Abstract
Pluripotent stem cells are being actively studied as a cell source for regenerating damaged liver. For long term survival of engrafting cells in the body, not only do the cells have to execute liver-specific function but also withstand the physical strains and invading pathogens. The cellular innate immune system orchestrated by the interferon (IFN) pathway provides the first line of defense against pathogens. The objective of this study is to assess the innate immune function as well as to systematically profile the IFN-induced genes during hepatic differentiation of pluripotent stem cells. To address this objective, we derived endodermal cells (day 5 post-differentiation), hepatoblast (day 15) and hepatocyte-like cells (day 21) from human embryonic stem cells (hESC). Day 5, 15 and 21 cells were stimulated with IFN-α and subjected to IFN pathway analysis. Transcriptome analysis was carried out by RNA sequencing. The results showed that the IFN-α treatment activated STAT-JAK pathway in differentiating cells. Transcriptome analysis indicated stage specific expression of classical and non-classical IFN-stimulated genes (ISGs). Subsequent validation confirmed the expression of novel ISGs including RASGRP3, CLMP and TRANK1 by differentiated hepatic cells upon IFN treatment. Hepatitis C virus replication in hESC-derived hepatic cells induced the expression of ISGs – LAMP3, ETV7, RASGRP3, and TRANK1. The hESC-derived hepatic cells contain intact innate system and can recognize invading pathogens. Besides assessing the tissue-specific functions for cell therapy applications, it may also be important to test the innate immune function of engrafting cells to ensure adequate defense against infections and improve graft survival.
Keywords: Pluripotent stem cells, endoderm, hepatocytes, hepatocyte-like cells, differentiation, interferon, innate immunity, ISG, interferon-stimulated genes
Introduction
Embryonic pluripotent stem cells are isolated from the inner cell mass of developing primordial embryo. These pluripotent stem cells have the ability to generate all cell types in the body. During tissue damage, resident stem and progenitor cells can be activated to repair and regenerate the damaged tissue. Transplantation of progenitor cells derived from pluripotent stem cells is a promising therapeutic option towards treating genetic, infectious and other degenerative disorders. Transplanted cells at the site of injury encounter the inflammatory niche comprised of cellular and soluble factors, including interferons (IFN). The engrafting cells respond to these stimuli by proliferation, differentiation or cell death. These cell types could also be exposed to infectious agents, such as pathogenic viruses and bacteria. In vitro experiments have shown that embryonic pluripotent stem cells can respond to type I (IFN-α and IFN-β) and II (IFN-γ) interferons that mediate the first line of defense against invading pathogens and generating specific immunity (Whyatt, Duwel et al. 1993; Drukker, Katz et al. 2002; Hong and Carmichael 2013). These observations are critical for understanding the innate immune function of pluripotent stem as well as progenitor cells and tissue rejection during cell therapy application.
Interferons are a diverse family of cytokines which possess antiviral and immunomodulatory properties (Isaacs and Lindenmann 1957; Stark, Kerr et al. 1998; Theofilopoulos, Baccala et al. 2005). These specialized proteins are produced by cells in response to viral infection. Cellular sensors such as retinoic acid inducible gene I (RIG-I or DDX58) and toll like receptors (TLR3, TLR7 and TLR9) can recognize pathogen associated molecular patterns (PAMPs) that can trigger an innate immune response (Thompson and Locarnini 2007). Viral specific DNA, RNA and proteins, and microbial products are sensed by TLR and RIG-I resulting in the phosphorylation and activation of interferon regulatory factor 3 (IRF3) and IRF7 (Diebold, Kaisho et al. 2004; Gale and Foy 2005; Perry, Chen et al. 2005). Phosphorylated IRFs and NF-κB translocate to the nucleus and transactivate the expression of IFN-α and IFN-β genes that are critical in the elimination of invading pathogens. By autocrine and paracrine means, the produced interferons serve as ligands for the Janus activated kinase (JAK) -signal transducer and activator of transcription (STAT) pathway (Horvath 2004). Type I interferons bind to the IFN alpha/beta receptors associated with two tyrosine kinases, JAK1 and TYK2 which phosphorylate STAT1 and STAT2 (Horvath 2004). Both STAT proteins bind with IRF9 generating the interferon stimulating gene 3 (ISGF3) complex (Kessler, Veals et al. 1990; Veals, Santa Maria et al. 1993; Darnell, Kerr et al. 1994; Li, Leung et al. 1996). This complex is translocated to the nucleus binding to the IFN-stimulated response element (ISRE) directing the expression of IFN stimulated genes (ISGs) and converting the host cell to an antiviral state (Muller, Steinhoff et al. 1994; Haque and Williams 1998; Liu, Sanchez et al. 2012). ISGs are involved in executing antiviral response against pathogen like hepatitis C virus (Saito, Owen et al. 2008). ISGs function in modulating various cellular processes, which includes altering the cell cycle, apoptosis, angiogenesis, metabolism and transcriptional regulation (Balachandran, Kim et al. 1998; Tan and Katze 1999; de Veer, Holko et al. 2001). There are over 300 canonical ISGs described and recently genetic screens have been used for identification of ISGs with antiviral functions (Schoggins, Wilson et al. 2011; Schoggins, MacDuff et al. 2014). Antiviral ISGs including GBP1, MXA, CNP, cGAS, TAP1, RIG-I, MDA5 (IFIH1), IRF1 and LGP2 (DHX58) are well characterized (Anderson, Carton et al. 1999; Kochs and Haller 1999; Yoneyama, Kikuchi et al. 2005; Schoggins, Wilson et al. 2011; Liu, Sanchez et al. 2012; Wilson, Schoggins et al. 2012; Schoggins, MacDuff et al. 2014). Due to its antiviral properties, IFN-α is a widely used therapeutic agent against HCV and hepatitis B virus (HBV) that cause hepatitis, fibrosis and cirrhosis (Schalm, Heathcote et al. 2000; Manns, McHutchison et al. 2001).
Pluripotent stem cell-derived endodermal cells and hepatic lineage cells are currently being tested for regenerating damaged liver in preclinical animal models (Duan, Catana et al. 2007; Liu, Kim et al. 2011; Chen, Tseng et al. 2012). During embryonic development, hepatic lineage cells originate from endoderm layer. Growth factors HGF, BMP, Wnt and FGF signaling by mesenchymal cells are critical for liver development (Schmidt, Bladt et al. 1995; Uehara, Minowa et al. 1995; Weinstein, Monga et al. 2001; Michalopoulos 2010). Using a combination of these growth factors, pluripotent stem cells can be differentiated into hepatocytes via endoderm (Cai, Zhao et al. 2007; Rashid, Corbineau et al. 2010; Chen, Tseng et al. 2012; Cheng, Ying et al. 2012). This in vitro platform is useful for understanding the differentiation stage-specific cellular perturbations for external stimuli. Our objective is to investigate the innate immune response exhibited by cells at different phases of hepatic differentiation upon exposure to interferon (IFN-α) stimuli. We used an unbiased discovery based approach to profile ISGs during various hepatic differentiation phases of pluripotent stem cells. We identified both canonical and non-canonical ISGs, including novel ISGs. Functional relevance of selected ISGs was verified during HCV replication in differentiated hepatic cells.
Materials and Methods
Cells
Human embryonic stem cell (hESC) line, WA09 (H9), was sourced from WiCell Research Institute, USA. The human induced pluripotent stem cell (iPSC) line 83i-CTRL was obtained from Cedars-Sinai Medical Center iPSC core facility and was described previously (Sareen, Ebert et al. 2012; Ren, Irudayam et al. 2015). The feeder-free hESCs were cultured using serum-free chemically-defined media, mTeSR1, (STEMCELL Technologies, Canada) with every day media change regimen. The HeLa cell line (ATCC, USA) and human liver cancer cell lines, HepG2 and Huh-7.5.1, were maintained using complete Dulbecco's modified Eagle's medium (DMEM) (Fisher Scientific). Complete DMEM was supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/ml), streptomycin (100 mg/ml), and 2 mM L-glutamine (Life Technologies). DMEM media for HepG2 and Huh-7.5.1 cells contained additional supplements, 10 mM Hepes and 10 mM nonessential amino acids. All the cell lines were maintained at 37°C with 5% CO2.
Differentiation of pluripotent stem cells into hepatic lineage cells
For in vitro differentiation, single cell preparation of the pluripotent stem cells was plated on a matrigel-coated 6-well plate, cultured at 37°C in 5% CO2. The cells were subjected to a 3 week hepatic differentiation protocol that consisted of three phases, including endoderm induction (day 1-5), hepatic specification (day 6-15) and hepatic maturation (day 16-21). The protocol could yield endoderm cells (day 5), hepatoblast (day 15) and hepatocyte-like cells (day 21). The cytokines were procured from Peprotech Inc., (Rocky Hill, NJ) unless otherwise mentioned. For differentiation we used a serum-free defined (SFD) media described by Cheng and colleagues (Cheng, Ying et al. 2012). SFD base media comprised of IMDM (75%), Ham's F12 (25%) growth media supplemented with N2 and B27 (without vitamin A) supplements (Life Technologies). For endoderm differentiation, Wnt 3A (40 ng/ml, R and D Systems) and Activin A (100 ng/ml) supplement was used in RPMI media (Life Technologies) for the first one day and the following four days the cells were treated with Activin A, VEGF (10 ng/ml), BMP4 (0.5 ng/ml) and bFGF (10 ng/ml) in SFD media. From day 6 onwards, the SFD media supplemented with BMP4 (50 ng/ml), VEGF (10 ng/ml), EGF (10 ng/ml), TGF-α (20 ng/ml), HGF (100 ng/ml), dexamethasone (1×10−7M; Sigma-Aldrich, St. Louis, MO), DMSO (1%, Sigma-Aldrich), ascorbic acid (50 μg/ml), and monothioglycerol (4.5×10−4 M) was used. From day 12 onwards, BMP4 and TGF-α were removed from the cocktail. For hepatocyte maturation phase, HGF, dexamethasone and oncostatin M (20 ng/ml) were included in the SFD media from day 16 onwards. At indicated time points, endoderm and hepatic lineage specific markers were assessed by immunocytochemistry (ICC), flow cytometry and reverse transcription-quantitative PCR (RT-qPCR).
IFN-α treatment
At day 5, day 15 and day 21 post-differentiation, the cells were stimulated with human interferon-alpha A (R and D Systems) at a concentration of 1000 IU or 5000 IU for 6 hours. Untreated cells were included as negative control. The 5000 IU IFN-α treated (D5, D15 and D21) and untreated (D5, D15 and D21) cell samples were collected for RNA sequencing analysis and gene expression studies. Experiments for RNA sequencing were done in duplicate with two independent biological repeats (n=2).
RNA sequencing sample preparation
Total RNA from cells was isolated using RNeasy Mini Kit (QIAGEN, USA). RNase-Free DNase treatment was performed on column to remove residual DNA. Twelve RNA samples were assessed for concentration and quality using the Thermo Scientific (Waltham, MA) NanoDrop 8000 Spectrophotometer. RNA sequencing libraries were constructed using Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA) per manufacturer's instructions. Briefly, total RNA with RNA integrity (RIN) scores of 9 or better using Agilent Bioanalyzer RNA 6000 nano kit were used for RNA library preparation. mRNA was then purified from one microgram of total RNA from each sample using oligo-dT attached magnetic beads with two rounds of purification. Subsequently, the mRNA was fragmented and primed for cDNA synthesis according to manufacturer's recommendations. RNA adapters and barcodes were ligated to cDNA to allow for clonal amplification and multiplexing on a single lane on a single end Illumina flow cell. Sequencing was done on an Illumina HiSeq 2500 to an average read depth of 26 million reads per sample and the minimum number of reads for a sample was at least 23 million reads.
Quality Assessment, Filtering and Alignment of RNA sequencing
Raw Illumina reads in FASTQ format were analyzed for quality control using RNASEQC (DeLuca, Levin et al. 2012). We assessed read depth, average read length, average coverage across the gene, number of genes identified, PCR duplication rate, ribosomal content, and exon/intron representation to confirm the quality of the library and sequencing. Reads were aligned to the UCSC human reference genome (hg19) using TOPHAT (Trapnell, Pachter et al. 2009). Gene expression was calculated using reference-guided transcript assembly using a gene transfer file (GTF) from UCSC genes and normalized quantification of reads as RPKM (# of Reads Per Kilobase of ORF per Million reads aligned) calculated with Cufflinks (Cufflinks 2014) and compiled into a gene counts table for all samples.
Gene Expression Filtering
Reads Per Kilobase of transcript per Million mapped reads (RPKM) were calculated for 51,057 genes. Raw FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) values less than 1.1 were considered poorly measured and therefore increased to a minimum threshold of 1.1 to prevent overestimating gene expression differences between groups of samples and to permit downstream analyses where data values may be log-transformed. 30,616 genes without significant expression (FPKM = 1.1) were removed from further analysis, leaving 20,441 genes. All gene expression values were log2 transformed.
Principle component analysis (PCA)
We first assessed the data in an unbiased way for evidence of biological or technical artifacts related to coordinated gene expression changes by calculating the principle components and visualizing the greatest three components of gene expression variance in a three-dimensional plot (Supplementary Fig. S1). In this unsupervised analysis, we identified the genes with the highest variance independent of sample annotation by first calculating the standard deviations for each of the remaining 20,441 genes among all samples. Genes were rank ordered by standard deviation and the top 1000 genes were used to calculate the top three principal components (PCA) and plotted using R (Vienna, Austria) version 3.0.2 package “rgl” (Adler and Murdoch 2014).
Transcriptome Analysis
Unique Genes Expressed
We next evaluated the data for differences in gene expression during differentiation time points (day 5, day 15 and day 21) or respective IFN-α treatment conditions. We first considered the number of genes expressed in each stage and defined a gene as well measured and expressed as having an average of at least a FPKM of five in biological replicates from 1 time point/treatment. This stringent cut-off of 5 FPKM represents as little as one transcript per cell (Kellis, Wold et al. 2014). Thus, genes with observations that averaged below 5 in a time point or treatment were considered not expressed. In this way, we proposed a relative number of unique genes that were expressed and shared among each stage illustrated in a Venn diagram. The Venn diagram was generated using program “Venny” (Oliveros 2007).
Differentially Expressed genes (DEG)
We next set out to characterize the global gene expression effects induced by differentiation or by IFN-α treatment. To do this, we selected only genes that had an FPKM of 5 or greater in at least half the samples of a comparison, and were defined as well measured and expressed for analysis. In this way, for example, we identified for 12,617 genes as expressed in at least two samples between day 5 and day 15 control samples and 11,009 genes between days 15 and 21.
A minimum of two fold change (the minimum fold change measurable by qPCR) and a false discovery rate of 10 percent (q<.1) were the minimum criteria for all supervised analysis to define DEG. Q values were calculated using two-tailed students T-Test and applied the Benjamini and Hochberg adjustment for multiple hypothesis testing. For example, we identified 359 genes that were significantly differentially expressed (fold change at least 2, FDR at most 0.1) and 359 significantly differentially expressed genes from day 15 and 21. We illustrated the gene expression differences with a one-way hierarchical clustering analysis. The gene co-expression correlations were calculated for all samples using “hclust, and plotted using “gplots” and “heatmaps.2”in R V2.14.2 (Warnes, Bolker et al. 2014). RNA Sequencing data is retrievable with GEO accession number GSE67848.
Pathway Analysis
Significantly expressed genes were assessed for pathway enrichment using Ingenuity Pathway Analysis (IPA 2014) (QIAGEN, Redwood City) and significantly enriched canonical pathways were defined as having a q-value <.1.
Reverse Transcription Quantitative PCR Analysis
Total RNA was extracted from cells using RNeasy Mini Kit (QIAGEN). As a control, human primary hepatocyte RNA from fetal liver was included. RNA samples were reverse-transcribed using SuperScript III Reverse Transcriptase kit (Life Technologies) as recommended by the manufactures using random primers. Gene expression was quantified using Platinum SYBR Green qPCR SuperMix-UDG with ROX Kit (Life Technologies) by the ViiA7 real-time PCR system (Applied Biosystems). The reaction for each gene expression was completed in triplicates and housekeeping human genes, PPIG (cyclophilin G) or GAPDH, were used as an internal reference control. The qPCR primer sequences are given in supplementary Table S1. The following conditions were used for cDNA amplification: 50°C for 2 min; 95°C for 2 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The Ct value of each gene is normalized to that of the housekeeping gene to provide comparison among various samples.
Flow Cytometry
At indicated time points, the differentiating cells were collected, membrane permeabilized using the Cytofix/Cytoperm kit ( BD Biosciences) and analyzed using antibodies against endoderm [SOX17 and CXCR4 (R & D Systems)], and hepatic markers [AFP (Dako, USA) and Albumin (Dako, USA)]. Cells were analyzed on a BD Fortessa (BD Biosciences) flow cytometer using FACSdiva software and FlowJo software (TreeStar Inc.).
Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde (Polysciences, Inc., Warrington, PA). Following three PBS washes, the cells were blocked with 10% fetal bovine serum, 3% BSA, 0.1% Triton-x 100 in PBS). Subsequently, the cells were incubated with human albumin goat polyclonal primary antibody (Bethyl Laboratories, Inc) and alpha-1 antitrypsin (AAT) mouse monoclonal primary antibody (Santa Cruz Biotechnology) at a dilution of 1:200 for overnight at 4°C. The secondary antibody, mouse anti-goat polyclonal antibody-FITC (Santa Cruz Biotechnology) or goat anti-mouse polyclonal antibody (Alexa fluor 594) (Life Technologies, USA) was added at 1:1000 dilutions and incubated for 1 hr at room temperature. Between antibody changes the cells were washed five times with PBS. The nuclei were stained with addition of Hoechst dye (Life Technologies, USA).
Western blotting
For Western blotting, the cell lysates were resolved by SDS-PAGE using 4-15% gradient gel (Bio-Rad) and transferred to a PVDF membrane using Trans-Blot turbo transfer system (Bio-Rad). The membranes were blocked (5% skim milk, 0.2% Tween-20 in PBS) for 1 hour at room temperature and probed with rabbit monoclonal antibodies STAT1, Phospho-STAT1 (Tyr701) and beta-actin (Cell Signaling Technology) and mouse monoclonal antibody NS3 [clone H23 (Abcam)]. Goat anti-rabbit or goat anti-mouse IgG conjugated with horseradish peroxidase (Amersham Pharmacia Biotech) secondary antibody was detected by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate kit, Thermo Scientific, USA).
Human apolipoprotein B-100 (ApoB-100) and triglycerides assays
24 hour spent culture media from differentiating cells (one million cells per well) at indicated time points (day 0, day 4, day 10 and day 15 post-differentiation) were collected. The amount of ApoB-100 (ng/ml) produced by the cells was measured by the Human Apolipoprotein B ELISA kit (MABTECH, Inc). Triglycerides (mg/ml) were measured using colorimetric assay (L-Type TG M, Wako Diagnostics). Fold change value to day 0 (undifferentiated cells) was obtained by calculating the ratio with that of each time point.
Hepatitis C virus (HCV) genomic RNA generation and transfection
An intragenotype 2a chimeric virus, having genomic regions from J6CF and JFH-1 strains was used in this study (Chu, Ren et al. 2013). Mutant HCV with catalytically inactive viral RNA polymerase NS5B (GDD amino acids replaced with AAG residues) was reported previously (Chu, Ren et al. 2013). T7-RNA polymerase-mediated in vitro transcription of HCV plasmid DNA template was described previously (Chu, Ren et al. 2013; Ren, Contreras et al. 2014). The pFNX-HCV plasmids containing HCV cDNA were linearized with XbaI restriction digestion and subjected to in vitro transcription (T7 Ribomax Express Large Scale RNA Production System - Promega Corporation, Madison, WI) and RNA purification (Qiagen RNeasy Mini Kit) as per manufacturer's recommendations. The hESC-derived hepatic cells (day 14 post-differentiation) or Huh-7.5.1 cells cultured in 12-well plates were used. HCV RNA (400 ng per well) was transfected into the cells (1 ×106 per well) using TransIT mRNA transfection reagent (Mirus Bio LLC., USA). 400 ng HCV RNA for 1 ×106 cells per well is non-cytotoxic as determined by viable cell count at 24 hour post transfection of hESC-derived hepatic cells during RNA dose optimization step. For mock control, the cells received only transfection reagent without HCV RNA. After 4 hours, the culture supernatant was replaced with fresh media. Samples were collected for RNA and protein at indicated time points. For interferon treatment, 1000 IU of IFN-α was added once to cells 2 hrs before transfection.
Statistical analysis
Error bars in the graph reflect the standard deviation. P-values were determined by the two-tailed student's t-test and significance was reported if p value is p<0.05 (*); p<0.001 (**); p<0.0001 (***).
Results
Derivation of hepatic cells from pluripotent stem cells
To assess the cellular response to type-1 interferon during various stages of hepatic differentiation, a three stage differentiation program was used. This protocol recapitulates embryonic endoderm induction, hepatic specification and hepatic maturation phases (Fig. 1A) (Cai et al., 2007; Rashid et al., 2010; Chen et al., 2012; Cheng et al., 2012). We utilized feeder-free H9 human embryonic stem cells. First, we evaluated the differentiation efficiency by investigating stage specific markers. At day 5, over 85% cells co-expressed CXCR4 and SOX17 proteins demonstrating a robust endoderm induction (Fig. 1B). Then, we evaluated hepatic markers by flow cytometry to provide a quantitative estimation of differentiation efficiency. At day 21, 85% of cells expressed liver markers albumin and alpha-fetoprotein (AFP) (Fig. 1C). Gene expression kinetics of albumin and AFP indicated that these markers are significantly upregulated in differentiating cells at day 15 and day 21 compared to that of hESCs (Fig. 1D). At day 15 to day 21, as the maturation progressed, a trend of AFP reduction with an increase in albumin expression was observed. Positive controls of human primary hepatocytes of fetal origin and liver cancer cell line HepG2 were included for liver marker analysis. The results indicated that our three-phase differentiation protocol yielded fetal stage immature hepatocyte-like cells. Furthermore, we tested the functionality of differentiating hepatic cells. The secretion of very low-density lipoprotein (VLDL) particles is a hallmark characteristic of hepatocytes. To test if the hepatic cells are competent for VLDL secretion, we measured triglycerides (Fig. 1E) and ApoB-100 (Fig. 1F) components of VLDL secreted into the culture media of one million cells for a 24 hour period. Our results from differentiating hESCs and iPSCs indicated that the hepatic lineage-specified cells at day 10 and 15 are capable of releasing VLDL particles, thus reproducing physiological function of liver. Additional liver marker, alpha-1 antitrypsin (AAT) co-expression with albumin was demonstrated in differentiated-hepatocyte-like cells by ICC (Fig. 1G).
Figure 1.
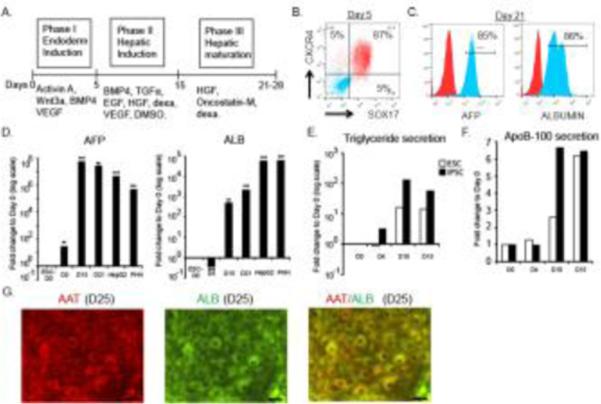
Differentiation of hESC to hepatic lineage cells and gene expression analysis. (A) Schematic diagram depicting the experimental protocol used for the differentiation of hESCs into hepatic lineage cells. Flow cytometry analysis of endoderm (B), and hepatic markers (C) by differentiating cells. In the histogram, the isotype l antibody stained cells are shown in red. (D) Kinetics of liver specific-marker gene expression by RT-qPCR analysis. Expression of each gene is normalized to housekeeping gene PPIG and compared to that of day 0 hESCs. Means with standard deviations are presented as bar graphs. HepG2 and primary human hepatocytes (PHH) from fetal liver are included as positive controls. (E and F) Functional analysis of hepatic cells derived from hESC and iPSC lines. VLDL components, triglycerides and ApoB-100 production by differentiating cells at indicated time points are shown in the graphs. (G) Immunocytochemistry of alpha 1 antitrypsin (AAT) and albumin expression by differentiating hepatocytes at day 25 (scale bar 10 μm).
We then assessed the differentiation-stage specific gene expression by RNA sequencing analysis. 11009 genes were expressed at least one time point (defined as an average of FPKM >5) (Fig 2A). We observed the most unique gene expression (2075 genes) in the earliest time point (i.e. the hESC-derived endodermal cells) (Fig 2A) and the largest number of differentially expressed genes (6165 genes) between the earliest time points (day 5 and day 15) (Fig 2B). On the contrary, we observed fewer differences in later stages, for example the shared gene profiles concurred with the differentiation states of the cells. Genes expressed in day 15 cells were highly similar to day 21 cells, specifically only 359 genes out of the 11009 expressed genes in day 15 and day 21 were uniquely expressed in either time points (Fig 2B). Furthermore, differentially expressed genes at the liver-specified time points (day 15 and day 21) demonstrate an enrichment of gene expression in liver specific metabolic pathways, including urea cycle, and prothrombin activation (Fig. 2C), while the cells at the earliest time point (day 5) expressed a higher proportion of genes involved in embryonic development, pattern formation, and in the BMP4 pathway all of which are consistent with the earliest phase of differentiation the endoderm. These data suggest the cells likely determined their endoderm development fate by day 5 and had differentiated into hepatic lineage by day 15 with little further differentiation occurring after day 15. List of differentially regulated genes are given in Supplementary Table S2. Taken together, we established a robust hepatic differentiation platform for evaluating type-1 IFN response pathway.
Figure 2.

Global transcriptional analysis of differentiating hESCs by RNA-sequencing. (A) Venn diagram shows the shared and unique genes expressed at various stages of differentiation. (B) Heat map of differentially expressed genes at day 5, 15 and 21 of differentiation (in duplicate; red-up regulated; green-down regulated). 2-way supervised Hierarchical clustering was performed using 6313 unique genes specifically identified for each time point for the combined dataset. From day 5 to day 15 cells most of the 6165 genes were down-regulated. Day 15 and Day 21 cells had only 359 differentially expressed genes. (C) Canonical pathways activated in hepatic lineage cells are shown (Day 5 =T1; Day 15 =T2; Day 21 =T3).
IFN-α treatment and transcriptome analysis
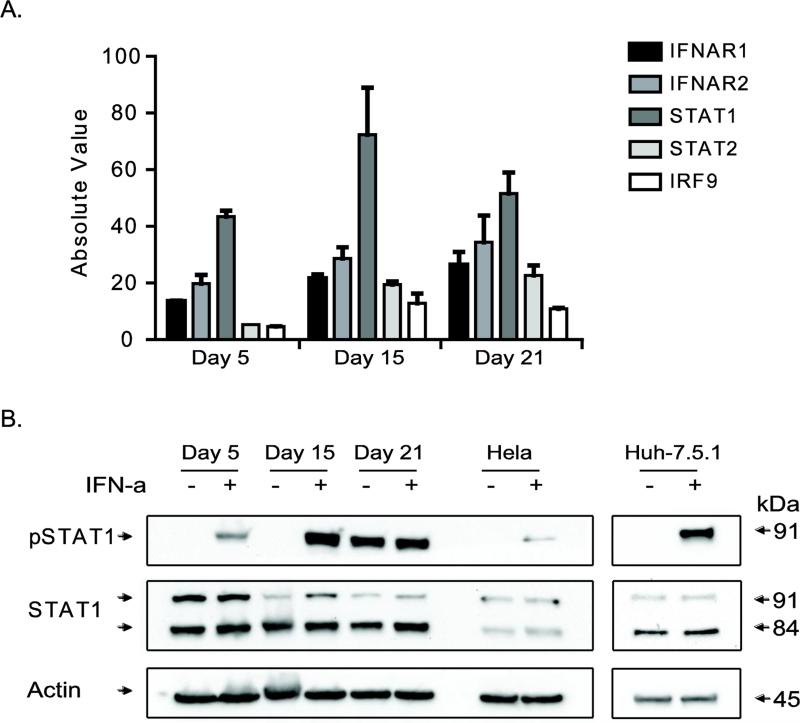
We chose cells from the endoderm phase (day 5) and the hepatic phases (days 15 and 21) for assessing response to IFN-α. Type-1 IFN exerts the biological effect by binding to IFN receptors and activating the JAK-STAT pathway. The differentiating cells expressed the JAK-STAT pathway genes, including Type I Interferon Receptor 1 (IFNAR1), IFNAR2, STAT1, STAT2 and IRF9 (Fig. 3A). Treatment of differentiating cells with IFN-α for 1 hr resulted in phosphorylation of STAT1, a key component of ISG3 transcription factor complex (Fig. 3B). Interestingly, the untreated day 21 cells had higher basal level of phosphorylated form of STAT1 which could be due to use of oncostatin M (OSM), an IL-6 family of cytokine that shares signaling pathway with STAT1. However, both IFN-α and OSM have distinct receptors and downstream target genes resulting in specific biological outcomes (Darnell, Kerr et al. 1994; Heinrich, Behrmann et al. 1998; Dey, Radhakrishnan et al. 2013). Our results indicated that the JAK-STAT pathway can be triggered by IFN-α in pluripotent stem cell-derived endoderm-hepatic lineage cells.
Figure 3.
Analysis of JAK/STAT1 pathway in differentiating cells. (A) Basal expression values of factors involved in mediating the IFN-signaling pathway are given in the bar graph. The absolute expression values were obtained from RNA-seq data of differentiating cells (without IFN-α treatment). (B) Analysis of STAT1 upon IFN-α treatment. Phosphorylated STAT1 (Tyr701) was detected following IFN-α addition to differentiating cells. Total STAT1 and actin was included as a loading control. HeLa cells and Huh-7.5.1 cells with or without IFN-α treatment were included as positive control. Note that the day 21 untreated cells have higher basal levels of pSTAT1.
To characterize the complete IFN response, we evaluated transcriptomic differences between IFN-α treated (5000 IU for 6 hours) and matched untreated control samples at three different time points (days 5, 15, and 21). First, we assessed whether the greatest amount of gene expression variation between samples was associated with biological conditions or another factor such as technical that could affect downstream analysis. To do this we calculated the top three principal components and plotted them in 3D space (Supplementary Fig. S1). The samples clearly clustered based on differentiation time points along with their respective IFN-treatment groups suggesting the largest gene expression differences were associated with treatment and time.
Next, we evaluated the ability of these cells especially the later-stage differentiated cells to respond to IFN-α treatment (Fig. 4). We observed that day 21 cells, a mature phase of differentiation, exhibited stronger response to INF-α compared to that of day 5 and 15 cells. 438 genes were significantly upregulated (q-value 0.2 with 2 fold change) between IFN-α treated and control day 21 samples (Fig. 4A). A list of these genes can be found in Supplementary Table S3. Comparative analysis of previously known canonical ISGs indicated that 83 of the 438 genes were shared (Supplementary Table S3). Data for these IFN induced 438 genes were filled in for day 15 IFN-α treated and control samples regardless of fold change or significance (Fig. 4A). 42 canonical ISGs were significantly changed in all three time points of differentiation upon IFN treatment (Fig. 4B). The pathway analysis of differentially regulated genes upon IFN treatment revealed the following innate immunity pathways: interferon signaling, activation of IRF by cytosolic pattern recognition receptors, role of pattern recognition receptors in recognition of bacteria and viruses, antigen presentation pathway, role of RIG1-like receptors in antiviral innate immunity, LPS/IL-1 mediated inhibition of RXR function and virus entry via endocytic pathways. The interferon signaling and virus entry via endocytic canonical pathways with enriched genes are shown in Supplementary Figure S2.
Figure 4.
Transcriptome analysis of IFN-α treated differentiating cells. (A) Heat maps show differentially regulated genes upon IFN-α treatment of differentiating cells at day 15 and day 21 time points. Green depicts down-regulated gene levels and red induced levels. Expression levels of each gene indicated by intensities are normalized in each row. Time points and genes are clustered according to their similarity in profile. The dendrograms show unique clusters of genes according to common functionality. (B) Heat map depicts the expression levels of 42 known canonical ISGs during differentiation upon IFN-α stimulation.
Validation of novel ISGs
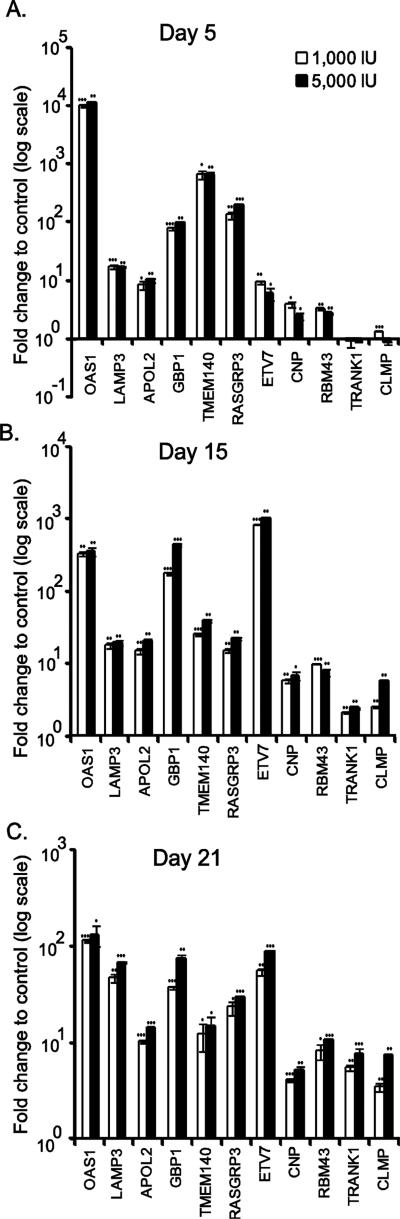
For confirmation, the IFN-α treatment (1000 IU and 5000 IU doses) experiment was repeated on differentiating hES cells to hepatic cells. Gene expression analyses were done using RT-qPCR. We selected both canonical ISGs and novel hits from the transcriptome analysis, which had a two-fold induction in at least two of the differentiation time points studied. The canonical ISGs tested include, OAS1, LAMP3, APOL2, TMEM140 and GBP1 (Fig. 5) as well as MX1, ISG15, HSH2D, and ZNFX1 (Supplementary Fig. S3). We also verified the induction of new candidate ISGs such as RASGRP3, TRANK1, and CLMP and less characterized ISGs like ETV7, CNP and RBM43. Consistent with the transcriptome profile, the canonical ISGs, MX1, ISG15, OAS1, HSH2D, and ZNFX1 had exhibited significant IFN-mediated stimulation in all three stages of cellular differentiation. The genes, LAMP3, TMEM140, GBP1, RBM43 and ETV7 with no significant change at day 5 due to high FDR ranking (Supplementary Table S2), had strong induction on all three time points as tested by RT-qPCR. RASGRP3 was verified as a new ISG in differentiating cells (Fig. 5) as well as in human liver cancer cell line Huh-7.5.1 and HeLa cells (Supplementary Fig. S3). Consistent with transcriptome analysis, the genes TRANK1 and CLMP exhibited insensitivity for IFN-α treatment at day 5, but were stimulated at day 15 and 21 post-differentiation. Induction level of APOL2 and CNP remained relatively similar in cells of three differentiation phases. Our result suggests that the validated genes exhibit differential induction pattern during endodermal and hepatic differentiation states.
Figure 5.
Validation of interferon induced genes profiled by RNA sequencing analysis. Day 5 (A), day 15 (B) and day 21 (C) cells treated with indicated doses of IFN-α was subjected to RT-qPCR analysis. Relative gene expression to untreated control cells was calculated and the mean values are shown in the graphs with standard deviations.
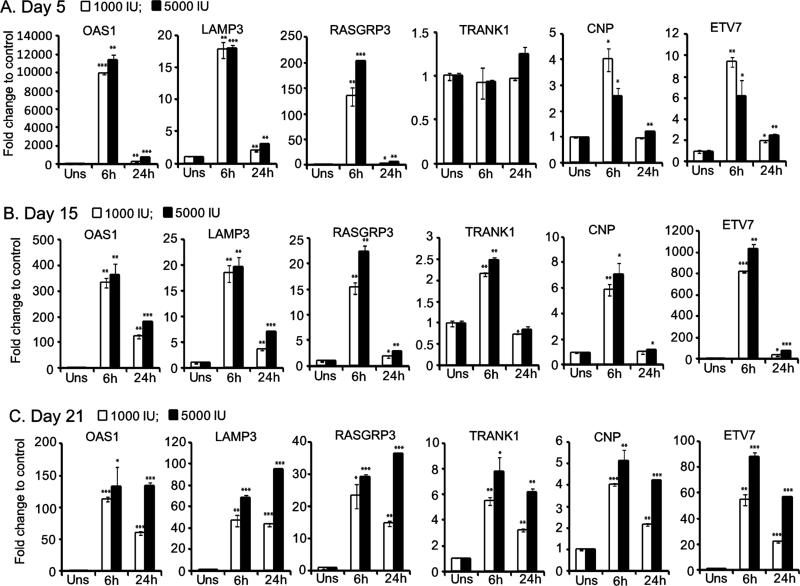
Expression kinetics analysis
In addition to the 6hr time point, we also tested the expression of selected ISGs at 24hr post-IFN treatment. We studied the expression kinetics of RASGRP3, TRANK1, CNP and ETV7. The OAS1 and LAMP3 genes were included as positive control. In general, these ISGs showed peak induction at 6 hrs post-IFN treatment (Fig. 6). At endoderm phase (day 5), a marginal induction was observed for TRANK1 in 5000 IU IFN treatment for 24 hrs. Since the genes were selected from the transcriptome data of 6 hr IFN treated samples, it was not surprising to observe peak induction kinetic of 6 hr. In conclusion, our data suggested that the above tested ISGs have immediate early expression kinetics.
Figure 6.
Expression kinetics of selected ISGs. Induction of ISGs RASGRP3, ETV7, CNP and TRANK1 are shown for day 5 (A), day 15 (B) and day 21 (C) post-differentiation of hESCs. The well characterized ISGs OAS1, and LAMP3 are included as positive control. Normalized gene expression values were used for calculating the fold change with that of unstimulated control and presented as graphs. All the genes but TRANK1 were significantly induced in 6h and 24h time points across differentiation phases.
Functional analysis of ISGs
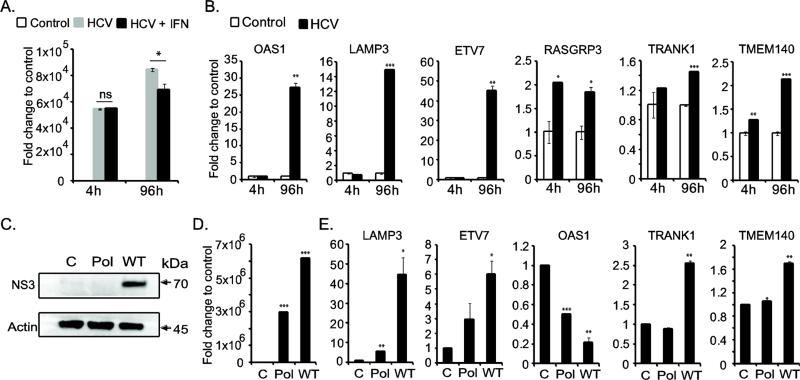
In order to determine the biological relevance of the newly profiled ISGs, we utilized a hepatitis C viral infection model. HCV is a major liver pathogen affecting human. Hepatic cells derived from pluripotent stem cells have been shown to be permissive for HCV replication (Schwartz, Trehan et al. 2012; Wu, Robotham et al. 2012). RNA sequencing analysis showed that the differentiating hepatic cells expressed various cell surface receptors for HCV entry, including CD81, claudin 1 (CLDN1), occludin (OCLN), low-density-lipoprotein receptor (LDLR) and scavenger receptor class B member 1 (SCARB1) (Supplementary Fig. S4) (Lindenbach and Rice 2013). The day 14 differentiated cells (hepatic-specified can support HCV growth) were transfected with HCV genomic RNA with or without IFN-α. The viral genome replication was tested using reverse transcription quantitative-PCR assay. An increase in HCV genome level was detected at 96hr post-transfection compared to that of 4hr input indicating establishment of viral infection (Fig. 7A). The HCV replication was significantly inhibited by IFN-α treatment indicating the triggering of anti-viral response by IFN-α. Furthermore, HCV replication strongly induced the ISGs – LAMP3, OAS1 and ETV7 (Fig. 7B). A marginal but significant upregulation of RASGRP3, TMEM140 and TRANK1 genes were also observed in HCV infected cells indicating an indirect or direct role of these genes in innate immune response. Furthermore, we carried out HCV replication experiment in human hepatoma cell line (Huh-7.5.1) as an additional control. The results indicated similar upregulation of IFN-stimulated genes with the exception of OAS1 (Fig. 7C, 7D and 7E). We also observed a mild induction of LAMP3 in HCV polymerase-null genomic RNA transfected cells suggesting that the innate immune system can be activated by viral PAMP in the non-functional genomic RNA to a lesser extend (Fig. 7E). The ISG induction by HCV replication in Huh-7.5.1 cells is likely through a RIG-I independent manner, as the parental clone of Huh-7.5.1 line lacks RIG-I (Sumpter, Loo et al. 2005). Taken together, the hepatic cells derived from H9 hESC retained an intact innate immune system that is capable of sensing viral RNA and mounting anti-viral response.
Figure 7.
Functional validation of ISGs during hepatitis C virus replication. (A) HCV genome level was quantified by RT-qPCR. 4 hour time point indicates the equal amount of transfected RNA in IFN-α untreated (HCV) and treated (HCV-IFN) cells. At 96 hour-post transfection, the HCV replication was enhanced, but was inhibited in IFN-α treated cells. (B) HCV replication resulting in upregulation of ISGs. (C) Western blot analysis of HCV NS3 protein production during viral replication. Protein lysates from mock transfected control cells, as well as HCV polymerase null (Pol) and HCV wild type (WT) genomic RNA transfected cells at 96 hour post-transfection were used. β-actin was included as loading control. (D) HCV replication level in Huh-7.5.1 cells is presented in the bar graph. The cells were transfected with 1 μg of viral genomic RNA. At 96 hour-post transfection, the HCV genome level was quantified and the fold change was calculated to that of control cells (mock transfected). (E) The ISGs induced in Huh-7.5.1 cells upon HCV replication at 96 hour-post transfection are shown in the bar graph.
Discussion
We utilized an unbiased RNA sequencing approach to profile and characterize IFN-stimulated genes during hepatic differentiation of pluripotent stem cells. Our transcriptome analysis mainly focused on protein-coding genes and approximately 20,441 genes were expressed. Upon INF-α treatment, about 438 genes were significantly induced at day 21 cells including 83 canonical ISGs. Transcription of 42 ISGs were significantly changed by IFN-α in all three differentiation time points. Our data suggested that the hESC-derived endoderm and hepatic cells can positively respond to IFN, albeit with differences in gene expression levels.
Murine embryonic stem cells have been shown to be responsive to IFN-α and IFN-β (Whyatt, Duwel et al. 1993). The undifferentiated hESC and iPSC showed an attenuated type I interferon response that correspond to high expression of SOCS1, a suppressor of cytokine signaling factor (Hong and Carmichael 2013). Here, we demonstrate that the endodermal phase cells are responsive to type I IFN. These in vitro observations suggest that during in utero the developing embryo can respond to the invading viral pathogens by up-regulating the innate defense system.
In the course throughout differentiation of hESCs, different cocktails of cytokines and growth factors were used to achieve each phase of hepatic lineage derivation (Fig. 1A). During IFN-α treatment, these cytokine-infused cocktails could influence the gene expression profile by altering the activation of respective signaling pathways. For instance, oncostatin M used during day 16 to 21 of differentiation can bind to heterodimeric cell surface receptors- gp130 and either leukemia inhibiting factor receptor (LIFR) or OSM receptor-beta (OSMR-beta) resulting in activation of JAK-STAT, MAPK, and PI3K/AKT pathways (Heinrich, Behrmann et al. 1998; Van Wagoner, Choi et al. 2000; Arita, South et al. 2008; Dey, Radhakrishnan et al. 2013). This variable was taken into account while calculating gene transcription specifically altered upon IFN treatment by comparing between IFN-α untreated (control) and treated cells of each endoderm, hepatoblast and immature hepatocyte phases. The canonical ISGs, including OAS1, ISG15 and MX1 were activated in all three phases; whereas, the new ISGs TRANK1 and CLMP had a phase-specific expression profile (Fig. 5). This observation could be biologically relevant as various cell or tissue types subjected to differential epigenetic control to accommodate specific functions.
Genes induced by IFN-α in various cell types have been profiled previously (Der, Zhou et al. 1998; Lanford, Guerra et al. 2006; Liu, Sanchez et al. 2012). Microarray analysis of primary human hepatocytes following IFN-α treatment revealed the induction of LAMP3, OAS1, STAT1, GBP1, and several other ISGs (Lanford, Guerra et al. 2006). For validation of RNA sequencing data, we specifically focused on genes activated by IFN-α. LAMP3 or CD208 regulates the transcription of anti-viral genes (Mohty, Vialle-Castellano et al. 2003; Mine, Shulzhenko et al. 2013). LAMP3 is involved in the replication of influenza virus and its expression is induced upon influenza viral infection (Zhou, Xue et al. 2011). In the present study, induction of LAMP3 upon HCV infection suggests that the hESC-derived hepatic cells have the ability to mount anti-viral response. LAMP3 can provide positive feedback signal to augment the IFN-α mediated anti-viral response.
ETV7 or TEL2, a transcription factor, may involve in hematopoietic development and oncogenic transformation (Potter, Buijs et al. 2000; Carella, Potter et al. 2006). ETV7 expression had been shown to be induced in IFN-α treated monocytes and HIV-1 infected cells (Rempel, Calosing et al. 2008). We observed that ETV7 was highly induced upon HCV replication. The role of ETV7 during viral infection or IFN-mediated antiviral activity is unknown, but may be attributed to regulating cell survival. RASGRP3 is a guanyl-nucleotide exchange factor which promotes the active GTP-bound form of Ras. A recent report observed that the RASGRP3 acts as a negative regulator of inflammatory response by macrophages through activating Rap1 (Tang, Chen et al. 2014). Induction of RASGRP3 during HCV infection may be a cellular response to mitigate inflammatory damage while mounting anti-viral defense. TRANK1 is a conserved gene in mammals with no known function. The protein contains tetratricopeptide and ankyrin repeats with a predicted hydrolase activity. Our results indicate that TRANK1 is a novel ISG. TRANK1 mechanism of action in the context of IFN-α signaling pathway and HCV infection requires further investigation.
Host-pathogen interaction is a balancing act as viruses evolved to have strategies to evade the host defense system. HCV NS3, core and NS5A proteins are involved in immune evasion by modulating specific and innate immune pathways (Basu, Meyer et al. 2001; Horner and Gale 2013). NS3 protease can effectively inactivate RIG-I by proteolysis, thus negating the RIG-I mediated sensing of HCV RNA and activation of downstream IRF-3 and NF-kB effectors (Korth and Katze 2000; Gale and Foy 2005). Besides IFN-α, IL28 or IFN-λ, a type III IFN has been shown to inhibit HCV replication by inducing ISGs (Thomas, Gonzalez et al. 2012; Zhou, Sun et al. 2014). Robust viral production is observed in sub-clones of Huh-7 cell line that lack the innate immune sensor RIG-I and Huh-7 based cell lines have been commonly used for HCV studies (Lindenbach, Evans et al. 2005; Sumpter, Loo et al. 2005; Wakita, Pietschmann et al. 2005; Pietschmann, Kaul et al. 2006; Chu, Ren et al. 2013). Pluripotent stem cell-derived hepatic cells is a useful model system for studying the interaction of innate immune factors with hepatic pathogens, including HCV, hepatitis B virus (HBV) and dengue virus. A previous study reported that the iPSC Hepatocytes-like cells (iHLC) in response to HCV infection up-regulated antiviral inflammatory genes (Schwartz, Trehan et al. 2012). To further improve the utility of human pluripotent stem cell lines for modeling patho-physiological processes of innate immune system, individual or combination of ISG knock-out cell lines could be established. Recent advances in gene editing technologies through designer nucleases, Zinc-finger nucleases (ZFN) and Transcription activator-like effector nucleases (TALENs) as well as CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system, facilitate precise editing of human genome by double-strand DNA breaks (Ding, Lee et al. 2013; Ding, Regan et al. 2013; Ran, Hsu et al. 2013).
Utilizing human pluripotent stem cell lines for modeling processes of innate immune system can expand beyond studying viral infection; furthermore, it can support human clinical trials by evaluating the cell candidate population for transplant by measuring the presence of an intact immune response to improve clinical outcome. For instance, retinal pigment epithelium cells derived from iPSCs are currently being evaluated in human clinical trials for an eye disorder and several other stem cell-based preclinical studies are poised for human testing (Trounson, Thakar et al. 2011; Riken 2013). Due to immuno-suppressive treatment following allogeneic cell transplant and natural exposure to pathogens in the environment, the recipients have the risk of complications from infectious diseases. To increase the chance of better therapeutic outcome, the cell candidate for transplant can be evaluated for presence of functional innate immune system.
Conclusion
Here, we describe the first comprehensive analysis of the type I IFN-induced gene profile of endoderm and hepatic lineage cells derived from pluripotent stem cells. The endodermal cells, hepatoblast and hepatocyte-like cells contained active type I IFN signaling pathway which exhibited differential responses to IFN-α treatment. We identified novel non-canonical ISGs such as RASGRP3, TRANK1 and CLMP. The hESC-derived hepatic cells infected with HCV responded with upregulation of innate immunity genes LAMP3, OAS1, ETV7, RASGRP3, and TRANK1 indicating that the novel ISGs may play a role in antiviral response. The transcriptome profile of various stages of hepatic differentiation in combination with IFN-α treatment is a valuable resource for understanding its basic biology as well as host-pathogen interactions.
Supplementary Material
Highlights.
Human embryonic stem cells (hESCs) were efficiently differentiated to endodermal and hepatic lineage cells.
Interferon-stimulated genes were profiled by RNA sequencing method at various stages of hepatic differentiation of hESCs.
Profiled canonical and novel ISGs had differentiation-stage specific expression pattern.
Confirmed the biological relevance of novel ISGs during hepatitis C virus infection of hESC-derived hepatic cells.
Acknowledgments
Huh-7.5.1 cell line was a gift from Francis V. Chisari (The Scripps Research Institute, USA). This work was funded by Cedars-Sinai Medical Center's Institutional Research Award and National Center for Advancing Translational Sciences, Grant UL1TR000124 to V.A and NIH R01DK090794 grant to S.W.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler D, Murdoch D. rgl: 3D visualization device system (OpenGL). R package version 0.94.1143. 2014 from https://r-forge.r-project.org/projects/rgl/
- Anderson SL, Carton JM, et al. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256(1):8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- Arita K, South AP, et al. Oncostatin M receptor-beta mutations underlie familial primary localized cutaneous amyloidosis. Am J Hum Genet. 2008;82(1):73–80. doi: 10.1016/j.ajhg.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Kim CN, et al. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17(23):6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Meyer K, et al. Hepatitis C virus core protein modulates the interferon- induced transacting factors of Jak/Stat signaling pathway but does not affect the activation of downstream IRF-1 or 561 gene. Virology. 2001;288(2):379–390. doi: 10.1006/viro.2001.1100. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhao Y, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45(5):1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- Carella C, Potter M, et al. The ETS factor TEL2 is a hematopoietic oncoprotein. Blood. 2006;107(3):1124–1132. doi: 10.1182/blood-2005-03-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Tseng CY, et al. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55(4):1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ying L, et al. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell. 2012;10(4):371–384. doi: 10.1016/j.stem.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Ren S, et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J Virol. 2013;87(10):5678–5696. doi: 10.1128/JVI.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cufflinks Cufflinks. 2014 from http://cole-trapnell-lab.github.io/cufflinks/
- Darnell JE, Jr., Kerr IM, et al. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der SD, Zhou A, et al. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey G, Radhakrishnan A, et al. Signaling network of Oncostatin M pathway. J Cell Commun Signal. 2013;7(2):103–108. doi: 10.1007/s12079-012-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Ding Q, Lee YK, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12(2):238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukker M, Katz G, et al. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci U S A. 2002;99(15):9864–9869. doi: 10.1073/pnas.142298299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Catana A, et al. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells. 2007;25(12):3058–3068. doi: 10.1634/stemcells.2007-0291. [DOI] [PubMed] [Google Scholar]
- Gale M, Jr., Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436(7053):939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Haque SJ, Williams BR. Signal transduction in the interferon system. Semin Oncol. 1998;25(1 Suppl 1):14–22. [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, et al. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334(Pt 2):297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong XX, Carmichael GG. Innate immunity in pluripotent human cells: attenuated response to interferon-beta. J Biol Chem. 2013;288(22):16196–16205. doi: 10.1074/jbc.M112.435461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Gale M., Jr. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19(7):879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM. The Jak-STAT pathway stimulated by interferon alpha or interferon beta. Sci STKE. 2004;2004(260):tr10. doi: 10.1126/stke.2602004tr10. [DOI] [PubMed] [Google Scholar]
- IPA. Ingenuity Pathway Analysis. 2014 from www.qiagen.com/ingenuity.
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147(927):258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- Kellis M, Wold B, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111(17):6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DS, Veals SA, et al. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4(10):1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- Kochs G, Haller O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci U S A. 1999;96(5):2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth MJ, Katze MG. Evading the interferon response: hepatitis C virus and the interferon-induced protein kinase, PKR. Curr Top Microbiol Immunol. 2000;242:197–224. doi: 10.1007/978-3-642-59605-6_10. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, et al. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43(5):961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- Li X, Leung S, et al. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271(10):5790–5794. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11(10):688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kim Y, et al. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3(82):82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Sanchez DJ, et al. Systematic identification of type I and type II interferon- induced antiviral factors. Proc Natl Acad Sci U S A. 2012;109(11):4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine KL, Shulzhenko N, et al. Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer. Nat Commun. 2013;4:1806. doi: 10.1038/ncomms2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohty M, Vialle-Castellano A, et al. IFN-alpha skews monocyte differentiation into Toll-like receptor 7-expressing dendritic cells with potent functional activities. J Immunol. 2003;171(7):3385–3393. doi: 10.4049/jimmunol.171.7.3385. [DOI] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 from http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Perry AK, Chen G, et al. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15(6):407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- Pietschmann T, Kaul A, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103(19):7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter MD, Buijs A, et al. Identification and characterization of a new human ETS- family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood. 2000;95(11):3341–3348. [PubMed] [Google Scholar]
- Ran FA, Hsu PD, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid ST, Corbineau S, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120(9):3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel H, Calosing C, et al. Sialoadhesin expressed on IFN-induced monocytes binds HIV-1 and enhances infectivity. PLoS One. 2008;3(4):e1967. doi: 10.1371/journal.pone.0001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Contreras D, et al. A protocol for analyzing hepatitis C virus replication. J Vis Exp. 2014;(88):e51362. doi: 10.3791/51362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Irudayam JI, et al. Bioartificial Liver Device Based on Induced Pluripotent Stem Cell-Derived Hepatocytes. Journal of Stem Cell Research & Therapy. 2015;05(02) [Google Scholar]
- Riken Pilot clinical study into iPS cell therapy for eye disease starts in Japan. 2013 [Google Scholar]
- Saito T, Owen DM, et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, Ebert AD, et al. Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS One. 2012;7(6):e39113. doi: 10.1371/journal.pone.0039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SW, Heathcote J, et al. Lamivudine and alpha interferon combination treatment of patients with chronic hepatitis B infection: a randomised trial. Gut. 2000;46(4):562–568. doi: 10.1136/gut.46.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Bladt F, et al. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373(6516):699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RE, Trehan K, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109(7):2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, et al. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr., Loo YM, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79(5):2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SL, Katze MG. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J Interferon Cytokine Res. 1999;19(6):543–554. doi: 10.1089/107999099313677. [DOI] [PubMed] [Google Scholar]
- Tang S, Chen T, et al. RasGRP3 limits Toll-like receptor-triggered inflammatory response in macrophages by activating Rap1 small GTPase. Nat Commun. 2014;5:4657. doi: 10.1038/ncomms5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, et al. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Thomas E, Gonzalez VD, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142(4):978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007;85(6):435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A, Thakar RG, et al. Clinical trials for stem cell therapies. BMC Med. 2011;9:52. doi: 10.1186/1741-7015-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara Y, Minowa O, et al. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373(6516):702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- Van Wagoner NJ, Choi C, et al. Oncostatin M regulation of interleukin-6 expression in astrocytes: biphasic regulation involving the mitogen-activated protein kinases ERK1/2 and p38. J Neurochem. 2000;75(2):563–575. doi: 10.1046/j.1471-4159.2000.0750563.x. [DOI] [PubMed] [Google Scholar]
- Veals SA, Santa Maria T, et al. Two domains of ISGF3 gamma that mediate protein- DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol Cell Biol. 1993;13(1):196–206. doi: 10.1128/mcb.13.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, et al. gplots: Various R programming tools for plotting data. R package version 2.14.2. 2014 [Google Scholar]
- Weinstein M, Monga SP, et al. Smad proteins and hepatocyte growth factor control parallel regulatory pathways that converge on beta1-integrin to promote normal liver development. Mol Cell Biol. 2001;21(15):5122–5131. doi: 10.1128/MCB.21.15.5122-5131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt LM, Duwel A, et al. The responsiveness of embryonic stem cells to alpha and beta interferons provides the basis of an inducible expression system for analysis of developmental control genes. Mol Cell Biol. 1993;13(12):7971–7976. doi: 10.1128/mcb.13.12.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SJ, Schoggins JW, et al. Inhibition of HIV-1 particle assembly by 2′,3′- cyclic-nucleotide 3′-phosphodiesterase. Cell Host Microbe. 2012;12(4):585–597. doi: 10.1016/j.chom.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Robotham JM, et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8(4):e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sun P, et al. Modulating innate immunity improves hepatitis C virus infection and replication in stem cell-derived hepatocytes. Stem Cell Reports. 2014;3(1):204–214. doi: 10.1016/j.stemcr.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Xue Q, et al. Lysosome-associated membrane glycoprotein 3 is involved in influenza A virus replication in human lung epithelial (A549) cells. Virol J. 2011;8:384. doi: 10.1186/1743-422X-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.