Abstract
Introduction
Chronically elevated circulating inflammatory markers are common in older persons but mechanisms are unclear. Many blood transcripts (>800 genes) are associated with interleukin-6 protein levels (IL6) independent of age. We aimed to identify gene transcripts statistically mediating, as drivers or responders, the increasing levels of IL6 protein in blood at older ages.
Methods
Blood derived in-vivo RNA from the Framingham Heart Study (FHS, n=2422, ages 40–92 yrs) and InCHIANTI study (n=694, ages 30–104 yrs), with Affymetrix and Illumina expression arrays respectively (>17,000 genes tested), were tested for statistical mediation of the age-IL6 association using resampling techniques, adjusted for confounders and multiple testing.
Results
In FHS, IL6 expression was not associated with IL6 protein levels in blood. 102 genes (0.6% of 17,324 expressed) statistically mediated the age-IL6 association of which 25 replicated in InCHIANTI (including 5 of the 10 largest effect genes). The largest effect gene (SLC4A10, coding for NCBE, a sodium bicarbonate transporter) mediated 19% (adjusted CI 8.9 to 34.1%) and replicated by PCR in InCHIANTI (n=194, 35.6% mediated, p=0.01). Other replicated mediators included PRF1 (perforin, a cytolytic protein in cytotoxic T lymphocytes and NK cells) and IL1B (Interleukin 1 beta): few other cytokines were significant mediators.
Conclusions
This transcriptome-wide study on human blood identified a small distinct set of genes that statistically mediate the age-IL6 association. Findings are robust across two cohorts and different expression technologies. Raised IL6 levels may not derive from circulating white cells in age related inflammation.
Keywords: Aging, Inflammation, Transcriptome, Blood, Human, Epidemiology
1. Introduction
Chronically elevated levels of pro-inflammatory biomarkers are a core feature of aging, and a risk factor for many diseases and adverse phenotypes that are frequent in older persons (De Martinis and others 2006). Increased levels of pro-inflammatory markers in blood and other tissues with aging are paralleled by a progressive decline in overall immune responsiveness (Franceschi and Campisi 2014).
Elevated Interleukin-6 (IL6) and C-reactive protein (CRP) have emerged as robust age-related risk factors for multiple adverse outcomes including several diseases, disability, cognitive impairment and death (Singh and Newman 2011). IL6 is a pleiotropic cytokine produced by several cell types, including immune cells, hepatocytes, vascular endothelial cells, adipocytes and skeletal muscle (Singh and Newman 2011). IL6 levels are often undetectable in young individuals but increase with advancing age, even in the absence of detectable causes, including cardiovascular risk factors and disease (Ferrucci and others 2005). High levels of IL6 predict all major age-associated diseases, physical and cognitive disability and mortality (Economos and others 2013; Ferrucci and others 1999; Heikkila and others 2008; Ridker and others 2000). The mechanisms that cause and sustain high levels of inflammatory markers in aging are largely unknown.
The overproduction of pro-inflammatory markers may occur in many sites beyond circulating leukocytes; candidates include tissue resident macrophages, adipocytes, endothelial cells, cells within atherosclerotic plaques and muscle cells (Howcroft and others 2013). The accumulation of cells expressing a senescence-associated secretory phenotype (Campisi 2013; Young and Narita 2009) in different tissues, possibly induced by age-related NF-kB dysregulation (Bektas and others 2013), may also be a possible cause. Data collected in animal models have provided inconsistent results and there is a scarcity of data in humans.
In a recent study, the authors determined the whole blood gene expression transcripts associated with IL6 levels in 2 human populations, independent of age (Lin and others 2014). That analysis identified 4139 genes that were significantly associated with interleukin-6 levels (FDR<0.05), independent of age, sex and blood cell components, of which 807 genes replicated in the smaller InCHIANTI cohort. Many of the top genes generally associated with blood IL6 protein levels (independent of age) are in inflammation-related pathways or erythrocyte function, including the JAK/Stat signalling pathway and interleukin-10 signalling pathway.
In the study presented here, we aimed to identify the gene transcripts in whole blood that might be specific to the chronic inflammation of aging. In particular, we hypothesized that genes with expression profiles that statistically mediate “age-related inflammation” – that account for a proportion of the statistical association between age and IL6, a proxy for age-related inflammation - may reflect the most relevant molecular pathways in aging and may help to identify the specific cell subtypes most closely involved. We therefore estimated the degree to which each whole blood gene transcript statistically explained the age-IL6 association, using mediation models in 2 independent community-based cohorts in a discovery (Framingham Heart Study, FHS) and replication (InCHIANTI) analysis. The cohorts utilize two different microarray platforms, allowing replication of findings robust to cohort and microarray differences.
2. Methods
Our study was performed in two independent and well-characterized human cohorts. The discovery cohort was the Framingham Heart Study (FHS) Offspring cohort, USA (Kannel and others 1979), with replication of the significant mediators in the InCHIANTI (Invecchiare in Chianti, aging in the Chianti area) study, a community-based cohort study of aging in Florence, Italy (Ferrucci and others 2000).
FHS participants targeted in this analysis were from the Offspring Cohort enrolled in 1971 as the offspring (and offspring spouses) of the Original FHS cohort. Offspring participants who attended examination 8 (2005–2008) and had blood-derived RNA prepared were included in this analysis (n=2,422, see Table 1 for further cohort details). This study was approved by the Institutional Review Boards at Boston University Medical Center, and all participants gave written informed consent (Feinleib and others 1975).
Table 1.
Summary statistics of the Framingham and InCHIANTI cohort characteristics
| Framingham Heart Study Offspring
| ||
|---|---|---|
| Gender | N | % |
| Males | 1,093 | 45.1 |
| Females | 1,329 | 54.9 |
| Age | ||
| 30–49 | 50 | 2.1 |
| 50–69 | 1508 | 62.3 |
| 70–89 | 859 | 35.5 |
| 90–104 | 5 | 0.2 |
| Mean (SD) | 2422 | 66.4 (9.0) |
| Tobacco Exposure | ||
| None | 775 | 32 |
| Former Smoker | 1444 | 59.6 |
| Current Smoker | 203 | 8.38 |
| N | Mean (SD) | |
| BMI (kg/m2) | 2422 | 28.5 (5.4) |
| Interleukin 6 (pg/mL) | 2422 | 2.7 (3.0) |
| Leukocyte Composition | ||
| Neutrophils | 2422 | 59.8 (7.9) |
| Lymphocytes | 2422 | 27.0 (7.5) |
| Monocytes | 2422 | 9.2 (1.9) |
| Eosinophils | 2422 | 3.3 (1.6) |
| Basophils | 2422 | 0.8 (0.2) |
| InCHIANTI
| ||
|---|---|---|
| Gender | N | % |
| Males | 313 | 45.1 |
| Females | 381 | 54.9 |
| Age | ||
| 30–49 | 88 | 12.7 |
| 50–69 | 100 | 14.4 |
| 70–89 | 477 | 68.7 |
| 90–104 | 29 | 4.2 |
| Mean (SD) | 694 | 72.2 (15.3) |
| Tobacco Exposure | ||
| None | 380 | 54.8 |
| Former Smoker | 240 | 34.6 |
| Current Smoker | 74 | 10.7 |
| N | Mean (SD) | |
| BMI (kg/m2) | 694 | 27.1 (4.3) |
| Interleukin 6 (pg/mL) | 694 | 3.8 (2.9) |
| Leukocyte Composition | ||
| Neutrophils | 694 | 57.5 (9.1) |
| Lymphocytes | 694 | 30.8 (8.7) |
| Monocytes | 694 | 8.0 (2.1) |
| Eosinophils | 694 | 3.2 (2.1) |
| Basophils | 694 | 0.6 (0.2) |
InCHIANTI participants were originally enrolled in 1998–2000, and were interviewed and examined every 3 years. Ethical approval was granted by the Instituto Nazionale Riposo e Cura Anziani Institutional Review Board in Italy. Participants gave informed consent to participate. RNA was available at wave 4 (year 9) of the study, with IL6 also measured at year 9. All the data required for the full analyses were available for 694 individuals (see Table 1 for further cohort details).
2.1 RNA collection and extraction
FHS
The methods for gene expression profiling were previously published (Joehanes and others 2013). Briefly, peripheral blood samples were extracted using the PAXgene Blood mRNA kit (PreAnalytiX, Hombrechtikon, Switzerland), and amplified by the WT-Ovation Pico RNA Amplification System (NuGEN, San Carlos, CA), according to manufacturer’s instructions. cDNA was then hybridized to the Human Exon 1.0 ST Array (Affymetrix, Inc., Santa Clara, CA) for quantification. The raw data were quantile-normalized and natural-log transformed, followed by summarization using Robust Multi-array Average (Irizarry and others 2003). The gene annotations were obtained from Affymetrix NetAffx Analysis Center (version 31). We excluded transcript clusters that were not mapped to RefSeq transcripts, resulting in 17,873 distinct transcripts (17,324 unique gene identifiers) for downstream analysis.
InCHIANTI
Peripheral blood samples were also extracted using the PAXgene Blood mRNA kit according to the manufacturer’s instructions (Debey-Pascher and others 2009), which preserves transcript expression levels as at the time of blood sampling. Samples were collected in 2008/9 (wave 4) from 733 participants. Whole genome expression profiling of the samples was conducted using the Illumina Human HT-12 microarray (Illumina, San Diego, USA) as previously described (Zeller and others 2010). Data processing was done using the Illumina and BeadStudio software (Illumina, San Diego, USA) as previously described (Zeller and others 2010). All microarray experiments and analyses complied with MIAME guidelines (Brazma and others 2001). Participants were excluded if mean signal intensities across all probes with p≤0.01 were >3 standard deviations from the cohort mean; probes with <5% of participants giving intensities with p≤0.01 different from background also were excluded, 3 further exclusions were made due to missing leukocyte data. More detailed methods have been previously published (Harries and others 2011). Data from 695 individuals and 16,571 (11,393 unique gene identifiers) probes passed our quality control process and went forward into our analyses. The expression data were first normalized using natural log transformation and standardized using z-scores.
2.2 Serum Protein Measures
FHS
The interleukin-6 concentration was assayed by the quantitative enzyme-linked immunosorbent assay according to the manufacturers’ protocols (R&D Systems, Minneapolis, MN, USA). The minimum detectable concentration was 0.039 pg/ml. Ten percent of measures were run in duplicate. Standard quality control was performed, and the mean intra-assay coefficient of variation was 4% and the inter-assay coefficient of variation was 3.7% (Fontes and others 2013) (http://www.framinghamheartstudy.org/researchers/description-data/vascular-manuals/offspring_exam8_omni1_exam3_marker_manual.pdf). No participants included in the analysis were below the minimum detectable concentration.
InCHIANTI
Serum IL6 was quantified using a quantitative sandwich enzyme-linked assay (BioSource Cytoscreen UltraSensitive kits, BioSource International Inc., Camarillo, CA, USA). The lowest detectable concentration was 0.10 pg/ml and the interassay coefficient of variation (CV) was 7%. No participants included in the analysis were below the minimum detectable concentration.
2.3 Blood Cell Count Measurement
FHS
For the Framingham Heart Study (FHS) Generation 2 (Offspring) participants included in this study, blood cell fractions (CBC) are predicted using a partial least square regression (PLS) as follows. Of 3,155 participants in FHS Generation 3 cohort (not included in this study), 2,280 who attended the second examination cycle were measured using a Coulter HmX hematology autoanalyzer and the standard protocol (Beckman Coulter Inc, Brea, CA, USA). Of these 2,280, two thirds were randomly chosen as a training set and the rest as the testing set. The PLS model was built on the training set only using PLS method with 10-fold cross validation. The resulting model is tested on the testing set and gave the following testing-set prediction accuracy R2: 0.41, 0.61, 0.83, 0.83, 0.81, 0.87, 0.34, and 0.25 for red blood cell count, white blood cell count, percent neutrophil, percent lymphocyte, percent monocyte, percent eosinophil, percent basophil, and platelet count, respectively. The resulting model was used to predict the CBC in all remaining participants with no CBC measure, including the Generation 2 participants included in this study. We performed a test to compare results with measured and predicted CBCs on the 2,280 participants with both and found no significant difference.
InCHIANTI
Assessments of the number of red blood cells, white blood cells, platelets, the hemoglobin concentration were performed through an automated system at the Laboratory of Clinical Chemistry and Microbiological Assays, SS. Annunziata Hospital, Azienda Sanitaria 10, Florence, Italy, using a Coulter LH 750 Hematology Autoanalyzer (Beckman Coulter Inc, Brea, CA, USA).
2.4 Statistical analyses
In all models using IL6 as the outcome we used natural log of the serum levels to reduce the skewness inherent to the measure.
Mediation analysis
Using the R statistical software application (v2.14.1) (R-Development-Core-Team 2011) and package ‘mediation’ (Imai and others 2010) (v4.2) we individually assessed each transcript’s mediation effect in a model using logged interleukin 6 serum levels as the dependent measure (or ‘treatment’ as it is referred to in the package), and age as the ‘exposure’. In each model (1 per gene) the mediating variable was the expression level of that particular gene. The common approach to testing the effect of a potential mediator is to adjust for the proposed mediator in a linear/logistic model, and determine by how much the effect size is changed. Whilst this tells the analyst whether an association is affected by a mediator, it does not provide an estimate of statistical significance or confidence intervals for the mediation effect; hence we are using the package ‘mediation’ as this re-samples the data to determine confidence intervals for the mediation estimate.
Using parametric linear regression models throughout, we modeled the mediating effects for each transcript on the association between IL6 and age. Each model was adjusted for the following independent variables; sex, technical covariates (including microarray batches and study site – these are specific to each cohort) and, leukocyte sub-type proportions. The cell type proportions included lymphocytes, monocytes, eosinophils and basophils, but neutrophil percentage was not included (as it was closely negatively associated with lymphocyte proportion), to avoid these measures totaling 100% and leaving no degrees of freedom in the model. To avoid issues of co-linearity with age in the regression models additional co-factors were investigated as sensitivity analyses only (see section 2.8), rather than in the discovery phase analysis. The package uses quasi-Bayesian resampling simulations to estimate the uncertainty of the effects derived from our parametric models. After exploration of the package it was decided that 10,000 simulations per model was sufficient to give relatively stable estimates whilst keeping computation time realistic.
Although the package provides a p-value estimate, this was not of high enough resolution to control for multiple testing and was susceptible to sample distribution biases (following correspondence with package authors – who agreed with the following solution). We have therefore used adjusted confidence-intervals (CIs) to reflect the number of tests performed, akin to the Bonferroni adjustment (Bonferroni 1936). In FHS the number of array probes (and thus models) available was 17,873, so the CI were set to 99.99972% (1-(0.05/17873)). Significant mediators are those with CIs that do not cross the null (0) value. When we refer to “genome-wide significant mediators”, or “significant mediators in FHS”, these were determined by this CI-adjusted (Bonferroni) method.
2.5 Concordance with InCHIANTI
Genes that mediate a proportion (≥5%) of the association between age and IL6 after adjustment for multiple-testing in FHS were then assessed in the InCHIANTI study. Each test was conducted as described in the previous section for the FHS, except 95% confidence intervals that did not cross the null (0) were considered significant replication (rather than the multiple-testing adjusted confidence intervals applied to the discovery analysis in the FHS cohort). We are therefore using a discovery/replication procedure, where any findings significant in the discovery analysis after adjustment for multiple testing are considered independent tests for replication, and thus we accepted p<0.05 (i.e. 95% confidence intervals) as significant.
2.6 InCHIANTI: PCR analysis of gene expression
A number of genes including IL6 and SLC4A10 did not have gene expression information available in the InCHIANTI microarray data. We selected 2 random subsamples of 200 InCHIANTI participants for additional analysis and validation using polymerase-chain-reaction (PCR) techniques. Subsets of the participants were chosen because these are historic samples and the RNA is a finite resource.
Firstly, PCR 383-well plates were used to assess the expression levels of SLC4A10, DAAM2 and FLT3; total RNA (45–85 ng) was reverse transcribed in 13-μL reactions using the Superscript III VILO kit (Invitrogen, Paisley, UK), according to the manufacturer’s instructions. Expression levels for each target transcript were then measured in triplicate 5ul reactions using 384-well plates on the ABI Prism 7900HT platform (Life Technologies, Foster City, CA, USA), using commercially available assays (DAAM2 assay Id Hs00322497_m1, FLT3 assay Id Hs00174690_m1, SLC4A10 assay Id Hs00222849_m1). Gene expression levels were calculated relative to the mean crossing point of the endogenous control genes IDH3B, GUSB and 18s, and normalized to the median DAAM2, FLT3 or SLC4A10 expression level across the sample.
Secondly, TaqMan Low Density Arrays were used to measure IL6 and CDKN2A expression data. Total RNA (30–170 ng) was reverse transcribed in 20-μL reactions using the Superscript III VILO kit (Invitrogen, Paisley, UK), according to the manufacturer’s instructions. Expression levels for each target transcript were then measured using the TaqMan Low Density Array approach on the ABI Prism 7900HT platform (Life Technologies, Foster City, CA, USA), using commercially available assays (IL6 assay Id Hs00985639_m1, CDKN2A assay Id Hs00923894_m1). Gene expression levels were calculated relative to the mean crossing point of the endogenous control genes GUSB and 18s, and normalized to the median IL6 or CDKN2A expression level across the 200 samples.
2.7 Mediation Analysis in InCHIANTI PCR Subset
To assess the statistical significance of the genes unavailable in the InCHIANTI microarray data but were selected for PCR analysis, we used the Sobel test, as it provides higher power in small samples sizes when compared to bootstrapping methods (Fritz and Mackinnon 2007).
Cluster analysis of significant mediators: principal components analysis (R package: psych) was performed to determine the correlation structure of the significant mediators in FHS. Default options were used (including “Varimax” rotation). The number of components to extract was determined by Scree analysis.
Pathways analysis of significant mediators
DAVID functional annotation tools (Huang da and others 2009) were used to test for enrichment of pathways or processes in the significant mediators identified by FHS.
2.8 Sensitivity Analyses
We ran the analysis using a binary phenotype of IL6 using an empirically determined clinically relevant cut-point (>3.5pg/mL) (Cesari and others 2004). To assess the effect of adjusting for whole white blood cell (WBC) count (number of cells in sample) in addition to the proportions of major white cell subtypes, we ran the analysis with and without WBC count.
We also investigated the effect of additional adjustments for potential confounding factors in the InCHIANTI cohort for the subset of replicated genes. The adjustments iteratively investigated were; waist circumference (continuous: cm), self-reported anti-inflammatory medication (binary: yes/no. Includes: “Cytokines and immunomodulators”, “Immunosuppressive agents”, “NSAID, FANS”, “Coxibs”), self-reported physical activity in the last year (categorical: “hardly any”, “mostly sitting/some walking”, “light exercise 2–4 hrs/week”, “moderate 1–2 hrs or light >4 hrs/wk”, “>3 hrs moderate exercise per week”), smoking status (binary: “ever smoked” vs. “never smoked”), and type-2 diabetes (binary: Fasting blood glucose>140 or glycosuria).
3. Results
3.1 Characteristics of the samples
The cohorts differ in size and age-distributions, but are otherwise broadly similar (Table 1). Overall, 2422 participants from FHS (age-range: 40–92 years) were eligible for the discovery analysis. 694 participants (age-range: 30–104 years) from the InCHIANTI study had complete data for the replication analysis.
3.2 Serum IL6 protein levels associated with age but not with leukocyte IL6 transcript abundance
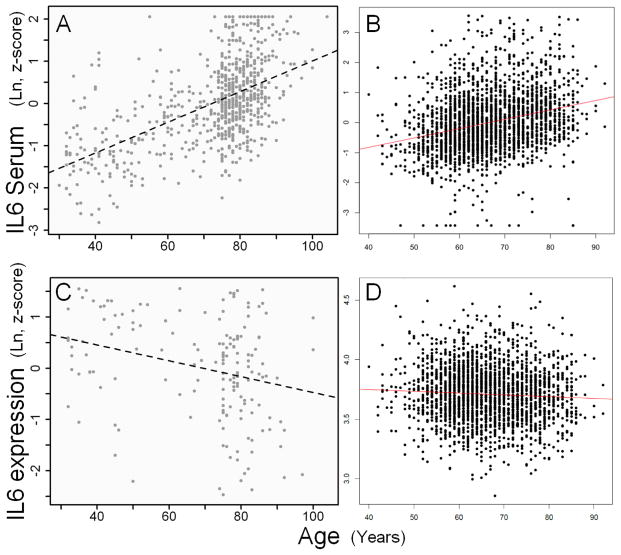
Interleukin-6 serum (protein) levels (IL6) were strongly positively associated with advancing age both in FHS (N=2422; unadjusted: beta=0.031, p=1×10−44) and InCHIANTI (N=694; unadjusted: beta=0.038, p=1 ×10−61, adjusted: beta=0.036, p= 1.1 ×10−36) (Figure 1 A and B). Expression of IL6 transcripts (from circulating blood leukocytes, see methods) was not associated with age in FHS and mildly negatively associated with age in InCHIANTI (Figure 1C and D) (FHS: n=2422, coefficient=0.013, p=0.12; InCHIANTI: n=186, coefficient=−4.7, p= 5.6 ×10−4). Surprisingly, IL6 transcript levels were not significantly correlated with IL6 protein levels in serum in either study (FHS: coefficient=0.0064, p= 0.26; InCHIANTI: coefficient=0.021, p=0.76).
Figure 1. The associations between interleukin-6 and age.
A) IL6 protein serum levels and age in 694 InCHIANTI samples. B) IL6 protein serum levels and age in 2422 FHS samples. C) IL6 gene expression and age in leukocytes in 186 InCHIANTI samples. D) IL6 gene expression and age in leukocytes in 2422 FHS samples.
3.3 Immune cell sub-type associations
In InCHIANTI, directly measured lymphocyte, neutrophil, monocyte and eosinophil percentages were each associated with higher mean age (Beta= −0.1, 0.09, 0.02, 0.02; P= 9×10−9, 1×10−4, 2×10−4, 3×10−3, respectively); lymphocyte, neutrophil and basophil percentages were associated with serum IL6 adjusting for age (Beta= −2.9, 2.8, −0.03; P= 4×10−14, 5×10−12, 1×10−3, respectively); lymphocyte and neutrophil percentages mediated the association between age and IL6 (proportion of effect mediated= 0.09, 0.06 respectively); the ratio between lymphocyte and neutrophil percentages also significantly mediated the age~IL6 association (proportion of effect mediated= 0.06).
3.4 A small set of transcripts partially mediate the serum IL6 – age association
Transcripts for 17,324 unique genes could be tested in FHS, of which 4693 (27.1%) were associated with age (FDR<0.05) and 4140 (23.9%) were associated with IL6 (Lin and others 2014). However, expression of only 102 (0.6% of the 17,324) genes significantly mediated a proportion of the association between age and IL6 (after adjustment for confounders – see methods - and multiple testing using 99.99972% confidence intervals, and after excluding 3 invalidly annotated transcripts, Supplementary Table 1). Twenty-nine genes mediated ≥5% of the association, with SLC4A10 (solute carrier family 4, sodium bicarbonate transporter) mediating 19% of the association (CI 8.9 to 34.1%) in FHS (Table 2).
Table 2.
Transcripts mediating ≥5% of the age-IL6 association in FHS, with array based replication estimates from the InCHIANTI study
| FHS | InCHIANTI | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| N | Entrez | Gene | Description | Prop ɫ | 99.9% CIs~ | Prop ɫ | 95% CIs | Sig. | PCR | ||
| 1 | 57282 | SLC4A10 | solute carrier family 4, sodium bicarbonate transporter | 0.191 | 0.089 | 0.341 | 0.356 | * | « | ||
| 2 | 2322 | FLT3 | fms-related tyrosine kinase | −0.099 | −0.211 | −0.04 | −0.01 | * | « | ||
| 3 | 9839 | ZEB2 | zinc finger E-box binding homeobox 2 | 0.098 | 0.032 | 0.192 | 0 | −0.008 | 0.008 | ||
| 4 | 8460 | TPST1 | tyrosylprotein sulfotransferase 1 | −0.09 | −0.182 | −0.036 | −0.025 | −0.07 | 0.012 | ||
| 5 | 1432 | MAPK14 | mitogen-activated protein kinase 1 | 0.086 | 0.03 | 0.172 | 0.001 | −0.01 | 0.015 | ||
| 6 | 3553 | IL1B | interleukin 1, beta | −0.081 | −0.171 | −0.032 | −0.052 | −0.093 | −0.023 | * | |
| 7 | 23500 | DAAM2 | dishevelled associated activator of morphogenesis 2 | −0.079 | −0.176 | −0.027 | −0.079 | * | « | ||
| 8 | 2153 | F5 | coagulation factor V (proaccelerin, labile factor) | 0.075 | 0.026 | 0.148 | 0.014 | −0.007 | 0.039 | ||
| 9 | 1462 | VCAN | versican | 0.073 | 0.024 | 0.15 | 0.043 | 0.018 | 0.076 | * | |
| 10 | 3820 | KLRB1 | killer cell lectin-like receptor subfamily B, member 1 | 0.068 | 0.019 | 0.135 | 0.016 | −0.007 | 0.043 | ||
| 11 | 5551 | PRF1 | perforin 1 (pore forming protein) | −0.064 | −0.14 | −0.001 | −0.057 | −0.104 | −0.021 | * | |
| 12 | 3267 | AGFG1 | ArfGAP with FG repeats 1 | 0.064 | 0.02 | 0.147 | −0.003 | −0.022 | 0.013 | ||
| 13 | 51099 | ABHD5 | abhydrolase domain containing 5 | −0.062 | −0.145 | −0.017 | 0.002 | −0.004 | 0.015 | ||
| 14 | 400360 | C15orf54 | chromosome 15 open reading frame 54 | 0.062 | 0.017 | 0.123 | |||||
| 15 | 116369 | SLC26A8 | solute carrier family 26, member 8 | 0.062 | 0.005 | 0.128 | 0.012 | −0.008 | 0.037 | ||
| 16 | 768211 | RELL1 | RELT-like 1 | −0.06 | −0.173 | −0.011 | |||||
| 17 | 26253 | CLEC4E | C-type lectin domain family 4, member E | −0.059 | −0.135 | −0.016 | −0.012 | −0.037 | 0.008 | ||
| 18 | 2204 | FCAR | Fc fragment of IgA, receptor for | 0.059 | 0.019 | 0.12 | 0.005 | −0.004 | 0.021 | ||
| 19 | 55350 | VNN3 | vanin 3 | −0.056 | −0.133 | −0.01 | −0.003 | −0.019 | 0.008 | ||
| 20 | 147947 | ZNF542 | zinc finger protein 542 | 0.054 | 0.016 | 0.115 | |||||
| 21 | 285533 | RNF175 | ring finger protein 175 | 0.053 | 0.012 | 0.125 | 0 | −0.006 | 0.005 | ||
| 22 | 330 | BIRC3 | baculoviral IAP repeat-containing 3 | −0.053 | −0.119 | −0.017 | −0.01 | −0.033 | 0.007 | ||
| 23 | 4609 | MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | −0.052 | −0.128 | 0 | −0.048 | −0.103 | 0 | ||
| 24 | 1374 | CPT1A | carnitine palmitoyltransferase 1A (liver) | 0.052 | 0.004 | 0.13 | −0.001 | −0.011 | 0.005 | ||
| 25 | 22901 | ARSG | arylsulfatase G | −0.052 | −0.124 | −0.012 | 0.001 | −0.007 | 0.012 | ||
| 26 | 317 | APAF1 | apoptotic peptidase activating factor 1 | −0.051 | −0.133 | −0.013 | 0.013 | −0 | 0.034 | ||
| 27 | 84255 | SLC37A3 | solute carrier family 37, member 3 | 0.051 | 0.014 | 0.109 | −0.004 | −0.019 | 0.004 | ||
| 28 | 55356 | SLC22A15 | solute carrier family 22, member 15 | −0.05 | −0.153 | −0.001 | 0.001 | −0.004 | 0.013 | ||
| 29 | 4050 | LTB | lymphotoxin beta (TNF superfamily, member 3) | −0.05 | −0.118 | −0.006 | −0.079 | −0.138 | −0.031 | * | |
Proportion of the IL6~age association mediated
Bonferroni-adjusted confidence intervals (ie. False-discovery-rate adjusted)
Sig. = significant at p<0.05 in InCHIANTI
PCR = not detected in Illumina; data from PCR in 200 InCHIANTI participants; no CI’s as Sobel method used due to small sample size
The InCHIANTI Illumina array provided data on n=11,393 genes, substantially fewer than the 17,324 measured in FHS. Data on 9,971 unique genes were available in both cohorts with an additional 7,345 measured in FHS only and 1,422 in InCHIANTI only. Of the 102 genes partial mediators of the age-IL6 association identified in FHS, 88 were available (Supplementary Table 1) in InCHIANTI and 22 of them significantly mediated the association of age with IL6 after adjustment for confounders (with 3 additionally validated by PCR, see below). Five of the ten most strongly significant mediators were replicated in InCHIANTI. Of the 29 genes mediating ≥5% in FHS (Table 2), 22 were also in the InCHIANTI microarray data. Significant associations in the same direction were found for IL1b, VCAN, PRF1 and LTB. No genes mediating ≥5% had significant associations in both cohorts but different directions of effect. Four of those mediating <5% were significant and with opposite directions of effect (Supplementary Table 1).
Three genes (SLC4A10, FLT3, DAAM2) with larger mediating effects (>7%) in the FHS analysis did not pass the background inclusion criteria in InCHIANTI microarray data. These were selected for confirmation by PCR in a random subset of the InCHIANTI sample. In 194 samples (after exclusions/data cleaning) there was statistically significant partial mediation of the association between age and IL6 for all 3 genes (SLC4A10 % mediated=35.6%, p=0.01; DAAM2 %=−7.92%, p=0.0001; FLT3 %=−1.03%, p=0.0002), although this small sample size of the replication makes comparisons of effect sizes difficult.
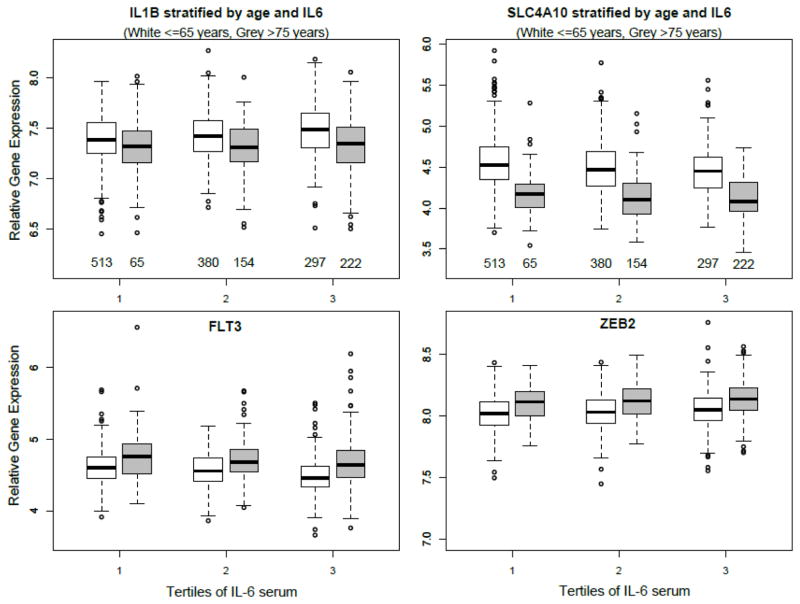
As noted previously, both positive and negative mediators were identified. Figure 2 shows boxplots of expression (in FHS) for 4 genes that have been stratified by age (young ≤65years, old >75) and IL6 expression (tertiles); IL1B (negative mediator), SLC4A10 (positive mediator), FLT3 (negative) and ZEB2 (positive). The plot illustrates that different mediators were positively or negatively associated with age and/or IL6. Figure 2 plots also show that SLC4A10 expression reduces with increasing IL6 concentration, and also has markedly lower expression in the older group in general.
Figure 2. The relationships between expression of four genes, IL6 protein, and age.
Boxplots of gene expression in the FHS cohort stratified by tertile of IL6 serum protein concentrations, split by age group; white boxes are individuals ≤65 years (mean age= 59.0 +/− 4.7 years), grey boxes are individuals >75 (mean age= 79.9 +/− 3.2 years).
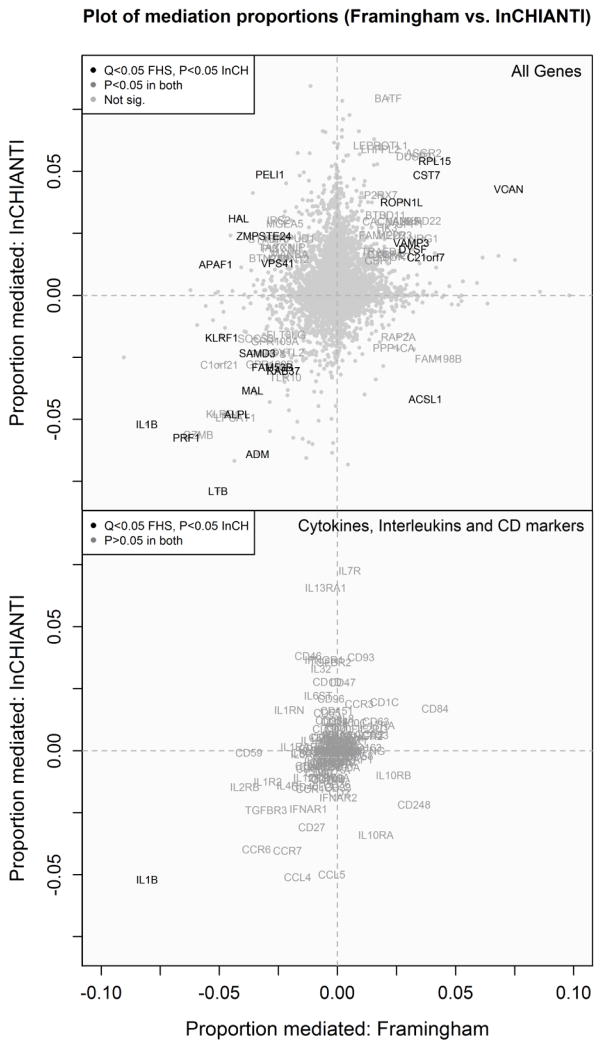
3.5 Consistent negative findings
Overall, very few interleukins or cytokines emerged as significant mediators (Figure 3) with all mediation effect sizes for these genes in FHS being <5%, with the sole exception of IL1B. Similarly there were few larger effect CD marker mediators (Cluster of Differentiation, used to identify and characterize leukocyte subtypes) other than notable exceptions with updated gene symbols, such as KLRB1 (CD161).
Figure 3. Comparing the mediation effects of genes between FHS and InCHIANTI.
Scatter plots of the proportion of the age/serum-IL6 association mediated by each gene expression transcript in FHS and InCHIANTI for (A) all genes, (B) cytokines, interleukin-related and CD (cluster of differentiation) molecules.
CDKN2A (p16INK4a) expression is increased in-vivo with circulating lymphocyte cell senescence (Liu and others 2011). In InCHIANTI, CDKN2A array expression was not above background, so TaqMan Low Density Array real-time PCR measured expression in a subsample. Expression of CDKN2A was not a significant mediator of the IL6-age association in either cohort.
3.6 Clustering and gene ontology enrichment of mediators
To determine which genes “cluster” together (have similar expression profiles) we used principal components analysis. Eighteen clusters were identified by the Scree analysis of the 102 significant mediators in FHS (Supplementary Figure 1). Of particular note, SLC4A10, the largest single mediator in FHS, only clusters strongly with KLRB1 (highlighted in yellow - also called CD161, which inhibits natural killer cell (NK) toxicity and is also expressed in some T cells). IL1B and ADM were also closely related.
Gene ontology enrichment analyses of the 102 FHS expression probes were explored in the DAVID bioinformatics tool. Only 1 KEGG pathway was significant in the functional annotation analysis; the NOD-like receptor signaling pathway (p=4.9 × 10−5, Benjamin-Hochberg multiple testing adjusted p=3.2 × 10−3). In an analysis restricted to the 7 replicated genes only, no output was returned by DAVID.
3.7 Sensitivity Analyses
To test the consistency of the results of the associations of age and genes with linear IL6 concentrations, we also performed an analysis using a binary IL6 trait as the outcome. The correlation between the proportions mediated by the genes was highly consistent, the R2 = 0.81 (Supplementary Figure 2).
Adjusting for WBC count slightly altered the ordering (by proportion mediated) in the list of significant mediators in FHS, however the final list of genes mediating >5% found to replicate in InCHIANTI did not change from the original list of 7.
Additional adjustment for potential confounding factors in the analyses of the 7 replicated genes in InCHIANTI found that all genes were still statistically significant mediators (p<0.05), with the exception of SLC4A10 which was no longer significant after adjustment for waist or physical activity (p=0.06 and p=0.08 respectively), although this may be due to statistical power in the small sample size (n=196). As expected, the exact effect estimates (the proportion of the IL6-age association mediated) varied between the models depending on the specific factor included (Supplementary Table 2).
4. Discussion
Our study presents the first larger-scale transcriptome-wide analysis of age-related inflammation in human blood. Very strong associations were present in both study cohorts between age and serum interleukin-6 (IL6) protein concentrations (Lin and others 2014). Examining statistical mediators of the age-IL6 association, we identified and independently replicated 7 partial mediators with effect sizes ≥5%, including one larger effect transcript, namely SLC4A10. Our results include known immune markers and several novel genes not previously linked to aging. These results are very different from the many associations with general IL6 levels independent of age (807 replicated genes) (Lin and others 2014) and represent a mediator signature specific to age-related inflammation.
Increased Interleukin 1 beta protein concentrations (IL1B), a cytokine involved in initiation of most inflammatory responses and a product of inflammasome activation, is a core feature of the pro-inflammatory state of aging (Didier and others 2012). IL1B expression emerged as the only replicated cytokine mediator in our analyses. The negative mediation value reflects IL1B expression increasing with IL6, but decreasing with age (as shown in Figure 2). Perforin (PRF1) is one of the main proteins of cytolytic granules and is a key effector molecule for T-cell and natural killer-cell-mediated cytolysis. Perforin expression per cell falls with advanced age (Rukavina and others 1998). A high proportion of CD8+ T cells express Granzyme B but not PRF1 in response to an influenza challenge, and the absence of PRF1 results in Granzyme B degradation of the extracellular matrix and inflammation (McElhaney and others 2012). Alterations to T-cell populations (and other immune cell types) with aging leads to reduced antibody response and increased incidence of infectious disease, contributing to age-related inflammation (Del Giudice and others 2014). Our finding of IL1B and perforin amongst the replicated mediators shows that our study was able to identify primary mechanisms involved in age-related chronic inflammation.
Our largest effect mediator SLC4A10 (19.1% in FHS, 35.6% in InCHIANTI) codes for NCBE (also called NBCn2) and is a sodium bicarbonate transporter, part of a gene family involved in intracellular acid-base homeostasis. SLC4A10 expression is highest in the brain, and SLC4A10 has been linked to autism, epilepsy and mental retardation (Parker and Boron 2013). There are little published data on SLC4A10 in the immune system, but the BioGPS database (Wu and others 2009) shows similar levels of expression in all studied leukocyte cell sub-types in humans, suggesting that SLC4A10 mechanisms may be widespread. Inflammation is associated with a local drop in pH resulting from infiltration of inflammatory cells, and extracellular acidosis in inflammation may be a ‘danger signal’ activating immune responses (Rajamaki and others 2013). Farwell and Taylor (Farwell and Taylor 2010) showed that a higher anion gap and lower bicarbonate level were associated with a higher leukocyte count and higher C-reactive protein level, in the NHANES study (Farwell and Taylor 2010). Clearly, much more work is needed to clarify the mechanisms of involvement of SLC4A10 in age-related inflammation.
FTL3 (also called CD135) emerged as the second largest effect gene in FHS (replicated in InCHIANTI), and plays a key role in lymphocyte (B and T cell) development but not for the development of other blood cells. The third larger effect gene, ZEB2, is mainly expressed in monocytes and is involved in epithelial-mesenchymal transition (ETM), which is characterized by the downregulation of cellular adhesion complexes and is a key step in cancer development (Gheldof and others 2012). The ZEB2 mediation effect was not significant in the InCHIANTI array data, but further work using gold standard lab methods is needed to be sure that the InCHIANTI estimate is not a ‘false negative’ due to differences in array technology or sample size.
An important finding of our study is that although we confirmed a strong positive relationship between serum IL6 levels and age, we found strikingly few interleukins, cytokines (including mRNA for IL6) or interferon gene transcripts mediating the age-IL6 association in the studied blood white cells (Figure 3), with the exception of IL1B, based on an analysis checking synonyms for these genes. These findings suggest that the primary origin of age-related inflammation, in particular the age-related increase in circulating IL6 protein, may not be circulating immune cells. However that we see a few specific gene expression mediators of age-related inflammation in whole blood suggests that the immune system responds to the age-associated increase in circulating IL6 in a specific way, distinct from the 807 genes seen in the analysis independent of age.
4.1 Limitations and Future Work
Although moderate (50%) replication of the most significant mediators was observed, there were many small effect genes that were not significant in the InCHIANTI cohort. Our study includes two separate expression arrays (plus three PCR validations), with the arrays having different gene coverage and sensitivities, imposing some limitations. However the alternate technologies do provide evidence for the robustness of our positive findings across our two study cohorts. The Affymetrix Human Exon 1.0 ST array (used by FHS) uses 25-mer probes, typically grouped in sets of four per exon, to estimate exon-specific levels of expression, while the (InCHIANTI) Illumina Human HT12v3 array uses 50-mer probes biased towards the 3′ end of mRNA transcripts to estimate whole-gene levels of expression (Ramasamy and others 2013). The latter array appears less sensitive in our blood derived RNA samples as far fewer gene expression measures passed quality control in InCHIANTI than FHS, and thus some of the genes that appeared not to replicate may be found in future studies. The analysis is cross-sectional, so more work will be needed to establish the direction of causation. The study subjects were of European origin, so further work will be needed to establish whether the results are applicable to other ethnic groups. The RNA samples were from whole blood containing a mixture of white cells with results reflecting overall expression patterns. Our results should be seen as starting points for establishing underlying mechanisms, including changes in cell subtype proportions not already adjusted for in our analyses.
Future work should include studies to establish the cellular subtype origins of the novel mediators reported, and their mechanistic roles. This should account for associations with age-related diseases; a recent study found that white-blood cell counts, but not IL6, is associated with cardiovascular diseases in >70 year-olds (Compté and others 2014). Analyses of gene expression changes with inflammation of aging in key subgroups may be helpful, especially identifying gender specific expression. As longitudinal follow-up becomes available, dynamic changes in expression and the predictive value of expression changes for age related health outcomes can be examined. Follow-up analyses, including analysis of C-reactive protein, are being initiated with collaborating cohorts.
4.2 Conclusions
To conclude, we have performed the first larger-scale transcriptome-wide analysis of age-related inflammation in two human population cohorts. A small set of genes appear to have partially mediated the age-IL6 association, several of which have not previously been linked to the pro-inflammatory state of aging. We have identified a novel larger effect partial mediator (SLC4A10, coding for NCBE), thought to be involved in pH homeostasis. The results are very different from the widespread associations with general IL6 levels independent of age, suggesting that age-related chronic inflammation has a discrete gene expression mediator signature. We did not find increased expression of IL6 with age in our blood derived data, suggesting that the raised IL6 protein concentrations in blood may be predominantly generated from other body compartments. Further work is needed to distinguish mediators of age-related inflammation from the much larger numbers of age correlated expression changes in blood.
Supplementary Material
Principal Components Analysis of 102 significant mediator gene transcripts in FHS
Sensitivity analysis of transcripts mediating the age binary IL6 (‘high’ defined as >3.5pg/mL) association
Full list of gene transcripts mediating the age IL6 association
Highlights.
First study of gene expression specific to age related inflammation in human blood
25 genes replicated across two cohorts (n=3116)
7 replicated genes mediated >5% of the age-IL6 association
IL1B, PRF1 and LTB are known immune markers now identified as age-IL6 mediators
SLC4A10 is largest (novel) mediator, may regulate pH homeostasis in white cells
Acknowledgments
Funding
FHS gene expression profiling was funded through the Division of Intramural Research (Principal Investigator, Daniel Levy), National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD. Measurement of interleukin-6 was funded through RO1 HL 064753, RO1 HL076784, and 1 R01 AG028321. The Framingham Heart Study is supported by contract N01-HC-25195. JMM was supported by RO1 AG29451.
This study was supported in part by the Intramural Research Program, National Institute on Aging. The analysis was generously supported by a Wellcome Trust Institutional Strategic Support Award (WT097835MF). WH was funded by the National Institute for Health Research (NIHR) Collaboration for Leadership in Applied Health Research and Care (CLAHRC) for the South West Peninsula. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health in England.
This work has made use of the resources provided by the University of Exeter Science Strategy and resulting Systems Biology initiative. Primarily these include high-performance computing facilities managed by Konrad Paszkiewicz and Robin Batten of the College of Environmental and Life Sciences and the University of Exeter IT service.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bektas A, Zhang Y, Wood WH, 3rd, Becker KG, Madara K, Ferrucci L, Sen R. Age-associated alterations in inducible gene transcription in human CD4+ T lymphocytes. Aging. 2013;5:18–36. doi: 10.18632/aging.100522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annual review of physiology. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari M, Penninx BW, Pahor M, Lauretani F, Corsi AM, Rhys Williams G, Guralnik JM, Ferrucci L. Inflammatory markers and physical performance in older persons: the InCHIANTI study. J Gerontol A Biol Sci Med Sci. 2004;59:242–248. doi: 10.1093/gerona/59.3.m242. [DOI] [PubMed] [Google Scholar]
- Compté N, Bailly B, De Breucker S, Goriely S, Pepersack T. Study of the association of total and differential white blood cell counts with geriatric conditions, cardiovascular diseases, low-grade inflammation and telomere length. Experimental Gerontology. 2014 doi: 10.1016/j.exger.2014.11.016. In press. [DOI] [PubMed] [Google Scholar]
- De Martinis M, Franceschi C, Monti D, Ginaldi L. Inflammation markers predicting frailty and mortality in the elderly. Experimental and molecular pathology. 2006;80:219–227. doi: 10.1016/j.yexmp.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Debey-Pascher S, Eggle D, Schultze JL. RNA stabilization of peripheral blood and profiling by bead chip analysis. Methods Mol Biol. 2009;496:175–210. doi: 10.1007/978-1-59745-553-4_13. [DOI] [PubMed] [Google Scholar]
- Del Giudice G, Weinberger B, Grubeck-Loebenstein B. Vaccines for the Elderly. Gerontology. 2014 doi: 10.1159/000366162. [DOI] [PubMed] [Google Scholar]
- Didier ES, Sugimoto C, Bowers LC, Khan IA, Kuroda MJ. Immune correlates of aging in outdoor-housed captive rhesus macaques (Macaca mulatta) Immunity & ageing: I & A. 2012;9:25. doi: 10.1186/1742-4933-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economos A, Wright CB, Moon YP, Rundek T, Rabbani L, Paik MC, Sacco RL, Elkind MS. Interleukin 6 plasma concentration associates with cognitive decline: the northern Manhattan study. Neuroepidemiology. 2013;40:253–259. doi: 10.1159/000343276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwell WR, Taylor EN. Serum anion gap, bicarbonate and biomarkers of inflammation in healthy individuals in a national survey. CMAJ: Canadian Medical Association journal = journal de l’Association medicale canadienne. 2010;182:137–141. doi: 10.1503/cmaj.090329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP The Framingham Offspring Study. Design and preliminary data. Preventive medicine. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, Guralnik JM. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. Journal of the American Geriatrics Society. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- Ferrucci L, Corsi A, Lauretani F, Bandinelli S, Bartali B, Taub DD, Guralnik JM, Longo DL. The origins of age-related proinflammatory state. Blood. 2005;105:2294–2299. doi: 10.1182/blood-2004-07-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Harris TB, Guralnik JM, Tracy RP, Corti MC, Cohen HJ, Penninx B, Pahor M, Wallace R, Havlik RJ. Serum IL-6 level and the development of disability in older persons. Journal of the American Geriatrics Society. 1999;47:639–646. doi: 10.1111/j.1532-5415.1999.tb01583.x. [DOI] [PubMed] [Google Scholar]
- Fontes JD, Yamamoto JF, Larson MG, Wang N, Dallmeier D, Rienstra M, Schnabel RB, Vasan RS, Keaney JF, Jr, Benjamin EJ. Clinical correlates of change in inflammatory biomarkers: The Framingham Heart Study. Atherosclerosis. 2013;228:217–223. doi: 10.1016/j.atherosclerosis.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. The journals of gerontology Series A, Biological sciences and medical sciences. 2014;69(Suppl 1):S4–9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- Fritz MS, Mackinnon DP. Required sample size to detect the mediated effect. Psychological science. 2007;18:233–239. doi: 10.1111/j.1467-9280.2007.01882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cellular and molecular life sciences: CMLS. 2012;69:2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, Yaghootkar H, Dutta A, Murray A, Frayling TM, Guralnik JM, Bandinelli S, Singleton A, Ferrucci L, Melzer D. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging cell. 2011;10:868–878. doi: 10.1111/j.1474-9726.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkila K, Ebrahim S, Lawlor DA. Systematic review of the association between circulating interleukin-6 (IL-6) and cancer. European journal of cancer. 2008;44:937–945. doi: 10.1016/j.ejca.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Howcroft TK, Campisi J, Louis GB, Smith MT, Wise B, Wyss-Coray T, Augustine AD, McElhaney JE, Kohanski R, Sierra F. The role of inflammation in age-related disease. Aging. 2013;5:84–93. doi: 10.18632/aging.100531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imai K, Keele L, Tingley D. A general approach to causal mediation analysis. Psychological methods. 2010;15:309–334. doi: 10.1037/a0020761. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Joehanes R, Ying S, Huan T, Johnson AD, Raghavachari N, Wang R, Liu P, Woodhouse KA, Sen SK, Tanriverdi K, Courchesne P, Freedman JE, O’Donnell CJ, Levy D, Munson PJ. Gene expression signatures of coronary heart disease. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:1418–1426. doi: 10.1161/ATVBAHA.112.301169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. American journal of epidemiology. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- Lin H, Joehanes R, Pilling LC, Dupuis J, Lunetta KL, Ying SX, Benjamin EJ, Hernandez D, Singleton A, Melzer D, Munson PJ, Levy D, Ferrucci L, Murabito JM. Whole Blood Gene Expression and Interleukin-6 Levels. Genomics. 2014 doi: 10.1016/j.ygeno.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Johnson SM, Fedoriw Y, Rogers AB, Yuan H, Krishnamurthy J, Sharpless NE. Expression of p16(INK4a) prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhaney JE, Zhou X, Talbot HK, Soethout E, Bleackley RC, Granville DJ, Pawelec G. The unmet need in the elderly: how immunosenescence, CMV infection, co-morbidities and frailty are a challenge for the development of more effective influenza vaccines. Vaccine. 2012;30:2060–2067. doi: 10.1016/j.vaccine.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Boron WF. The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiological reviews. 2013;93:803–959. doi: 10.1152/physrev.00023.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R-Development-Core-Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- Rajamaki K, Nordstrom T, Nurmi K, Akerman KE, Kovanen PT, Oorni K, Eklund KK. Extracellular acidosis is a novel danger signal alerting innate immunity via the NLRP3 inflammasome. The Journal of biological chemistry. 2013;288:13410–13419. doi: 10.1074/jbc.M112.426254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Gibbs JR, Dillman A, Hernandez DG, Arepalli S, Walker R, Smith C, Ilori GP, Shabalin AA, Li Y, Singleton AB, Cookson MR, Nabec, Hardy J, Ukbec, Ryten M, Weale ME. Resolving the polymorphism-in-probe problem is critical for correct interpretation of expression QTL studies. Nucleic acids research. 2013;41:e88. doi: 10.1093/nar/gkt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Rifai N, Stampfer MJ, Hennekens CH. Plasma concentration of interleukin-6 and the risk of future myocardial infarction among apparently healthy men. Circulation. 2000;101:1767–1772. doi: 10.1161/01.cir.101.15.1767. [DOI] [PubMed] [Google Scholar]
- Rukavina D, Laskarin G, Rubesa G, Strbo N, Bedenicki I, Manestar D, Glavas M, Christmas SE, Podack ER. Age-related decline of perforin expression in human cytotoxic T lymphocytes and natural killer cells. Blood. 1998;92:2410–2420. [PubMed] [Google Scholar]
- Singh T, Newman AB. Inflammatory markers in population studies of aging. Ageing research reviews. 2011;10:319–329. doi: 10.1016/j.arr.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Narita M. SASP reflects senescence. EMBO reports. 2009;10:228–230. doi: 10.1038/embor.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hubner N, Tregouet D, Munzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PloS one. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal Components Analysis of 102 significant mediator gene transcripts in FHS
Sensitivity analysis of transcripts mediating the age binary IL6 (‘high’ defined as >3.5pg/mL) association
Full list of gene transcripts mediating the age IL6 association