70.0 (14/20) | 1.00 |
| III-IV | 9 | 66.7 (6/9) | 9 | 66.7 (6/9) | ||
| AFP | ||||||
| ≥ 200 ng/mL | 7 | 42.9 (3/7) | 1.00 | 7 | 71.4 (5/7) | 1.00 |
| < 200 ng/mL | 22 | 45.5 (10/22) | 22 | 68.2 (15/22) | ||
AFP: α-fetoprotein.
The correlations between the clinicopathologic characteristics of liver cancer patients and the expression of PRDX1 mRNA in their tumors were also compared (Figure 1C). The levels of PRDX1 mRNA were higher in males than females (P < 0.001). The correlation between PRDX1 mRNA and other features, such as age, differentiation degree, vascular invasion, Child-Pugh classification, TNM staging, hepatic fibrosis degree, serum AFP levels, and hepatic inflammation in adjacent liver tissue, was not observed.
Expression and localization of PRDX1 protein in liver cancer cell lines
In addition to whole cell lysates, cytosol, membrane/organelle, and nuclear protein fractions were extracted from liver cancer cells (HepG2, Hep3B, SK-HEP-1, Bel-7404, SMMC-7721, and HLE) to enhance the visibility of moderate- and low-abundance proteins. PRDX1 was expressed in the whole lysates of all six cells, represented by multiple different bands (Figure 2). Additionally, the subcellular protein analysis showed that PRDX1 in the cytosol fraction existed near the theoretical molecular weight form (22 kDa), whereas two higher molecular weight bands, approximately 35 and 50 kDa, were present in the membrane/organelle and nuclear fractions, especially the membrane/organelle fraction (Figure 2). Thus, PRDX1 protein was ubiquitously distributed in the liver cancer cells, and it existed as multiple forms in the membrane/organelle and nuclear fractions.
Figure 2.
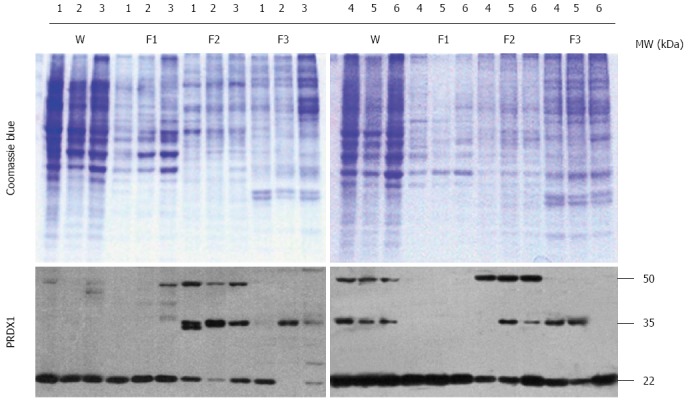
Western blotting analysis of peroxiredoxin 1 protein in liver cancer cells. The codes of liver cancer cells: 1: HepG2; 2: Hep3B; 3: SK-HEP-1; 4: Bel-7404; 5: SMMC-7721; 6: HLE. W: Whole lysates; F1: Cytosol fraction; F2: Membrane/organelle fraction; F3: Nucleus fraction. The upper panel is the Coomassie Blue stained SDS-PAGE gel, and the lower panel is the Western blotting of PRDX1. PRDX1: Peroxiredoxin 1.
Expression of PRDX1 protein in human liver cancer samples
To confirm the observation in liver cancer cells, the expression of PRDX1 in liver cancer patients was analyzed using Western blotting. We found that PRDX1 was detected as two bands with different molecular weight, the theoretical 22 kDa band and the higher 50 kDa band. Compared with the non-tumorous corresponding tissues, the theoretical PRDX1 band was increased in 48.3% (14/29) of neoplastic liver tissues, whereas the higher 50 kDa form was downregulated in 69.0% (20/29) of tumor tissues (Figure 3). The total intensity of PRDX1 was downregulated in 18 out of 29 tumor tissues (62.1%, 18/29).
Figure 3.
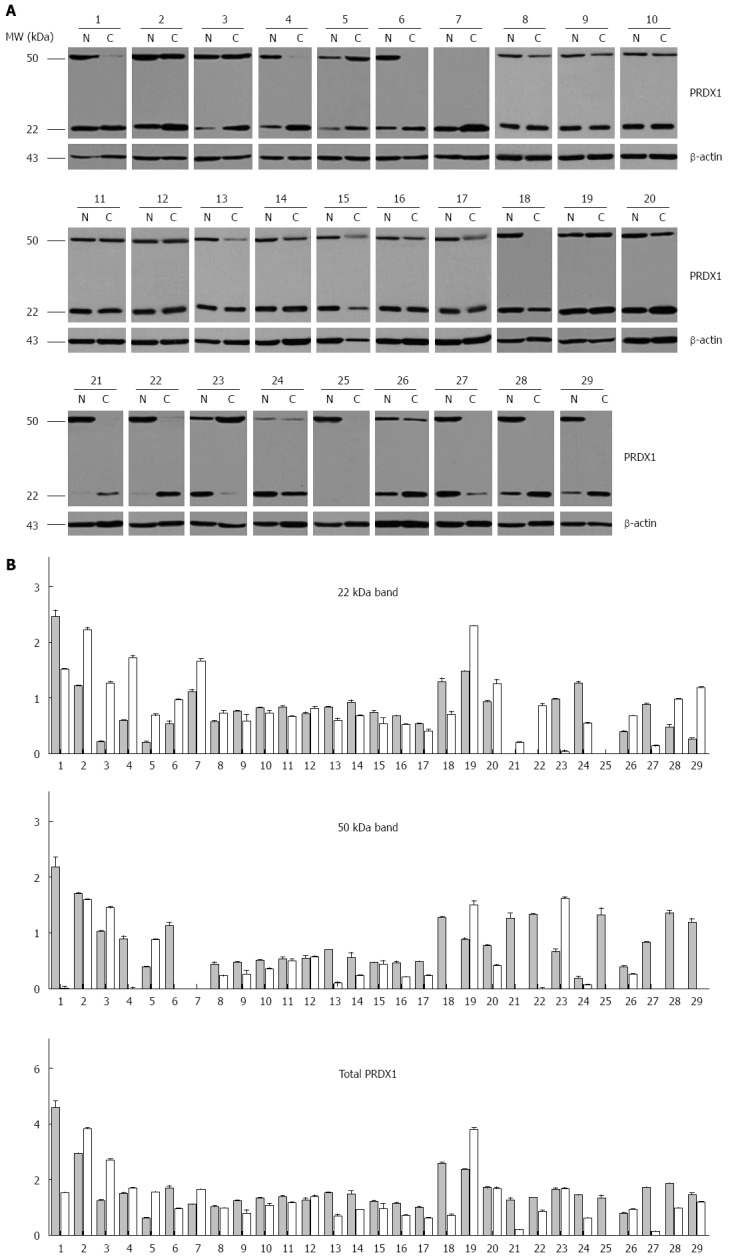
Expression of peroxiredoxin 1 in human liver cancer specimens. A: Western blot analysis of tumor (C) and matching adjacent non-tumor liver tissues (N) of 29 patients. β-actin protein levels are shown as a loading control. The patients were coded from 1 to 29; B: Densitometric analysis of 29 hepatocellular carcinoma cases. The black and gray bars represent the relative band intensity of peroxiredoxin 1 (PRDX1) in non-tumor or tumor tissues, showing the ratio between 22 kDa, 50 kDa or total PRDX1 and β-actin. Each data point represents the mean ± SD derived from three independent experiments.
The clinical association trend for PRDX1 expression was observed in these 29 patients. We only found that downregulation of the 50 kDa band was greater in younger patients (< 60 years old). However, there was no correlation between PRDX1 protein and gender, tumor size, differentiation, TNM staging, or serum AFP levels.
Bioinformatic prediction of mechanisms involved in the formation of high molecular weight PRDX1
The theoretical molecular weight of PRDX1 is approximately 22 kDa. Our Western blot analysis, with reducing SDS-PAGE with 10% β-mercaptoethanol, revealed two extra high molecular weight bands near 35 and 50 kDa, especially in subcellular fractions. Therefore, we suspected that PRDX1 possessed some covalent modifications that were increasing the molecular weight. The prediction of SUMOplot tool identified two consensus sequence of SUMOylation with high probability and four sites with low probability in PRDX1 (Figure 4A). The molecular weight of SUMO1 is near 12 kDa, thus we supposed that the higher molecular weight bands of PRDX1 were due to its SUMOylation.
Figure 4.
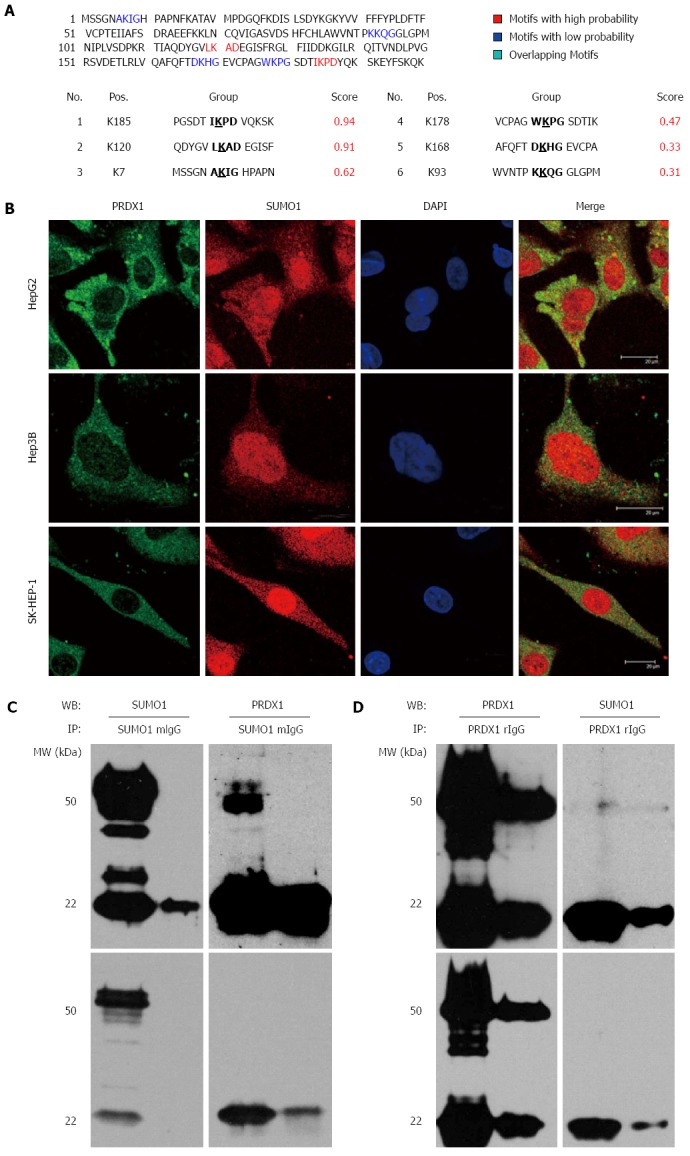
Peroxiredoxin 1 might be sumoylated in liver cancer cells. A: The bioinformatic prediction of PRDX1 using SUMOplot tool; B: Immunofluorescence staining visualized under a confocal microscope illustrating the co-localization of PRDX1 and SUMO1 proteins in the cytoplasm of three liver cancer cells. C, D: Co-immunoprecipitation of PRDX1 with SUMO1 in HepG2 cell extract; C: Lysates were subjected to immunoprecipitation (IP) with anti-SUMO1 antibody, followed by Western blotting (WB) with anti-PRDX1 and anti-SUMO1 to detect sumoylated PRDX1; D: Lysates were subjected to IP with anti-PRDX1 antibody, followed by WB with anti-SUMO1 and anti-PRDX1 to detect sumoylated PRDX1. The upper and lower panels are the results of dark and light exposure by Western blotting.
PRDX1 might be sumoylated in liver cancer cells
To investigate whether PRDX1 might be sumoylated, we determined the colocalization of PRDX1 and SUMO1 in three liver cancer cells, HepG2, Hep3B, and SK-HEP-1. We found that green endogenous PRDX1 was partially co-localized with the red SUMO1 molecules in the cytoplasm, according to immunofluorescence and confocal microscopy analysis (Figure 4B).
Furthermore, a co-immunoprecipitation assay was performed. HepG2 cells were lysed in the presence of N-ethylmaleimide, which inhibits SUMO-specific proteases, and were immunoprecipitated with an anti-PRDX1 or anti-SUMO1 antibody. As shown in Figure 4C, when the anti-SUMO1 antibody was used to precipitate SUMO1 interacting proteins, compared with non-immune IgG control, the theoretical and higher molecular weight bands of PRDX1 could be detected. Conversely, when the PRDX1 and its interacting proteins were enriched by anti-PRDX1 antibody, a weak band, about 50 kDa, was also recognized by anti-SUMO1 antibody (Figure 4D). Unfortunately, due to the low molecular weight characteristics of SUMO1, it was not detected in our Western blot system. These results indicated that PRDX1 and SUMO1 interacted with one another and suggested that PRDX1 might undergo SUMOylation in liver cancer cells.
DISCUSSION
In the present study, we found PRDX1 mRNA was upregulated in the tumor tissues of liver cancer in a large sample size. Increased PRDX1 was associated with male gender and shorter overall survival. Western blotting revealed that PRDX1 had two bands on SDS-PAGE gel (about 35 and 50 kDa) that were higher molecular weight than the theoretical molecular weight (22 kDa). These higher molecular weight bands mainly existed in the membrane/organelle and nuclear fractions. Subsequent immunofluorescence and co-immunoprecipitation assays hinted that the higher molecular weight bands might be due to SUMOylation of PRDX1.
In addition to its peroxide detoxifying function, PRDX1 interacts and regulates the activity of several vital proteins. For example, PRDX1 can bind the SH3 domain of c-Abl, Myc Box II domain of c-Myc, C2 domain of PTEN, androgen receptor, apoptosis signal-regulating kinase 1, mammalian ste20-like kinase 1 to modulate their activities[13,17,27,31-34]. Moreover, PRDX1 promotes tumor development and progression through Toll-like receptor 4 and mammalian target of rapamycin/p70S6K pathways and tumor growth factor β1-induced epithelial-mesenchymal transition[14,15,35]. Meanwhile, PRDX1 can act as a chaperone to enhance the transactivation potential of NF-κB in estrogen receptor negative breast cancer cells[36]. Recently, PRDX1 was found to bind RNA and serve as a transcription anti-terminator to enhance the survival of cells exposed to cold stress[37].
In tumors, hypoxia or ROS can induce the expression of PRDX1[38-41]. Although PRDX1 was shown to be overexpressed in most of tumors, because of its complex functions, its clinical significance was dependent on tumor type. For example, PRDX1 was a favorable prognostic factor in esophageal squamous cell carcinoma, breast cancer, bladder cancer, and cholangiocarcinoma[42-45], whereas an opposite role was attributed to PRDX1 in squamous cell carcinoma of the tongue, gallbladder cancer, ovarian serous carcinomas, lung cancer, pancreatic cancer, and liver cancer[18-22,29].
Our study confirmed that PRDX1 mRNA was upregulated in liver cancer tumor tissues, and its high levels were associated with shorter overall survival time. However, according to our large sample size, we did not observe the correlation between PRDX1 and the recurrence-free survival of patients, as reported previously[29]. Moreover, the expression of PRDX1 was higher in male patients. A previous study also found that expression levels of PRDX1 were relatively high in hepatitis C virus-related hepatocellular carcinoma samples from men[46], concordant with our results. It was known that liver cancer has a high male-to-female incidence rate ratio of 2-4:1[1], and men with liver cancer tend to have a more invasive phenotype and shorter survival[47]. Thus, the prognostic value of PRDX1 mRNA might relate to the gender-disparity of liver cancer.
At the protein level, we found that PRDX1 had three different molecular weight forms in liver cancer cells, and the higher molecular weight bands were mainly distributed in the cell organelles and nucleus. They were also detected in the tissue lysates. Meanwhile, the higher molecular weight bands were downregulated in tumor tissues, whereas the theoretical molecular weight band was upregulated. Our Western blot system is reducing SDS-PAGE including 10% β-mercaptoethanol; therefore, the dimer cannot explain the phenomenon. We predicted that the shift in molecular weight was due to a covalent modification rather than a disulfide bond for PRDX1 in liver cancer. To support this hypothesis, we performed the bioinformatic prediction, immunofluorescence, and co-immunoprecipitation assay, and the putative modification forms of PRDX1 by SUMO were confirmed.
SUMOs are ubiquitin-like polypeptides that covalently conjugate to proteins in an ATP-dependent enzymatic cascade that resembles ubiquitylation[48]. Hundreds of proteins can be modified by SUMOs, including oxidative stress-related proteins. Hydrogen peroxide enhances the global protein SUMO conjugation profile and induces a reversible blockade of SUMO proteases sentrin-specific protease 1[49,50]. ROS induces a rapid de-SUMOylation of transcription factors c-Fos and c-Jun, resulting in stimulation of their activity and activation of numerous anti-oxidant proteins[51,52]. Thus, it seems that SUMOylation is a fine sensor for ROS and participates in anti-oxidative responses and ROS-dependent cell death[51].
It is known that the activity of PRDX1 can be regulated by some post-translational modifications. For example, phosphorylation of PRDX1 on Thr90 or Ser32 reduced its peroxidase activity[53-55]. Acetylation of PRDX1 on Lys197 increased its reducing activity[56]. Glutathionylation of PRDX1 at Cys52, Cys83, and Cys173 inactivated its molecular chaperone function[57]. Similarly, consequences of SUMOylation can also modulate the functions and activities of target proteins. It is widely accepted that SUMOylation can mask the interaction surface, induce conformational changes, and create SUMO-dependent interaction with downstream effectors[48]. However, due to low steady-state levels of endogenous protein modification and isopeptidase activity in nondenaturing lysates, detection and analysis of SUMOylation are challenged. Therefore, the SUMOylation of PRDXs had not been reported yet. To increase the visibility of sumoylated PRDX1, we added 10 mmol/L N-ethylmaleimide to our cell lysate buffer to stable SUMO conjugates, leading to the discovery of the higher molecular weight bands of PRDX1. It was known that PRDX1 exhibits both nuclear and cytoplasmic localization in cells[58]. However, PRDX1 has no nuclear localization signals (NLS) as predicted by PredictNLS (https://www.predictprotein.org/) and cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/) bioinformatics tools. One of the roles of SUMOylation is as a molecular switch to control the nuclear localization[59]; therefore, the SUMOylation of PRDX1 might provide a possible mechanism to localize PRDX1 to the nucleus. Furthermore, the downregulation of sumoylated PRDX1 might be involved in hepatocarcinogenesis. The confirmation and functions of sumoylated PRDX1 are continuing to be explored in our laboratory.
In conclusion, our results demonstrated upregulation of PRDX1 mRNA in liver cancer is an independent poor prognostic factor for overall survival. PRDX1 protein might modified by SUMO in liver cancer cells to form higher molecular weight bands. The sumoylated PRDX1 protein was downregulated in tumor tissues, suggesting its specific functions may be distinct from the un-modified forms in hepatocarcinogenesis. Overall, PRDX1 acts as an “oncogene” in liver cancer cells. It may be a useful prognostic marker and a promising molecular target for the therapeutic intervention of liver cancer.
COMMENTS
Background
Liver cancer is the second leading cause of cancer death worldwide. The exposures to common etiological factors generally lead to extensive oxidative stress and the promotion of hepatocarcinogenesis via oxidative DNA, protein, and lipid damage. In normal cells, several antioxidant enzymes and non-enzymatic molecules participate in the detoxification of intracellular reactive oxygen species. Among them, peroxiredoxin 1 (PRDX1) catalyzes the reduction of hydrogen peroxide, organic hydroperoxides, and peroxynitrite.
Research frontiers
As a major hydroperoxide scavenging enzyme in cytoplasm, PRDX1 is considered a tumor suppressor gene. However, PRDX1 recently was found to act in a manner independent of its anti-oxidative function. It also regulates cell proliferation, differentiation, apoptosis, migration, angiogenesis, and radio-chemosensitivity. Therefore, PRDX1 plays a dual role in tumorigenesis.
Innovations and breakthroughs
In liver cancer, the overexpression of PRDX1 had been observed in limited clinical specimens. In this study, the authors investigated the expression characteristics of PRDX1 mRNA and protein in liver cancer using RNA sequencing dataset and Western blotting. In addition, the subcellular distribution and a putative post-translational modification form of PRDX1 were explored.
Applications
PRDX1 was overexpressed in the tumor tissues of liver cancer and was shown to serve as an independent unfavorable prognostic factor for overall survival. PRDX1 protein might be modified by small ubiquitin-like modifier (SUMO) in liver cancer cells to form higher molecular weight isoforms. PRDX1 may be a useful prognostic marker for liver cancer, and it is a promising molecular target for the therapeutic intervention of liver cancer.
Terminology
PRDXs use thioredoxin as the electron donor to catalyze the reduction of hydrogen peroxide, organic hydroperoxides, and peroxynitrite. Mammalian cells express six isoforms of PRDXs, which are classified into three subfamilies based on the location or absence of the essential catalytic cysteine residue. PRDX1 is the most abundant and ubiquitously distributed isoform.
Peer-review
This manuscript is well written and suggests that PRDX1 overexpressed in the tumor tissue of liver cancer may be a poor prognostic factor for overall survival. In addition, it might be modified by SUMO to play specific roles in hepatocarcinogenesis. However, as the authors point out, SUMOylation of PRDX1 was not confirmed and its function in hepatocarcinogenesis remains unclear. Further studies are needed to confirm this idea.
Footnotes
Supported by State Key Project for Infectious Diseases, No. 2013ZX10002009 and No. 2012ZX10002-017; State Key Project for Basic Research, No. 2014CBA02001 and No. 2014CBA02002; National High-tech R and D Program, No. 2012AA020206; and Natural Science Foundation of China, No. 81071789 and No. 81321091.
Institutional review board statement: All routine colonic biopsy specimens and blood samples from the patients were taken after informed consent and ethical permission was obtained for participation in the study.
Conflict-of-interest statement: To the best of our knowledge, no conflict of interest exists.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: April 22, 2015
First decision: June 19, 2015
Article in press: August 31, 2015
P- Reviewer: Yu DY S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Liu XM
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Kumar M, Zhao X, Wang XW. Molecular carcinogenesis of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: one step closer to personalized medicine? Cell Biosci. 2011;1:5. doi: 10.1186/2045-3701-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 6.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 7.Rhee SG, Woo HA, Kil IS, Bae SH. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 9.D’Autréaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 10.Neumann CA, Cao J, Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8:4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarvis RM, Hughes SM, Ledgerwood EC. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic Biol Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Turner-Ivey B, Manevich Y, Schulte J, Kistner-Griffin E, Jezierska-Drutel A, Liu Y, Neumann CA. Role for Prdx1 as a specific sensor in redox-regulated senescence in breast cancer. Oncogene. 2013;32:5302–5314. doi: 10.1038/onc.2012.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morinaka A, Funato Y, Uesugi K, Miki H. Oligomeric peroxiredoxin-I is an essential intermediate for p53 to activate MST1 kinase and apoptosis. Oncogene. 2011;30:4208–4218. doi: 10.1038/onc.2011.139. [DOI] [PubMed] [Google Scholar]
- 14.Riddell JR, Bshara W, Moser MT, Spernyak JA, Foster BA, Gollnick SO. Peroxiredoxin 1 controls prostate cancer growth through Toll-like receptor 4-dependent regulation of tumor vasculature. Cancer Res. 2011;71:1637–1646. doi: 10.1158/0008-5472.CAN-10-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong F, Hou G, Liu H, Zhang M. Peroxiredoxin 1 promotes tumorigenesis through regulating the activity of mTOR/p70S6K pathway in esophageal squamous cell carcinoma. Med Oncol. 2015;32:455. doi: 10.1007/s12032-014-0455-0. [DOI] [PubMed] [Google Scholar]
- 16.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 17.Cao J, Schulte J, Knight A, Leslie NR, Zagozdzon A, Bronson R, Manevich Y, Beeson C, Neumann CA. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28:1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagawa T, Omura K, Harada H, Ishii T, Uwayama J, Nakaso K, Iwasa S, Koyama Y, Onizawa K, Yusa H, et al. Peroxiredoxin I expression in tongue squamous cell carcinomas as involved in tumor recurrence. Int J Oral Maxillofac Surg. 2005;34:915–920. doi: 10.1016/j.ijom.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Cai CY, Zhai LL, Wu Y, Tang ZG. Expression and clinical value of peroxiredoxin-1 in patients with pancreatic cancer. Eur J Surg Oncol. 2015;41:228–235. doi: 10.1016/j.ejso.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Yang ZL, Ren X, Zou Q, Yuan Y, Liang L, Chen M, Chen S. ILK and PRDX1 are prognostic markers in squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Tumour Biol. 2013;34:359–368. doi: 10.1007/s13277-012-0557-2. [DOI] [PubMed] [Google Scholar]
- 21.Chung KH, Lee DH, Kim Y, Kim TH, Huh JH, Chung SG, Lee S, Lee C, Ko JJ, An HJ. Proteomic identification of overexpressed PRDX 1 and its clinical implications in ovarian carcinoma. J Proteome Res. 2010;9:451–457. doi: 10.1021/pr900811x. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Bogner PN, Baek SH, Ramnath N, Liang P, Kim HR, Andrews C, Park YM. Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clin Cancer Res. 2008;14:2326–2333. doi: 10.1158/1078-0432.CCR-07-4457. [DOI] [PubMed] [Google Scholar]
- 23.Dittmann LM, Danner A, Gronych J, Wolter M, Stühler K, Grzendowski M, Becker N, Bageritz J, Goidts V, Toedt G, et al. Downregulation of PRDX1 by promoter hypermethylation is frequent in 1p/19q-deleted oligodendroglial tumours and increases radio- and chemosensitivity of Hs683 glioma cells in vitro. Oncogene. 2012;31:3409–3418. doi: 10.1038/onc.2011.513. [DOI] [PubMed] [Google Scholar]
- 24.Hwang KE, Park DS, Kim YS, Kim BR, Park SN, Lee MK, Park SH, Yoon KH, Jeong ET, Kim HR. Prx1 modulates the chemosensitivity of lung cancer to docetaxel through suppression of FOXO1-induced apoptosis. Int J Oncol. 2013;43:72–78. doi: 10.3892/ijo.2013.1918. [DOI] [PubMed] [Google Scholar]
- 25.Poschmann G, Grzendowski M, Stefanski A, Bruns E, Meyer HE, Stühler K. Redox proteomics reveal stress responsive proteins linking peroxiredoxin-1 status in glioma to chemosensitivity and oxidative stress. Biochim Biophys Acta. 2015;1854:624–631. doi: 10.1016/j.bbapap.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Xie B, Li X, Chen Y, Xu Y, Xu-Welliver M, Zou L. Downregulation of peroxiredoxin-1 by β-elemene enhances the radiosensitivity of lung adenocarcinoma xenografts. Oncol Rep. 2015;33:1427–1433. doi: 10.3892/or.2015.3732. [DOI] [PubMed] [Google Scholar]
- 27.Park SY, Yu X, Ip C, Mohler JL, Bogner PN, Park YM. Peroxiredoxin 1 interacts with androgen receptor and enhances its transactivation. Cancer Res. 2007;67:9294–9303. doi: 10.1158/0008-5472.CAN-07-0651. [DOI] [PubMed] [Google Scholar]
- 28.Song IS, Kim SU, Oh NS, Kim J, Yu DY, Huang SM, Kim JM, Lee DS, Kim NS. Peroxiredoxin I contributes to TRAIL resistance through suppression of redox-sensitive caspase activation in human hepatoma cells. Carcinogenesis. 2009;30:1106–1114. doi: 10.1093/carcin/bgp104. [DOI] [PubMed] [Google Scholar]
- 29.Sun QK, Zhu JY, Wang W, Lv Y, Zhou HC, Yu JH, Xu GL, Ma JL, Zhong W, Jia WD. Diagnostic and prognostic significance of peroxiredoxin 1 expression in human hepatocellular carcinoma. Med Oncol. 2014;31:786. doi: 10.1007/s12032-013-0786-2. [DOI] [PubMed] [Google Scholar]
- 30.Aguilar-Melero P, Prieto-Álamo MJ, Jurado J, Holmgren A, Pueyo C. Proteomics in HepG2 hepatocarcinoma cells with stably silenced expression of PRDX1. J Proteomics. 2013;79:161–171. doi: 10.1016/j.jprot.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappaB activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- 32.Mu ZM, Yin XY, Prochownik EV. Pag, a putative tumor suppressor, interacts with the Myc Box II domain of c-Myc and selectively alters its biological function and target gene expression. J Biol Chem. 2002;277:43175–43184. doi: 10.1074/jbc.M206066200. [DOI] [PubMed] [Google Scholar]
- 33.Chhipa RR, Lee KS, Onate S, Wu Y, Ip C. Prx1 enhances androgen receptor function in prostate cancer cells by increasing receptor affinity to dihydrotestosterone. Mol Cancer Res. 2009;7:1543–1552. doi: 10.1158/1541-7786.MCR-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SY, Kim TJ, Lee KY. A novel function of peroxiredoxin 1 (Prx-1) in apoptosis signal-regulating kinase 1 (ASK1)-mediated signaling pathway. FEBS Lett. 2008;582:1913–1918. doi: 10.1016/j.febslet.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Ha B, Kim EK, Kim JH, Lee HN, Lee KO, Lee SY, Jang HH. Human peroxiredoxin 1 modulates TGF-β1-induced epithelial-mesenchymal transition through its peroxidase activity. Biochem Biophys Res Commun. 2012;421:33–37. doi: 10.1016/j.bbrc.2012.03.103. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, He S, Sun JM, Delcuve GP, Davie JR. Selective association of peroxiredoxin 1 with genomic DNA and COX-2 upstream promoter elements in estrogen receptor negative breast cancer cells. Mol Biol Cell. 2010;21:2987–2995. doi: 10.1091/mbc.E10-02-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, Lee JM, Lee HN, Kim EK, Ha B, Ahn SM, Jang HH, Lee SY. RNA-binding properties and RNA chaperone activity of human peroxiredoxin 1. Biochem Biophys Res Commun. 2012;425:730–734. doi: 10.1016/j.bbrc.2012.07.142. [DOI] [PubMed] [Google Scholar]
- 38.Kim HJ, Chae HZ, Kim YJ, Kim YH, Hwangs TS, Park EM, Park YM. Preferential elevation of Prx I and Trx expression in lung cancer cells following hypoxia and in human lung cancer tissues. Cell Biol Toxicol. 2003;19:285–298. doi: 10.1023/b:cbto.0000004952.07979.3d. [DOI] [PubMed] [Google Scholar]
- 39.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M, Hou M, Ge L, Miao C, Zhang J, Jing X, Shi N, Chen T, Tang X. Induction of peroxiredoxin 1 by hypoxia regulates heme oxygenase-1 via NF-κB in oral cancer. PLoS One. 2014;9:e105994. doi: 10.1371/journal.pone.0105994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiota M, Izumi H, Miyamoto N, Onitsuka T, Kashiwagi E, Kidani A, Hirano G, Takahashi M, Ono M, Kuwano M, et al. Ets regulates peroxiredoxin1 and 5 expressions through their interaction with the high-mobility group protein B1. Cancer Sci. 2008;99:1950–1959. doi: 10.1111/j.1349-7006.2008.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshino I, Matsubara H, Akutsu Y, Nishimori T, Yoneyama Y, Murakami K, Sakata H, Matsushita K, Ochiai T. Tumor suppressor Prdx1 is a prognostic factor in esophageal squamous cell carcinoma patients. Oncol Rep. 2007;18:867–871. [PubMed] [Google Scholar]
- 43.O’Leary PC, Terrile M, Bajor M, Gaj P, Hennessy BT, Mills GB, Zagozdzon A, O’Connor DP, Brennan DJ, Connor K, et al. Peroxiredoxin-1 protects estrogen receptor α from oxidative stress-induced suppression and is a protein biomarker of favorable prognosis in breast cancer. Breast Cancer Res. 2014;16:R79. doi: 10.1186/bcr3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan C, Cha EJ, Lee HL, Han KH, Lee KM, Kim WJ. Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J Urol. 2006;175:1512–1516. doi: 10.1016/S0022-5347(05)00659-2. [DOI] [PubMed] [Google Scholar]
- 45.Yonglitthipagon P, Pairojkul C, Chamgramol Y, Loukas A, Mulvenna J, Bethony J, Bhudhisawasdi V, Sripa B. Prognostic significance of peroxiredoxin 1 and ezrin-radixin-moesin-binding phosphoprotein 50 in cholangiocarcinoma. Hum Pathol. 2012;43:1719–1730. doi: 10.1016/j.humpath.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takemoto N, Iizuka N, Yamada-Okabe H, Hamada K, Tamesa T, Okada T, Hashimoto K, Sakamoto K, Takashima M, Miyamoto T, et al. Sex-based molecular profiling of hepatitis C virus-related hepatocellular carcinoma. Int J Oncol. 2005;26:673–678. [PubMed] [Google Scholar]
- 47.Dohmen K, Shigematsu H, Irie K, Ishibashi H. Longer survival in female than male with hepatocellular carcinoma. J Gastroenterol Hepatol. 2003;18:267–272. doi: 10.1046/j.1440-1746.2003.02936.x. [DOI] [PubMed] [Google Scholar]
- 48.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 49.Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, Au SW. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 2008;22:127–137. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- 50.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 51.Feligioni M, Nisticò R. SUMO: a (oxidative) stressed protein. Neuromolecular Med. 2013;15:707–719. doi: 10.1007/s12017-013-8266-6. [DOI] [PubMed] [Google Scholar]
- 52.Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang TS, Jeong W, Choi SY, Yu S, Kang SW, Rhee SG. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J Biol Chem. 2002;277:25370–25376. doi: 10.1074/jbc.M110432200. [DOI] [PubMed] [Google Scholar]
- 54.Zykova TA, Zhu F, Vakorina TI, Zhang J, Higgins LA, Urusova DV, Bode AM, Dong Z. T-LAK cell-originated protein kinase (TOPK) phosphorylation of Prx1 at Ser-32 prevents UVB-induced apoptosis in RPMI7951 melanoma cells through the regulation of Prx1 peroxidase activity. J Biol Chem. 2010;285:29138–29146. doi: 10.1074/jbc.M110.135905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawat SJ, Creasy CL, Peterson JR, Chernoff J. The tumor suppressor Mst1 promotes changes in the cellular redox state by phosphorylation and inactivation of peroxiredoxin-1 protein. J Biol Chem. 2013;288:8762–8771. doi: 10.1074/jbc.M112.414524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parmigiani RB, Xu WS, Venta-Perez G, Erdjument-Bromage H, Yaneva M, Tempst P, Marks PA. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci USA. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JW, Piszczek G, Rhee SG, Chock PB. Glutathionylation of peroxiredoxin I induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry. 2011;50:3204–3210. doi: 10.1021/bi101373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen ST, Van Etten RA. The PAG gene product, a stress-induced protein with antioxidant properties, is an Abl SH3-binding protein and a physiological inhibitor of c-Abl tyrosine kinase activity. Genes Dev. 1997;11:2456–2467. doi: 10.1101/gad.11.19.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu GH, Gerace L. Sumoylation regulates nuclear localization of lipin-1alpha in neuronal cells. PLoS One. 2009;4:e7031. doi: 10.1371/journal.pone.0007031. [DOI] [PMC free article] [PubMed] [Google Scholar]


