Abstract
Urothelial cell carcinoma of the bladder (UCCB) is the most common form of bladder cancer and it is estimated that ~15,000 people in the US succumbed to this disease in 2013. Bladder cancer treatment options are limited and research to understand the molecular mechanisms of this disease is needed to design novel therapeutic strategies. Recent studies have shown that microRNAs play pivotal roles in the progression of cancer. MiR-148a has been shown to serve as a tumor suppressor in cancers of the prostate, colon, and liver, but its role in bladder cancer has never been elucidated. Here we show that miR-148a is down-regulated in UCCB cell lines. We demonstrate that overexpression of miR-148a leads to reduced cell viability through an increase in apoptosis rather than an inhibition of proliferation. We additionally show that miR-148a exerts this effect partially by attenuating expression of DNA methyltransferase 1 (DNMT1). Finally, our studies demonstrate that treating cells with both miR-148a and either cisplatin or doxorubicin is either additive or synergistic in causing apoptosis. These data taken together suggest that miR-148a is a tumor suppressor in UCCB and could potentially serve as a novel therapeutic for this malignancy.
Keywords: bladder cancer, apoptosis, miR-148a, cisplatin, cell cycle
Introduction
Bladder cancer in the US is the fourth most common type of cancer in men and the ninth in women and it was estimated that ~150,000 people died worldwide from bladder cancer in 2008 [1,2]. Approximately 90% of all bladder cancer cases are classified as urothelial cell carcinomas (UCC) which are derived from epithelial cells lining the interior of the organ [3]. Therapeutic options for UCC of the bladder (UCCB) include surgical resection and the GC (gemcitabine and cisplatin) and MVAC (methotrexate, vinblastine, doxorubicin, and cisplatin) chemotherapy regimens, but disease recurrence leading to death is common [4,5]. Little progress has been made in the development of new therapies and survival has improved little in the last 20 years [6]. Identifying alternative treatments is critical for prolonging survival.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (18-24 nucleotides in length) that primarily function to negatively regulate gene expression by promoting mRNA degradation or repressing translation [7]. Recent evidence has demonstrated that miRNAs play a role in the etiology and progression of bladder cancer by affecting several facets of the disease including proliferation, apoptosis, and migration. They can serve as either oncogenes or tumor suppressors depending on context and targets. MiR-708 was shown to be oncogenic in bladder cancer by inhibiting apoptosis via targeting caspase-2 [8]. MiR-182-5p was also shown to be oncogenic in bladder cancer by targeting RECK and Smad4 leading to increased proliferation, migration, and invasion [9]. A tumor suppressive role was discovered for the p53 target miR-34a due to its ability to sensitize bladder cancer cells to cisplatin treatment [10]. MiR-409-3p was shown to inhibit migration and invasion by targeting c-Met [11].
MiR-148a plays the role of tumor suppressor in several contexts. MiR-148a was shown to suppress the epithelial to mesenchymal transition and reduce metastasis in non-small cell lung cancer and gastric cancer respectively by targeting Rock1 [12,13]. MiR-148a was shown to inhibit the epithelial to mesenchymal transition in hepatocellular carcinoma cells as well [14]. In hepatitis B associated hepatocellular carcinoma, miR-148a was shown to be a p53 target and its re-introduction into cultured liver tumor cells resulted in decreased proliferation [15]. In pancreatic and colorectal cancers, miR-148a has been shown to induce apoptosis by targeting Bcl-2 [16,17]. MiR-148a down-regulation by hypermethylation has been associated with metastasis and reintroduction of this miRNA into cells with epigenetic inactivation reduced motility, tumor growth and inhibited metastasis in a SIHN-011B head and neck cancer xenograft model [18]. Further studies on colon cancer that focused on expression of circulating miRNAs revealed that decreased circulating levels of miR-148a correlated with disease recurrence [19]. More recent studies report that miR-148a was one of the miRNAs that were down regulated in triple negative breast cancer cell lines and this was associated with deregulation of the E2F pathway [20]. Studies in prostate cancer have also demonstrated that miR-148a levels were decreased in metastatic prostate cancer cell lines and this was implicated in drug resistance [21]. The role of miR-148a in bladder cancer has never been investigated.
DNA methylation is an epigenetic modification that plays an essential role in the control of gene expression and is integral to many processes including cellular differentiation, development, and tumorigenesis [22]. DNA methyltransferase 1 (DNMT1) is a major and ubiquitously expressed methyltransferase whose expression is regulated at multiple levels [23]. Deregulation of DNMT1 expression and the accompanying disruption of DNA methylation patterns are associated with many cancers, making DNMT1 an attractive therapeutic target [24]. Studies have revealed that DNMT1 is up-regulated in bladder cancer [25]. However, the role of DNMT1 in bladder cancer cell survival is controversial. One study reported that silencing of DNMT1 by shRNA in the T24 UCCB cell line led to decreased proliferation and increased apoptosis [26]. A conflicting report indicated that T24 cells depleted of DNMT1 exhibited no growth defects [27]. Recent studies have suggested that miR-148a serves as a tumor suppressor by targeting DNMT1 [28]. In gastric cancer cells, miR-148a overexpression was shown to suppress cellular proliferation and also to downregulate DNMT1 [29]. Whether the miR-148a/DNMT1 expression axis plays a role in bladder cancer proliferation or survival is unknown.
In this study, we characterize the expression of miR-148a in UCCB cell lines and assess its role in proliferation and induction of apoptosis. We found that miR-148a is down-regulated in several cell lines when compared to immortalized urothelial cells. Overexpression of miR-148a in vitro reduced cell viability by promoting apoptosis. Furthermore, miR-148a decreased DNMT1 expression and we demonstrate that the effects observed following miR-148a overexpression are partially due to DNMT1 depletion. Lastly, we show that induction of cell death can be enhanced by combining miR-148a with chemotherapeutic agents.
Materials and Methods
Cell Culture, Plasmids, siRNA, and Transfections
SV-HUC-1, T24, TCCSUP, J82, and UM-UC-3 cells were generous gifts from Drs. Sweeney and De Vere White (UC Davis). All cell lines were cultured in 10% FBS RPMI-1640 supplemented with glutamine and both penicillin and streptomycin at 37C and 5% CO2. All siRNA transfections were carried out using Lipofectamine RNAiMAX (Invitrogen) at an oligonucleotide concentration of 50nM unless otherwise specified. The following oligonucleotides were used for transfection; miR-148a mimic and anti-miR-148a (Qiagen), control non-targeting oligonucleotide and DNMT1 siRNA (Dharmacon). The following drugs were used for treatments; cisplatin and doxorubicin (Fisher Scientific). DNMT1-PCDNA3.1 was created by inserting DNMT1 sequence excised from an LZRS vector (LZRS-DNMT1 was a gift from Paul Khavari (Addgene plasmid # 24952)) into the multiple cloning site of PCDNA3.1. UM-UC-3 cells were transfected using Lipofectamine 3000 with empty vector or DNMT1 containing vector and subjected to G418 selection.
RNA Extraction and Quantitative PCR
Total RNA containing miRNAs was extracted using the mirVana miRNA Isolation Kit (Ambion) and reverse transcribed into cDNA using the miScript II RT Kit (Qiagen). qPCR was performed using the miScript PCR kit (Qiagen) on a ViiA 7 Real Time PCR System (Applied Biosystems, Grand Island, NY). Expression of miR-148a was normalized to RNU6-2_1 and was analyzed using the efficiency corrected ΔCt method. Primer assays; RNU6-2_1 and miR-148a (Qiagen).
Proliferation Assays
For miR-148a titration curves, cells were plated at 10,000-20,000 cells/well in a 24 well plate and transfected 24 hours later with 0, 10, 25, or 50nM miR-148a mimic and/or a balancing concentration of a control oligonucleotide. Proliferation was assessed 72 hours later using Cell Counting Kit – 8 (CCK-8) (Dojindo). All conditions were performed in triplicate. Data is displayed as the mean +/− standard deviation.
For experiments assessing the growth of cells transfected with DNMT1 siRNA, cells were plated in a 24 well plate at 10,000-20,000 cells/well and transfected 24 hours later with a control oligonucleotide or DNMT1 targeting siRNA. Proliferation was assessed 48 and 96 hours later using CCK-8. All conditions were performed in triplicate. Data is displayed as the mean +/− standard deviation.
For quantitative studies using chemotherapeutic drugs in conjunction with either miR-148a mimic, anti-miR-148a, or DNMT1 targeting siRNA, cells were plated 24 hours prior to transfection at 10,000-20,000 cells/well. 24 hours post transfection, cells were treated with either cisplatin or doxorubicin at 0.5μM or 0.05μM respectively. Proliferation was assessed 48 hours later using CCK-8. All conditions were performed in triplicate. Data are displayed as the mean +/− standard deviation.
For qualitative studies assessing cell growth visually, T24 and UM-UC-3 cells were plated at 100,000 cells/35mm dish and treated with miR-148a mimic or a control oligonucleotide and either cisplatin or doxorubicin in the same manner as above. Cell growth was assessed 48 hours after drug treatment via staining with crystal violet. Images were taken using an Alpha Innotech MultiImage II system (San Jose, CA).
Colony Formation Assay
T24 and UM-UC-3 cells were plated in a 6 well plate at 1500 cells/well. The following day, cells were transfected with control oligonucleotide or miR-148a mimic in triplicate. T24 cells were allowed to grow for 10 days and UM-UC-3 cells were allowed to grow for 12 days post transfection. Colonies were then stained using crystal violet and photographed using an Alpha Innotech MultiImage II system (San Jose, CA). For the rescue experiments using DNMT1 overexpressing cells, we plated 2000 cells/well and cultured for 11 days. All other conditions and analyses were as described above.
Flow Cytometry
T24 and UM-UC-3 cells were transfected with either a control oligonucleotide or miR-148a mimic or DNMT1 targeting siRNA in triplicate or quadruplicate. 72 hours later, cells were harvested and either ethanol fixed and propidium iodide (PI) stained (Sigma-Aldrich) for cell cycle analysis, or were Annexin V-FITC and PI stained (eBioscience) for analysis of apoptosis. All flow cytometry was carried out on a FACS Calibur flow cytometer (BD, Franklin Lakes, NJ). Cell cycle data was analyzed using ModFit software and apoptosis data was analyzed using CellQuest software. Data is displayed as the mean +/− standard deviation.
Western Blot
Whole cell lysates were subjected to SDS-PAGE and transferred to 0.2μm nitrocellulose membranes. Membranes were blocked in 5% milk in PBS-T and left to incubate with primary antibody overnight at 4C. The following day, membranes were incubated with secondary antibody conjugated to HRP and development was carried out using the WesternBright Sirius Kit (Advansta) and visualized with a SRX-101A developer (Konica Minolta, Wayne, NJ) or a c-Digit digital scanner (LI-COR, Lincoln, Nebraska). Loading was assessed either by actin, tubulin, or Ponceau S. The following antibodies were used: cleaved caspase-3 and PARP/cleaved PARP (Cell Signaling), Bcl-2, DNMT1, and actin (Santa Cruz Biotechnology), Tubulin (Thermo Scientific).
Statistics
For all assays, a two tailed two sample equal variance student's t-test was used to assess differences between samples. A p<0.05 was accepted as significant.
Results
MiR-148a is down-regulated in UCCB cell lines
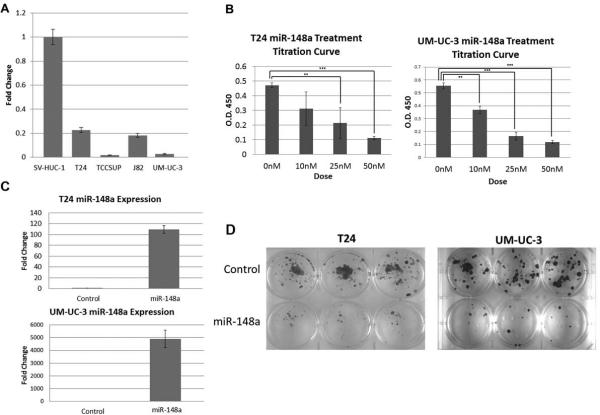
Multiple groups have reported that miR-148a functions as a tumor suppressor. In this study, we asked if expression of miR-148a is altered in bladder tumor-derived cell lines when compared to immortalized control cells. Using qPCR, we assessed the expression of miR-148a in the UCCB cell lines T24, TCCSUP, J82, and UM-UC-3 and in the immortalized urothelial cell line SV-HUC-1 (Fig. 1A). All of the tumor-derived lines have a ≥3 fold decrease in the levels of miR-148a in comparison to the immortalized control line, SV-HUC-1. We could further stratify the cancer lines into two classes based on relative expression; T24 and J82 have significantly more miR-148a than TCCSUP and UM-UC-3. These data demonstrate that miR-148a expression is both down-regulated and variable in UCCB. This analysis suggests that miR-148a may function as a tumor suppressor in this context.
Figure 1. miR-148a expression and effects in UCCB cell lines.
A. qPCR was performed on four UCCB cell lines (T24, TCCSUP, J82, and UM-UC-3) and the immortalized urothelial cell line SV-HUC-1 to assess the relative expression of miR-148a. B. Proliferation was assessed in T24 and UM-UC-3 cells via titration curves with treatments of 0, 10, 25, and 50nM of either miR-148a mimic and/or a balancing amount of control oligonucleotide. C. qPCR was used to assess expression of miR-148a in T24 and UM-UC-3 cells following 50nM transfection with the miR-148a mimic. D. Colony formation assays were performed on T24 and UM-UC-3 cells treated with either miR-148a mimic or a control oligonucleotide. ** denotes p-value ≤ 0.01, *** denotes p-value ≤ 0.001.
MiR-148a overexpression attenuates viability in T24 and UM-UC-3 cells
Based upon our results from the expression analysis of miR-148a in UCCB cell lines, we hypothesized that re-expression of miR-148a might lead to an inhibition of cellular growth. The T24 and UM-UC-3 cell lines were chosen for subsequent functional studies as they represent the two classes of relative expression. To determine if miR-148a overexpression attenuates cellular viability, both cell lines were transfected with 0, 10, 25, or 50nM miR-148a mimic and dose response was assessed by proliferation assays (Fig. 1B). As hypothesized, we found miR-148a overexpression results in decreased cell growth and that this effect was dose dependent. Since the 50nM dose was most effective, all subsequent studies used this dose. Using qPCR, we were able to confirm overexpression of miR-148a (Fig. 1C). Further studies of cellular viability using colony formation assays also demonstrated a strong attenuation of cellular growth in cells treated with miR-148a mimic (Fig. 1D). These results taken together demonstrate that miR-148a exerts a growth inhibitory effect in UCCB cells.
MiR-148a does not attenuate cellular viability via cell cycle inhibition
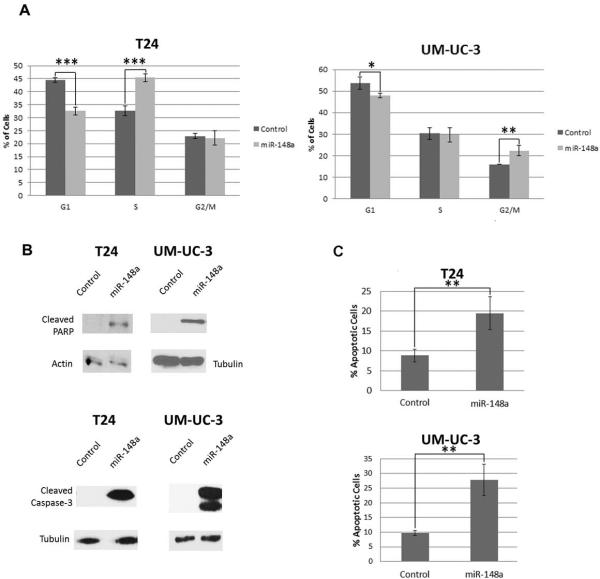
The above data suggests that miR-148a acts as a tumor suppressor in the context of UCCB but does not differentiate between cell cycle inhibition and increased apoptosis. To address this question, flow cytometry was used to detect cell cycle changes of T24 and UM-UC-3 cells treated either with the miR-148a mimic or a control oligonucleotide (Fig. 2A). In T24 cells, there was no statistically significant G1 or G2/M arrest that could account for the observed decrease in viability of miR-148a transfected cells. Surprisingly, there was an increase in S-phase. In UM-UC-3 cells overexpressing miR-148a, there was no change in S-phase and similar to T24, no G1 arrest. However, in UM-UC-3 cells, there was a statistically significant increase in the G2/M population of miR-148a mimic treated cells. Although this could lead to some proliferative inhibition, these results taken together suggest that cell cycle retardation is not the primary mechanism by which miR-148a overexpression inhibits cellular viability.
Figure 2. Effects of miR-148a mimic treatment on cell cycle and apoptosis in T24 and UM-UC-3 cells.
A. Cell cycle analysis by PI staining and flow cytometry was performed on miR-148a or control oligonucleotide treated T24 cells and UM-UC-3 cells. B. Whole cell lysates from T24 and UM-UC-3 cells treated with either miR-148a mimic or a control oligonucleotide were used in western blot analysis of cleaved caspase-3 and cleaved PARP. Either actin or tubulin served as loading controls. C. Annexin-V staining coupled to flow cytometry was performed to further assess induction of apoptosis in response to overexpression of miR-148a in T24 and UM-UC-3 cells. * denotes p-value ≤ 0.05, ** denotes p-value ≤ 0.01, *** denotes p-value ≤ 0.001.
MiR-148a overexpression induces apoptosis in T24 and UM-UC-3 cells
Based on the results from our cell cycle analysis, we hypothesized that overexpression of miR-148a induces apoptosis. Western blot analysis of cleaved caspase-3 and cleaved PARP demonstrated that treatment of cells with miR-148a mimic activates apoptosis (Fig. 2B). Flow cytometry coupled to annexin-V staining was used to confirm the results. As anticipated, treatment of T24 and UM-UC-3 cells with miR-148a mimic induced apoptosis (Fig. 2C). These data argue that the decrease in viability of bladder cancer cells observed following overexpression of miR-148a is predominately dependent on activation of apoptosis.
MiR-148a inhibits UCCB cell viability in part by targeting DNMT1
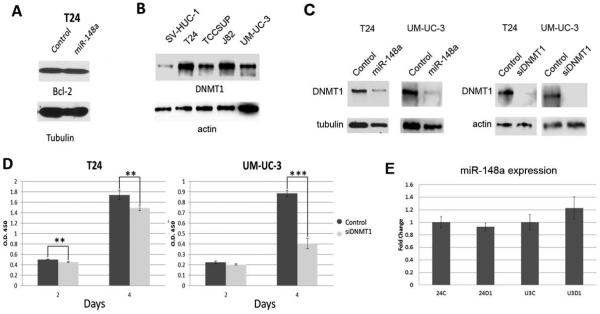
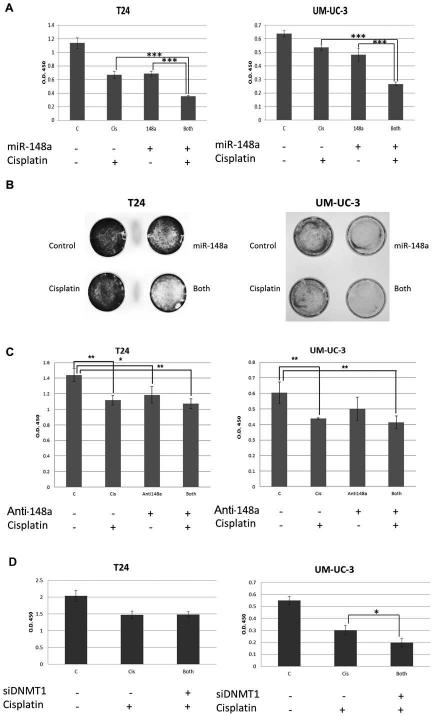
We next explored the mechanism by which overexpression of miR-148a induces apoptosis in UCCB cell lines. Previous studies in pancreatic and colorectal cancer cells reported that miR-148a induced apoptosis by targeting the anti-apoptotic oncogene Bcl-2. However, we did not observe changes in Bcl-2 levels when miR-148a was overexpressed in T24 cells (Fig. 3A). It has also been shown that miR-148a can target DNMT1. Given that DNMT1 is thought to be an oncogene in bladder cancer, we first asked in DNMT1 is overexpressed in our UCCB cell line context (Fig. 3B). We found that DNMT1 protein was overexpressed to varying degrees in all four lines assayed versus SV-HUC-1. We next assessed whether the miR-148a/DNMT1 axis was mediating our observed results. Western blots of whole cell lysates from T24 and UM-UC-3 cells treated with either miR-148a mimic or a control oligonucleotide showed reduced DNMT1 levels demonstrating that in the context of UCCB, DNMT1 is a miR-148a target (Fig. 3C). Using an siRNA against DNMT1, we asked if DNMT1 knockdown could phenocopy the results attained in the miR-148a studies (Fig.3C and D). Proliferation assays indicated that DNMT1 knockdown leads to a decrease in viability in both cell lines. However, the result in UM-UC-3 was far more pronounced indicating that DNMT1 is more important for survival in this cellular context than it is in T24 cells. Additionally, we asked whether there was a negative feedback loop between DNMT1 and miR-148a. Previous studies have shown that miR-148a is methylated in cancer [18]. Using qPCR, we demonstrated that miR-148a levels do not change in response to DNMT1 knockdown suggesting methylation may not be involved in miR-148a down-regulation in UCCB (Fig. 3E).
Figure 3. Assessment of DNMT1 as a downstream target of miR-148a.
A. Whole cell lysates from T24 cells treated either with miR-148a mimic or control oligonucleotide were used for western blot analysis of Bcl-2. Tubulin served as loading control. B. Whole cell lysates from SV-HUC-1, T24, TCCSUP, J82, and UM-UC-3 cells were used for western blot analysis of DNMT1. Actin served as loading control. C. Whole cell lysates from T24 and UM-UC-3 cells treated with either miR-148a mimic, DNMT1 targeting siRNA, or a control oligonucleotide were used for western blots for DNMT1. Tubulin or actin served as loading controls. D. Proliferation was assessed in T24 and UM-UC-3 cells treated with either DNMT1 targeting siRNA or a control oligonucleotide. ** denotes p-value ≤ 0.01, *** denotes p-value ≤ 0.001. E. qPCR was used to assess miR-148a expression in response to either control oligonucleotide (C) or DNMT1 targeting siRNA (D1) in both T24 (24) and UM-UC-3 (U3) cells.
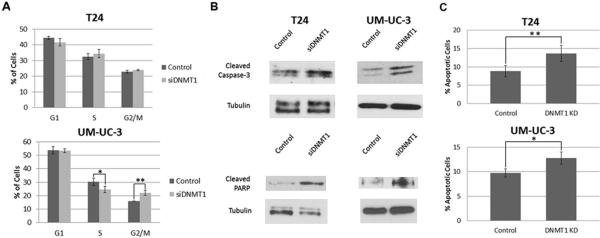
We next further assessed whether the effects of DNMT1 knockdown on viability were mediated by cell cycle inhibition or induction of apoptosis (Fig. 4). Flow cytometry was used to analyze the cell cycle in cells treated either with DNMT1 targeting siRNA or a control oligonucleotide (Fig. 4A). Similar to miR-148a overexpression, DNMT1 knockdown causes no change and a small increase in the G2/M population in T24 and UM-UC-3 cells respectively. Unlike the miR-148a overexpression experiments, we found no significant change in the G1 populations. However, there was a statistically significant decrease in S-phase in UM-UC-3 cells following DNMT1knockdown. In UM-UC-3, these results suggest that there is a modest retardation of the cell cycle. However, together these results suggested that decreased DNMT1 expression inhibits cellular viability predominantly through an increase in apoptosis. An analysis of apoptosis revealed that attenuation of DNMT1 leads to increased caspase-3 cleavage and cleavage of PARP (Fig. 4B). To further verify the induction of apoptosis, T24 and UM-UC-3 cells treated with either siRNA targeting DNMT1 or a control oligonucleotide were subjected to annexin-V staining and flow cytometry (Fig. 4C). This analysis confirmed that decreased DNMT1 expression promoted apoptosis.
Figure 4. Effect of DNMT1 knockdown on cell cycle and apoptosis.
A. Cell cycle analysis by PI staining and flow cytometry was performed on DNMT1 siRNA treated or control oligonucleotide treated T24 cells and UM-UC-3 cells. B. Whole cell lysates from T24 and UM-UC-3 cells treated with either siRNA targeting DNMT1 or a control oligonucleotide were used for western blots for cleaved caspase-3 and cleaved PARP. Tubulin served as a loading control. C. Annexin-V staining coupled to flow cytometry was used to assess apoptosis on T24 and UM-UC-3 control oligonucleotide treated cells and DNMT1 targeting siRNA treated cells * denotes p-value ≤0.05, ** denotes p-value ≤ 0.01.
We next performed a rescue experiment to deteremine if overexpression of DNMT1 abrogates the effects of miR-148a treatment. We focused this study on UM-UC-3 because DNMT1 seemed most important for survival in this cell type. We generated stable DNMT1 overexpressing UM-UC-3 cells. UM-UC-3 cells harboring the control PCDNA3.1 vector were generated to serve as a control. Western blot analysis confirmed DNMT1overexpression (Fig. 5A). Western blots for cleaved PARP demonstrated that DNMT1 overexpression completely abolishes increased PARP cleavage indicating a rescue from miR-148a- induced apoptosis (Fig. 5B). We further assessed growth of these cells in response to miR-148a treatment via a colony formation assay (Fig. 5C). Our data demonstrate that viability is restored in response to miR-148a overexpression when DNMT1 is also overexpressed. However, we note that the effects of miR-148a on control cell viability are not as pronounced as previously shown. We hypothesize that high levels of manipulation and stress from cloning altered the phenotype of these cells making them more resistant than their wild-type counterparts.
Figure 5. Overexpression of DNMT1 attenuates effect of miR-148a treatment.
A. Whole cell lysates from DNMT1 overexpressing and control empty vector UM-UC-3 cells were used for western blots to demonstrate overexpression of DNMT1. Tubulin served as a loading control. B. Whole cell lysates from DNMT1 overexpressing and control empty vector UM-UC-3 cells treated either with control oligonucleotide or miR-148a mimic were used for western blots of PARP. Tubulin served as a loading control. C. DNMT1 overexpressing and control empty vector UM-UC-3 cells treated either with control oligonucleotide or miR-148a mimic were used for colony formation assays to demonstrate the effect of DNMT1 overexpression on miR-148a induced reduction of viability. PCDNA3.1 = control empty vector.
Taken together, these results indicate that DNMT1 is a miR-148a target in UCCB and that this pathway is partly responsible for the observed decrease in viability through an induction of apoptosis. However, because the DNMT1 data do not completely phenocopy the miR-148a data, we conclude that miR-148a has additional targets in this context.
MiR-148a used in conjunction with cisplatin or doxorubicin leads to an additive or synergistic induction of apoptosis in T24 and UM-UC-3 cells
Based on the above results, we asked whether miR-148a overexpression could cooperate with standard of care chemotherapeutics to further decrease cellular viability. We used two types of proliferation assays (one quantitative and one qualitative) where T24 and UM-UC-3 cells were treated either with miR-148a mimic, low dose of cisplatin, or both (Fig. 6). While treatment of the cells with each agent individually led to significant reductions in viability, the inhibition seen by combined treatment was additive in T24 and synergistic in UM-UC-3 cells (Fig. 6A and B). We next assessed whether we could protect against cisplatin treatment using an antagomir against miR-148a to reduce miR-148a expression (Fig. 6C). Surprisingly, our results demonstrated that anti-miR-148a has a slight anti-proliferative effect in T24 and no effect in UM-UC-3. We hypothesize that miR-148a levels are already so low in these cells that further reduction has no major effect. Accordingly, no protective effect was observed combining cisplatin with anti-miR-148a. A recent report suggested that DNMT1 expression plays a role in cisplatin sensitivity in ovarian cancer [30]. Therefore, we asked if this pathway was mediating the observed effect when combining miR-148a with cisplatin. We found that in UM-UC-3 cells, the effect of decreased DNMT1 expression and cisplatin treatment was additive (Fig. 6D). However, the same effect was not observed for T24 cells. These data suggest again that the effects of miR-148a are context dependent and that additional miR-148a targets contribute to the pro-apoptotic effect of miR-148a.
Figure 6. Effect of combining miR-148a with cisplatin in T24 and UM-UC-3 cells.
A. T24 and UM-UC-3 cells were transfected with either control oligonucleotide or miR-148a mimic and 24 hours post transfection were treated with either 0.5μM cisplatin or vehicle. Cell viability was assessed 48 hours post drug treatment using the CCK-8 reagent. B. T24 and UM-UC-3 cells were plated in 35mm dishes and were subjected to identical treatments as described in A. 48 hours post drug treatment, cells were stained with crystal violet and images were taken. C. T24 and UM-UC-3 cells were transfected with either control oligonucleotide or anti-miR-148a mimic and 24 hours post transfection were treated with either 0.5μM cisplatin or vehicle. Cell viability was assessed 48 hours post drug treatment using the CCK-8 reagent. D. T24 and UM-UC-3 cells were transfected with either control oligonucleotide or DNMT1 targeting siRNA and 24 hours post transfection were treated with either 0.5uM cisplatin or vehicle. Cell viability was assessed 48 hours post drug treatment using the CCK-8 reagent. * denotes p-value ≤ 0.05, ** denotes p-value ≤ 0.01, *** denotes p-value ≤ 0.001.
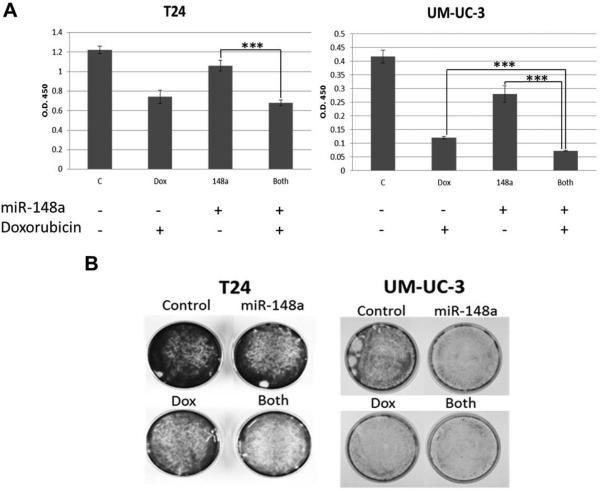
Combination of miR-148a with doxorubicin was also assessed using the same approach described for cisplatin (Fig. 7A and B). The qualitative assay argued that miR-148a could cooperate with doxorubicin in reducing cellular viability in both lines. However, the quantitative assays indicated an additive effect of the two agents for UM-UC-3 only, while the assay failed to demonstrate statistical significance in T24 cells.
Figure 7. Effect of combining miR-148a with doxorubicin in T24 and UM-UC-3 cells.
A. T24 and UM-UC-3 cells were transfected with either control oligonucleotide or miR-148a mimic and 24 hours post transfection were treated with either 0.05μM doxorubicin or vehicle. Cell viability was assessed 48 hours post drug treatment using the CCK-8 reagent. B. T24 and UM-UC-3 cells were plated in 35mm dishes and were subjected to identical treatments as described in A. 48 hours post drug treatment, cells were stained with crystal violet and images were taken. *** denotes p-value ≤ 0.001.
Discussion
Evasion of cellular death and uncontrolled cellular proliferation are two of the hallmarks of cancer and multiple studies have focused on the role miRNAs play in regulating these processes [31]. MiR-148a has been shown to be deregulated in different tumors but the downstream targets of this miRNA differ and it is unclear how miR-148a modulates cellular responses in varying cellular contexts. Many studies point to a tumor suppressor role for miR-148a. In our present study, we demonstrate that miR-148a expression is lower in UCCB cell lines versus non-transformed cells and also acts as a tumor suppressor by potently inducing apoptosis. Our results suggest that repression of miR-148a in bladder cancer cells promotes cell survival and thus miR-148a could serve as a potential therapeutic.
Moreover, our study demonstrates that miR-148a leads to induction of apoptosis in part by down-regulating DNMT1, an epigenetic modifier. Modulation of DNMT1 levels would globally modify transcription, where a decrease in DNMT1 would lead to a de-repression of silenced genes. The overall effect of such wide-spread transcriptional changes would be cell context dependent. Previous studies have investigated the role of DNMT1 in bladder tumorigenesis. Wu et al showed that DNMT1 expression was increased in bladder cancers and correlated with stage of the disease [25]. Furthermore, inhibition of DNMT1 resulted in decreased cellular viability [25]. Additional studies indicated that decreased expression of DNMT1 in T24 cells inhibited cellular proliferation and increased apoptosis [26]. Using a canine model of bladder cancer, Dhawan et al demonstrated that targeting DNMT1 may be a viable treatment option and provided an animal model with which to translate future discoveries [32]. However, the mechanism(s) that lead to increased DNMT1 expression in bladder tumors have not been identified. Our study suggests that one mechanism which contributes to elevated DNMT1 levels in bladder tumors is a deregulation of miR-148a and further supports targeting of DNMT1 as a therapeutic strategy.
The current 5-year survival rate for muscle invasive UCCB is low and there are few treatment options [10]. Cisplatin, a potent chemotherapeutic agent, has been shown to have limited efficacy in the treatment of this malignancy. The results presented here indicate that while cisplatin and miR-148a independently induce apoptosis, the effect of combining the two agents is additive in T24 cells and synergistic in UM-UC-3 cells resulting in significantly increased cell death. The combined treatment of cells with miR-148a and doxorubicin also enhanced apoptosis in T24 and UM-UC-3 cells compared to treatment with a single agent but the results were less impressive suggesting that the complementary effect of miR-148a was specific to the chemotherapeutic agent. Our results demonstrate that miR-148a has potential as a novel agent for treatment of UCCB and that combination therapy with specific chemotherapeutics could be used as potent therapeutic regimens.
Our work additionally points to the potential increased utility inherent in miRNAs as therapies over targeting individual oncoproteins/tumor suppressor proteins alone. We show that using siRNA to specifically target DNMT1 leads to only a partial phenocopy of the results obtained using miR-148a mimic. Thus, although DNMT1 is targeted by miR-148a in this context, it is not the only target. Use of a molecule with broader rather than restricted specificity may be a better option for promoting cell death. MiRNAs have the ability to regulate several different mRNA targets simultaneously and because of this, they are key regulators of gene networks. This provides a method to tailor de-regulated expression of several transcripts with a single molecule.
In conclusion, we have elucidated a novel pathway regulating cell death in UCCB. Our study highlights the potential utility of miR-148a as a novel therapeutic for the treatment of UCCB.
Acknowledgments
Grant Support:
U.S Department of Veterans Affairs, Office of Research and Development, VA MERIT awards BX00004000 and BX001079 to PMG and MM, respectively, National Cancer Institute CA133209 (PMG), and part by Grant Number T32-GM008799 from NIH-NIGMS. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NIGMS, NIH, VA or the US government.
Abbreviation
- DNMT1
DNA methyltransferase 1
- UCC
urothelial cell carcinoma
- qPCR
quantitative PCR
- PARP
poly ADP ribose polymerase
- FBS
fetal bovine serum
- SDS-PAGE
sodium dodecyl sulfate- polyacrylamide gel electrophoresis
- PBS-T
phosphate buffered saline-Tween 20
- PI
propidium iodide
References
- 1.Yafi FA, North S, Kassouf W. First- and second-line therapy for metastatic urothelial carcinoma of the bladder. Curr Oncol. 2011;18(1):e25–34. doi: 10.3747/co.v18i1.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Amsellem-Ouazana D, Bieche I, Tozlu S, Botto H, Debre B, Lidereau R. Gene expression profiling of ERBB receptors and ligands in human transitional cell carcinoma of the bladder. J Urol. 2006;175(3 Pt 1):1127–1132. doi: 10.1016/S0022-5347(05)00317-4. [DOI] [PubMed] [Google Scholar]
- 4.Pliarchopoulou K, Laschos K, Pectasides D. Current chemotherapeutic options for the treatment of advanced bladder cancer: a review. Urol Oncol. 2013;31(3):294–302. doi: 10.1016/j.urolonc.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Kim JJ. Recent advances in treatment of advanced urothelial carcinoma. Curr Urol Rep. 2012;13(2):147–152. doi: 10.1007/s11934-012-0238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdoorn BP, Kessler ER, Flaig TW. Targeted therapy in advanced urothelial carcinoma. Oncology (Williston Park) 2013;27(3):219–226. [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Song T, Zhang X, Zhang L, et al. miR-708 promotes the development of bladder carcinoma via direct repression of Caspase-2. J Cancer Res Clin Oncol. 2013;139(7):1189–1198. doi: 10.1007/s00432-013-1392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirata H, Ueno K, Shahryari V, et al. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012;7(11):e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinall RL, Ripoll AZ, Wang S, Pan CX, deVere White RW. MiR-34a chemosensitizes bladder cancer cells to cisplatin treatment regardless of p53-Rb pathway status. Int J Cancer. 2012;130(11):2526–2538. doi: 10.1002/ijc.26256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Chen H, Lin Y, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36(1):62–68. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Song Y, Wang Y, Luo J, Yu W. MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem. 2013;380(1-2):277–282. doi: 10.1007/s11010-013-1682-y. [DOI] [PubMed] [Google Scholar]
- 13.Zheng B, Liang L, Wang C, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17(24):7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 14.Yan H, Dong X, Zhong X, et al. Inhibitions of epithelial to mesenchymal transition and cancer stem cells-like properties are involved in miR-148a-mediated anti-metastasis of hepatocellular carcinoma. Mol Carcinog. 2013 doi: 10.1002/mc.22064. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Fan Z, Kang L, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123(2):630–645. doi: 10.1172/JCI64265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Li M, Zang W, et al. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. 2013 doi: 10.1007/s13277-013-1115-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Li Y, Huang Q, et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011;18(11):1702–1710. doi: 10.1038/cdd.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shivapurkar N, Weiner LM, Marshall JL, et al. Recurrence of Early Stage Colon Cancer Predicted by Expression Pattern of Circulating microRNAs. PLoS One. 2014;9(1):e84686. doi: 10.1371/journal.pone.0084686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aydogdu E, Katchy A, Tsouko E, et al. MicroRNA-regulated gene networks during mammary cell differentiation are associated with breast cancer. Carcinogenesis. 2012;33(8):1502–1511. doi: 10.1093/carcin/bgs161. [DOI] [PubMed] [Google Scholar]
- 21.Fujita Y, Kojima K, Ohhashi R, et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285(25):19076–19084. doi: 10.1074/jbc.M109.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kar S, Deb M, Sengupta D, et al. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics. 2012;7(9):994–1007. doi: 10.4161/epi.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin W, Leonhardt H, Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus. 2011;2(5):392–402. doi: 10.4161/nucl.2.5.17928. [DOI] [PubMed] [Google Scholar]
- 24.Singh V, Sharma P, Capalash N. DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer. Curr Cancer Drug Targets. 2013;13(4):379–399. doi: 10.2174/15680096113139990077. [DOI] [PubMed] [Google Scholar]
- 25.Wu CT, Wu CF, Lu CH, et al. Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer. 2011;117(22):5221–5233. doi: 10.1002/cncr.26150. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Zeng F, Peng S, Zhu C, Li H, Wang L. Effects on biological behavior of bladder carcinoma T24 cells via silencing DNMT1 and/or DNMT3b with shRNA in vitro. J Huazhong Univ Sci Technolog Med Sci. 2009;29(2):215–219. doi: 10.1007/s11596-009-0216-z. [DOI] [PubMed] [Google Scholar]
- 27.Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 2006;66(2):729–735. doi: 10.1158/0008-5472.CAN-05-1537. [DOI] [PubMed] [Google Scholar]
- 28.Gailhouste L, Gomez-Santos L, Hagiwara K, et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58(3):1153–1165. doi: 10.1002/hep.26422. [DOI] [PubMed] [Google Scholar]
- 29.Zhu A, Xia J, Zuo J, et al. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med Oncol. 2012;29(4):2701–2709. doi: 10.1007/s12032-011-0134-3. [DOI] [PubMed] [Google Scholar]
- 30.Xiang Y, Ma N, Wang D, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2013 doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Dhawan D, Ramos-Vara JA, Hahn NM, et al. DNMT1: An emerging target in the treatment of invasive urinary bladder cancer. Urol Oncol. 2012 doi: 10.1016/j.urolonc.2012.03.015. [DOI] [PubMed] [Google Scholar]