Abstract
A defining feature of viral central nervous system (CNS) infection is the rapid onset of severe neuroinflammation. However, the mechanisms underlying glial responses to replicative neurotropic viruses are only now becoming apparent with the discovery of a number of cytosolic sensors for viral nucleic acids. We have described the expression by murine and human glial cells of two disparate pattern recognition receptors, retinoic acid inducible gene-I (RIG-I) and DNA-dependent activator of interferon regulatory factors (DAI), receptors for viral RNA and DNA moieties, respectively. In the present study, we demonstrate the functional significance of RIG-I expression in primary murine microglia and astrocytes. Our data indicate that murine glial immune responses to a model neurotropic RNA virus, vesicular stomatitis virus, are RIG-I dependent and independent of levels of DAI expression or RNA polymerase III activity. In contrast, maximal glial inflammatory and antiviral responses to the DNA virus herpes simplex virus-1 (HSV-1) are dependent on the expression of both RIG-I and DAI, and require RNA polymerase III activity. These findings indicate that the RNA sensor, RIG-I, acts in parallel with DAI in an RNA polymerase III-dependent manner to initiate glial responses to HSV-1. We therefore suggest that RIG-I plays a significant role in the detection of both RNA and DNA pathogens by microglia and astrocytes.
Keywords: RLR, microglia, astrocytes, neuroinflammation, innate immunity
INTRODUCTION
RNA and DNA virus infections of the central nervous system (CNS) are known to broadly trigger microglial and astrocyte activation and the release of mediators that can promote innate and adaptive immune responses. Such responses can limit viral replication and dissemination leading to infection resolution. However, a defining feature of viral CNS infection is the rapid onset of severe neuroinflammation, and overzealous glial responses associated with significant neurological damage or even death (as reviewed in Furr and Marriott, 2012). Pathogens as dissimilar as the DNA virus, herpes simplex virus (HSV)-I, and the single-stranded RNA virus, West Nile Virus (WNV), induce the production of proinflammatory and chemotactic molecules by microglia (Aravalli et al, 2005; Cheeran et al, 2005; Furr et al, 2011; Lokensgard et al, 2001; van Marle et al, 2007). In addition, we have demonstrated the ability of both microglia and astrocytes to respond to other RNA and DNA viruses, including vesicular stomatitis virus (VSV), Sendai virus and murine gammaherpesvirus (MHV)-68 by producing inflammatory mediators such as IL-6 and TNF-α (Chauhan et al, 2010; Furr et al, 2008; 2010; Rasley et al, 2004). However, the mechanisms underlying glial responses to neurotropic viruses are only now becoming apparent with the discovery of a variety of cell surface and cytosolic molecules that serve as sensors for viral components.
Host cells principally sense viral pathogens by their nucleic acid genomes, typically as a result of viral transcription and/or their replicative activity. Two disparate pattern recognition receptors (PRRs), retinoic acid inducible gene (RIG)-I and DNA-dependent activator of interferon regulatory factors (DAI), have been identified and serve as sensors for distinct viral classes. RIG-I senses the 5′-triphosphate moiety of negative-sense single-stranded (ss) RNA viruses (Hornung et al, 2006; Pichlmair et al, 2006) or short blunt-ended double-stranded (ds) RNA (Takahasi et al, 2008). Ligation of RIG-I by these moieties triggers activation of the NF-kB and/or interferon regulatory factor (IRF3) pathway(s) and the subsequent production of proinflammatory cytokines and type I interferons (IFNs), respectively. DNA is also a potent activator of innate immunity (Stetson and Medzhitov, 2006) and DAI has been shown to bind synthetic dsDNA, B-DNA, to similarly initiate NF-kB and IRF3-mediated cytokine production (Ishii et al, 2006; Takaoka et al, 2007). We have demonstrated that mouse microglia and astrocytes express RIG-I (Furr et al, 2008) and shown that this viral sensor plays a key role in human astrocyte responses to the neurotropic ssRNA virus, VSV (Furr et al, 2010). Furthermore, we have established that murine glia functionally express DAI and demonstrated that this cytosolic DNA sensor plays a significant role in glial inflammatory and neurotoxic responses to HSV-1 (Furr et al, 2011).
Interestingly, some studies have suggested that RIG-I may also play a role in the recognition of DNA pathogens via the activity of RNA polymerase III (Ablasser et al, 2009; Chiu et al, 2009). In this pathway, AT-rich dsDNA serves as a template for RNA polymerase III and is transcribed into RNA containing a 5′-triphosphate, a ligand for RIG-I. Activation of RIG-I by dsDNA has been shown to induce type I IFN production and NF-kB activation indicating this pathway represents another mechanism by which cells sense viral DNA (Ablasser et al, 2009; Chiu et al, 2009). In the present study, we demonstrate that RIG-I is functional in murine microglia and astrocytes, and show that this molecule is essential for inflammatory glial responses to VSV. Furthermore, we have determined that this viral sensor and RNA polymerase III are also important components in inflammatory and antiviral glial responses to HSV-1 and MHV-68, suggesting a key role for RIG-I in both RNA and DNA virus-associated neuroinflammation.
METHODS
Isolation of neonatal brain microglia and astrocytes
Primary neonatal murine microglia and astrocytes were isolated according to a modified protocol to that previously described by our laboratory (Bowman et al, 2003; Chauhan et al, 2010; Furr et al, 2008; 2010; 2011; Rasley et al, 2004). Briefly, six to eight neonatal C57Bl/6J mouse brains per preparation were dissected free of meninges and large blood vessels and finely minced with sterile surgical scissors. The minced tissue was then forced through a wire screen and briefly incubated with 0.25% trypsin-1 mM EDTA in serum free RPMI 1640 medium for 5 minutes. The cell suspension was then washed and this mixed glial culture was maintained in RPMI 1640 containing 10% fetal bovine serum (FBS) and gentamicin for 2 weeks.
Astrocytes were isolated from mixed glial cultures by mild trypsinization (0.25% trypsin-1 mM EDTA for 20 minutes) in the absence of FBS as previously described (Saura et al, 2003). The remaining intact layer of adherent cells was demonstrated to be >98% microglia by immunohistochemical staining for the microglial surface marker CD11b (Saura et al, 2003) and the isolated astrocytes were determined to be >96% pure based on morphological characteristics and the expression of the astrocyte marker glial fibrillary acidic protein (GFAP) as determined by immunofluorescence microscopy. The microglia were maintained for 1 week in RPMI 1640 with 10% FBS and 20% conditioned medium from LADMAC cells (ATCC number CRL-2420), a murine monocyte-like cell line that secretes colony stimulating factor-1 (CSF-1), while astrocytes were cultured in RPMI 1640 containing 10% FBS. All studies were performed in accordance with relevant federal guidelines and institutional policies regarding the use of animals for research purposes.
Source and Propagation of Cell Lines
EOC 13.31 (ATCC® CRL-2468), an immortalized murine microglia cell line, was purchased from American Type Culture Collection (Manassas, VA). These cells were derived from C3H/HeJ mice and so are TLR4 defective. The lack of this receptor would not be anticipated to effect cell responses to VSV or viral nucleic acids, and there is no data to support the notion that TLR4 mediates microglial responses to HSV-1. Cells were maintained in RPMI 1640 supplemented with 10% FBS, gentamicin, and 20% LADMAC conditioned media. EOC 13.31 cells have been classified as microglia due to the presence of CD11b and other microglia-specific cell surface markers (Walker et al, 1995).
Preparation of viral stocks and in vitro infection of glial cells
Wild type VSV (Indiana Strain), VSV-ΔM51-GFP, herpes simplex virus-1 (HSV-1; MacIntyre strain, ATCC®, VR-539), and murine gammaherpesvirus-68 (MHV-68) were used in this study. VSV-ΔM51-GFP was kindly provided by Dr. Valery Grdzelishvili (University of North Carolina at Charlotte) and was generated by the insertion of the green fluorescent protein (GFP) open reading frame at position 5 of the viral genome and a methionine deletion in the M protein as previously described (Wollmann et al, 2010). Wild type and mutant VSV viral stocks were prepared by infecting monolayer cultures of baby hamster kidney epithelial cells (BHK-21; ATCC®, CCL-10) as we have described previously (Chauhan et al, 2010; Furr et al, 2008; 2010). BHK-21 cells were maintained in DMEM and were infected with VSV at a low multiplicity of infection (MOI) of 0.05 plaque-forming units (PFU) per cell and incubated for 48 hours at 37°C in SFM4MegaVir protein-free medium (Thermo Scientific Hyclone, Waltham, MA). The cell-free medium containing released virus was collected, centrifuged to remove unwanted cellular debris, and the concentration of infectious viral particles was quantified using a standard plaque assay of serial dilutions of VSV on BHK-21 cells at 37°C. It should be noted that the MOI values are relative and calculated based on the concentration of infectious VSV particles determined on BHK cells supporting robust virus replication. It is therefore likely that the actual MOI values for microglia and/or astrocytes were actually lower than indicated.
HSV-1 viral stocks were prepared by infecting monolayer cultures of African green monkey kidney cells (Vero; ATCC, CCL-81) with HSV-1 (MacIntyre strain from a patient with encephalitis; ATCC, VR-539) at a low MOI of 0.05 PFU per cell and incubated for 48 hours at 37°C in SFM4MegaVir protein-free medium as we have described previously (Furr et al, 2011). MHV-68 viral stocks were prepared by infecting monolayer cultures of BHK-21 cells at a low viral MOI of 0.1 PFU per cell as we have previously described (Furr et al, 2011). Cells were removed with trypsin and pulse sonicated (Vibra Cell; Sonics & Materials Inc., Newtown, CT) to release intact virions. The sonicated material was centrifuged to remove unwanted cellular debris. The concentration of infectious viral particles in the cell-free supernatant was quantified using a standard plaque assay of serial dilutions of HSV-1 on Vero cells or MHV-68 on NIH-3T3 cells at 37°C, and glial cells were exposed to infectious viral particles at the MOIs indicated.
Microglia and astrocytes were infected with VSV, HSV-1, or MHV-68 at MOIs between 0.01 and 10 PFU per cell as indicated and the viruses were allowed to adsorb for 1 hour prior to washing to remove non-adherent viral particles. Cultures were maintained for the indicated time periods prior to collection of culture supernatants, preparation of whole cell protein isolates, or RNA isolation.
In vitro stimulation of glial cells with RIG-I or DAI ligands
Glial cells were transfected with the RIG-I ligands 5′-triphosphate single-stranded RNA (5′ppp-ssRNA) or 5′ triphosphate double stranded RNA (5′ppp-dsRNA; Invivogen, San Diego, CA), or a synthetic double stranded B-DNA analog poly(deoxyadenylic-deoxythymidylic) acid sodium salt (Poly(dA:dT; Invivogen) that is a DAI specific ligand.
To generate 5′ppp-ssRNA, we created a linear template for run-off in vitro transcription by digesting the pGEM-SeV-NP plasmid (previously described in (Curran and Kalokofsky, 1991) with EcoRV. EcoRV cuts pGEM-SeV-NP at a single site allowing a 342 base RNA to be generated when using the SP6 promoter. RNA was generated using the MAXIscript In Vitro Transcription Kit (Life Technologies, Grand Island, NY) at 37°C for 2.5 hours in a final volume of 200 ul containing approximately 1 ug of linearized pGEM-SeV-NP, all four nucleoside triphosphates, transcription buffer, and 200 U of SP6 enzyme mix. RNA was then incubated at 37°C for 20 minutes in the presence of 10 U Turbo DNase (Life Technologies) to remove the plasmid template. Following addition of EDTA (35 mM), the samples were phenol/chloroform extracted and ethanol precipitated. Precipitated RNA was resuspended in RNase free water to a final concentration of 200–250 ng/ul. The final RNA products were analyzed by electrophoresis on a 2.5% agarose gel. 5′ppp-ssRNA was transfected into murine glial cells using FuGENE HD transfection reagent (Promega) at the indicated concentrations for one hour. For comparison purposes, cells were exposed to transfection reagent alone.
B-DNA or 5′ppp-dsRNA was introduced into microglia and astrocytes at the indicated concentrations using FuGENE HD (Promega) or LyoVec™ (Invivogen) transfection reagents according to the manufacturer’s instructions. At the indicated time points following transfection, RNA was isolated and culture supernatants were collected for analysis.
Western blot analyses
Western blot analyses for the presence of RIG-1, DAI, phosphorylated IRF3 (pIRF3), and the HSV-1 tegument protein, VP16, in glial cells were performed as described previously by our laboratory (Bowman et al, 2003). After incubation with a rabbit polyclonal antibody against mouse and human RIG-1 (Abgent, San Diego, CA) or DAI (Abcam, Cambridge, MA), a purified rabbit monoclonal antibody specific for IRF-3 that has been phosphorylated at Ser396 (Cell Signaling Technology), or a purified mouse monoclonal antibody directed against HSV-1 and HSV-2 VP16 (Abcam) for 24 hours at 4°C, blots were washed and incubated in the presence of appropriate horseradish peroxidase-conjugated secondary antibodies. Bound enzyme was detected with the Super Signal system (Thermo Scientific, Rockford, IL). To assess total protein loading in each well, immunoblots were reprobed with a goat anti-mouse β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoblots shown are representative of at least three separate experiments. ImageJ software (National Institutes of Health) was used for densitometric analysis and pIRF3 levels are reported relative to the levels of total IRF expression normalized to β-actin levels.
Quantification of IL-6, TNF-α, and IFN-β in glial cell culture supernatants
Specific capture ELISAs were performed to quantify concentrations of murine IL-6, TNF-α, and IFN-β. Commercially available ELISA kits were used to measure TNF-α and IFN-β secretion (R&D Systems, Minneapolis, MN and BioLegend, San Diego, CA, respectively) while murine IL-6 secretion was measured using a rat anti-mouse IL-6 capture antibody (Clone MP5-20F3) and a biotinylated rat anti-mouse IL-6 detection antibody (Clone MP5-C2311) (BD Biosciences, San Jose, CA). Bound antibody was detected by addition of streptavidin-horseradish peroxidase (BD Biosciences). After addition of TMB substrate and H2SO4 stop solution, absorbances were measured at 450 nm. A standard curve was constructed using varying dilutions of recombinant cytokines (BD Biosciences) and the cytokine content of culture supernatants determined by extrapolation of absorbances to the standard curve.
Semi-quantitative and quantitative reverse transcribed PCR
Total cellular RNA was isolated from microglia and astrocytes with Trizol reagent (Life Technologies) and reverse transcribed as previously described (Bowman et al, 2003). Semi-quantitative polymerase chain reactions (PCR) were performed to determine the expression of mRNA encoding murine IFN-β and the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). In addition, quantitative real-time PCR was also performed to determine the level of expression of mRNA encoding IFN-β, the MHV-68 gene product ORF65, and GAPDH using the SYBR Green approach on a 7500 Fast Real-Time PCR machine (Life Technologies) as described previously (Chauhan et al. 2008). Positive and negative strand PCR primers used, respectively, were 5′-TTGTGCTTCTCCACTACAGC-3′ and 5′-CTGTAAGTCTGTTAATGAAG-3′ to amplify mRNA encoding murine IFN-β, 5′-ATGCTCCAGAAGAGGAAGGGACAC-3′ and 5′-TTGGCAAAGACCCAGAAGAAGCC-3′ to amplify mRNA encoding ORF65, and 5′-CCATCACCATCTTCCAGGAGCGAG-3′ and 5′-CACAGTCTTCTGGGTGGCAGTGAT-3′ to amplify mRNA encoding murine GAPDH. Primers were designed using Primer-BLAST (National Center for Biotechnology Information, Bethesda, MD) based on their location in different exons of the genomic sequence and their lack of significant homology to sequences present in GenBank (National Center for Biotechnology Information, Bethesda, MD). The identity of the PCR amplified fragments was verified by size comparison with DNA standards (Promega). All RNA expression levels are reported as relative levels normalized to the expression of the housekeeping gene GADPH determined in parallel PCR reactions.
siRNA-mediated knockdown of RIG-I and DAI
RIG-I and/or DAI expression in murine microglia and astrocytes was knocked down by transfection with siRNA targeting each as we have previously described (Furr et al, 2010; 2011). Three validated Stealth RNAi™ siRNA duplexes targeting murine RIG-I and three targeting DAI, as well as universal negative control siRNA not homologous to anything in the vertebrate transcriptome, were purchased from Life Technologies. Glial cells were transfected with each siRNA duplex individually or the three in concert using FuGENE HD transfection reagent (Promega) according to the manufacturer’s instructions. In some studies, double knockdown was achieved in glial cells by transfecting with both RIG-I and DAI siRNA duplexes. Antibiotic-free media was replaced with complete media at 6 hours following transfection. At 72 hours after transfection, whole cell lysates were collected for immunoblot analyses to confirm RIG-I and/or DAI expression knockdown. Targeted knockdown of RIG-I and DAI reduced expression of these proteins by 72.7 +/− 2.7% and 79.6 +/− 5.5% SEM, respectively, based on three independent experiments (p < 0.05) consistent with previous results from our laboratory (Furr et al, 2010; 2011).
RNA polymerase III inhibition
Glial cells were pre-treated with the RNA polymerase inhibitors Tagetin™ (10 U for 8 hours; Epicentre Biotechnologies, Madison, WI) or InSolution™ RNA Polymerase III Inhibitor (25 uM for 24 hours; Calbiochem, San Diego, CA) prior to viral challenge or siRNA tranfection.
Fluorescence Measurements
VSV-ΔM51-GFP GFP fluorescence levels were measured using a CytoFluor multi-well plate reader (Life Technologies) with the following parameters: excitation filter of 450/50 nm, emission filter of 530/25 nm and gain = 50 (Moerdyk-Schauwecker et al, 2013).
Statistical analyses
Results of the present studies were tested statistically by ANOVA and Tukey’s post hoc test using commercially available software (GraphPad Prism v5.03, GraphPad Software Inc. La Jolla, CA). In all experiments, results were considered statistically significant when a P-value of less than 0.05 was obtained.
RESULTS
Viral RNA and DNA motifs induce RIG-I expression by primary murine glial cells
We have previously demonstrated that primary murine microglia and astrocytes constitutively express RIG-I and we have shown that such expression is upregulated following RNA virus infection (Furr et al, 2008). To begin to establish the functional status of RIG-I in murine glia we have determined whether the expression of this viral sensor is altered following cytosolic exposure to uncapped 5′ppp-ssRNA, a specific RIG-I ligand (Hornung et al, 2006; Saito and Gale, 2008; Takahasi et al, 2008). As shown in Figure 1A, intracellular 5′ppp-ssRNA administration elicits marked elevations in RIG-I protein levels in microglia and modest increases in astrocytes over levels seen in resting cells, implying that this receptor is functional in both cell types.
FIGURE 1.
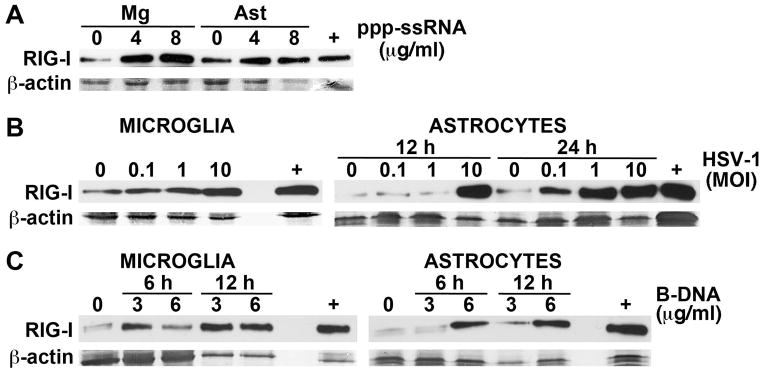
RIG-I protein expression in cultured primary murine astrocytes and microglia is upregulated following exposure to viral RNA and DNA motifs. Panel A: Cultured primary microglia (Mg) or astrocytes (Ast) were treated with transfection reagent alone (0) or transfected with uncapped 5′ppp-ssRNA (4 or 8 ug/ml). At 6 hrs following transfection RIG-I expression was assessed in whole cell protein isolates by immunoblot analysis. Panel B: Primary microglia and astrocytes were uninfected (0) or infected with HSV-1 (MOI of 0.1, 1, or 10). At 12 hrs following infection RIG-I expression was assessed in microglial whole cell protein isolates by immunoblot analysis and such expression in astrocytes was similarly assessed at 12 and 24 hrs. Panel C: Cells were treated with transfection reagent alone (0) or transfected with B-DNA (3 or 6 ug/ml). At 6 and 12 hrs following transfection RIG-I expression was assessed in whole cell protein isolates by immunoblot analysis. For comparison purposes, RIG-I protein expression in a similar number of resting HeLa cells is shown (+). Representative immunoblots are shown from one of three independent experiments.
To determine whether RIG-I expression in glial cells is sensitive to the presence of intracellular viral DNA motifs we have assessed RIG-I protein levels in microglia and astrocytes following infection with the DNA virus HSV-1. As shown in Figure 1B, RIG-I levels were significantly enhanced in microglia as early as 12 hours post-infection. HSV-1 challenge also elevated RIG-I expression in murine astrocytes, albeit with slower kinetics of induction, with significant upregulation at all MOIs used at 24 hours following infection (Figure 1B). Finally, we have assessed the effect of viral DNA motifs on RIG-I expression by primary murine glial cells. B-DNA is a double-stranded DNA in its canonical B helical form, and is a putative ligand of DAI (Takaoka et al, 2007; Wang et al, 2008). As shown in Figure 1C, intracellular B-DNA administration elicited rapid and marked elevations in RIG-I protein expression by both microglia and astrocytes. Together, these data indicate that RIG-I expression is not only auto-induced by its ligand, but can also be upregulated by the presence of intracellular viral DNA motifs.
A specific RIG-I ligand rapidly induces inflammatory mediator production by glial cells
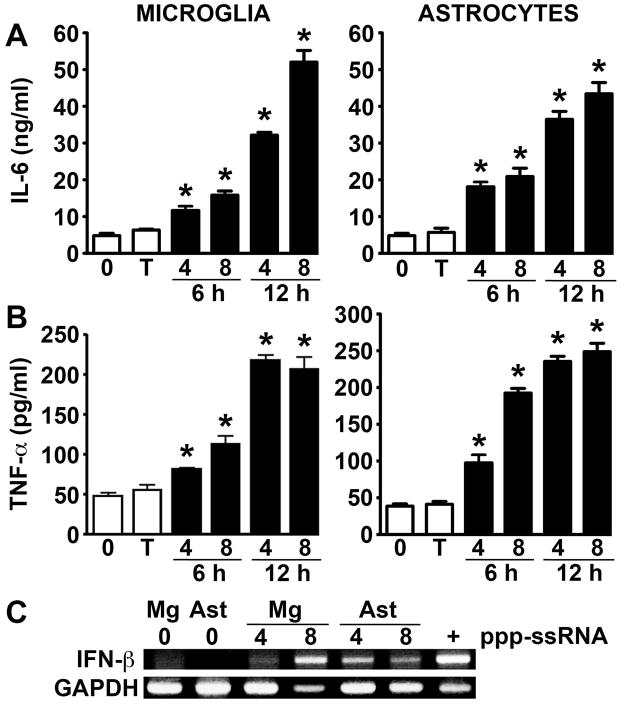
To more directly assess the functional role of RIG-I in primary murine glia, we have evaluated the sensitivity of microglia and astrocytes to intracellular administration of 5′ppp-ssRNA. As shown in Figure 2, this specific RIG-I ligand rapidly induces release of the key inflammatory cytokines IL-6 and TNF-α by microglia and astrocytes in both a dose and time dependent manner. Furthermore, we have also determined that introduction of 5′ppp-ssRNA similarly induces expression of mRNA encoding IFN-β by primary glia (Figure 2C), and the release of this antiviral type I IFN in a microglial cell line (1792 +/− 11 pg/ml versus undetectable levels in cells exposed to transfection reagent alone; n = 3, p < 0.05) and primary murine astrocytes (1804 +/− 19 pg/ml versus undetectable levels in cells exposed to transfection reagent alone; n = 3, p < 0.05) at 24 hrs following administration of 5′ppp-ssRNA (4 ug/ml). As such, the ability of 5′ppp-ssRNA to upregulate RIG-I expression and induce antiviral and inflammatory cytokine expression demonstrates the functional status of this cytosolic sensor in primary murine glial cells.
FIGURE 2.
Intracellular administration of the RIG-I specific ligand ppp-ssRNA induces type-I interferon and inflammatory cytokine production by murine microglia and astrocytes. Cells were untreated (0), treated with transfection reagent alone (T), or transfected with uncapped 5′ppp-ssRNA (4 or 8 ug/ml; black bars). At 6 and 12 hrs following transfection IL-6 (Panel A) and TNF-α (Panel B) levels were assessed in culture supernatants by specific capture ELISA. Data are expressed as the mean +/− SEM of a representative experiment, performed in triplicate, of studies that were performed three independent times. An asterisk indicates a statistically significant difference from cells treated with transfection reagent alone (p < 0.05). Expression of mRNA encoding IFN-β was determined in glial cells at 4 and 8 hrs by RT-PCR following transfection with the RIG-I ligand (Panel C). For comparison purposes, IFN-β mRNA expression in a similar number of activated murine macrophages is shown (+) and the results shown are representative of three independent experiments.
RIG-I knockdown attenuates VSV-induced inflammatory cytokine production by primary glia
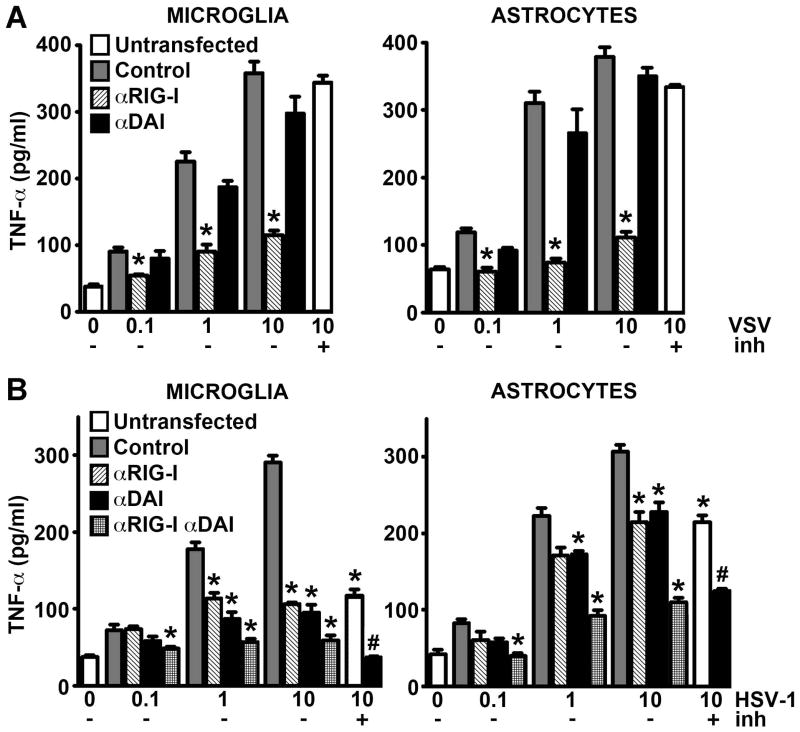
To confirm the functional status of RIG-I in murine glia and to begin to assess the relative importance of this sensor in glial responses to RNA viruses, we have determined the effect of siRNA-mediated RIG-I knockdown on inflammatory cytokine production by microglia and astrocytes following infection with the RNA virus, VSV. In agreement to our previous findings in human astrocytes (Furr et al, 2010), VSV-induced TNF-α production by murine astrocytes was almost totally abolished following RIG-I knockdown relative to cells transfected with scrambled control RNA (Figure 3A). This effect was not observed in astrocytes transfected with siRNA directed against the putative cytosolic viral DNA sensor, DAI (Figure 3A), despite effective DAI expression knockdown (data not shown). Interestingly, similar results were obtained in primary murine microglia where RIG-I, but not DAI, knockdown markedly reduced VSV-induced cytokine production (Figure 3A). These results indicate that the viral RNA sensor RIG-I, but not the DNA sensing molecule DAI, is essential for the detection of, and/or initiation of cellular responses to, this neurotropic RNA virus by primary murine glia.
FIGURE 3.
RIG-I is required for inflammatory glial responses to VSV while both RIG-I and DAI are required for maximal responses to HSV-1. Microglia and astrocytes were untransfected or transfected with scrambled control RNA (Control), or siRNA directed against RIG-I (αRIG-I) or DAI (αDAI). At 72 hrs following transfection cells were infected with VSV (Panel A; MOI of 0.1, 1, or 10) or HSV-1 (Panel B; MOI of 0.1, 1, or 10) and TNF-α levels were assessed in culture supernatants by specific capture ELISA at 12 hrs (Panel A) or 24 hrs (Panel B) following infection. The time points were selected based on the ability of VSV and HSV-1 to elicit maximal cytokine responses at each time point. Some untransfected cells were also pretreated with a RNA polymerase III inhibitor (10 U) for 8 hrs prior to infection with VSV (MOI of 10) or HSV-1 (MOI of 10) and assessment of TNF-α production at 12 hrs (Panel A) or 24 hrs (Panel B) post infection (+inh). Data are expressed as the mean +/− SEM of a representative experiment, performed in triplicate, of studies that were performed two independent times. An asterisk indicates a statistically significant difference from cells transfected with scrambled control RNA (p < 0.05) and a pound symbol indicates a statistically significant difference between RNA polymerase III inhibitor treated cells that were transfected with scrambled control RNA or siRNA directed against DAI (p < 0.05).
RIG-1 and DAI are both involved in glial responses to the DNA virus HSV-1
To investigate whether RIG-I also plays a role in the detection of viral DNA and to begin to determine the relative importance of this innate immune sensor in microglial and astrocyte responses to DNA viruses, we have assessed the effect of RIG-I knockdown on inflammatory cytokine production by microglia and astrocytes following infection with the DNA virus HSV-1. Consistent with our previous studies (Furr et al, 2011), knockdown of the DNA sensor, DAI, significantly reduced HSV-1 associated cytokine production by both microglia and astrocytes, with TNF-α production being attenuated by approximately 67% and 30%, respectively, at the highest viral MOI used (Figure 3B). Surprisingly, RIG-I knockdown achieved similar reductions in HSV-1-induced TNF-α release by both glial cell types (Figure 3B). Furthermore, transfection with siRNAs directed against both DAI and RIG-I reduced microglial TNF-α production following HSV-1 infection by approximately 80% to levels that were statistically indistinguishable from uninfected cells (Figure 3B), while cytokine production by similarly treated astrocytes was also reduced albeit to a lesser extent (60%). Taken together, these data indicate that RIG-I, like DAI, plays a significant role in glial responses to DNA viruses.
RNA polymerase III is essential for DNA virus recognition by RIG-I in murine glial cells
To begin to determine the mechanisms underlying the involvement of RIG-I in glial responses to this DNA virus we have assessed the role of RNA polymerase III. As shown in Figure 3B, pretreatment of cells with a selective RNA polymerase III inhibitor significantly reduced HSV-1-mediated TNF-α production by microglia and astrocytes and did so to the same extent as RIG-I knockdown (approximately 62% and 30%, respectively). This finding was in contrast to parallel studies in which inhibitor pretreatment failed to significantly affect VSV-induced cytokine production by either cell type (Figure 3A; +inh). Importantly, microglial and astrocyte responses to HSV-1 were reduced by approximately 85% and 50%, respectively, when the RNA polymerase III inhibitor was used in combination with DAI knockdown, levels of inhibition that were comparable to those seen in cells following knockdown of both DAI and RIG-I (Figure 3B; +inh). As such, these findings suggest that the RNA sensor, RIG-I, acts in parallel with DAI in a RNA polymerase III-dependent manner to initiate inflammatory glial responses to DNA viruses.
RNA polymerase III is essential for maximal anti-viral glial responses by murine glial cells to DNA viruses
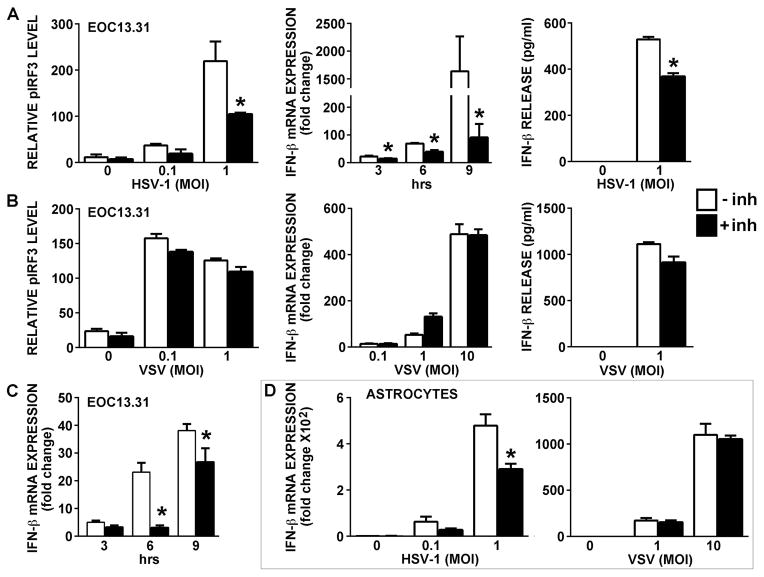
To determine whether RNA polymerase III activity is required for glial antiviral responses, EOC13.31 microglia-like cells or primary astrocytes were pretreated with a selective RNA polymerase III inhibitor prior to challenge with neurotropic DNA or RNA viruses. As shown in Figure 4, exposure of microglial cells to either HSV-1 or VSV significantly increased pIRF3 levels and resulted in robust IFN-β mRNA expression and protein production (Figures 4A and 4B). This response was not limited to neurotropic DNA viruses as the lymphotropic herpesvirus MHV-68 was also capable of inducing IFN-β mRNA expression in microglial cells (Figure 4C). HSV-1 and VSV also induced such responses in primary murine astrocytes (Figure 5D). Importantly, pharmacological inhibition of RNA polymerase III markedly attenuated microglial type I interferon responses to the DNA viruses HSV-1 (Figure 4A) and MHV-68 (Figure 4C), but did not significantly affect an elevation in pIRF3 levels or IFN-β production induced by the RNA virus, VSV (Figure 4B). Similarly, the HSV-1-induced increase in IFN-β mRNA expression in astrocytes was significantly attenuated following RNA polymerase III inhibition while responses of this cell type to VSV were unaffected (Figure 4D).
FIGURE 4.
RNA polymerase III activity is required for maximal type-I IFN responses by glia to the DNA viruses HSV-1 and MHV-68 but not the RNA virus VSV. EOC13.31 microglia (Panels A–C), or primary astrocytes (Panel D) were untreated (open bars) or pretreated with a RNA polymerase III inhibitor (25 uM) for 24 hrs (black bars) prior to being uninfected (0) or infected with HSV-1 (MOI of 0.1 or 1; Panels A and D), VSV (MOI of 0.1, 1, or 10 as indicated in Panels B and D), or MHV-68 (MOI of 1; Panel C). At various times following infection, pIRF3 levels were determined by immunoblot analysis (24 hrs), relative expression of mRNA encoding IFN-β was determined by qPCR (3, 6, and 9 hrs; 6 hr time point shown in Panel B), and IFN-β protein release was assessed by specific capture ELISA (24 hrs). Data are expressed as the mean +/− SEM of a representative experiment, performed in triplicate, of studies that were performed three independent times. An asterisk indicates a statistically significant effect of RNA polymerase III inhibition on glial responses.
FIGURE 5.
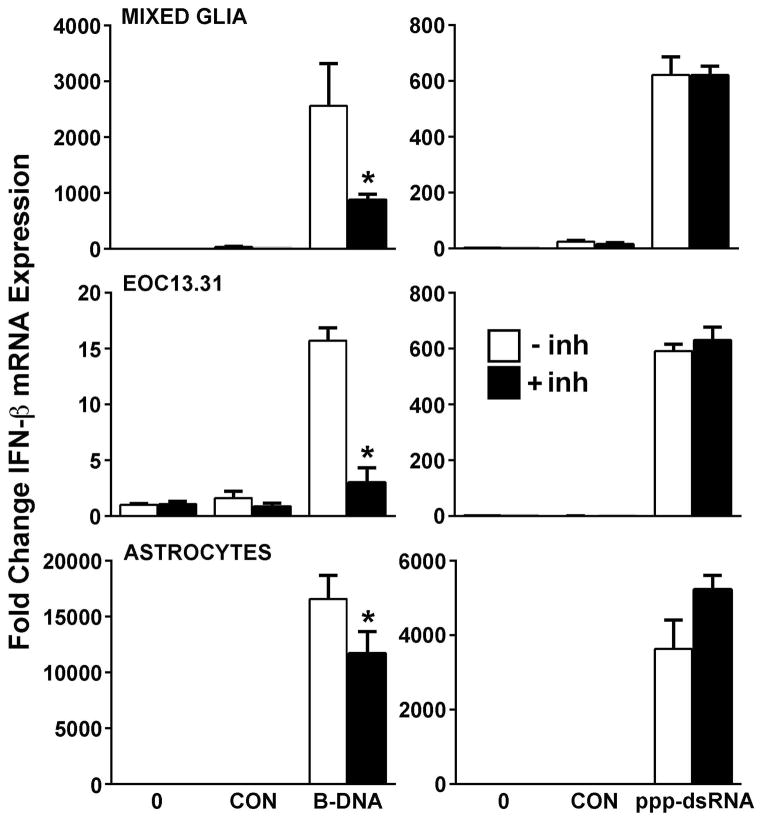
RNA polymerase III activity is required for maximal glial responses to a DAI ligand but not a RIG-I agonist. Mixed glial cells, EOC13.31 microglia-like cells, or primary murine astrocytes were untreated (open bars) or pretreated with a RNA polymerase III inhibitor (25 uM) for 24 hrs (black bars) prior to being untransfected (0), treated with transfection reagent alone (CON), or transfected with uncapped 5′ppp-dsRNA (4 ug/ml) or B-DNA (3 ug/ml). At 24 hrs following transfection, expression of mRNA encoding IFN-β was determined by qPCR. Data are expressed as the mean +/− SEM of a representative experiment, performed in triplicate, of studies that were performed three independent times. An asterisk indicates a statistically significant effect of RNA polymerase III inhibition on IFN-β expression.
To confirm the specificity of the effect of pharmacological RNA polymerase III inhibitors on viral DNA motif-induced type I interferon responses, we have assessed their effect on glial IFN-β production induced by specific DNA and RNA ligands for DAI and RIG-I, respectively. As shown in Figure 5, the intracellular introduction of the DAI ligand B-DNA or the RIG-I ligand 5′ppp-dsRNA elicited a marked increase in the expression of mRNA encoding IFN-β in cultures of primary mixed glial cells, isolated primary astrocytes, or EOC13.31 microglia-like cells. Importantly, prior treatment with a RNA polymerase III inhibitor significantly attenuated the responses of mixed glia, astrocytes, and microglia to B-DNA, but failed to inhibit responses induced by uncapped 5′ppp-dsRNA (Figure 5). This effect was also observed in the IFN-β protein secretion by EOC 13.31 cells at 24 hrs (76 +/− 7 pg/ml in control B-DNA transfected cells versus 26 +/− 8 pg/ml in B-DNA challenged cells pretreated with the RNA polymerase III inhibitor; n = 3, p < 0.0001) and primary astrocytes at 6 hrs (1210 +/− 57 pg/ml in control B-DNA treated cells versus 262 +/− 37 pg/ml in B-DNA challenged cells pretreated with the RNA polymerase III inhibitor; n = 3, p < 0.0001) following intracellular administration (3 ug/ml), while responses to uncapped 5′ppp-dsRNA (4 ug/ml) were unaffected (data not shown).
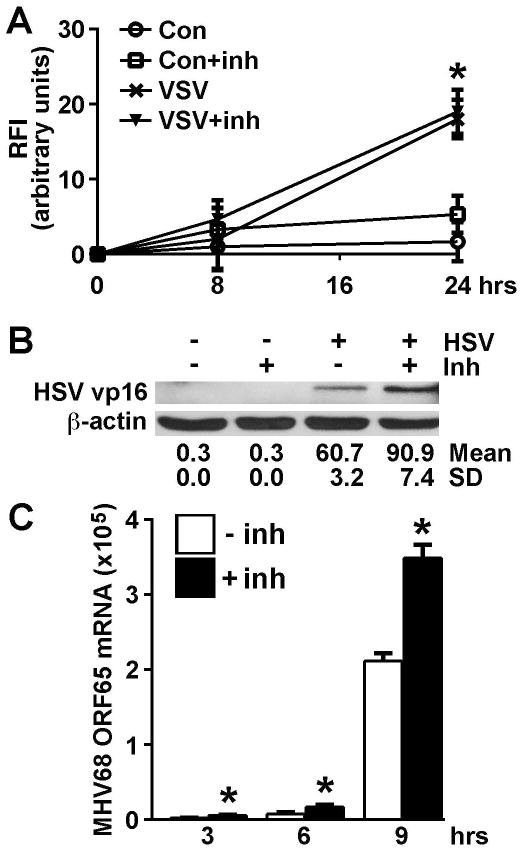
Finally, we have performed experiments to determine whether RNA polymerase III activity is essential for glial responses that limit viral burden. EOC13.31 microglia-like cells were untreated or pretreated with an RNA polymerase III inhibitor for 24 hrs prior to challenge with GFP-expressing VSV, wild type HSV-1, or wild type MHV-68. As shown in Figure 6A, RNA polymerase III inhibition does not influence intracellular GFP-VSV virion production as assessed by relative fluorescence intensity (RFI). In contrast, inhibition of RNA polymerase III activity significantly increased intracellular levels of the HSV-1 tegument protein VP16 (Figure 6B) and the expression of mRNA encoding the MHV-68 product ORF65 (Figure 6C). Together, these data support the contention that RNA polymerase III activity is required to limit DNA, but not RNA, virus product levels.
FIGURE 6.
RNA polymerase III activity limits HSV-1 and MHV-68 protein expression but does not effect VSV virion levels. EOC13.31 microglia-like cells were untreated or pretreated with the RNA polymerase III inhibitor (25 uM) for 24 hrs prior to being uninfected or infected with GFP-expressing VSV (MOI of 1; Panel A), HSV-1 (MOI of 1; Panel B), or MHV-68 (MOI of 1; Panel C). At the indicated times following infection, levels of VSV-GFP expression were determined (Panel A) according to relative fluorescence intensity (RFI), HSV-1 VP16 levels were determined by immunoblot analysis, and relative mRNA expression of the MHV-68 product ORF65 was determined by qPCR. Data are expressed as the mean +/− SEM of a representative experiment, performed in triplicate, of studies that were performed two independent times. An asterisk indicates a statistically significant effect of RNA polymerase III inhibition on glial responses. A representative immunoblot is shown from one of two separate experiments and the average band densities +/− SD is indicated.
DISCUSSION
It has become increasingly apparent that glial cells possess intracellular sensors that can detect compromise of the cytosolic compartment. Most RNA viruses replicate in the cytoplasm and therefore allow detection of viral replicative motifs by RIG-I-like receptors (RLRs) present in the host cell. Indeed, we have shown that microglia and astrocytes are permissive for VSV infection and that productive replication is required for inflammatory mediator production by these cells (Chauhan et al, 2010). Importantly, we have described the robust constitutive expression of RIG-I and another RLR double stranded RNA helicase, melanoma differentiation-associated protein (MDA5), by murine glial cells (Furr et al, 2008). Furthermore, we have previously demonstrated that human astrocytes also express RIG-I and that expression levels of this cytosolic viral RNA sensor increase in murine and human glial cells following challenge with the model neurotropic RNA virus, VSV (Furr et al, 2008; 2010). In the present study, we show that intracellular administration of an in vitro generated 5′ triphosphorylated RNA ligand for RIG-I (Furr et al, 2010; Saito and Gale, 2008; Takahasi et al, 2008) induces RIG-I expression in primary murine microglia and astrocytes. This result is consistent with our previous findings in human astrocytes (Furr et al, 2010), and more recent studies showing the ability of Japanese encephalitis virus (JEV) and Chikungunya virus to elevate RIG-I expression in BV-2 microglia-like cells (Jiang et al, 2014) and U87MG astrocytoma cells (Abraham et al, 2013), respectively. As such, these findings support the functional status of RIG-I in both glial cell types in mice. In addition, the inducible nature of such expression by intact viral particles and a specific RIG-I ligand suggests the possibility that glial cells could become sensitized to the presence of intracellular viral moieties that are produced during viral replication in a feed-forward manner.
Perhaps more importantly, the present study demonstrates that this RLR plays an essential role in the inflammatory and antiviral immune responses of murine astrocytes and microglia to RNA virus infection. Similar to our findings in human astrocytes (Furr et al, 2010), intracellular delivery of a 5′triphosphorylated RNA ligand into primary murine astrocytes initiates increases in type I IFN mRNA expression and robust inflammatory cytokine production by this major glial cell type. Significantly, our results indicate that intracellular delivery of this RIG-I ligand is also a potent stimulus for the expression of the inflammatory mediators IL-6 and TNF-α and type I interferon production by primary microglia. Finally, we have confirmed that RIG-I plays a critical role in the immune responses of microglia and astrocytes to a model neurotropic rhabdovirus by demonstrating that RIG-I knockdown specifically and almost totally abolishes VSV-induced cytokine production by either of these CNS cell types. This finding is consistent with several studies in glial cell lines demonstrating that inflammatory cytokine production by JEV infected BV-2 microglia-like cells (Jiang et al, 2014) and poly I:C-mediated chemokine release by U373MG human astrocytoma cells (Yoshida et al, 2007; Imaizumi et at 2014) is decreased following RIG-I knockdown. Furthermore, it is in agreement with the observations that a putative RLR inhibitor can attenuate RNA ligand-mediated astrocyte activation (de Rivero Vaccari et al, 2012) while RIG-I knockdown leads to increased levels of JEV virus proliferation in BV-2 microglia-like cells (Jiang et al, 2014).
In contrast to RNA viruses, many DNA viruses are known to replicate within the nucleus. In addition, specific recognition of viral DNA motifs is further complicated by the apparent detection by DAI of a structure common to both self and non-self DNA. This suggests that discrimination between these types of DNA is based on its subcellular localization and may be a function of the amount and length of the DNA present in the cytosol rather than a specific chemical feature of the ligand (as reviewed in (Keating et al, 2011)). However, a study has shown that HSV-1 DNA, dislocated from the viral capsid, is readily detectable in the cytoplasm of infected primary macrophages and monocyte/macrophage cell lines (Unterholzner et al, 2010) suggesting that viral DNA can be detected by receptors present in the cytosol. The present study and our previous work support such a hypothesis since intracellular administration of the putative DAI ligand B-DNA is a potent stimulus for inflammatory cytokine production (Furr et al, 2011) and type I IFN (Figure 5) expression by murine glial cells. Furthermore, our demonstration that DAI knockdown significantly reduces HSV-1-induced cytokine production by microglia and astrocytes (Furr et al, 2011 and Figure 3) indicates that this cytosolic viral DNA sensor plays a significant role in the recognition of DNA viruses.
However, RIG-I may also contribute to type I IFN and inflammatory cytokine production by cells in response to DNA virus challenge via the actions of DNA-dependent RNA polymerase III. In this model, AT-rich double-stranded DNA serves as a template for RNA polymerase III and is transcribed as RNA containing a 5′-triphosphate cap, a ligand for RIG-I. Such a hypothesis is supported by the demonstration that cell responses to the gammaherpesvirus, Epstein-Barr virus, are attenuated following RNA polymerase III knockdown or treatment with pharmacological inhibitors of this enzyme (Ablasser et al, 2009; Chiu et al, 2009). The present finding that the DNA virus HSV-1, as well as a synthetic DNA ligand for DAI, can up-regulate RIG-I expression by both astrocytes and microglia provides circumstantial evidence for the involvement of RIG-I in the perception of DNA pathogens. However, more direct evidence comes from the demonstration that both RIG-I knockdown and RNA polymerase III inhibition significantly attenuates inflammatory cytokine and type I IFN production by microglia and astrocytes in response to the DNA viruses HSV-1 and MHV-68, and a synthetic DAI ligand. Furthermore, we provide evidence that such a mechanism can contribute to the control of viral replication within glial cells by demonstrating that the pharmacological inhibition of RNA polymerase III results in the elevated expression of DNA virus products in infected microglia-like cells, but has no effect on VSV product levels. Together, these data provide the first evidence that transcription of cytosolic viral DNA by RNA polymerase III and subsequent RNA recognition by RIG-I could serve as a mechanism for the perception of DNA viruses by CNS cells.
The present finding that both RIG-I and DAI are necessary to mount maximal glial cytokine responses to DNA pathogens is consistent with the observation that DAI knockdown reduces, but does not abolish, glial responses to HSV-1 (Figure 3 and Furr et al, 2011). While the ability of a specific RNA polymerase III inhibitor to attenuate cytokine production by HSV-1 infected or B-DNA challenged glial cells provides strong support for the notion that RIG-I detects RNA moieties transcribed from cytosolic viral DNA, we cannot discount the possibility that DAI and RIG-I signaling pathways interact in other ways in microglia or astrocytes. For example, the adaptor molecule STING has been shown to play a role in both RIG-I and DAI signaling pathways (Ishikawa and Barber, 2008). As such, the possibility exists that HSV-1-initiated RIG-I and DAI signaling pathways converge at this component to potentiate cellular activation. Alternatively, it is conceivable that RIG-I and DAI interact either directly or via STING to promote glial responses. In support of this possibility, immunoprecipitation studies indicate that STING associates with RIG-I complexes in close proximity to endoplasmic reticulum associated-mitochondria (Dixit et al, 2010; Ishikawa and Barber, 2009). However, it remains to be determined whether DAI associates with RIG-I, STING, or other RIG-I signaling components in glial cells. Finally, TLR3 and its associated adaptor molecule TRIF, have been associated with the control of HSV-1 in the CNS of human patients and mice (Conrady et al, 2013; Zhang et al, 2007), and inflammatory processes that lead to CNS pathology following HSV-1 infection have been reported to be associated with TLR2 (Kurt-Jones et al, 2004). While these studies appear to contrast with our findings, a possible explanation may lie in the ability of PRRs to act in a cooperative manner to promote glial inflammatory responses (as discussed in (Furr and Marriott, 2012)). Clearly, further work will be required to resolve these issues.
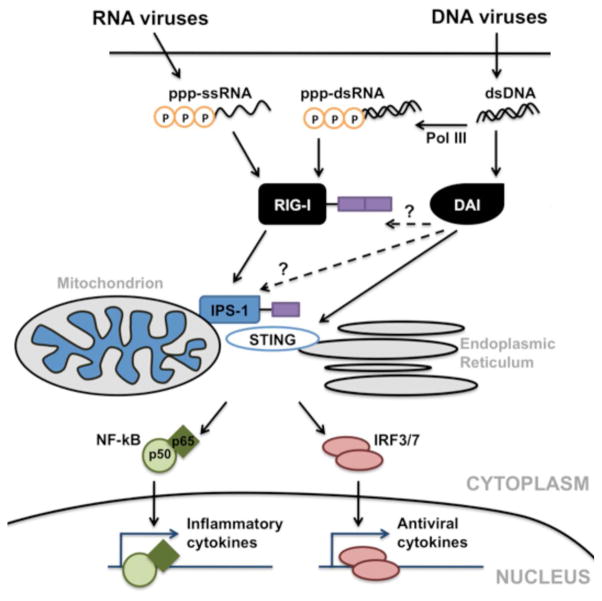
Taken in concert, the present study illustrates the functional significance of RIG-I expression in murine glial cells. Our data show that murine microglial and astrocyte responses to the RNA virus, VSV, are RIG-I dependent but independent of the expression of DAI or RNA polymerase III activity. In contrast, maximal glial inflammatory and antiviral responses to HSV-1 are dependent on the expression of both RIG-I and DAI, and require RNA polymerase III activity. Based upon our results we propose the model shown in Figure 7. We suggest that neurotropic single-stranded RNA viruses, such as VSV, infect microglia and astrocytes and replicate within them generating phosphorylated single stranded RNA (5′ppp-ssRNA) in the cytoplasm that acts as a ligand for RIG-I. RIG-I then associates with IPS-1, and perhaps STING, which subsequently activates NF-kB, IRF3, and IRF7. These transcription factors translocate to the nucleus and initiate the production of inflammatory mediators including TNF-α, IL-6, and type I IFNs. In contrast, neurotropic double-stranded DNA viruses, such as HSV-1, infect glial cells and replicate within them generating genomic DNA. Viral DNA is recognized by DAI which activates downstream effector molecules including IPS-1 and STING, subsequently activating NF-kB, IRF3, and IRF7. In addition, RNA polymerase III (Pol III) in the cytosol transcribes viral DNA into dsRNA containing 5′-triphosphate (5′ppp-dsRNA) that is recognized by RIG-I and similarly initiates transcription factor activation via IPS-1 and STING. Upon release, cytokines such as TNF-α, and IL-6 would be anticipated to promote inflammation, increase blood-brain barrier permeability, and facilitate leukocyte recruitment into the CNS, while type I IFNs function to limit viral replication. As such, these studies suggest that RIG-I plays an important role in the perception of both RNA and DNA viral pathogens by microglia and astrocytes. However, it remains to be determined whether RIG-I-mediated glial responses are, on balance, protective or contribute to the rapid and potentially lethal inflammation associated with such CNS pathogens.
FIGURE 7.
Proposed mechanisms by which murine glia recognize neurotropic RNA and DNA viruses to initiate cytokine production. Replicating RNA viruses generate phosphorylated single stranded RNA (5′ppp-ssRNA) that act as a ligand for RIG-I. RIG-I associates with IPS-1, and perhaps STING, which subsequently activates NF-kB, IRF3, and IRF7. These transcription factors translocate to the nucleus and initiate the production of inflammatory mediators including TNF-α, IL-6, and type I IFNs. Replicating DNA viruses generate genomic DNA that serves as a ligand for DAI. DAI then associates with IPS-1 and STING, subsequently activating NF-kB and IRF3/7. In addition, RNA polymerase III (Pol III) in the cytosol transcribes viral DNA into dsRNA containing 5′-triphosphate (ppp-dsRNA) that is also recognized by RIG-I and similarly initiates transcription factor activation via IPS-1 and STING.
Acknowledgments
This work was supported by grant NS050325 to IM from the National Institutes of Health.
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS CONTRIBUTIONS
EKC and SRF isolated and cultured CNS cells, prepared viral stocks and determined viral titers, carried out the in vitro experiments, performed data analysis, and drafted the manuscript. IM conceived the study, contributed to the experimental design, and edited the final manuscript. All authors read and approved the final version of the manuscript.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham R, Mudaliar P, Padmanabhan A, Sreekumar E. Induction of cytopathogenicity in human glioblastoma cells by chikungunya virus. PLoS One. 2013;8:e75854. doi: 10.1371/journal.pone.0075854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravalli RN, Hu S, Rowen TN, Palmquist JM, Lokensgard JR. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J Immunol. 2005;175:4189–4193. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- Bowman CC, Rasley A, Tranguch SL, Marriott I. Cultured astrocytes express toll-like receptors for bacterial products. Glia. 2003;43:281–291. doi: 10.1002/glia.10256. [DOI] [PubMed] [Google Scholar]
- Chauhan VS, Furr SR, Sterka DG, Jr, Nelson DA, Moerdyk-Schauwecker M, Marriott I, Grdzelishvili VZ. Vesicular stomatitis virus infects resident cells of the central nervous system and induces replication-dependent inflammatory responses. Virology. 2010;400:187–196. doi: 10.1016/j.virol.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Chauhan VS, Sterka DG, Jr, Gray DL, Bost KL, Marriott I. Neurogenic exacerbation of microglial and astrocyte responses to Neisseria meningitidis and Borrelia burgdorferi. J Immunol. 2008;180:8241–8249. doi: 10.4049/jimmunol.180.12.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeran MC, Hu S, Sheng WS, Rashid A, Peterson PK, Lokensgard JR. Differential responses of human brain cells to West Nile virus infection. J Neurovirol. 2005;11:512–524. doi: 10.1080/13550280500384982. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady CD, Zheng M, van Rooijen N, Drevets DA, Royer D, Alleman A, Carr DJ. Microglia and a functional type I IFN pathway are required to counter HSV-1-driven brain lateral ventricle enlargement and encephalitis. J Immunol. 2013;190:2807–2817. doi: 10.4049/jimmunol.1203265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JA, Kolakofsky D. Rescue of a Sendai virus DI genome by other parainfluenza viruses: implications for genome replication. Virology. 1991;182:168–176. doi: 10.1016/0042-6822(91)90660-4. [DOI] [PubMed] [Google Scholar]
- de Rivero Vaccari JP, Minkiewicz J, Wang X, De Rivero Vaccari JC, German R, Marcillo AE, Dietrich WD, Keane RW. Astrogliosis involves activation of retinoic acid-inducible gene-like signaling in the innate immune response after spinal cord injury. Glia. 2012;60:414–421. doi: 10.1002/glia.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Chauhan VS, Moerdyk-Schauwecker MJ, Marriott I. A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J Neuroinflammation. 2011;8:99. doi: 10.1186/1742-2094-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Chauhan VS, Sterka D, Jr, Grdzelishvili V, Marriott I. Characterization of retinoic acid-inducible gene-I expression in primary murine glia following exposure to vesicular stomatitis virus. J Neurovirol. 2008;14:503–513. doi: 10.1080/13550280802337217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Marriott I. Viral CNS infections: role of glial pattern recognition receptors in neuroinflammation. Front Microbiol. 2012;3:201. doi: 10.3389/fmicb.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Moerdyk-Schauwecker M, Grdzelishvili VZ, Marriott I. RIG-I mediates nonsegmented negative-sense RNA virus-induced inflammatory immune responses of primary human astrocytes. Glia. 2010;58:1620–1629. doi: 10.1002/glia.21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Numata A, Yano C, Yoshida H, Meng P, Hayakari R, Xing F, Wang L, Matsumiya T, Tanji K, Tatsuta T, Murakami M, Tanaka H. ISG54 and ISG56 are induced by TLR3 signaling in U373MG human astrocytoma cells: possible involvement in CXCL10 expression. Neurosci Res. 2014;84:34–42. doi: 10.1016/j.neures.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Ye J, Zhu B, Song Y, Chen H, Cao S. Roles of TLR3 and RIG-I in mediating the inflammatory response in mouse microglia following Japanese encephalitis virus infection. J Immunol Res. 2014;2014:787023. doi: 10.1155/2014/787023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, Bronson R, Arnold MM, Knipe DM, Finberg RW. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc Natl Acad Sci USA. 2004;101:1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokensgard JR, Hu S, Sheng W, vanOijen M, Cox D, Cheeran MC, Peterson PK. Robust expression of TNF-alpha, IL-1beta, RANTES, and IP-10 by human microglial cells during nonproductive infection with herpes simplex virus. J Neurovirol. 2001;7:208–219. doi: 10.1080/13550280152403254. [DOI] [PubMed] [Google Scholar]
- Moerdyk-Schauwecker M, Shah NR, Murphy AM, Hastie E, Mukherjee P, Grdzelishvili VZ. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: role of type I interferon signaling. Virology. 2013;436:221–34. doi: 10.1016/j.virol.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Rasley A, Bost KL, Marriott I. Murine gammaherpesvirus-68 elicits robust levels of interleukin-12 p40, but not interleukin-12 p70 production, by murine microglia and astrocytes. J Neurovirol. 2004;10:171–180. doi: 10.1080/13550280490444119. [DOI] [PubMed] [Google Scholar]
- Saito T, Gale M., Jr Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med. 2008;205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura J, Tusell JM, Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi M, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Hondra K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–506. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G, Antony J, Ostermann H, Dunham C, Hunt T, Halliday W, Maingat F, Urbanowski MD, Hobman T, Peeling J, Power C. West Nile virus-induced neuroinflammation: glial infection and capsid protein-mediated neurovirulence. J Virol. 2007;81:10933–10949. doi: 10.1128/JVI.02422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker WS, Gatewood J, Olivas E, Askew D, Havenith CE. Mouse microglial cell lines differing in constitutive and interferon-gamma-inducible antigen-presenting activities for naïve and memory CD4+ and CD8+ T cells. J Neuroimmunol. 1995;63:163–174. doi: 10.1016/0165-5728(95)00146-8. [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann G, Rogulin V, Simon I, Rose JK, van den Pol AN. Some attenuated variants of vesicular stomatitis virus show enhanced oncolytic activity against human glioblastoma cells relative to normal brain cells. J Virol. 2010;84:1563–73. doi: 10.1128/JVI.02040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Imaizumi T, Lee SJ, Tanji K, Sakaki H, Matsumiya T, Ishikawa A, Taima K, Yuzawa E, Mori F, Wakabayashi K, Kimura H, Satoh K. Retinoic acid-inducible gene-I mediates RANTES/CCL5 expression in U373MG human astrocytoma cells stimulated with double-stranded RNA. Neurosci Res. 2007;58:199–206. doi: 10.1016/j.neures.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Zhang SY, Jouanguy E, Sancho-Shimizu V, von Bernuth H, Yang K, Abel L, Picard C, Puel A, Casanova JL. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol Rev. 2007;220:225–236. doi: 10.1111/j.1600-065X.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]