Abstract
Background
Cognitive dysfunction in schizophrenia is associated with a lower density of dendritic spines on deep layer 3 pyramidal cells in the dorsolateral prefrontal cortex (DLPFC). These alterations appear to reflect dysregulation of the actin cytoskeleton required for spine formation and maintenance. Consistent with this idea, altered expression of genes in the CDC42 (cell division cycle 42)-CDC42 effector protein signaling pathway, a key organizer of the actin cytoskeleton, was previously reported in DLPFC gray matter from subjects with schizophrenia. Here, we examined the integrity in schizophrenia of the CDC42-PAK-LIMK signaling pathway in a layer- and cell type-specific fashion in DLPFC deep layer 3.
Methods
Using laser microdissection, we collected samples of DLPFC deep layer 3 from 56 matched pairs of schizophrenia and comparison subjects and measured levels of CDC42-PAK-LIMK pathway mRNAs by qPCR. These same transcripts were also quantified by microarray in samples of individually microdissected deep layer 3 pyramidal cells from a subset of the same subjects and from antipsychotic-exposed monkeys.
Results
Relative to comparison subjects, CDC42EP4, LIMK1, LIMK2, ARHGDIA and PAK3 mRNA levels were significantly up-regulated in schizophrenia subjects in both laminar and cellular samples. In contrast, CDC42 and PAK1 mRNA levels were significantly down-regulated specifically in deep layer 3 pyramidal cells. These differences were not attributable to psychotropic medications or other co-morbid factors.
Conclusions
Findings from the present and prior studies converge on synergistic alterations in CDC42 signaling pathway that could destabilize actin dynamics and produce spine deficits preferentially in deep layer 3 pyramidal cells in schizophrenia.
Keywords: CDC42, dendritic spine, prefrontal cortex, schizophrenia, actin cytoskeleton, mRNA
Introduction
Cognitive deficits, such as impairments in working memory, represent a core feature of schizophrenia (1) and these impairments appear to reflect altered circuitry in the dorsolateral prefrontal cortex (DLPFC;(2)). In particular, convergent lines of evidence from postmortem (3), neuroimaging (4) and pharmacological studies (5) implicate aberrant glutamate neurotransmission in cortical dysfunction in schizophrenia (6–8). Pyramidal cells, the principal source of cortical glutamate neurotransmission, exhibit lower dendritic spine density (9–12), shorter dendritic arbors (10) and smaller somal volumes (13–16) in subjects with schizophrenia. These morphological aberrations appear to have laminar specificity as smaller pyramidal cell volumes and lower dendritic spine density were observed in deep layer 3 but not in layers 5 or 6 (15,17,18). Importantly, none of these findings appeared to be attributable to antipsychotic medications or other co-morbid factors or potential confounds (19) and therefore likely reflect the underlying disease process.
Previous postmortem studies suggest that these morphological alterations in layer 3 pyramidal cells may be the consequence of disturbed expression of genes that regulate the actin cytoskeleton (20,21), which plays a critical role in dendritic spine formation and maintenance (22–25). For example, transcript levels of the Rho GTPase CDC42 (cell division cycle 42) are lower in DLPFC gray matter in subjects with schizophrenia and are positively correlated with layer 3 spine density measures (20). The specificity of the spine density decrement on layer 3 pyramidal neurons may reflect the laminar-specific expression of certain molecules that interact with CDC42. Indeed, CDC42 effector proteins (CDC42EP) are preferentially expressed in layers 2–3 of the human DLPFC (26) and CDC42EP3 mRNA expression was reported to be up-regulated in schizophrenia (21). The combination of lower CDC42 and elevated CDC42EP3 was hypothesized to alter the integrity of the barrier formed by the septin family of proteins in the spine neck (27), rendering it less permeable to the influx of postsynaptic molecules that are necessary for spine plasticity in response to glutamate stimulation (21).
However, these previous studies did not examine CDC42-related gene expression specifically in deep layer 3 or in pyramidal cells in this location. In addition, they did not address how altered CDC42 signaling could disrupt the regulation of the assembly and disassembly of actin filaments through cofilin, a family of actin-binding proteins (Figure 1A). The activity of cofilin is downstream of a signaling pathway that involves the interaction between CDC42 and the p21-activated serine/threonine protein kinases (PAK) family of proteins (28,29). These proteins are activated upon binding of the GTP-bound forms of CDC42 and activate (among other targets) the LIM domain-containing serine/threonine protein kinases (LIMK1 and LIMK2) (30,31) which in turn regulate the actin-depolymerizing activity of cofilin (32–34). Moreover, the activity of CDC42 is regulated by guanine nucleotide dissociation inhibitors such as ARHGDIA (Figure 1A), a class of molecules that inhibit the substitution of GDP for GTP, thereby suppressing GTPase activity (35). Therefore, multiple regulators in the CDC42-PAK-LIMK pathway appear to be crucial for the actin cytoskeleton to maintain the stability of spine structure.
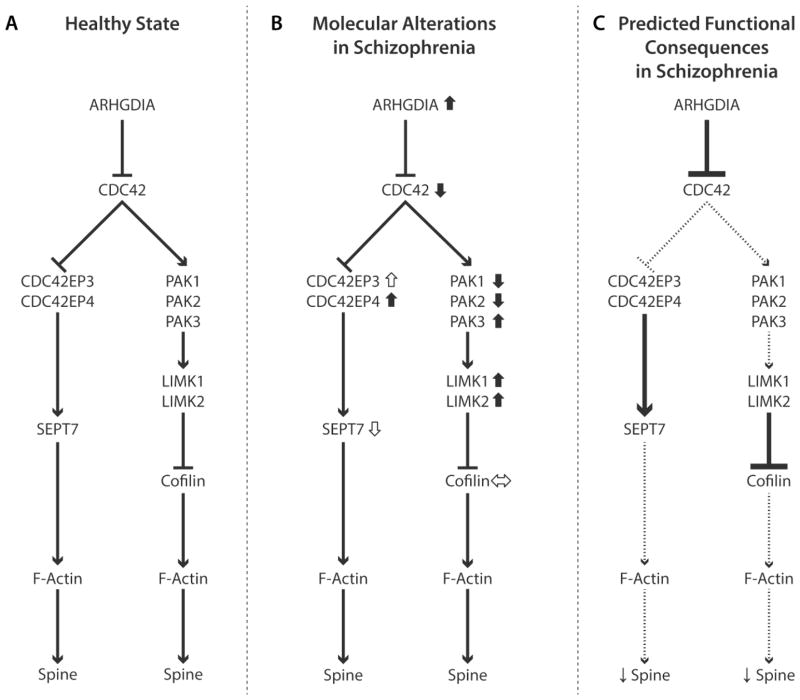
Figure 1. Schematic diagrams of CDC42-CDC42EP and CDC42-PAK-LIMK signaling pathways and their proposed roles in spine deficits in schizophrenia.
(A) CDC42 signaling pathways that regulate the contribution of F-actin to dendritic spine structure. The activity of CDC42 is inhibited by ARHGDIA, a guanine nucleotide dissociation inhibitor that suppresses intrinsic GTPase activity. CDC42-CDC42EP pathway: Activated CDC42 inhibits CDC42 effector proteins (CDC42EP), which dissociate the complex of septin filaments consolidated by SEPT7 in the spine neck. This opening of the septin barrier permits an influx of molecules from the parent dendrite that facilitate F-actin mediated growth of spines in response to excitatory inputs. CDC42-PAK-LIMK pathway: CDC42 activates p21-activated serine/threonine protein kinases (PAK), which in turn activate LIM domain-containing serine/threonine protein kinases (LIMK). Activation of this cascade inhibits the cofilin family of actin depolymerizing proteins that regulate the turnover of F-actin required for structural stability of spines. Arrows indicate activation and blunted lines indicate inhibition of each target. (B) Subjects with schizophrenia exhibit up- or down-regulation for multiple components of the CDC42-CDC42EP and CDC42-PAK-LIMK pathways in DLPFC deep layer 3 pyramidal cells. Solid short arrows next to transcript indicates reported evidence in this paper; open short arrows indicate previously reported evidence (21, 72). (C) Predicted functional consequences in schizophrenia of altered CDC42 signaling pathways in DLPFC deep layer 3 pyramidal neurons. Higher levels of ARHGDIA would directly inhibit the activation of CDC42 holding it in an inactive GDP-bound state. CDC42-CDC42EP pathway: The effect of higher levels of ARHGDIA would be amplified by the combination of lower levels of CDC42 and higher levels of CDC42EPs, impairing the transient opening of the septin barrier in spine necks in response to excitatory inputs, and thereby suppressing the influx of molecules into the spine head required for spine growth and maintenance (21). CDC42-PAK-LIMK pathway: The combination of higher levels of ARHGDIA mRNA, lower levels of CDC42 mRNAs and lower levels of PAK mRNAs (Given the much higher expression levels of PAK1 than PAK3, the down-regulation of PAK1 and PAK2 is predicted to have the dominant effect.) would all converge to increase phosphorylation (inactivation) of cofilin family proteins and suppress actin depolymerization, resulting in F-actin destabilization and spine loss. The up-regulated levels of LIMK1/2 and PAK3 may represent compensatory, but inadequate, responses to mitigate the negative impact on F-actin dynamics of the upstream molecular pathology in this pathway.
To determine if alterations in the CDC42-PAK-LIMK signaling pathway could contribute to the spine deficits on deep layer 3 pyramidal cells in schizophrenia, we conducted targeted gene expression analyses at laminar and cellular levels of specificity. We used laser microdissection to collect samples of DLPFC deep layer 3 from 56 matched pairs of schizophrenia and comparison subjects, and of individual deep layer 3 pyramidal cells in a subset of these subjects, and measured the expression levels of CDC42-related mRNAs in these samples using real-time quantitative polymerase chain reaction (RT-qPCR) and microarray, respectively.
Methods and Materials
Human subjects
Human brain specimens (N=112) were obtained during routine autopsies conducted at the Allegheny County Medical Examiner’s Office (Pittsburgh, PA, USA) following consent obtained from the next-of-kin. An independent committee of research clinicians made consensus DSM-IV (36) diagnoses, or confirmed the absence of a psychiatric diagnosis, for each subject using structured interviews with family members and review of prior medical records (37). To control for experimental variance, each subject with schizophrenia (n=34) or schizoaffective disorder (n=22) was matched to one healthy comparison subject for sex and as closely as possible for age (Table S1 contains individual subject details). As in prior studies (38–42), we consider schizoaffective disorder to be a variant of schizophrenia based on the DSM-IV requirement for the class A criteria of schizophrenia to be present, in the absence of mood symptoms. Samples from both subjects in a pair were processed together throughout all stages of the study. Subject groups (Table 1) did not differ in mean age, postmortem interval (PMI), RNA integrity number (RIN), tissue storage time at −80°C or race. Brain pH significantly differed between groups (t=2.51; df=55, p=0.015), although the mean difference between groups was very small (0.1 pH unit). Every subject had RIN ≥ 7.0 indicating an excellent quality of total RNA. All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research.
Table 1.
Characteristics of human subjects
| Characteristic | Quantitative PCR
|
Microarray
|
||||
|---|---|---|---|---|---|---|
| Comparison | Schizophrenia | Comparison | Schizophrenia | |||
| Number | 56 | 56 | 36 | 36 | ||
| Sex | 43 M, 13 F | 43 M, 13 F | 27 M, 9 F | 27 M, 9 F | ||
| Race | 47 W, 9 B | 37 W, 19 B | 30 W, 6 B | 24 W, 12 B | ||
| Age (years) | 48.8 ± 13.7 | 47.6 ± 12.5 | 48.1 ± 13.0 | 46.9 ± 12.4 | ||
| Postmortem Interval (hours) | 18.8 ± 5.6 | 19.3 ± 8.6 | 17.6 ± 6.1 | 18.0 ± 8.8 | ||
| RNA Integrity Number | 8.1 ± 0.6 | 8.1 ± 0.6 | 8.3 ± 0.6 | 8.2 ± 0.6 | ||
| Brain pH | 6.7 ± 0.2 | 6.6 ± 0.3 | 6.7 ± 0.2 | 6.6 ± 0.4 | ||
| Storage Time (months) | 116.4 ± 55.4 | 114.2 ± 60.5 | 122.2 ± 49.8 | 125.7 ± 53.1 | ||
|
| ||||||
| Comorbid factor | Yes | No | Yes | No | ||
|
| ||||||
| Schizoaffective Disorder | - | 22 | 34 | - | 13 | 23 |
| History of Substance Abuse | - | 38 | 18 | - | 23 | 13 |
| Nicotine ATOD | - | 38 | 12 | - | 23 | 7 |
| Antidepressants ATOD | - | 26 | 30 | - | 15 | 21 |
| Benzo/VPA ATOD | - | 25 | 31 | - | 14 | 22 |
| Antipsychotics ATOD | - | 49 | 7 | - | 31 | 5 |
| Death by Suicide | - | 15 | 39 | - | 10 | 26 |
Age, PMI, RIN and Brain pH are represented as mean ± SD.
M, Male; F, Female; W, White; B, Black. For the schizophrenia subject groups, the table indicates the number of subjects for the potential confounding variables such as, diagnosis of schizoaffective disorder; history of substance dependence or abuse; nicotine use at the time of death (ATOD); use of antipsychotics, antidepressants, or Benzo/VPA ATOD; or death by suicide. Benzo/VPA, benzodiazepines or valproic acid.
Laser microdissection procedure
We conducted two studies at different levels of resolution: 1) cortical layer-specific measures of gene expression in strips of tissue containing only DLPFC deep layer 3 (N=56 pairs; Table 1) and 2) cell-type specific measures of gene expression in DLPFC deep layer 3 pyramidal cells (N=36 pairs; Table 1).
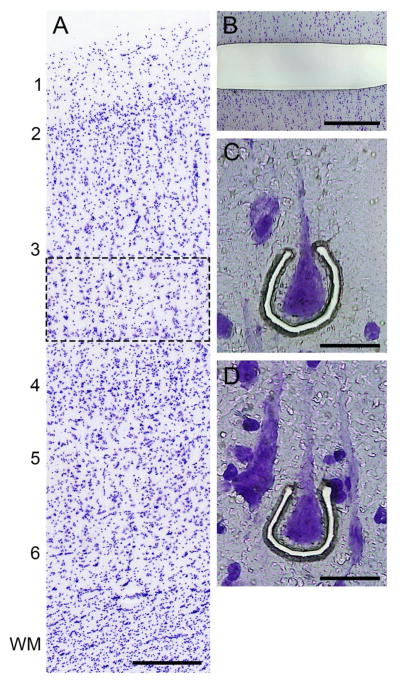
For both studies, the right hemisphere of each brain was blocked coronally, frozen and stored at −80°C (43). Cryostat sections (12 μm) were cut and thaw-mounted onto glass PEN membrane slides (Leica Microsystems, Bannockburn, IL) that had been UV-treated at 254nm for 30 minutes. The sections were dried and stored at −80 °C. On the day of the microdissection, sections were stained for Nissl substance with thionin (Figure 2A). DLPFC layer 3 was identified using a Leica microdissection system (LMD 6500; ×5 objective) in portions of the section cut perpendicular to the pial surface. For the laminar microdissections, strips (~10 million μm2) containing deep layer 3 (defined as the zone from the layer 3–4 border to 35% of the distance from the pial surface to the layer 6-white matter border) in DLPFC area 9 were collected from each subject using a 5× objective (Figure 2B). In nearby tissue sections, individual deep layer 3 pyramidal cells (~200 pyramidal cells per subject), identified based on their characteristic somal morphology and the presence of a prominent apical dendrite directed radially toward the pia mater (Figure 2C,D), were captured using a 40× objective as previously described (44).
Figure 2. Laser microdissection approach.
(A) Nissl stain of human DLPFC showing the location (dashed rectangle) of deep layer 3 which was sampled. Calibration bar equals 300 μm. (B) Nissl stained section after removal of strip of deep layer 3 by laser microdissection. Calibration bar equals 700 μm. (C and D) Representative images of individual deep layer 3 pyramidal cells being captured by laser microdissection in DLPFC deep layer 3. Calibration bar equals 30 μm. In panel A, numbers indicate cortical layers and WM indicates white matter.
qPCR analyses
For each sample, RNA was extracted and purified using the QIAGEN RNeasy Plus Micro Kit. Total RNA was converted to cDNA using the qScriptTM cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD). Forward and reverse primers were designed for each target mRNA to generate PCR amplicons of 85–120 base pairs. The specificity and efficiency of qPCR amplification for each target mRNA was demonstrated by: 1) high amplification efficiency (>92%) across a wide range of cDNA dilutions (Supplemental Table S3) and 2) single products demonstrated in dissociation curve analysis.
Three internal reference transcripts (Beta-Actin [ACTB], Cyclophilin A [PPIA] and guanine nucleotide binding protein [GNAS]), selected based on their stable expression across the subjects in this cohort regardless of diagnosis (45), were used to normalize data. Transcript expression levels of CDC42-related mRNAs (ARHGDIA, CDC42, CDC42EP4, PAK1, PAK2, PAK3, LIMK1 and LIMK2) were quantified using qPCR using Power SYBR green dye (Applied Biosystems, Foster City, CA) and ViiATM 7 Real-Time PCR system (Life Technologies, Carlsbad, CA). The cDNA samples from three matched pairs of schizophrenia and control subjects were processed together on the same 384-well qPCR plate with four replicates per primer set.
Microarray analyses
We used a microarray approach to quantify cellular expression of CDC42-related mRNAs in deep layer 3 pyramidal cells as previously described (44). For transcriptome analysis, the RNA was extracted using the QIAGEN RNeasy Plus Micro Kit. The extracted RNA was transcribed into cDNA and subjected to a single round of amplification using the Ovation Pico WTA System (Nugen Technologies, San Carlos, CA). After amplification, the cDNA was labeled using the Encore Biotin module and loaded on an Affymetric GeneChip® HT HG-U133+ PM Array Plate designed to assess expression levels of transcripts in the human genome (Affymetrix, Santa Clara, CA).
Antipsychotic-exposed monkeys
Experimentally naïve, male, macaque monkeys (Macaca fascicularis) received twice daily oral doses of olanzapine, haloperidol or placebo (n=6 monkeys per group) for 17–27 months (46). The doses of each antipsychotic medications produced trough serum levels in the therapeutic range for the treatment of schizophrenia (47). One monkey from each of the three groups was euthanized on the same day (48) and pyramidal cells captured (49) and subjected to microanalysis (See Supplemental Methods).
Data analysis and statistics
qPCR analyses
The comparative threshold cycle (CT) method was used in which transcript levels are normalized to the geometric mean of the three reference genes (ACTB, PPIA and GNAS). The difference in cycle threshold for each transcript was assessed by deducting the mean cycle threshold for the three reference genes from the cycle threshold of the target transcript. Since the difference in cycle threshold (dCT) represents the log2-transformed expression ratio of each target transcript to the geometric mean of the three reference genes, the relative expression ratio of each target transcript is determined as 2−dCT (37). We performed two analyses of covariance (ANCOVA) models on the expression ratio data for each transcript. Because subjects were selected and processed as pairs, the first paired ANCOVA model included mRNA level as the dependent variable, diagnostic group as the main effect, subject pair as a blocking factor, and tissue storage time, brain pH and RIN as covariates. Subject pairing may be considered an attempt to balance diagnostic groups for sex and age, and to account for the parallel processing of tissue samples from a pair, and thus to not be a true statistical paired design. Consequently, a second unpaired ANCOVA model was performed that included all covariates (i.e., age, sex, postmortem interval, storage time, RIN and pH). All statistical tests were conducted with α-level= 0.05.
We also assessed the potential influence of other factors that are frequently co-morbid with the diagnosis of schizophrenia using ANCOVA models. For these analyses, we compared subjects with schizophrenia using each variable (sex; diagnosis of schizoaffective disorder; history of substance dependence or abuse; nicotine use at the time of death; use of antipsychotics, antidepressants, or benzodiazepines and/or sodium valproate at the time of death; or death by suicide) as the main effect and age, tissue storage time, brain pH, PMI, and RIN as covariates.
Reported ANCOVA statistics include only those covariates that were statistically significant. As a result, the reported degrees of freedom vary across analyses.
Microarray analyses
The probe sets were filtered and paired t-tests were performed using the Random Intercept Model with Bayesian Information Criterion variable selection (50). Differentially expressed gene discovery was conducted using meta-analysis and an adaptively weighted (AW) Fisher’s method (51) was applied. Meta-analyzed p-values from AW were then adjusted by the Benjamini-Hochberg procedure for multiple comparisons to control false discovery rate (44,52).
Antipsychotic-exposed monkey analyses
An ANCOVA model with the level of pre-specified mRNAs as the dependent variable, treatment group as the main effect, and triad as a blocking factor was employed. For each transcript of interest, the values of all probe sets targeting that transcript were averaged within each animal.
Results
Expression of CDC42-related mRNAs in DLPFC deep layer 3
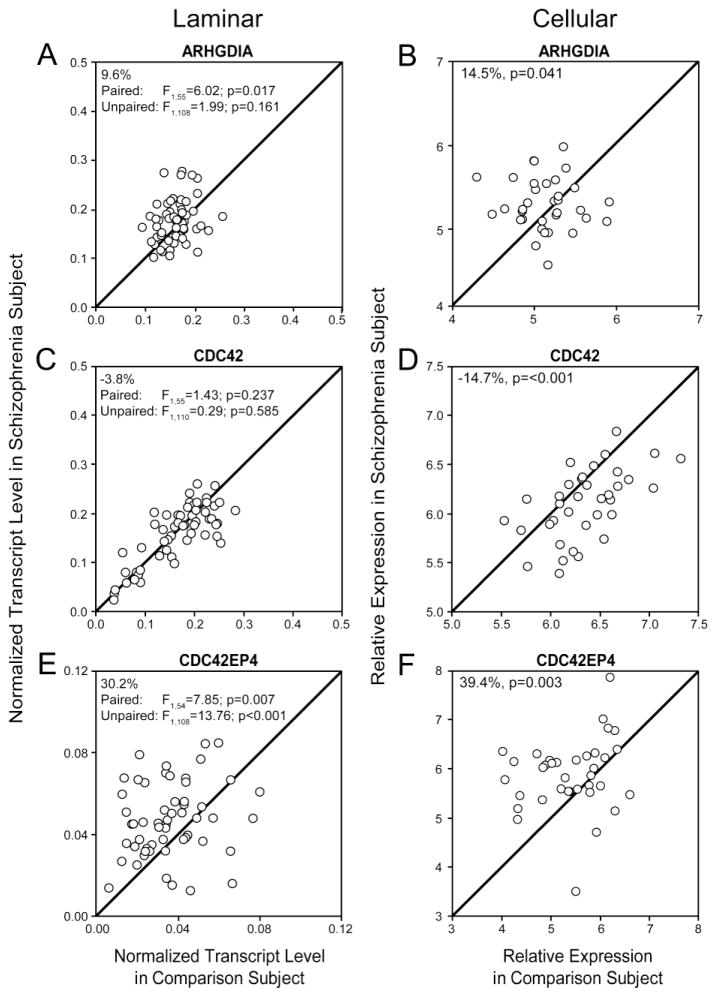
In subjects with schizophrenia, mean mRNA levels of ARHGDIA, a common upstream regulator of pathways related to CDC42 signaling (Figure 1B), were higher in tissue homogenates of DLPFC deep layer 3 from the 56 subject pairs (+9.6%; paired: F1,55=6.02, p=0.017; unpaired: F1,108=1.99, p=0.161; Figure 3A) and in deep layer 3 pyramidal cells from the 36 subject pairs (+14.5%; p=0.041; Figure 3B). Although mean CDC42 mRNA levels were not significantly lower (−3.8%; paired: F1,55=1.43, p=0.237; unpaired: F1,110=0.29, p=0.585; Figure 3C) in the deep layer 3 tissue homogenates, they were significantly lower (−14.7%; p<0.001; Figure 3D) in DLPFC deep layer 3 pyramidal cells. Consistent with prior studies (21) of the CDC42-CDC42EP filament pathway (Figure 1B), mean CDC42EP4 transcript levels were significantly higher in the subjects with schizophrenia in DLPFC deep layer 3 tissue homogenates (+30.2%; paired: F1,54=7.85, p=0.007; unpaired: F1,108=13.76, p<0.001; Figure 3E) and pyramidal cells (+39.4%; p=0.003; Figure 3F).
Figure 3.
(A,B) Rho GDP dissociation inhibitor encoded by ARHGDIA (C,D) Cell division cycle 42 (CDC42), (E,F) CDC42 Effector Protein 4 (CDC42EP4) mRNA levels in DLPFC deep layer 3 tissue homogenates (A,C,E) and DLPFC deep layer 3 pyramidal cells (B,D,F) from schizophrenia and comparison subjects. Scatter plots show the transcript levels for each matched pair of a comparison and schizophrenia subject. Values above the unity line reflect pairs in which transcript levels are higher in the schizophrenia subject relative to the comparison subject. Values below the unity line reflect pairs in which transcript levels are lower in the schizophrenia subject relative to the comparison subject.
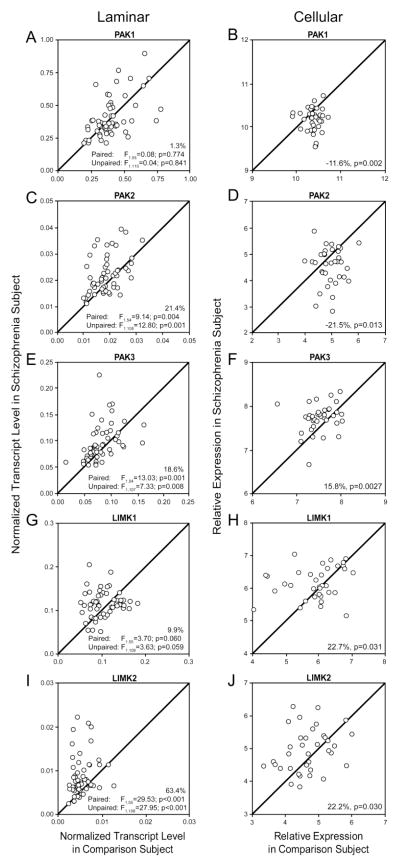
To interrogate the integrity of the CDC42-PAK-LIMK signaling pathway that regulates the assembly and disassembly of actin filaments through cofilin, we evaluated the expression level of several members of this pathway (Figure 1B). Mean transcript levels for PAK1 did not differ between groups in tissue homogenates of DLPFC deep layer 3 (paired: F1,55=0.08, p=0.774; unpaired: F1,110=0.04, p=0.841; Figure 4A) but were significantly lower in pyramidal cell samples from schizophrenia subjects (−11.6%; p=0.002, Figure 4B). In contrast, in the subjects with schizophrenia mean transcript levels were higher for PAK2 in tissue homogenates of DLPFC deep layer 3 (+21.4%; paired: F1,54=9.14, p=0.004; unpaired: F1,108=12.80, p=0.001; Figure 4C) but were significantly lower in deep layer 3 pyramidal cells (−21.5%; p=0.013, Figure 4D). This discrepancy between layer-specific and cell type-specific measures for PAK2 could represent opposing patterns of change in pyramidal cells relative to other cell types (i.e., interneurons and glial cells) in the illness. Furthermore, the difference in sample size between qPCR (N=56 pairs) and microarray (N=36 pairs) analyses do not account for the marked difference in expression as the mean transcript levels were higher for PAK2 (+24.2%) in tissue homogenates from the same 36 pairs used for microarray analyses. Mean transcript levels for PAK3 were significantly higher in both tissue homogenates of DLPFC deep layer 3 (+18.6%; paired: F1,54=13.03, p=0.001; unpaired: F1,107=7.33, p=0.008; Figure 4E) and pyramidal cells (+15.8%; p=0.0027; Figure 4F). Finally, in the subjects with schizophrenia mean transcript levels for LIMK1 were modestly higher in deep layer 3 tissue homogenates (+9.9%; paired: F1,55=3.70, p=0.060; unpaired: F1,109=3.63, p=0.059; Figure 4G) and were significantly higher in pyramidal cells (+22.7%; p=0.031; Figure 4H). Consistent with these changes, mean transcript levels for LIMK2 were significantly higher in both deep layer 3 tissue homogenates (+63.4%; paired: F1,55=29.53, p<0.001; unpaired: F1,108=27.95, p<0.001; Figure 4I) and pyramidal cells (+22.2%; p=0.030; Figure 4J).
Figure 4.
(A,B) p21 protein (CDC42/RAC)-activated kinase 1 (PAK1), (C,D) p21 protein (CDC42/RAC)-activated kinase 2 (PAK2), (E,F) p21 protein (CDC42/RAC)-activated kinase 3 (PAK3), (G,H) LIM domain kinase 1 (LIMK1), (I,J) LIM domain kinase 2 (LIMK2) mRNA levels in DLPFC deep layer 3 tissue homogenates (A,C,E,G,I) and DLPFC deep layer 3 pyramidal cells (B,D,F,H,J) from schizophrenia and comparison subjects. Scatter plots show the transcript levels for each matched pair of a comparison and schizophrenia subject. Values above the unity line reflect pairs in which transcript levels are higher in the schizophrenia subject relative to the comparison subject. Values below the unity line reflect pairs in which transcript levels are lower in the schizophrenia subject relative to the comparison subject.
Effects of psychotropic medications and other confounding variables
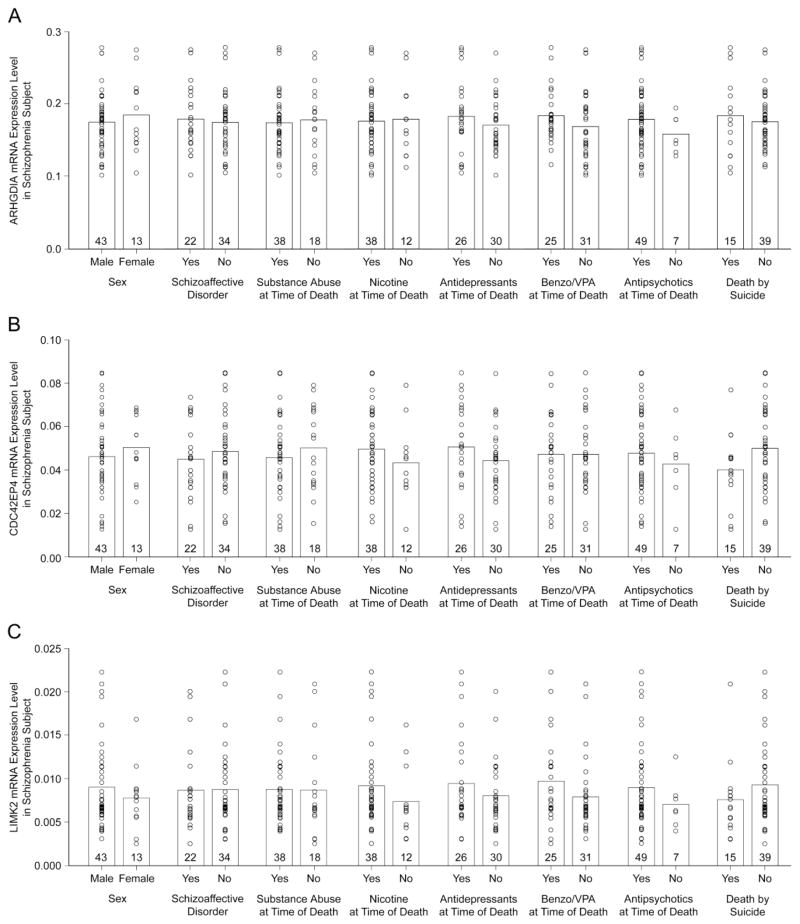
For the transcripts that were significantly altered in the same direction in both the deep layer 3 tissue homogenates and pyramidal cell samples from the subjects with schizophrenia, we evaluated the effect of potential confounding variables in the qPCR data. In the 56 schizophrenia subjects, levels of ARHGDIA (Figure 5A), CDC42EP4 (Figure 5B), LIMK2 (Figure 5C) mRNAs did not differ as a function of sex, diagnosis of schizoaffective disorder; history of substance dependence or abuse; nicotine use at the time of death; use of antipsychotics, antidepressants, or benzodiazepines and/or sodium valproate at the time of death; or death by suicide (all F1,48≤2.39; all p≥0.13). Similar results (all F1,48≤1.78; all≥p 0.19) were found for the other transcripts (i.e., PAK2, PAK3) that were significantly altered by RT-PCR.
Figure 5.
The effect of co-morbid factors on (A) Rho GDP dissociation inhibitor encoded by ARHGDIA (B) CDC42 Effector protein 4 and (C) LIM domain kinase 2 mRNA expression levels in subjects with schizophrenia in DLPFC deep layer 3 tissue homogenates. For each panel, the circles represent mRNA expression levels for individual schizophrenia subjects by qPCR and the bars represent mean mRNA levels for the indicated group. Numbers at the bottom of bars indicate the number of subjects with schizophrenia per group. None of these confounding variables were significant for any of the transcripts (all F1,48≤2.39, all p≥0.13). Two subjects had an undetermined manner of death and were not included in the death by suicide analysis. Six subjects had unknown nicotine use at the time of death and were not included in the nicotine analysis. Benzo/VPA, benzodiazepines or valproic acid.
In order to interrogate the potential effect of long-term exposure to typical or atypical antipsychotics, we also evaluated CDC42-related mRNAs in DLPFC deep layer 3 pyramidal cells from monkeys chronically exposed to olanzapine, haloperidol or placebo. Levels of CDC42-related mRNAs did not significantly differ among these three groups of monkeys (Table 2). Serum levels of antipsychotic medications were not significantly correlated with any gene expression measures (all lrl<0.75; all p>0.10).
Table 2.
Summary of differences by transcript in antipsychotic-exposed monkeys
| Transcript | Mean Expression ± Standard Deviation
|
ANCOVA | ||
|---|---|---|---|---|
| Placebo | Olanzapine | Haloperidol | ||
| ARHGDIA | 8.2 ± 0.2 | 8.1 ± 0.2 | 8.1 ± 0.3 | F2,10=0.09, p=0.91 |
| CDC42 | 9.9 ± 0.2 | 9.8 ± 0.2 | 10.0 ± 0.1 | F2,10=3.43, p=0.08 |
| CDC42EP4 | 5.7 ± 0.5 | 5.2 ± 0.3 | 5.4 ± 0.4 | F2,10=3.02, p=0.09 |
| PAK1 | 10.0 ± 0.1 | 10.2 ± 0.1 | 10.1 ± 0.1 | F2,10=2.51, p=0.13 |
| PAK2 | 6.4 ± 0.4 | 6.4 ± 0.3 | 6.6 ± 0.3 | F2,10=0.59, p=0.57 |
| PAK3 | 10.5 ± 0.2 | 10.7 ± 0.4 | 10.4 ± 0.2 | F2,10=3.21, p=0.09 |
| LIMK1 | 8.9 ± 0.4 | 8.9 ± 0.3 | 8.8 ± 0.3 | F2,10=0.95, p=0.42 |
| LIMK2 | 3.7 ± 0.2 | 3.7 ± 0.3 | 3.7 ± 0.2 | F2,10=0.03, p=0.97 |
Discussion
In this study, we found that subjects with schizophrenia exhibit altered gene expression, both up- and down-regulation, for components of the CDC42-PAK-LIMK pathway in DLPFC deep layer 3 tissue homogenates and pyramidal cells (Figure 1B, right side). In addition, we confirmed and extended to the cell type-specific level, earlier findings of altered expression in the CDC42-CDC42EP pathway (Figure 1B, left side). The levels of some transcripts (i.e., CDC42EP4, ARHGDIA, PAK3, LIMK1 and LIMK2) were significantly altered in schizophrenia, with the same direction and similar magnitude of difference from comparison subjects, in both layer- and pyramidal cell samples, as measured with qPCR or microarray analyses, respectively. In contrast, the levels of other transcripts (i.e., CDC42, PAK1) were significantly altered only in deep layer 3 pyramidal cell samples, but showed the same direction of change in the layer 3 tissue homogenates, suggesting that these alterations are specific to or at least enriched in, pyramidal cells. These transcript alterations may reflect the disease process in schizophrenia as none of these changes were attributable to antipsychotic medications or other factors frequently co-morbid with schizophrenia. Our findings support the notion that altered signaling in the CDC42-PAK-LIMK pathway could perturb the regulation of the assembly and disassembly of actin filaments through cofilin, and in concert with alterations in the CDC42-CDC42EP pathway, could contribute to the lower density of dendritic spines that is most pronounced in deep layer 3 pyramidal cells in the DLPFC of subjects with schizophrenia.
Differences in layer-specific vs. cell type-specific pathology in schizophrenia
The assessment of molecular pathology at both laminar and cellular levels of resolution revealed differences in gene expression in schizophrenia that might be cell type-specific. For example, disease-related differences in the levels of CDC42 and PAK1 mRNAs detected in DLPFC deep layer 3 pyramidal neurons, but not in tissue homogenates from the same laminar location in the same subjects, support the idea that these findings are pyramidal cell-specific. Thus, measures of gene expression in gray matter or even a specific cortical layer may obscure findings that are cell type-specific. Consistent with this idea, recent transcriptome analyses revealed substantial differential expression in schizophrenia of transcripts related to mitochondrial function in deep layer 3 pyramidal neurons and of transcripts related to the ubiquitin-proteasome system in layer 5 pyramidal cells (44), alterations that were not detected in layer-specific transcriptome studies (26). Similarly, previous studies in total gray matter tissue homogenates revealed a modest reduction (−6%) in levels of GABAA α1 subunit mRNA (53), whereas dual-label in situ hybridization studies showed that mean GABAA α1 subunit mRNA expression was significantly 40% lower in deep layer 3 pyramidal cells, but was not altered in interneurons in the same layer (54). Additionally, recent findings using laser microdissection to dissect neuronal populations in the thalamus from subjects with schizophrenia revealed lower expression of transcripts encoding glutamate receptor subunits and components of the postsynaptic scaffold in relay neurons but not in a mixed population of glial cells and interneurons (55). Thus, gene expression analyses at the level of individual cell types reveal distinctive alterations that are essential to understanding molecular pathology in the context of the neural circuits formed by different classes of neurons.
Contribution of CDC42-related signaling to dendritic spine abnormalities in DLPFC layer 3 pyramidal cells
Signaling through CDC42 pathways regulates the polymerization of the actin cytoskeleton and thus is essential for the maturation of filopodia into spines and for the maintenance of a normal complement of dendritic spines. In combination, findings of the present and prior studies suggest at least three different patterns of molecular disturbances in CDC42 signaling that could contribute to spine deficits preferentially on deep layer 3 pyramidal cells in schizophrenia.
First, our findings highlight a schizophrenia-related alteration in a regulatory component of the Rho family of GTPases, including CDC42. Like other members of this family, CDC42 cycles between an active-GTP bound state and an inactive GDP-bound state (56). The activity of these GTPases is regulated by guanine nucleotide dissociation inhibitors, such as ARHGDIA, which interacts with CDC42 (57), suppresses the exchange of GDP for GTP and renders CDC42 inactive (58). Thus, our findings of higher expression levels of ARHGDIA in deep layer 3 pyramidal cells (Figure 1B) suggest that CDC42 is more likely to be held in an inactive-GDP bound state in subjects with schizophrenia. In combination with lower levels of CDC42 (Figure 1B), the capacity of both the CDC42-CDC42EP and CDC42-PAK-LIMK signaling pathways to modulate the actin filaments that form the structural framework of dendritic spines would be predicted to be impaired in deep layer 3 pyramidal neurons in schizophrenia (Figure 1C).
Second, the prominence of dendritic spine abnormalities in deep layer 3 pyramidal neurons of subjects with schizophrenia has been proposed to be the consequence of altered signaling through molecules that are expressed in a layer-specific fashion (21). For example, CDC42 effector protein (CDC42EP) mRNAs are preferentially expressed in layer 3 of the human DLPFC (26). We previously demonstrated up-regulation of CDC42EP3 in layer 3 of subjects with schizophrenia (21), and here show that CDC42EP4 is up-regulated in layer 3 pyramidal cells. According to the previously proposed model for the CDC42-CDC42EP pathway (Figure 1A), the transient activation of CDC42 that normally occurs in individual spines following glutamate stimulation disrupts the CDC42EP-mediated assembly of the septin barrier consolidated by SEPT7 in the spine neck, enabling entrance into the spine head of the postsynaptic molecules, second messengers and cytoskeleton proteins necessary for F-actin mediated growth of spines and synaptic potentiation (21). In schizophrenia, the effect of higher levels of ARHGDIA would be amplified by the combination of lower levels of CDC42 and higher levels of CDC42EPs (Figure 1B) which impairs the opening of the septin barrier in response to glutamate stimulation, and thereby inhibits the influx of molecules into the spine head required for spine growth/maintenance, contributing ultimately to spine loss (Figure 1C).
Third, in this study we identified altered expression of several genes in the CDC42-PAK-LIMK pathway. In this pathway, activated, GTP-bound CDC42 activates the PAK proteins which in turn activate LIMK proteins. This signaling cascade inhibits the cofilin family of actin depolymerizing proteins that regulate the assembly and disassembly of F-actin required for structural stability of spines (59–62) (Figure 1A). Our findings from deep layer 3 pyramidal cells indicate that schizophrenia is associated with lower expression of PAK1/2 proteins (Figure 1B) which would further reduce activity in the CDC42-PAK-LIMK pathway due to the combination of elevated expression of ARHGDIA and lower expression of CDC42 and contribute to spine deficits. Consistent with this interpretation, down-regulation of CDC42 and PAK1 proteins results in impaired long-term maintenance of spines (63), and over-expression in vitro of dominant negative forms of PAK1 reduces spine density (64,65). Interestingly, a recent study demonstrated that pharmacological manipulation of downstream signaling components of the actin cytoskeleton such as the PAK proteins ameliorated synaptic deficits induced by DISC1 knockdown. Although this finding suggests that PAK proteins might serve as therapeutic targets, future experiments need to delineate the pattern of alterations in multiple signaling cascades in subjects with schizophrenia that can converge on these downstream signaling components (66). In contrast, we found higher expression levels of LIMK1/2 and PAK3 in deep layer 3 pyramidal cells. We interpret this finding as a compensatory, but inadequate, response to the multiple upstream alterations that increase phosphorylation (inactivation) of cofilin family proteins and suppress actin depolymerization, resulting in F-actin destabilization and a reduction in spine number on deep layer 3 pyramidal cells (Figure 1C).
In concert, these findings are consistent with other data suggesting that alterations in deep layer 3 pyramidal cells are “upstream” in the disease process of schizophrenia (44,67) or perhaps even directly related to genetic risk for the illness. For example, recent findings highlight the CDC42-PAK-LIMK regulatory network as a pathogenetic factor in actin cytoskeleton dysregulation in schizophrenia (68). In addition, de novo mutations in proteins that regulate actin filament dynamics are preferentially found in individuals with schizophrenia (69), and copy number variations at the 15q11.2 locus implicate genes such as CYFIP1 that regulate dendritic complexity and spine actin dynamics (70,71).
Conclusion
In concert with previous findings of alterations in other mediators (e.g., RhoA, Duo, Reelin, DISC1) of spine morphogenesis in schizophrenia (20,21,72–79), the findings of our study suggest that aberrant CDC42 signaling through two different pathways might represent a molecular pathology that converges on dendritic spine deficits specifically in deep layer 3 pyramidal cells (10,17) in subjects with schizophrenia. These observations support the notion that the actin cytoskeleton is dysregulated in schizophrenia. Because both spines and the axon terminals that innervate them are rich in actin, this dysregulation could result in lower excitatory drive to DLPFC deep layer 3 pyramidal cells (67) and a reduced need for energy production in these neurons (44).
Supplementary Material
Acknowledgments
The authors thank Mary L. Brady, Carol Sue Johnston, Mary Ann Kelly, Kelly Rogers, Kiley Laing (all University of Pittsburgh) and Aiqing He and Amy Truong (both Bristol-Myers Squibb) for excellent technical assistance. This work was supported by National Institutes of Health Grants MH103204 (DAL) and MH043784 (DAL) and a grant from Bristol-Myers Squibb.
Footnotes
Financial Disclosures
David A. Lewis currently receives investigator-initiated research support from Bristol-Myers Squibb and Pfizer and in 2012–2014 served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals, and Sunovion. The other authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70:1107–1112. doi: 10.1001/jamapsychiatry.2013.155. [DOI] [PubMed] [Google Scholar]
- 2.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 3.Kristiansen LV, Huerta I, Beneyto M, Meador-Woodruff JH. NMDA receptors and schizophrenia. Curr Opin Pharmacol. 2007;7:48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Merritt K, McGuire P, Egerton A. Relationship between Glutamate Dysfunction and Symptoms and Cognitive Function in Psychosis. Front Psychiatry. 2013;4:151. doi: 10.3389/fpsyt.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Javitt DC, Zukin SR. Recent advances in the phencyclidine model of schizophrenia. Am J Psychiatry. 1991;148:1301–1308. doi: 10.1176/ajp.148.10.1301. [DOI] [PubMed] [Google Scholar]
- 6.Coyle JT. The GABA-glutamate connection in schizophrenia: which is the proximate cause? Biochem Pharmacol. 2004;68:1507–1514. doi: 10.1016/j.bcp.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 7.Konradi C, Heckers S. Molecular aspects of glutamate dysregulation: implications for schizophrenia and its treatment. Pharmacol Ther. 2003;97:153–179. doi: 10.1016/s0163-7258(02)00328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moghaddam B. Targeting metabotropic glutamate receptors for treatment of the cognitive symptoms of schizophrenia. Psychopharmacology (Berl) 2004;174:39–44. doi: 10.1007/s00213-004-1792-z. [DOI] [PubMed] [Google Scholar]
- 9.Garey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM, et al. Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry. 1998;65:446–453. doi: 10.1136/jnnp.65.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 11.Konopaske GT, Lange N, Coyle JT, Benes FM. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71:1323–1331. doi: 10.1001/jamapsychiatry.2014.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweet RA, Henteleff RA, Zhang W, Sampson AR, Lewis DA. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology. 2009;34:374–389. doi: 10.1038/npp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold SE, Franz BR, Gur RC, Gur RE, Shapiro RM, Moberg PJ, et al. Smaller neuron size in schizophrenia in hippocampal subfields that mediate cortical-hippocampal interactions. Am J Psychiatry. 1995;152:738–748. doi: 10.1176/ajp.152.5.738. [DOI] [PubMed] [Google Scholar]
- 14.Pierri JN, Volk CL, Auh S, Sampson A, Lewis DA. Decreased somal size of deep layer 3 pyramidal neurons in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2001;58:466–473. doi: 10.1001/archpsyc.58.5.466. [DOI] [PubMed] [Google Scholar]
- 15.Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatry. 1998;55:215–224. doi: 10.1001/archpsyc.55.3.215. [DOI] [PubMed] [Google Scholar]
- 16.Sweet RA, Pierri JN, Auh S, Sampson AR, Lewis DA. Reduced pyramidal cell somal volume in auditory association cortex of subjects with schizophrenia. Neuropsychopharmacology. 2003;28:599–609. doi: 10.1038/sj.npp.1300120. [DOI] [PubMed] [Google Scholar]
- 17.Kolluri N, Sun Z, Sampson AR, Lewis DA. Lamina-specific reductions in dendritic spine density in the prefrontal cortex of subjects with schizophrenia. Am J Psychiatry. 2005;162:1200–1202. doi: 10.1176/appi.ajp.162.6.1200. [DOI] [PubMed] [Google Scholar]
- 18.Sweet RA, Bergen SE, Sun Z, Sampson AR, Pierri JN, Lewis DA. Pyramidal cell size reduction in schizophrenia: evidence for involvement of auditory feedforward circuits. Biol Psychiatry. 2004;55:1128–1137. doi: 10.1016/j.biopsych.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 21.Ide M, Lewis DA. Altered cortical CDC42 signaling pathways in schizophrenia: implications for dendritic spine deficits. Biol Psychiatry. 2010;68:25–32. doi: 10.1016/j.biopsych.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonhoeffer T, Yuste R. Spine motility. Phenomenology, mechanisms, and function. Neuron. 2002;35:1019–1027. doi: 10.1016/s0896-6273(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 23.Koleske AJ. Molecular mechanisms of dendrite stability. Nat Rev Neurosci. 2013;14:536–550. doi: 10.1038/nrn3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negishi M, Katoh H. Rho family GTPases and dendrite plasticity. Neuroscientist. 2005;11:187–191. doi: 10.1177/1073858404268768. [DOI] [PubMed] [Google Scholar]
- 25.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Arion D, Unger T, Lewis DA, Mirnics K. Molecular markers distinguishing supragranular and infragranular layers in the human prefrontal cortex. Eur J Neurosci. 2007;25:1843–1854. doi: 10.1111/j.1460-9568.2007.05396.x. [DOI] [PubMed] [Google Scholar]
- 27.Joberty G, Perlungher RR, Sheffield PJ, Kinoshita M, Noda M, Haystead T, et al. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat Cell Biol. 2001;3:861–866. doi: 10.1038/ncb1001-861. [DOI] [PubMed] [Google Scholar]
- 28.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 29.Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 31.Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell. 2002;9:73–83. doi: 10.1016/s1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 32.Chen TJ, Gehler S, Shaw AE, Bamburg JR, Letourneau PC. Cdc42 participates in the regulation of ADF/cofilin and retinal growth cone filopodia by brain derived neurotrophic factor. J Neurobiol. 2006;66:103–114. doi: 10.1002/neu.20204. [DOI] [PubMed] [Google Scholar]
- 33.Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumi T, Matsumoto K, Nakamura T. Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase. J Biol Chem. 2001;276:670–676. doi: 10.1074/jbc.M007074200. [DOI] [PubMed] [Google Scholar]
- 35.Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64:58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- 36.Psychiatric Association A. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- 37.Volk DW, Eggan SM, Lewis DA. Alterations in metabotropic glutamate receptor 1alpha and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. Am J Psychiatry. 2010;167:1489–1498. doi: 10.1176/appi.ajp.2010.10030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, et al. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eggan SM, Lazarus MS, Stoyak SR, Volk DW, Glausier JR, Huang ZJ, et al. Cortical glutamic acid decarboxylase 67 deficiency results in lower cannabinoid 1 receptor messenger RNA expression: implications for schizophrenia. Biol Psychiatry. 2012;71:114–119. doi: 10.1016/j.biopsych.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimoto S, Bazmi HH, Lewis DA. Lower expression of glutamic acid decarboxylase 67 in the prefrontal cortex in schizophrenia: contribution of altered regulation by Zif268. Am J Psychiatry. 2014;171:969–978. doi: 10.1176/appi.ajp.2014.14010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto T, Bazmi HH, Mirnics K, Wu Q, Sampson AR, Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 44.Arion D, Corradi JP, Shaowu T, Datta D, Boothe F, He A, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Molecular Psychiatry. 2014 doi: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volk DW, Chitrapu A, Edelson JR, Lewis DA. Chemokine receptors and cortical interneuron dysfunction in schizophrenia. Schizophr Res. 2014 doi: 10.1016/j.schres.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorph-Petersen KA, Pierri JN, Perel JM, Sun Z, Sampson AR, Lewis DA. The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharmacology. 2005;30:1649–1661. doi: 10.1038/sj.npp.1300710. [DOI] [PubMed] [Google Scholar]
- 47.Volk DW, Matsubara T, Li S, Sengupta EJ, Georgiev D, Minabe Y, et al. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169:1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datta D, Arion D, Lewis DA. Developmental Expression Patterns of GABAA Receptor Subunits in Layer 3 and 5 Pyramidal Cells of Monkey Prefrontal Cortex. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Lin Y, Song C, Sibille E, Tseng GC. Detecting disease-associated genes with confounding variable adjustment and the impact on genomic meta-analysis: with application to major depressive disorder. BMC Bioinformatics. 2012;13:52. doi: 10.1186/1471-2105-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Tseng GC. An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. The Annals of Applied Statistics. 2011;5:994–1019. [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 53.Hoftman GD, Volk DW, Bazmi HH, Li S, Sampson AR, Lewis DA. Altered Cortical Expression of GABA-Related Genes in Schizophrenia: Illness Progression vs Developmental Disturbance. Schizophr Bull. 2015;41:180–191. doi: 10.1093/schbul/sbt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glausier JR, Lewis DA. Selective pyramidal cell reduction of GABA(A) receptor alpha1 subunit messenger RNA expression in schizophrenia. Neuropsychopharmacology. 2011;36:2103–2110. doi: 10.1038/npp.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sodhi MS, Simmons M, McCullumsmith R, Haroutunian V, Meador-Woodruff JH. Glutamatergic gene expression is specifically reduced in thalamocortical projecting relay neurons in schizophrenia. Biol Psychiatry. 2011;70:646–654. doi: 10.1016/j.biopsych.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 57.Gorvel JP, Chang TC, Boretto J, Azuma T, Chavrier P. Differential properties of D4/LyGDI versus RhoGDI: phosphorylation and rho GTPase selectivity. FEBS Lett. 1998;422:269–273. doi: 10.1016/s0014-5793(98)00020-9. [DOI] [PubMed] [Google Scholar]
- 58.Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 59.Chia PH, Li P, Shen K. Cell biology in neuroscience: cellular and molecular mechanisms underlying presynapse formation. J Cell Biol. 2013;203:11–22. doi: 10.1083/jcb.201307020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ouyang Y, Wong M, Capani F, Rensing N, Lee CS, Liu Q, et al. Transient decrease in F-actin may be necessary for translocation of proteins into dendritic spines. Eur J Neurosci. 2005;22:2995–3005. doi: 10.1111/j.1460-9568.2005.04521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott RW, Olson MF. LIM kinases: function, regulation and association with human disease. J Mol Med (Berl) 2007;85:555–568. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 62.Sumi T, Matsumoto K, Takai Y, Nakamura T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J Cell Biol. 1999;147:1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayashi K, Ohshima T, Hashimoto M, Mikoshiba K. Pak1 regulates dendritic branching and spine formation. Dev Neurobiol. 2007;67:655–669. doi: 10.1002/dneu.20363. [DOI] [PubMed] [Google Scholar]
- 65.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayashi-Takagi A, Araki Y, Nakamura M, Vollrath B, Duron SG, Yan Z, et al. PAKs inhibitors ameliorate schizophrenia-associated dendritic spine deterioration in vitro and in vivo during late adolescence. Proc Natl Acad Sci U S A. 2014;111:6461–6466. doi: 10.1073/pnas.1321109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pathania M, Davenport EC, Muir J, Sheehan DF, Lopez-Domenech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubio MD, Haroutunian V, Meador-Woodruff JH. Abnormalities of the Duo/Ras-related C3 botulinum toxin substrate 1/p21-activated kinase 1 pathway drive myosin light chain phosphorylation in frontal cortex in schizophrenia. Biol Psychiatry. 2012;71:906–914. doi: 10.1016/j.biopsych.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lipska BK, Mitkus SN, Mathew SV, Fatula R, Hyde TM, Weinberger DR, et al. Functional genomics in postmortem human brain: abnormalities in a DISC1 molecular pathway in schizophrenia. Dialogues Clin Neurosci. 2006;8:353–357. doi: 10.31887/DCNS.2006.8.3/blipska. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lipska BK, Peters T, Hyde TM, Halim N, Horowitz C, Mitkus S, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 75.Arnsten AF, Wang MJ, Paspalas CD. Neuromodulation of thought: flexibilities and vulnerabilities in prefrontal cortical network synapses. Neuron. 2012;76:223–239. doi: 10.1016/j.neuron.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erdely HA, Tamminga CA, Roberts RC, Vogel MW. Regional alterations in RGS4 protein in schizophrenia. Synapse. 2006;59:472–479. doi: 10.1002/syn.20265. [DOI] [PubMed] [Google Scholar]
- 78.Guillozet-Bongaarts AL, Hyde TM, Dalley RA, Hawrylycz MJ, Henry A, Hof PR, et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2014;19:478–485. doi: 10.1038/mp.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.