Abstract
IQGAPs are scaffolding proteins that regulate actin assembly, exocyst function, cell motility, morphogenesis, adhesion and division. Vertebrates express 3 family members: IQGAP1, IQGAP2 and IQGAP3. IQGAP1 is known to stimulate nucleation of branched actin filaments through N-WASP and the Arp2/3 complex following direct binding to cytoplasmic tails of ligand-activated growth factor receptors, including EGFR, VEGFR2 and FGFR1. By contrast, little is known about functions of IQGAP2 or IQGAP3. Using in situ hybridization on whole mount zebrafish (Danio rerio) embryos, we show that IQGAP1 and IQGAP2 are associated with discrete tissues and organs, while IQGAP3 is mainly expressed in proliferative cells throughout embryonic and larval development. Morpholino knockdowns of IQGAP1 and IQGAP2 have little effect on embryo morphology while loss of function of IQGAP3 affects both cell proliferation and cell motility. IQGAP3 morphant phenotypes are similar to those resulting from overexpression of dominant negative forms of Ras or of Fibroblast Growth Factor Receptor 1 (FGFR1), suggesting that IQGAP3 plays a role in FGFR1-Ras-ERK signaling. In support of this hypothesis, dominant negative forms of FGFR1 or Ras could be rescued by co-injection of zebrafish IQGAP3 mRNA, strongly suggesting that IQGAP3 acts as a downstream regulator of the FGFR1-Ras signaling pathway.
Keywords: IQGAP3, Danio rerio, embryonic development, cell proliferation, cell motility, convergent extension
Introduction
IQGAPs are evolutionary conserved from yeast to humans. This scaffolding protein family regulates a broad range of cellular events including cell adhesion, cell migration and cytokinesis. In vertebrates, three family members, IQGAP1, IQGAP2 and IQGAP3, have been identified [Brill et al. 1996; Wang et al. 2007; Weissbach et al. 1994]. Human IQGAPs share uniform domain organization, with an N-terminal calponin homology domain (CHD), a series of repeated coiled coil domains (IQ repeats), a WW domain with two conserved tryptophans, multiple IQ motifs that bind calmodulin and other calcium-binding EF hand proteins, a GAP-related domain (GRD) and a RasGAP-related C-terminal domain (CTD). This richness of structural features allows IQGAPs to regulate a series of important cellular processes through direct binding to over 100 proteins, including F-actin, small Rho family GTPases (Rac1 and Cdc42), N-WASP, ERK 1/2, MEK 1/2, β-catenin, E-cadherin, calmodulin and CLIP 170 [Bensenor et al. 2007; Briggs et al. 2002; Brown and Sacks 2006; Erickson et al. 1997; Hart et al. 1996; Ho et al. 1999; Kuroda et al. 1998; Le Clainche et al. 2007; Li et al. 1999; Roy et al. 2004; Roy et al. 2005; Wang et al. 2007].
While vertebrate IQGAP proteins are 60–70% identical to each other in amino acid sequence, they display different expression patterns. In male rats, IQGAP1 is expressed in most tissues, IQGAP2 has a somewhat more resticted distribution, and IQGAP3 is limited to brain, lung, spleen and testis [Wang et al. 2007]. Each of the IQGAPs serves distinct functions in regulation of cell motility, morphogenesis, cell adhesion and cell proliferation. IQGAP1 is the best studied member in the family. Frequently concentrated in the cell cortex, IQGAP1 mediates cytoskeletal rearrangement through the regulation of actin filaments and microtubules [Bensenor et al. 2007; Fukata et al. 2002], and stimulates branched actin filament nucleation by activating the Arp 2/3 complex through N-WASP [Bensenor et al. 2007; Le Clainche et al. 2007]. This is triggered by direct binding of IQGAP1 to cytoplasmic tails of ligand-activated growth factor receptors, including EGFR [Blagoev et al. 2003], VEGFR2 [Yamaoka-Tojo et al. 2004] and FGFR1 [Bensenor et al. 2007]. We previously reported that FGF2 stimulates binding of IQGAP1 to FGFR1 and promotes the protrusion of lamellipodia, which bridges the signaling transduction pathway to cell motility [Bensenor et al. 2007]. IQGAP2 has been identified as a potent tumor suppressor in hepatocellular carcinoma [Gnatenko et al. 2013; Schmidt et al. 2008]. Discovered most recently, IQGAP3 is reportedly required for neurite outgrowth [Wang et al. 2007]. Accumulating at the distal region of axons in hippocampal neurons, IQGAP3 links the activation of Rac1 and Cdc42 with cytoskeletal structures during neuronal morphogenesis [Wang et al. 2007]. In addition, IQGAP3 expression is increased in highly proliferative hepatocytes in mouse and in human lung cancer cells, and the protein promotes cell growth and metastasis by regulating the Ras/ERK signaling cascade as a consequence of its direct binding to Ras [Kunimoto et al. 2009; Nojima et al. 2008; Skawran et al. 2008; Yang et al. 2014].
In the present study, we determined and compared the mRNA expression patterns during embryonic and early larval development for the three IQGAP genes present in the zebrafish genome (zIQGAPs), and analyzed the function of zIQGAP3 at early developmental stages. We found that zIQGAP1 and zIQGAP2 are expressed within specific tissues and organs, while zIQGAP3 is highly expressed in proliferative cells irrespective of their locations. While knockdown of zIQGAP1 and zIQGAP2 have little effect on embryonic development, we found that zIQGAP3 reduction affects cell proliferation, as well as cell motility. This results in strongly altered embryo morphology with phenotypes reminiscent of loss of function of FGFR1-Ras-ERK signaling. We found that zIQGAP3 expression can rescue the phenotype caused by dominant negative forms of FGFR1 or Ras, thereby implicating zIQGAP3 as a downstream regulator of the FGFR1-Ras signaling pathway.
Results
Molecular cloning and expression of IQGAP genes in zebrafish
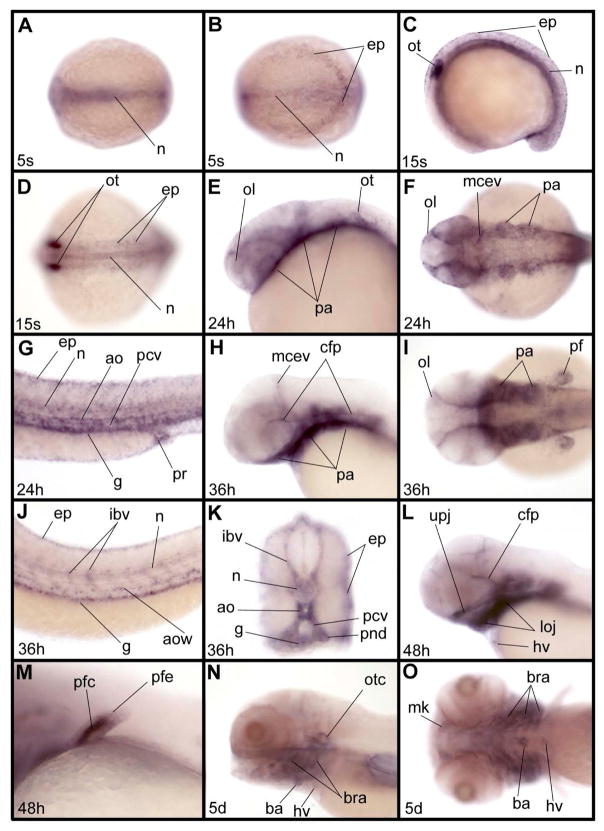
Full length cDNAs coding for the three zIQGAP proteins have been isolated and sequenced (Supplementary Fig. 1). Sequence analysis reveals the presence of the protein domains previously described for members of the IQGAP protein family (Fig. 1). As shown in Supplementary Table 1, we found that amino acid sequences of zIQGAPs 1, 2 and 3 share high similarity with each other (66–73%), as well as with their human homologs (62–85%). Expression of zIQGAP1, 2 and 3 genes were analyzed by whole mount in situ hybridization during zebrafish embryonic and early larva stages. zIQGAP1 transcripts were not observed at blastula or gastrula stages, and they appear first at the early somitogenesis stage in the posterior epidermis, as well as in the axial mesoderm, including the notochord (Fig. 2A, B). At middle of somitogenesis, expression was observed in the same tissues and a strong accumulation of transcripts was observed in the otic placodes (Fig. 2C, D). At 24 and 36 hours post fertilization (hpf) transcripts were mainly observed in pharyngeal arches, in part of the vasculature, including axial vasculature (aorta and posterior cardinal vein), intersegmental blood vessels and midcerebral vein, otic and olfactory vesicles, pectoral fins, gut and pronephric ducts (Fig. 2E-K). Transient expression of zIQGAP1 was also observed in the cephalic floor plate (Fig. 2H). At 48 hpf, zIQGAP1 transcripts were found in the cartilage of upper and lower jaws (Fig. 2L) as well as of the pectoral fins (Fig. 2M). Weak expression of zIQGAP1 was also observed in the heart valve (Fig. 2L). At larval stage (5 day post fertilization, or dpf) expression was restricted to the perichondrium of the chondrocranium, and to the pharyngeal and branchial arches (Fig. 2N, O).
Fig. 1.
Domain structures of zIQGAPs. Schematic of the three IQGAP proteins of Danio rerio displaying positions of the different conserved domains: CHD (calponin homology domain –mediates actin-binding and binds calponin); IQ repeats (repeated coiled-coil domains), WW (double tryptophan-containing protein-protein interaction domain that associates with proline-rich regions of other proteins), IQ motifs (calmodulin-binding), GRD (GAP-related domain - similar to the catalytic regions of Ras GTPase-activating proteins) and RasGAP C (RasGAP C-terminus-like domain important for binding β-catenin and E-cadherin).
Fig. 2.
Expression pattern of zIQGAP1 mRNA during embryonic and larval development. (A, B) Expression at the 5-somite (5s) stage in the notochord (n) and in posterior epidermis (ep) in dorsal view of the trunk (A) and of the posterior part of the embryo (B). (C, D) At the 15- somite stage (15s) additional expression is observed in otic vesicles (ot): lateral (C) and dorsal (D) views of trunk. (E–G) At 24 hpf (24h), expression is observed in olfactory vesicle (ol), pharyngeal arches (pa), middle cerebral vein (mcev), epidermis, notochord, aorta (ao), posterior cardinal vein (pcv), gut (g) and proctodeum (pr). E, lateral view of the head. F, dorsal view of the head. G, lateral view of the trunk. (H–K) At 36 hpf (36h), (H, I) in the anterior part of the embryo additional expression is observed in the cephalic floor plate (cfp) and in the pectoral fin. H, lateral view of head. I, dorsal view of head. In the trunk (J, K) zIQGAP1 is observed in epidermis, intersegmental blood vessels (ibv), notochord, aorta wall (aow), posterior cardinal vein (pcv) and gut (g). K is a thick cross section of the truncal region shown in J. (L, M) At 48 hpf (48h), expression is observed in the upper jaw (upj) and lower jaw (loj), in cephalic floor plate, as well as in the heart valve (hv) in the head region in lateral view. In the pectoral fin (M) expression is strong in cartilage (pfc) and weaker in the epidermis (pfe). (N, O) At 5 dpf (5d) expression is observed in the otic capsule (otc), perichondrium around the pharyngeal arch, and neurocranial cartilages, such as Meckel cartilage (m), branchial arches (bra), heart valve and bulbus arteriosus (ba). N, lateral view of head. O, ventral view of head. Embryos were presented anterior to the left except for the cross section in K.
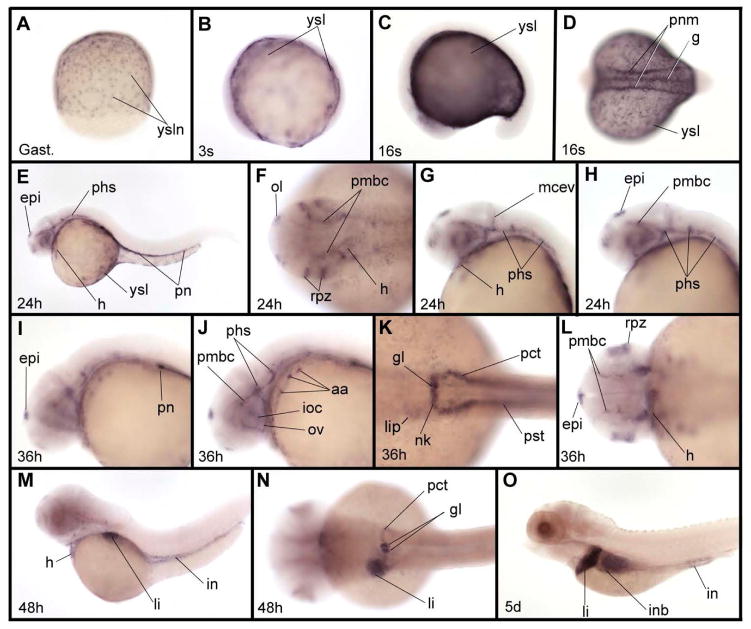
Expression was still observed in the heart valve at 5 dpf, when transcripts also became evident in the bulbus arteriosus (Fig. 2N, O). Altogether, zIQGAP1 mRNA expression is spatially restricted during embryonic development. Expression of zIQGAP2 begins during gastrulation and its transcripts were first observed in the yolk syncytial layer (ysl), accumulating in ysl nuclei (ysln, Fig. 3A). Throughout somitogenesis, strong expression in ysl occurs (Fig. 3 B-D), with additional expression in the developing gut and pronephric mesoderm starting around the middle of somitogenesis (Fig. 3D). At 24 and 36 hpf, the ysl expression of zIQGAP2 was strongly decreased, while the pronephric expression was maintained and was observed in the whole pronephric system from its most anterior part (the glomerulus) to its most posterior component (the pronephric ducts) [Wingert et al. 2007]. At these stages, additional expression territories were observed. zIQGAP2 transcripts were found in the cardiovascular system, including heart and part of the cephalic vasculature (Fig. 3G-J, L), the epiphysis, the olfactory placodes and the retina proliferative zone. We noticed that expression in the epiphysis was bilaterally asymmetrical with strong accumulation of transcripts on the left side (Fig. 3L). Expression here likely occurred in precursors of the parapineal gland, an organ that mediates left right asymmetry in the zebrafish diencephalon [Gamse et al. 2003], and appeared later in development on the left side of the epiphysis. At 36 hpf, weak expression was detected in the liver primordium (Fig. 3K) and increased at later stages. This region became the main expression domain of zIQGAP2 at 48 hpf and during early larval stages (Fig. 3M-O). Expression of zIQGAP2 in the heart, still observable at 48 hpf, was absent at 5 dpf while transcripts accumulated strongly in pronephros glomeruli (Fig. 3N), and appeared both in intestine and in intestinal bulb (Fig.3M-O).
Fig. 3.
Expression pattern of zIQGAP2 mRNA during embryonic and larval development. (A) zIQGAP2 transcripts accumulate in nuclei of the yolk syncytial layer (ysl) at gastrula stage (Gast.). (B) At early somitogenesis (3-somite stage, or 3s) expression is observed in the whole ysl. (C, D) At the middle of somitogenesis (16-somite stage, or 16s) transcripts were found in the ysl, gut and pronephric mesoderm (pnm). (E–H) At 24 hpf (24h) additional expression is found in the epiphysis (epi), heart (h), olfactory vesicles (ol), retina proliferative zone (rpz), and a subpopulation of cephalic blood vessels that includes the primary head sinus (phs), middle cerebral vein (mcev) and primordial midbrain channel (pmbc). In the trunk, expression is observed in the pronephros (pn). (I–L) At 36 hpf (36h) additional expression in cephalic blood vessels were observed in aortic arches (aa), optic artery (ov) and inner optic circle (ioc). In trunk, expression is observed in all derivatives of the pronephric mesoderm, including the glomeruli (g), neck (nk), proximal convoluted tubule (pct) and proximal straight tubule (pst). Expression in the epiphysis shown in (L) is much stronger on the left side. (M, N) At 48 hpf (48h) zIQGAP2 transcripts were observed in heart, liver (li) and intestine (in), as well as in pronephros tissues, including glomeruli, neck and proximal convoluted tubule. (O) At 5 dpf (5d) transcripts were observed in pronephros, liver, intestinal bulb (inb) and intestine. Embryos were shown anterior to the left and dorsal to the top in lateral views (C, E, G–J, M, O), and anterior to the left in dorsal views (B, D, F, K–L, N). Embryo in A is in dorsal view, animal pole to the top.
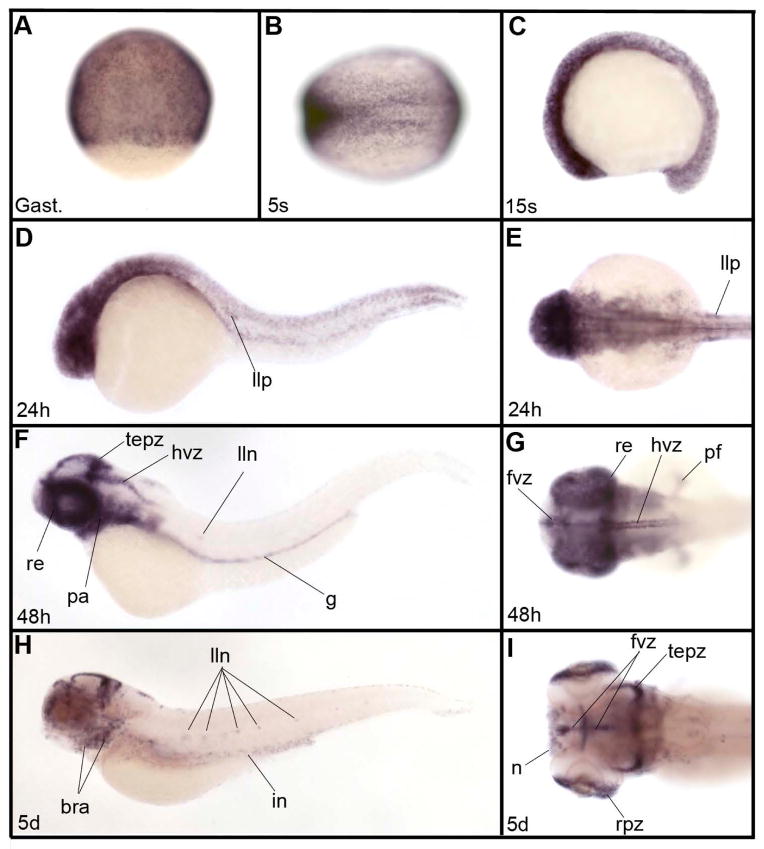
Finally, zIQGAP3 was already expressed at early developmental stages (Fig. 4A-D) with a salt and pepper pattern characteristic of cell proliferation genes, such as cyclin E2, H2A histone family member X, cell division cycle 6 homolog, cell division cycle 20 homolog, ubiquitin-conjugating enzyme E2S or polo-like kinase 1 (http://zfin.org gene expression section). Expression of zIQGAP3 in proliferative cells was observed throughout embryonic and larval development, in particular in the brain ventricular zone, tectum proliferative zone and proliferative zone of the retina. We also observed expression in territories that are not obviously related to cell proliferation, such as the posterior lateral line primordium (Fig. 4E) and lateral line neuromasts (Fig. 4F, H), nose (Fig. 4I), pectoral fins (Fig. 4G), pharyngeal and branchial arches (Fig. 4F-H), and gut (Fig. 4F, H).
Fig. 4.
Expression pattern of zIQGAP3 mRNA during embryonic and larval development. (A) Gastrula stage (Gast.). (B) Early somitogenesis (3-somite stage – 3s). (C) Middle of somitogenesis (15-somite stage or 15s). (DE) At 24hpf (24h) in addition to expression in proliferative cells, zIQGAP3 transcripts were detected in the lateral line primordium (llp). (E) Dorsal view of the head region. (F, G) At 48 hpf (48h), zIQGAP3 transcripts were observed in retina (re), forebrain ventricular zone (fvz), tectum proliferative zone (tepz), hindbrain ventricular zone (hvz), lateral line neuromasts (lln), pharyngeal arches (pa) gut (g) and pectoral fin (pf). (G) Dorsal view of head. (H, I) At 5 dpf (5d) zIQGAP3 expression is strong in proliferative cells of the brain (forebrain ventricular zone, tectum proliferative zone) and of the eye (retina proliferative zone, rpz). Transcripts are also observed in branchial arches (bra), nose (n), lateral line neuromasts and intestine (in). (I) Dorsal view of head region in H. Notice the expression in nose (n), rpz, tepz and fvz. Embryos in C, D, F and H are in lateral view dorsal to the top. Embryos in B, E, G and I are in dorsal view, anterior to the left. Embryo in A is in dorsal view, animal pole to the top.
Function of IQGAP proteins in embryonic development
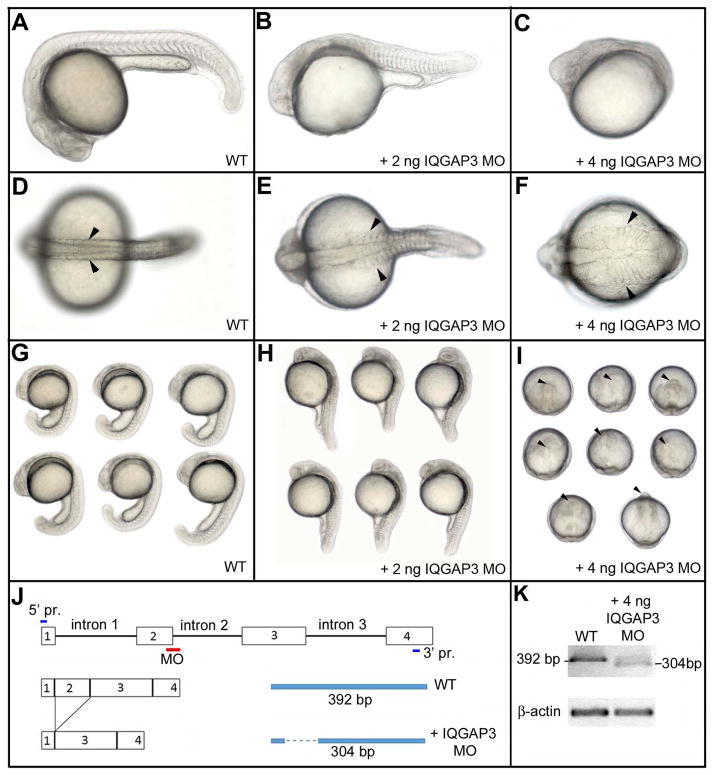
We investigated the function of the three zIQGAPs by performing loss of function experiments using a morpholino (MO) knockdown (KD) strategy. KD of zIQGAP1 and zIQGAP2 causes only a weak effect on global embryo morphology (Supplementary Fig. 2). In contrast, loss of function of zIQGAP3 strongly affects embryonic development (Fig. 5A-I). KD of zIQGAP3 was achieved by injection of a splice interfering MO targeting the 5 prime donor splice site of intron 2 (Fig. 5J). Hybridization of this MO to its target sequence on zIQGAP3 pre-mRNA results in an abnormal splicing characterized by the junction of exon 1 and exon 3 (Fig. 5J), leading to a frame shift and a premature stop codon. The resulting zIQGAP3 peptide translated from this abnormally spliced transcript is very short (16 AA instead of 1,639 AA, Supplementary Fig. 3) and is therefore presumed to be inactive. zIQGAP3 morphant showed dose-dependent phenotypes (Fig. 5). For a partial KD (injection of 2 ng zIQGAP3 MO) embryos displayed defects similar to those observed for mutants affecting convergent extension movements at gastrula stage [Marlow et al. 1998], with an abnormally short and wide embryonic axis. At 20 hpf, somites were enlarged (Fig. 5E) and did not display the characteristic chevron shape observed in wild type (WT) embryos (Fig. 5A, B). Strong morphant phenotypes were observed after injection of 4 ng of zIQGAP3 MO with an even more extreme posterior truncation, resulting in embryos restricted to head and anterior trunk (Figs. 5C, F, I). This morphant phenotype can be rescued, in a dose dependent manner, by co-injection of 0.25 to 1 ng of full length zIQGAP3 mRNA (Fig. 6) demonstrating the specificity of this MO KD. Following injection of zIQGAP3 MO, we investigated the formation of mesodermal tissues at gastrulation (Supplementary Fig. 4). We found that mesoderm induction, involution and regionalization occurs normally in zIQGAP3 morphants and that these embryos do not display early patterning defects but are likely to be defective in cell movements during gastrulation.
Fig. 5.
zIQGAP3 MO knockdown phenotypes. At 24hpf, compared to WT (A, D, G), zIQGAP3 morphant embryos obtained by injection of 2 ng (B, E, H; n = 271) or 4 ng (C, F, I; n = 350) of zIQGAP3 MO display a wider (arrowheads) and posteriorly truncated embryonic axis. To prevent cell death resulting from P53 activation (Robu et al. 2007) 4 ng of P53 MO was coinjected with IQGAP3 MO. Arrowheads in I indicate the anterior tip of the embryonic axis. Embryos are in lateral (A–C, G, H) or dorsal view (D–F), with anterior to the left. Groups of embryos (G–I) are in lateral (G, H) or dorsal view (I), with anterior on the top. (J) Schematic presenting the binding site of zIQGAP3 MO and its interfering mechanism by removing exon 2 of zIQGAP3 pre-mRNA. (K) WT and truncated pre-mRNA covering first four exons were amplified by RT-PCR and analyzed to confirm the interfered splicing of zIQGAP3 mRNA. Notice the loss of WT mRNA band and the appearance of a smaller band representing the truncated mRNA (which lacks exon 2 due to MO-driven splicing interference).
Fig. 6.
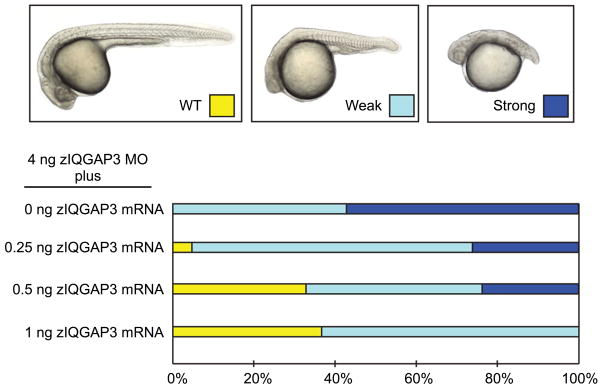
Rescue of zIQGAP3 MO KD. Graphs indicate the percentage of different phenotypes displayed on top (WT, yellow; weak phenotype, light blue and strong phenotype, dark blue) in zIQGAP3 MO KD (injection of 4 ng zIQGAP3 MO; n = 153), and in rescue experiments in which 0.25 ng (n = 128), 0.5 ng (n = 147) or 1 ng (n = 173) of zIQGAP3 mRNA was coinjected with the zIQGAP3 MO.
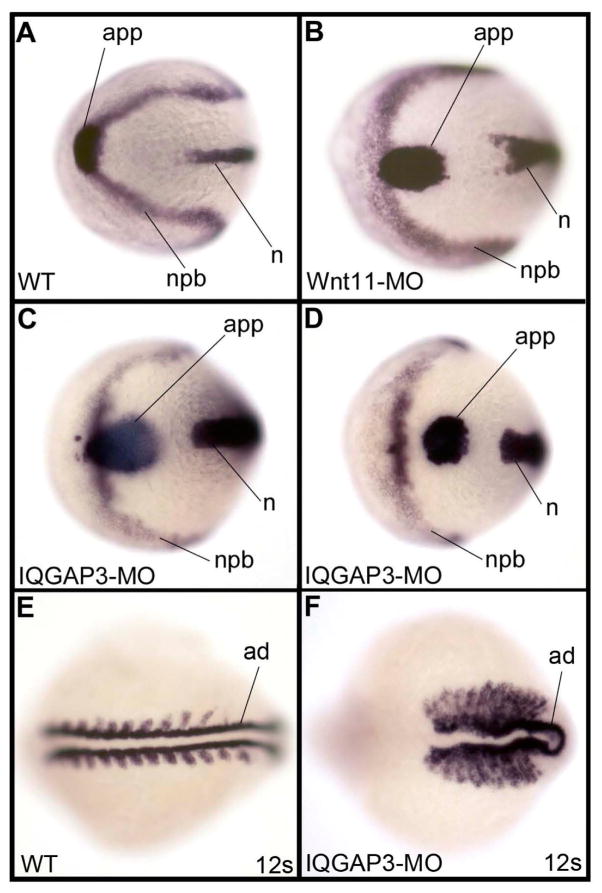
The effect of zIQGAP3 KD on cell movements was investigated by examining whether convergent extension is already altered at the gastrula stage. Accordingly, we compared the embryonic axes of WT gastrulae to those of embryos that were injected with MOs for zIQGAP3 or Wnt11, the latter of which is known to be required for convergent extension movements at gastrula stage [Heisenberg et al. 2000]. As shown in Fig. 7, at early somitogenesis zIQGAP3 morphants displayed abnormally wide notochord and neural plate, and the anterior prechoral plate was located posteriorly to the anterior neural-non neural boundary. This is very similar to defects due to loss of function of Wnt11 (Fig. 7B). Later in development, somites of zIQGAP3 morphant embryos were extremely elongated in their mediolateral dimension and were composed entirely of border cells with no internal cells, similar to what was observed in strong mutants affecting convergence and extension movements at gastrula stage, such as in trilobite/knypek double mutants [Henry et al. 2000].
Fig. 7.
Effect of zIQGAP3 KD on convergent extension movements. (A–D) In situ hybridization at the 3-somite stage with the combination of hgg1 (hatching gland gene 1, a marker for anterior prechordal plate – app), dlx3 (distal-less homeobox3, a marker of neural plate border – npb) and ntl (notail, a marker of the notochord – n) in a WT embryo (A), and embryos injected with Wnt11 MO (n = 105) (B) or zIQGAP3 MO (n = 121) (C,D). Compared to WT, Wnt11 and IQGAP3 morphants display a wider neural plate and notochord, and an anterior prechordal plate posterior to the neural plate border. These different defects signify disruption of the convergent extension movements occurring at gastrula stage. (E–F) In situ hybridization with myod1 (myogenic differentiation 1), which labels the adaxial cells (ad) as well as the border of the somites in (E) WT and (F) IQGAP3 morphant embryos at the 12-somite stage (n = 108). Embryos are presented dorsal view (head region), anterior to the left.
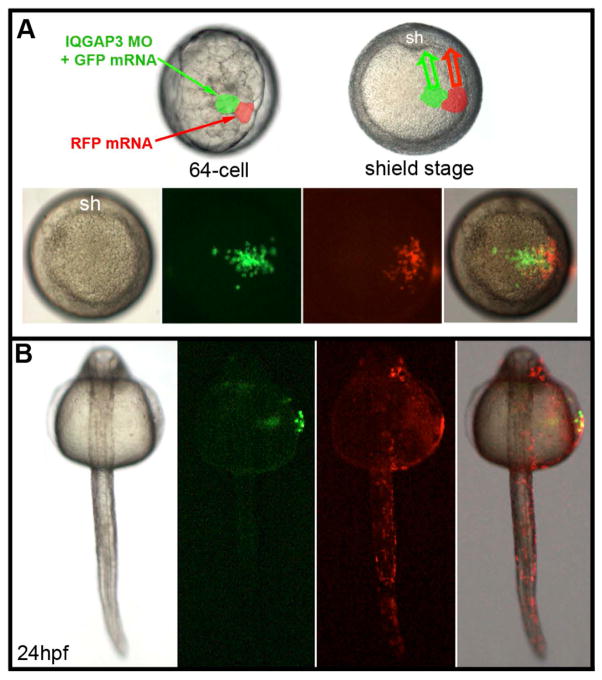
The zIQGAP3 morphant phenotype may result from abnormal cell movements, impairment of cell proliferation during embryogenesis, or both. To distinguish among these alternative hypotheses, we performed IQGAP3 loss of function analysis in a subset of embryonic cells by injecting zIQGAP3 MO together with GFP mRNA into one blastomere of the presumptive ectoderm at the 64-cell stage and, as a control, mRNA coding for RFP was injected into a neighboring blastomere of the presumptive ectoderm (Fig. 8A). At early gastrula stage (8hpf), embryos were screened for similar-size GFP and RFP clones at the same location (either dorsal, lateral or at animal pole) (Fig. 8A). At 24 hpf, the position and size of both zIQGAP3 morphant cells labeled in green and of wild-type control cells labeled in red were observed by fluorescence microscopy. Compared to the number of WT cells (labeled by RFP), the number of zIQGAP3 KD cells (labeled by GFP) was greatly reduced, strongly suggesting that IQGAP3 is essential for cell proliferation. In addition, IQGAP3 morphant cells were observed in a lateral position, staying on top of the yolk, far from the embryonic axis, while the wild-type cells converged toward the midline and populated the embryo along its entire length (Fig. 8B). These results indicate that zIQGAP3 acts cell autonomously and that morphant cells are defective in cell motility.
Fig. 8.
zIQGAP3 KD affects both cell proliferation and cell movements. (A) Top panel, schematic of the clonal analysis experiment. At the 64-cell stage, injection of 100 pg zIQGAP3 MO and 100 pg GFP mRNA mixture in one animal pole blastomere, and of 100 pg RFP mRNA in another, adjacent, animal pole blastomere (left, side view, center animal pole view) result in formation of two clones of cells (green for morphant cells and red for WT cells) at the beginning of gastrulation (shield stage, right). These cells converge (arrows) toward the dorsal side of the embryo (shield: sh). Bottom panel, live imaging in animal pole view at the onset of gastrulation, with bright field, GFP, RFP and merged channels (from left to right), indicating the positions of labeled clones at the onset of gastrulation. (B) Dorsal view of a typical embryo (n = 48) at 24 hpf (anterior to the top), with bright field, GFP, RFP and merged channels (from left to right). zIQGAP3 MO potently inhibited cell proliferation (very small population of GFP-labeled morphant cells compared to RFP-labeled WT cells) and cell movements (GFP-labelled morphant cells stayed in lateral position, on top of the yolk, while RFP-labeled WT cells converged toward the embryo midline and populated the entire length of the embryonic axis down to the tail region).
zIQGAP3 functions downstream of the Ras signaling pathway during embryonic development
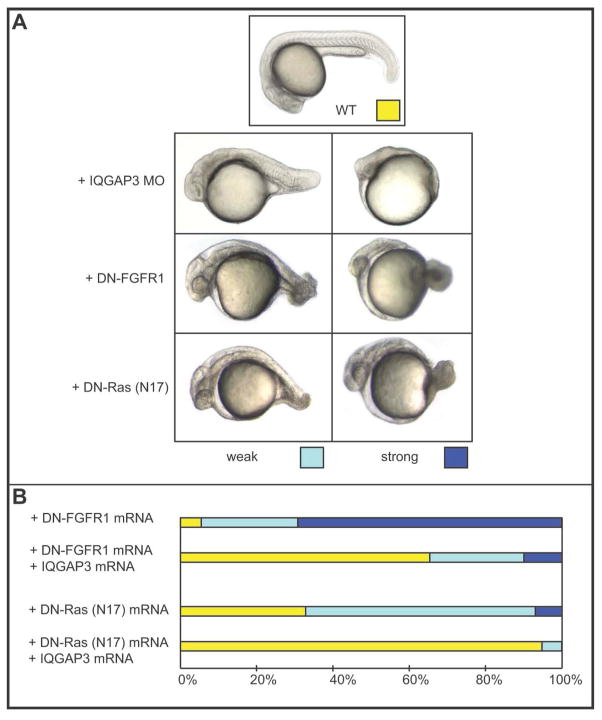
Mammalian IQGAP1 has been shown to serve as an important scaffold for the Ras/MAPK cascade, and to interact directly with FGFR1, K-Ras, Raf and MEK [Banon-Rodriguez et al. 2014; Jameson et al. 2013; McNulty et al. 2011; Ren et al. 2007; Ren et al. 2008; Roy et al. 2004; Roy et al. 2005]. To examine whether zIQGAP3 has a similar function we compared the phenotype of its morphants to those resulting from impairment of the FGFR1-Ras pathway. To do so, we overexpressed the dominant negative forms of FGFR1 (TR-FGFR1) and of Ras (DN-Ras; the TN17 mutant) [Furthauer et al. 2004; Umbhauer et al. 2000]. As shown in Fig. 9A, at 24 hpf, downregulation of the FGFR1-Ras pathway resulted in strong posterior truncation of the embryos, a phenotype verysimilar to zIQGAP3 morphants. We then investigated the possible involvement of zIQGAP3 in that signaling pathway through its gain of function in embryos overexpressing TR-FGFR1 or DN-Ras. We found that injection of mRNA coding for zIQGAP3 efficiently rescues the phenotypes resulting from the downregulation of the FGFR1-Ras pathway (Fig. 9B). zIQGAP3 is therefore an essential factor that acts as a downstream regulator of the Ras signaling pathway.
Fig. 9.
zIQGAP3 acts downstream to FGFR1 and Ras. (A) Phenotypes of zIQGAP3 morphants and of embryos overexpressing DN-FGFR1 and DN-Ras compared to a WT embryo at the same developmental stage. A WT embryo is shown on the top (yellow), weak phenotypes on bottom left (light blue) and strong phenotypes on bottom right (dark blue). (B) Percentage of the different phenotypes observed in the population of embryos injected with DN-FGFR1 mRNA (500 pg; n = 101) or DN-Ras mRNA (200 pg; n = 144) alone, or in combination with 1 ng of zIQGAP3 mRNA. The phenotypic rescue by coinjection of zIQGAP3 mRNA with either DN-FGFR1 (n = 112) or DN-Ras (n = 112) indicates that zIQGAP3 acts downstream of FGFR1 and Ras.
Discussion
Building on a prior description of IQGAP1 and IQGAP2 localization in developing Xenopus laevis embryos [Yamashiro et al. 2003] we now present the first functional comparison of all three vertebrate IQGAPs during early development. Using zebrafish (Danio rerio) as a model system, zIQGAP1 and zIQGAP2 were found to be expressed at the mRNA level in specific tissues and organs at specific times during embryogenesis and larval stages. zIQGAP1 is strongly expressed in the ear, in the vasculature and in cartilages, while a high level of zIQGAP2 expression is found in the ysl, in endodermal derivatives (liver, intestinal bulb, intestine) and in the pronephros. Surprisingly, only a subset of embryonic cells express these zIQGAP genes and no obvious correlation can be made between the observed expression territories and a particular physiological function (such a cell adhesion or cell motility) or type of tissue (such as epithelia or mesenchyme). While zIQGAP1 and zIQGAP2 mRNAs are associated with specific tissues and organs, the distribution of mIQGAP3 mRNA is largely confined to regions of high cellular proliferation, regardless of location. Furthermore, while KD of either zIQGAP1 or zIQGAP2 yielded minimal effects on development, zIQGAP3 KD dramatically interfered with normal embryogenesis. zIQGAP3, but not zIQGAP1 or zIQGAP2, therefore performs essential roles during early development. The impaired development caused by zIQGAP3 reduction was apparently due to two distinct effects: inhibition of both cell proliferation and cell motility (Figs. 7–8). Although IQGAP1 has also been implicated in regulation of cellular motility [Bensenor et al. 2007; Le Clainche et al. 2007; Yamaoka-Tojo et al. 2004], this study reinforces a prior report that IQGAP3 is unique among the IQGAPs as a protein that also regulates cell proliferation [Nojima et al. 2008].
It is likely that the role of IQGAP3 in cell proliferation relates to its direct binding of Ras, which is not known to bind either IQGAP1 or IQGAP2 [Nojima et al. 2008]. Although numerous cell surface receptors signal through Ras to drive cell replication, FGFR1 is especially important for activating Ras during early development. It is therefore noteworthy that expressing DN forms of FGFR1 or Ras yielded morphological phenotypes very similar to those caused by zIQGAP3 KD, and that the mutant developmental phenotype could be rescued by expression of zIQGAP3 (Fig. 9). These results indicate that zIQGAP3 functions downstream of FGFR1 and Ras as an essential protein for cell proliferation during zebrafish embryogenesis. How zIQGAP3 is involved in cell proliferation is unknown, but it is noteworthy that mammalian IQGAP1 binds mTORC1 [Tekletsadik et al. 2012], a multiprotein complex that regulates many fundamental aspects of cell behavior, including progression through the cell cycle and cell division [Zoncu et al. 2011]. It is therefore tempting to speculate that IQGAP3, including the version expressed in zebrafish, responds to Ras activation by promoting mTORC1-dependent cell proliferation.
The high structural similarity of zIQGAP3 to zIQGAP1 probably underlies the impaired motility of cells injected with zIQGAP3 MO. Binding to ligand-activated cell surface receptors, such as VEGFR2 and FGFR1, activates mammalian IQGAP1 for stimulation of branched actin filament assembly, which leads directly to plasma membrane protrusion and cell motility [Bensenor et al. 2007; Le Clainche et al. 2007; Yamaoka-Tojo et al. 2004]. IQGAP1 further regulates cell migration through capture of microtubules via CLIP-170 and APC [Fukata et al. 2002; Watanabe et al. 2004]. Although there have not been any reports that IQGAP3 binds to cell surface receptors, cofactors for branched actin filament formation, such as the Arp2/3 complex and N-WASP, or microtubule end binding proteins, like CLIP-170 and APC, the numerous functional motifs shared by zIQGAP1 and zIQGAP3 (Fig. 1) make it likely that the two proteins perform similar functions in the context of cell migration.
It has been reported that in Xenopus laevis, IQGAP1 regulates the nuclear localization of β-catenin [Goto et al. 2013]. That finding, along with the structural similarity between zIQGAP1 and zIQGAP3, suggests that the impaired cell migration caused by zIQGAP3 KD is an indirect effect of interfering with the canonical Wnt/β-catenin signaling pathway. However, expression of sp5l, a direct Wnt/β-catenin target gene [Thorpe et al. 2005], is not affected in IQGAP3 morphants (Supplementary Fig. 5). It is therefore unlikely that canonical Wnt/β-catenin signaling is regulated by IQGAP3.
Materials and Methods
Cloning of zIQGAP genes
Total RNA was extracted from 24-hour old embryos using RNA Stat-60 reagent (IsoTex Diagnostics, Inc.) and single strand cDNA was synthesized by RT-PCR using SuperScriptII Reverse Transcriptase (Invitrogen Cat #18064-014). Open reading frames for zIQGAP1, zIQGAP2 and zIQGAP3 were isolated by RT-PCR using respective primers (5' BamHI primer: 5'-TATCGCGGATCCATGTCAACTTCAGA-3'; (3253-3280) reverse primer: 5'-ATATGTCGACTGGGTCGGTTTTAATGTT-3'; 5’AgeI primer: 5'-TTATAACCGGTTACTATTTT-CCGTAGAACTT-3'; 3' AgeI (4987-5002) primer: 5'-TTTTACCGGTGTCTGCAACAGTCTG-3' for zIQGAP1; ClaI5’primer: 5’-AAACCCTATCGATAATGGTGAAGCCTCA-3’; Cla5’ UTR (71-83) primer: 5’-CCCATCG-ATAGTCAAAAAGTCC-3’; XhoI (3127-3151) primer: 5’-ATCAAATCTAAGGTGGATCATATCC-3’; 3277-3298 reverse primer: 5’-GATTAGTGTTGATGCCCAGGTT-3’; 3’ AgeI 3-UTR (5105-5123) primer: 5’-ATATTACCGGTAAGATGATTATTTTCCATA-3’for zIQGAP2; HindIII-XhoI-5’PCR primer: 5’-TTTAAA-AAGCTTCTCGAGAACGTACAGAGATG-3’; AgeI-EcoRI-3’primer: 5’-AAATTTACCGGTGAATTCCTTC-TTAAAGAACTTCTTG T-3’ for zIQGAP3). cDNA sequences of all three zIQGAPs have been submitted to GenBank, and their accession numbers are KP862587 for zIQGAP1, KP862588 for zIQGAP2 and KP862589 for zIQGAP3.
For subcloning of full length zIQGAP3 into pCS2+ vector, cDNA of zIQGAP3 was amplified as FLN-zQGAP3 (#1-1096bp) (by Hind III XhoI 5’ primer: 5’-TTTAAAAAGCTTCTCGAGAACGTA CAGAGATG-3’and 3’ 1096 KpnI primer:5’-TCTGCACTTAGCTGCTCCAGGTA-3’) and FLC-zIQGAP3 (#1096-4950bp) (by 5’-1096 KpnI primer: 5’-ATCACGCATCCTGGTACCTGGA-3’ and AgeI EcoRI 3’ primer: 5’-AAATTTACC-GGTGAATTCCTTCTTAAAGAACTTCTTGT-3’). pCS2+ vector was modified at multiple cloning sites (5’ modification primer, 5’-GATCCCCTCGAGATCGATTCGAATTCAAGGCCTACCGGTCCCCGCGGTGTA-CAAGTAAT-3’and 3’ modification primer, 5’-CTAGATTACTTGTACACCGCGGGGACCGGTAGGCCTT-GAATTCGAATCGATCTCGAGCG-3’). Digested FLN-zIQGAP3 (by XhoI and KpnI) and FLC-zQGAP3(KpnI and EcoRI) were ligated and inserted into digested pCS2+ vector (by XhoI and EcoRI) by T4DNA ligase (NEB, Cat. #M0202).
In Situ Hybridization
PCR-amplified sequences of genes of interest were used as templates for the synthesis of antisense RNA probes, labeled with digoxigenin-linked nucleotides. Embryos were fixed and permeabilized before being soaked in the digoxigenin-labeled probes. After washing away non-hybridized probes, digoxigenin-labeled hybrids were detected by immunohistochemistry using an alkaline phosphatase-conjugated antibody against digoxigenin and a chromogenic substrate. The detailed protocol for in situ hybridizations on whole-mount zebrafish embryos used in this study has been described previously [Thisse and Thisse 2014; Thisse and Thisse 2008]. The cDNA sequence for zIQGAP2 (clone fc20f09) was amplified using the two primers: 5'-GGATCCATTAACCCTCACTAAA-GGGAAGAGCTATGACGTCGCAT-3' and 5'-TAATACGACTCACTATAGGG-3', and an antisense probe was synthesized using T3 RNA polymerase. The probe used for zIQGAP3 corresponds to the full length cDNA sequence inserted into the pCS2+ vector linearized by digestion with HindIII and antisense RNA synthesis using the T3 RNA polymerase. zIQGAP1 (clone im:7152854). dlx3, hgg1, lbx2, msgn1, ntl, sp5l and twist1a digoxigenin labeled antisense RNA probes were synthesized as described (http://zfin.org). Because the twist1a probe includes sequence coding for the basic helix-loop-helix DNA binding domain, it cross hybridized with twist2 (a gene specifically expressed in the notochord).
In vitro mRNA synthesis and injections
For mRNA synthesis, cDNAs were cloned into a pCS2+ vector, subsequently linearized with NotI, and transcribed using SP6 RNA polymerase using the mMESSAGEmMACHINE kit from Ambion. DNA constructs used for overexpression of DN forms of zebrafish FGFR1 (TR-FGFR1) and Ras (DN-Ras; the T17N mutant) were used as reported previously [Furthauer et al. 2004; Umbhauer et al. 2000].
Morpholino sequence and preparation
The antisense morpholino oligonucleotide (5’-TGTGTGTGTACCTTTTGGCCTCCTC-3’) interfering with zIQGAP3 gene splicing (targeting the splice donor site of IQGAP3 intron 2) was synthesized by Gene Tools, LLC. Morpholinos were resuspended in water as a 4 mM stock solution and diluted in 0.2% Phenol Red and 0.1 M KCl before further dilution to the indicated concentrations.
Embryo injections
MOs and mRNAs were diluted in 0.2% Phenol Red, 0.1 M KCl. Embryos were dechorionated at the one-cell stage using Pronase E and and 1 nl of mRNA solution was injected into the yolk of 1 or 2-cell stage embryos using an Eppendorf 5426 microinjector. MO KDs of zIQGAP3 were performed by injecting 2–4 ng of splice interfering MO. Overexpression experiments were performed by injection of in vitro synthesized mRNA of zIQGAP3 (1 ng), DN-FGFR1 (500 pg) or DN-Ras (200 pg). In rescue experiments of zIQGAP3 morphant, a mixture of zIQGAP3 MO (4 ng) and zIQGAP3 mRNA (0.25-1 ng) was injected at the 1-cell stage and the resulting phenotypes were compared to phenotypes resulting from injection of 4 ng of IQGAP3 MO alone. For clonal analyses a mixture containing 100 pg of zIQGAP3 MO and 100 pg of GFP mRNA was injected into one animal pole blastomere at the 64-cell stage and the behavior of morphant cells was compared to that of wild-type cells injected with 100 pg of RFP mRNA. All injection experiments have been performed at least three times on the total number of embryos listed in the relevant figure legends.
Supplementary Material
Acknowledgments
This work has been supported by funds from the University of Virginia (CT; BT) and by the NIH grant NS051746 to GSB.
Footnotes
None of the authors have any conflicts of interest.
References
- Banon-Rodriguez I, Galvez-Santisteban M, Vergarajauregui S, Bosch M, Borreguero-Pascual A, Martin-Belmonte F. EGFR controls IQGAP basolateral membrane localization and mitotic spindle orientation during epithelial morphogenesis. EMBO J. 2014;33(2):129–45. doi: 10.1002/embj.201385946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensenor LB, Kan HM, Wang N, Wallrabe H, Davidson LA, Cai Y, Schafer DA, Bloom GS. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J Cell Sci. 2007;120(Pt 4):658–69. doi: 10.1242/jcs.03376. [DOI] [PubMed] [Google Scholar]
- Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nature Biotechnol. 2003;21(3):315–8. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Li Z, Sacks DB. IQGAP1-mediated stimulation of transcriptional co-activation by beta-catenin is modulated by calmodulin. J Biol Chem. 2002;277(9):7453–65. doi: 10.1074/jbc.M104315200. [DOI] [PubMed] [Google Scholar]
- Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, Bernards A, Snijders AJ. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16(9):4869–78. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 2006;16(5):242–249. doi: 10.1016/j.tcb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cerione RA, Hart MJ. Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J Biol Chem. 1997;272(39):24443–7. doi: 10.1074/jbc.272.39.24443. [DOI] [PubMed] [Google Scholar]
- Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 Capture Microtubules through IQGAP1 and CLIP-170. Cell. 2002;109(7):873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Van Celst J, Thisse C, Thisse B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development. 2004;131(12):2853–64. doi: 10.1242/dev.01156. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130(6):1059–68. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Gnatenko DV, Xu X, Zhu W, Schmidt VA. Transcript profiling identifies iqgap2(−/−) mouse as a model for advanced human hepatocellular carcinoma. PLoS One. 2013;8(8):e71826. doi: 10.1371/journal.pone.0071826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Sato A, Adachi S, Iemura S, Natsume T, Shibuya H. IQGAP1 protein regulates nuclear localization of beta-catenin via importin-beta5 protein in Wnt signaling. J Biol Chem. 2013;288(51):36351–60. doi: 10.1074/jbc.M113.520528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15(12):2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, Stemple DL, Smith JC, Wilson SW. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature. 2000;405(6782):76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- Henry CA, Hall LA, Burr Hille M, Solnica-Krezel L, Cooper MS. Somites in zebrafish doubly mutant for knypek and trilobite form without internal mesenchymal cells or compaction. Curr Biol. 2000;10(17):1063–6. doi: 10.1016/s0960-9822(00)00677-1. [DOI] [PubMed] [Google Scholar]
- Ho Y-D, Joyal JL, Li Z, Sacks DB. IQGAP1 Integrates Ca2+/Calmodulin and Cdc42 Signaling. J Biol Chem. 1999;274(1):464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- Jameson KL, Mazur PK, Zehnder AM, Zhang J, Zarnegar B, Sage J, Khavari PA. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nature Med. 2013;19(5):626–30. doi: 10.1038/nm.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K, Nojima H, Yamazaki Y, Yoshikawa T, Okanoue T, Tsukita S. Involvement of IQGAP3, a regulator of Ras/ERK-related cascade, in hepatocyte proliferation in mouse liver regeneration and development. J Cell Physiol. 2009;220(3):621–631. doi: 10.1002/jcp.21798. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science. 1998;281(5378):832–5. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282(1):426–35. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- Li Z, Kim SH, Higgins JM, Brenner MB, Sacks DB. IQGAP1 and calmodulin modulate E-cadherin function. J Biol Chem. 1999;274(53):37885–92. doi: 10.1074/jbc.274.53.37885. [DOI] [PubMed] [Google Scholar]
- Marlow F, Zwartkruis F, Malicki J, Neuhauss SC, Abbas L, Weaver M, Driever W, Solnica-Krezel L. Functional interactions of genes mediating convergent extension, knypek and trilobite, during the partitioning of the eye primordium in zebrafish. Dev Biol. 1998;203(2):382–99. doi: 10.1006/dbio.1998.9032. [DOI] [PubMed] [Google Scholar]
- McNulty DE, Li Z, White CD, Sacks DB, Annan RS. MAPK scaffold IQGAP1 binds the EGF receptor and modulates its activation. J Biol Chem. 2011;286(17):15010–21. doi: 10.1074/jbc.M111.227694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Adachi M, Matsui T, Okawa K, Tsukita S. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nature Cell Biol. 2008;10(8):971–8. doi: 10.1038/ncb1757. [DOI] [PubMed] [Google Scholar]
- Ren JG, Li Z, Sacks DB. IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci USA. 2007;104(25):10465–9. doi: 10.1073/pnas.0611308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren JG, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and B-Raf signaling. J Biol Chem. 2008;283(34):22972–82. doi: 10.1074/jbc.M804626200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, Li Z, Sacks DB. IQGAP1 Binds ERK2 and Modulates Its Activity. J Biol Chem. 2004;279(17):17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- Roy M, Li Z, Sacks DB. IQGAP1 Is a Scaffold for Mitogen-Activated Protein Kinase Signaling. Mol and Cell Biol. 2005;25(18):7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt VA, Chiariello CS, Capilla E, Miller F, Bahou WF. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol. 2008;28(5):1489–502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skawran B, Steinemann D, Weigmann A, Flemming P, Becker T, Flik J, Kreipe H, Schlegelberger B, Wilkens L. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol. 2008;21(5):505–516. doi: 10.1038/modpathol.3800998. [DOI] [PubMed] [Google Scholar]
- Tekletsadik YK, Sonn R, Osman MA. A conserved role of IQGAP1 in regulating TOR complex 1. J Cell Sci. 2012;125(Pt 8):2041–52. doi: 10.1242/jcs.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Thisse C. In situ hybridization on whole-mount zebrafish embryos and young larvae. Meth Mol Biol. 2014;1211:53–67. doi: 10.1007/978-1-4939-1459-3_5. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Weidinger G, Moon RT. Wnt/beta-catenin regulation of the Sp1-related transcription factor sp5l promotes tail development in zebrafish. Development. 2005;132(8):1763–72. doi: 10.1242/dev.01733. [DOI] [PubMed] [Google Scholar]
- Umbhauer M, Penzo-Mendez A, Clavilier L, Boucaut J, Riou J. Signaling specificities of fibroblast growth factor receptors in early Xenopus embryo. J Cell Sci. 2000;113( Pt 16):2865–75. doi: 10.1242/jcs.113.16.2865. [DOI] [PubMed] [Google Scholar]
- Wang S, Watanabe T, Noritake J, Fukata M, Yoshimura T, Itoh N, Harada T, Nakagawa M, Matsuura Y, Arimura N, et al. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J Cell Sci. 2007;120(Pt 4):567–77. doi: 10.1242/jcs.03356. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7(6):871–83. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan YX, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269(32):20517–21. [PubMed] [Google Scholar]
- Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, McMahon AP, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3(10):1922–38. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, Dikalov SI, Chen YE, Tojo T, Fukai T, Fujimoto M, Patrushev NA, Wang N, et al. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species--dependent endothelial migration and proliferation. Circ Res. 2004;95(3):276–83. doi: 10.1161/01.RES.0000136522.58649.60. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Noguchi T, Mabuchi I. Localization of two IQGAPs in cultured cells and early embryos of Xenopus laevis. Cell Motil Cytoskeleton. 2003;55(1):36–50. doi: 10.1002/cm.10109. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhao W, Xu QW, Wang XS, Zhang Y, Zhang J. IQGAP3 promotes EGFR-ERK signaling and the growth and metastasis of lung cancer cells. PLoS One. 2014;9(5):e97578. doi: 10.1371/journal.pone.0097578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.