Abstract
Purpose
It is well established that breast cancer stem cells (BCSCs) play an essential role in tumor invasion for both local and distant metastasis. The aim of this study was to establish whether BCSCs could act as a prognostic and clinical marker.
Methods
We analyzed tumor tissues from 161 breast cancer patients. Dual immunohistochemistry and immunofluorescence were performed on all the slides, and we analyzed the relationship between EpCAM-/CD49f+ tumor cells and key clinical and prognostic factors.
Results
Univariate survival analysis using the Kaplan-Meier method showed that the presence of EpCAM-/CD49f+ tumor cells in breast cancer was significantly associated with shorter disease-free survival (DFS) and overall survival (OS). Multivariate analysis showed that the presence of EpCAM-/CD49f+ cells was associated with shorter DFS (p=0.010; hazard ratio [HR], 2.070) and OS (p=0.002; HR, 3.235). Tumors containing EpCAM-/CD49f+ cells were also more likely to metastasize after initial surgery (p=0.048).
Conclusion
Our study suggests that breast tumors containing EpCAM-/CD49f+ cells are more likely to undergo distant metastasis after initial surgery and are associated with a shorter DFS and OS.
Keywords: Breast neoplasms, Neoplasm metastasis, Neoplastic stem cells, Prognosis
INTRODUCTION
One of the most important challenges in the treatment of breast cancer patients is recurrence and metastasis after initial surgery, and an early assessment of metastatic risk and the likelihood of treatment response are important in individualized therapy. Evidence from both in vitro and in vivo experiments suggests that breast tumors are highly heterogeneous, and that cancer cells can have diverse phenotypes and properties [1]. As certain subpopulations of cells might have a particularly high potential to cause recurrence and distant metastasis, elucidating their properties could provide a better understanding of these processes.
One possible explanation for the heterogeneous but hierarchical state of breast tumors is the cancer stem cell theory, which suggests that only a subgroup of breast cancer cells have tumor-initiating ability, and these are often referred to as breast cancer stem cells (BCSCs). Cancer stem cells have been identified in almost all cancers [2,3,4,5,6,7,8,9], and have been found to enhance tumorigenicity, self-renewal, and multilineage differentiation capabilities. These features allow de novo tumor formation through the recapitulation of the whole tumor population from a single cancer stem cell [10]. In addition, BCSCs more frequently undergo epithelial to mesenchymal transition (EMT) [11], and have an innate resistance to cytotoxic treatment [12]. Their very high malignant potential makes it difficult to completely eradicate residual cancer cells during treatment, and they are now usually considered a major source of recurrence and distant metastasis [11]. Clinical biomarkers for these cells may have clinical, prognostic, and therapeutic significance.
Many potential molecular phenotypes of BCSCs have been reported, the first of which was CD44+/CD24-/low/ESA+ together with negative staining for CD2, CD3, CD10, CD16, CD18, CD31, CD64, and CD140b (lineage-) in 2003 by Al-Hajj [2]. As few as 200 of these cells were able to form tumors after xenotransplantation into nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice, whereas the transplantation of thousands of other malignant epithelial cells did not result in tumor growth [13]. CD49f and epithelial cell adhesion molecule (EpCAM, also referred to as CD326 or epithelial-specific antigen) have been widely used as differentiation markers of normal and tumor gland cells [14]. The mammary regeneration capacity of EpCAMlow/CD49f+ cells was demonstrated by a transplantation study using NOD/SCID mice, and its clonogenic activity was confirmed by in vitro studies [15]. While an EpCAMhigh/CD49f- phenotype represents differentiated luminal cells, the EpCAMlow/CD49f+ subpopulation was enriched for bipotent mammary stem cell (MaSC) progenitors [15].
As the EpCAMlow/CD49f+ subpopulation is enriched for MaSC progenitors and shows mammary regeneration capacity in NOD/SCID mice, these cells are considered to have the capacity to differentiate into both mature luminal and myoepithelial cells. In this study, we labeled EpCAM-/CD49f+ cancer cells and evaluated their clinical, pathological, and therapeutic significance.
METHODS
Patients and sample preparation
We studied 161 randomized breast tumor tissues samples from a cohort of 276 patients who were diagnosed with breast cancer, and who underwent surgery to have the tumor removed in the West China Hospital between 2006 and 2009. Disease-free survival (DFS) and overall survival (OS) were defined as the time between the initial surgery and local or distant metastatic relapse, and between surgery and death, respectively. We prepared tissue microarray (TMA) cores 1.5 mm in diameter from the formalin-fixed paraffin-embedded samples. Two cores from each tumor were incorporated into the array. Approval for the study was granted by the Ethics Committee of West China Hospital (number: 2013-191).
Dual immunohistochemistry and immunofluorescence staining
Dual immunohistochemistry (IHC) and immunofluorescence (IF) staining were performed as previously described [13]. IHC was performed using antibodies against EpCAM (TA310957, 1:300; OriGene, Rockville, USA) and CD49f (TA506627, 1:150; OriGene) with an Autostainer with the EnVision G|2 Double Stain System ((DAKO company, Code K5361, Glostrup, Denmark) Rabbit/Mouse (DAB+/Permanent Red) according to the manufacturer's instructions. EpCAM was detected with Permanent Red and CD49f was detected using diaminobenzidene (DAB).
For IF staining, primary antibodies against EpCAM (TA310957, 1:300; OriGene) and CD49f (TA506627, 1:150; OriGene) were used. Secondary antibodies with coordinate species conjugated to either Alexafuor 488 (4412S, 1:200; CST, Boston, USA) or 546 (21206, 1:200; Invitrogen, Life Technologies, Eugene, USA) were used for detection. Nuclei were identified by staining with 4', 6-diamidino-2-phenylindole (DAPI, D9542-5MG, 1:10,000; Sigma-Aldrich, Carlsbad, USA). Finally, the slides were washed and covered with mounting media. Images were captured with a Zeiss LSM laser microscope.
In invasive ductal carcinoma (IDC) samples, EpCAM-positive cells were defined as those that stained positively with permanent red at any intensity. CD49f-positive cells were defined as those that were DAB positive at any intensity. The proportion of EpCAM-/CD49f+ tumor cells was determined as the percentage of cells negative for permanent red staining but positive for DAB staining. Likewise, the percentage of EpCAM+/CD49f+ cells and EpCAM+/CD49f- cells were estimated. For IF staining, EpCAM was stained with a secondary antibody conjugated with Alexa Fluor 546 (red) and CD49f was recognized by green fluorescence.
Tumor tissue histological analysis
Hematoxylin and eosin (H&E) and IHC slides were assessed by pathologists at West China Hospital. The epithelial growth factor receptor, cytokeratin 5/6 (CK5/6), estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) status of each tumor was obtained from the patient's pathology report. HER2 staining was analyzed according to the American Society of Clinical Oncology guidelines.
CD49f staining was mainly cytoplasmic, and dual IHC staining and counting was performed as described for CD24 [16]. The definitions used for the different breast cancer molecular subtypes were as follows: luminal A like (ER positive [ER+] and/or PR positive [PR+], and HER2 negative [HER2-]); luminal B like (ER+ and/or PR+, HER2+); basal-like (ER-, PR-, HER2-, cytokeratin 5/6 positive, and/or HER1+); HER2+/ER- (ER-, PR-, HER2+), and unclassified (negative for all five markers).
Statistical analysis
Associations between the presence of different EpCAM/CD49f phenotypes and clinical variables as well as breast cancer subgroups were assessed using the Pearson chi-square and Fisher exact test. The Kaplan-Meier method was used to estimate DFS and OS, and the log-rank test was used to compare survival between groups. All tests were two-sided and p<0.05 was considered statistically significant. Multivariate survival analysis was performed using the Cox proportional hazard model. All statistical tests were carried out using SPSS version 16.0 software (SPSS Inc., Chicago, USA), except for Kaplan-Meier survival curves, which were generated using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, USA).
RESULTS
Prevalence of EpCAM-/CD49f+ tumor cells
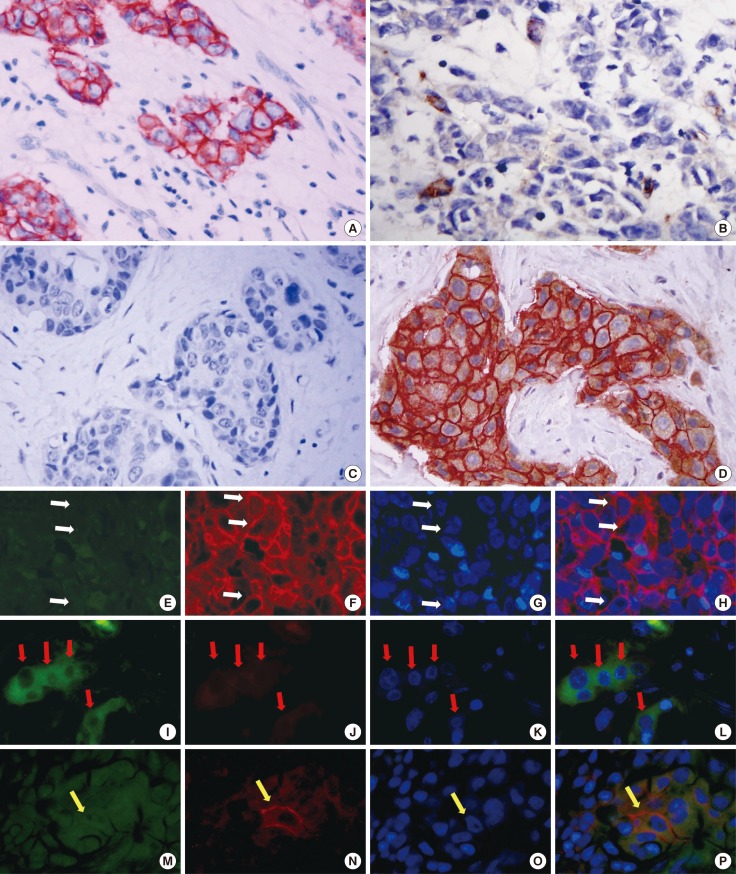
To identify putative BCSCs, we performed dual-IHC (Figure 1A-D) and IF staining (Figure 1E-P) on the TMA samples, in addition to single IHC and IF staining (using only one antibody) as a quality control. In breast tumor cells, EpCAM was predominantly expressed on the membrane, whereas CD49f was mainly present in cytoplasm, and both proteins showed distinct expression patterns in inter- and intratumor areas. Some tumor cells only expressed EpCAM (Figure 1A and Figure 1E-H, white arrow), some only expressed CD49f (Figure 1B and Figure 1I-L, red arrow), some expressed neither protein (Figure 1C), and some expressed both proteins (Figure 1D and Figure 1M-P, yellow arrow). EpCAM/CD49f expression was assessed in tumor samples from all 161 breast cancer patients. The proportion of tumor cells that were EpCAM-/CD49f+ ranged between 0% and 80% (data not shown).
Figure 1. Dual-immunohistochemistry (IHC) and immunofluorescence (IF) staining on tissue microarray (TMA) slides. Dual-IHC and IF staining with primary antibodies of EpCAM and CD49f were performed on TMA tissue. (A) Patients only contain EpCAM-positive cancer cells (membrane); (B) Patient show only CD49f-positive cancer cells (cytoplasm); (C) Case express none or both (D) biomarkers. In IF staining (E-P), green signal shows the expression of CD49f, red signal shows EpCAM with DAPI staining for nuclei. (E-H) One case that tumor cell expression only EpCAM on the membrane (white arrows); (I-L) One case that tumor cell express only CD49f (red arrow); (M-P) One case with tumor cells express both (yellow arrow) (for both IHC and IF, magnification is actually ×400).
Baseline clinical characteristics
Samples from only 161 of the 276 patients (58%) could be incorporated into the TMA for dual IHC and IF staining. Owing to insufficient clinical data and incomplete follow-up records, the clinicopathological characteristics of only 150 patients could be analyzed, all of whom were women, with a mean age of 49.7±9.05 years (range, 29-77 years). The majority of these patients (148/150, 98.7%) were diagnosed with IDC. The median follow-up time was 73.2 months, and the median DFS was 64.8 months. The significance of clinicopathological characteristics and prognostic factors was assessed using the Kaplan-Meier analysis and the log-rank test. As excepted, age (DFS, p=0.038), Histologic grade (DFS, p=0.014; OS, p=0.043), tumor size (DFS, p=0.022), nodule status (DFS, p<0.001; OS, p<0.001), PR status (DFS, p=0.014; OS, p=0.043), distant metastasis (DFS, p<0.001; OS, p<0.001), and recurrence (DFS, p<0.001; OS, p<0.001) were significant prognostic factors for a poor clinical outcome (Table 1).
Table 1. Baseline clinical characteristics.
| Characteristic | No. (%)* | DFS | OS | ||
|---|---|---|---|---|---|
| Log-rank | p-value | Log-rank | p-value | ||
| Histology | 150 | 0.001 | 0.981 | 0.652 | 0.419 |
| IDC | 148 (98.7) | ||||
| Others | 2 (3.3) | ||||
| Age (yr) | 150 | 4.327 | 0.038 | 2.908 | 0.088 |
| <45 | 48 (32.0) | ||||
| ≥45 | 102 (68.0) | ||||
| Histologic grade | 150 | 8.567 | 0.014 | 6.259 | 0.043 |
| I | 3 (2.0) | ||||
| II | 51 (34.0) | ||||
| III | 96 (64.0) | ||||
| T staging | 150 | 11.667 | 0.022 | 3.347 | 0.501 |
| Tis-1 | 39 (26.0) | ||||
| T2 | 84 (56.0) | ||||
| T3 | 19 (12.7) | ||||
| T4 | 11 (5.3) | ||||
| Nodal status | 150 | 20.582 | <0.001 | 22.502 | <0.001 |
| N0 | 61 (40.7) | ||||
| N1 | 38 (25.3) | ||||
| N2 | 31 (20.1) | ||||
| N3 | 20 (13.3) | ||||
| Metastasis | 150 | 0.503 | 0.478 | 0.391 | 0.532 |
| M0 | 145 (96.1) | ||||
| M1 | 5 (3.9) | ||||
| Menopause | 150 | 0.011 | 0.916 | 0.708 | 0.400 |
| Yes | 79 (52.7) | ||||
| No | 71 (47.3) | ||||
| ER status | 149 | 0.594 | 0.441 | 1.066 | 0.302 |
| Positive | 97 (65.3) | ||||
| Negative | 52 (34.7) | ||||
| PR status | 149 | 7.176 | 0.007 | 4.252 | 0.039 |
| Positive | 53 (35.5) | ||||
| Negative | 96 (64.5) | ||||
| HER2 status | 149 | 5.610 | 0.132 | 7.586 | 0.055 |
| Positive | 22 (17.1) | ||||
| Negative | 97 (75.2) | ||||
| Distant metastasis | 150 | 66.310 | <0.001 | 156.664 | <0.001 |
| Yes | 51 (33.4) | ||||
| No | 99 (66.6) | ||||
| Recurrence | 150 | 16.707 | <0.001 | 38.615 | <0.001 |
| Yes | 11 (8.3) | ||||
| No | 139 (91.7) | ||||
DFS=disease-free survival; OS=overall survival; IDC=invasive ductal carcinoma; ER=estrogen receptor; PR=progesterone receptor; HER2=human epithelial growth factor receptor 2.
*Number differences reflect missing data.
Relationship between the presence of EpCAM-/CD49f+ tumor cells and histopathological characteristics
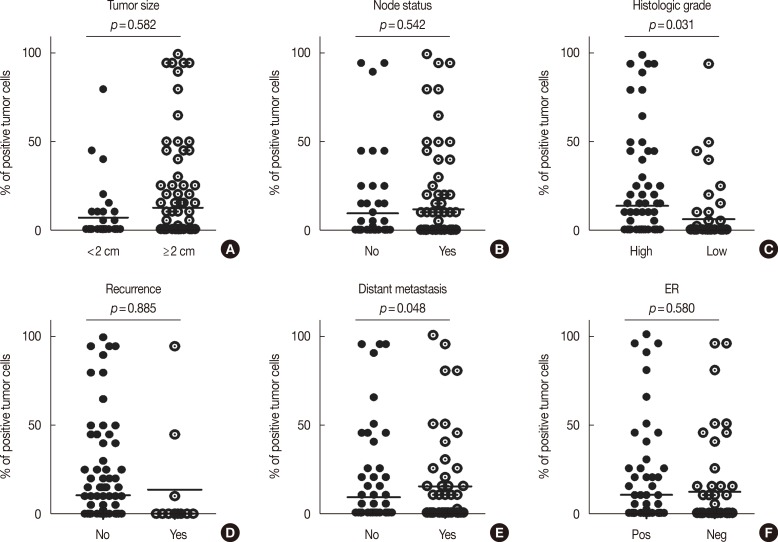
We also evaluated the relationship between EpCAM-/CD49f+ cells in IDC with specific tumor histopathological characteristics such as age, grade, tumor size, nodal status, ER status, PR status, distant metastasis, and recurrence (Figure 2). The presence of EpCAM-/CD49f+ cancer cells in the tumor was significantly associated with a higher tumor grade (p=0.031) (Figure 2C), and a higher probability of distant metastasis (p=0.048) (Figure 2E). Tumor size, node status, recurrence, and ER and PR expression status were not associated with the presence of EpCAM-/CD49f+ cancer cells in the tumor. We also assessed the relationship between the presence of EpCAM/CD49f cells in tumors and the breast cancer molecular subtype (as classified by IHC), although no significant associations were found (data not shown).
Figure 2. Prevalence of EpCAM-/CD49f+ with histopathologic characteristics. Analyzing of 150 breast cancer samples stratified to tumor size (A), node status (B), tumor grade (C; low=grade I and II, high=grade III), recurrence (D), distant metastasis (E), and estrogen receptor (ER) expression (F). Tumor containing EpCAM-/CD49f+ are associated with higher tumor grade (C; p=0.031) and distant metastasis (E; p=0.048). Chi-square exact and Fisher exact test were performed.
EpCAM-/CD49f+ tumor cells and breast cancer prognosis
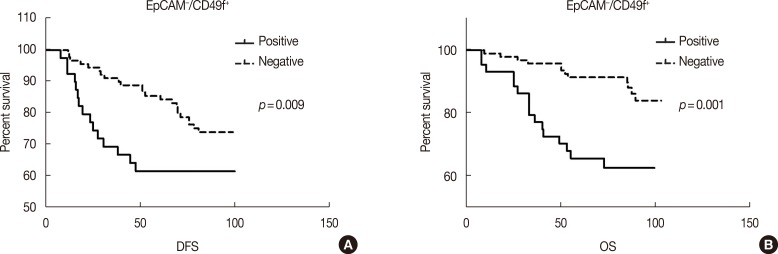
To determine whether the presence of EpCAM-/CD49f+ cells in breast tumors was associated with disease progression (recurrence and distant metastasis) and patient survival, we performed univariate and multivariate survival analyses on the entire cohort. In univariate survival analysis the presence of EpCAM-/CD49f+ tumor cells had a significant, negative association with both DFS and OS (p=0.009 and p=0.001, respectively) (Supplementary Table 1, available online, and Figure 3). EpCAM+/CD49f-, EpCAM+/CD49f+ and EpCAM-/CD49f- cells were evaluated as a control, and showed no significant association with DFS or OS in univariate survival analysis (Supplementary Table 1, Supplementary Figure 1, available online).
Figure 3. Univariate survival analysis of EpCAM-/CD49f+ with disease-free survival (DFS) and overall survival (OS). All the patients' tumor samples were stained with EpCAM and CD49f. (A, B) The present of EpCAM-/CD49f+ tumor cells have significant negative association with both DFS and OS (EpCAM-/CD49f+: pDFS=0.009; pOS=0.001).
In the multivariate analysis using Cox proportional hazard models, age, tumor grade, tumor size, and nodal, ER, PR, and HER2 status were included as covariates for the presence of EpCAM/CD49f cells. EpCAM-/CD49f+ cells were predictive of shorter DFS (p=0.010; hazard ratio [HR], 2.070; 95% confidence interval [CI], 1.188-3.608) and OS (p=0.020; HR, 3.235; 95% CI, 1.551-6.748) (Supplementary Table 2, available online). EpCAM+/CD49f+, EpCAM+/CD49f-, and EpCAM-/CD49f- cells were analyzed as a control. Neither EpCAM+/CD49f- nor EpCAM-/CD49f- cells were prognostic factors for DFS and OS, although the presence of EpCAM+/CD49f+ cells was associated with a shorter OS.
DISCUSSION
Metastasis is the main cause of breast cancer treatment failure and death [10], and there has been extensive research aimed at understanding the mechanism of metastasis. This has allowed more accurate prognoses and more individualized treatments. As breast cancer is highly heterogeneous at both the histological and molecular level, different cancer cells within the same tumor may have distinct characteristics [17]. The specific properties of a tumor, for example EMT, invasion, growth, and local and distant metastasis, might largely depend on tumor cell subpopulations with high EMT activity, a rapid cell cycle, and invasive characteristics.
It is now well established that integrins are involved in cell adhesion, migration, polarity, survival, growth, and death [18,19]. CD49f (integrin α6), in particular, has been associated with breast cancer aggressiveness in many studies [20,21]. CD49f expression is also increased in other malignancies, such as melanoma, esophageal squamous cell carcinoma, head and neck carcinoma, and prostate cancer [22,23,24,25]. Haraguchi et al. [26] reported that CD49f positive cells could initiate colon tumor growth. In our multivariate analysis of data including age, tumor grade, and tumor size, and nodal, ER, PR, and HER2 status, the presence of EpCAM-/CD49f+ cells had significant prognostic value for OS.
EpCAM is a 30- to 40-kDa type I membrane glycoprotein. It is expressed in a variety of human epithelial tissues, cancers, and progenitor and stem cells, and functions as a homotypic intercellular adhesion molecule. In tumor tissues, it is reported that the adhesive properties of EpCAM might prevent metastasis because intercellular adhesion should limit the ability of cells to migrate [27]. EpCAM has also been identified as a marker for cancer-initiating stem cells, which makes it an even more interesting target for cancer therapy [28]. CD49f and EpCAM expression is frequently used to identify the development of normal gland cells [14]. Previous reports indicated that luminal cells express EpCAM, and that differentiated luminal cells have an EpCAMhigh/CD49f- phenotype. While the EpCAMhigh/CD49f+ cell population contains "luminal progenitors," the EpCAM-/low/CD49f+ phenotype characterizes MaSC progenitors. This subgroup can differentiate into both mature luminal and myoepithelial cells. Thus, we propose that EpCAM-/low/CD49f+ cells may be enriched in BCSC populations, and hence, the EpCAM-/low/CD49f+ phenotype is a characteristic of cancer stem cells in the tumor, and may also predict metastasis and have prognostic and therapeutic value [15]. Treatment resistance is an important challenge in breast cancer. Putative cancer stem cells are widely accepted as mediators of resistance to clinical treatment [29], and likewise, BCSCs are reported to have an innate resistance to cytotoxic agents and other clinical treatments [12]. We therefore speculate that the putative BCSCs may act as a source of cells that prevents the complete eradication of tumors by standard anticancer therapies. More importantly still, BCSCs may have significant prognostic value and help guide personalized treatment plans.
In conclusion, we identified EpCAM-/low/CD49f+ cells within BCSC populations and assessed their relationship with clinicopathological parameters and clinical outcomes in breast cancer patients. Our findings suggests that breast tumors containing EpCAM-/CD49f+ or CD49f+ cancer cells are associated with a higher probability of distant metastasis after initial surgery, and poor clinical outcomes with respect to both DFS and OS.
Footnotes
This work was funded by grants from the National Natural Science Foundation of China (31000601, 81200461), and Young Investigator Scholar in Sichuan University (2012SCU04A14).
CONFLICT OF INTEREST: The authors declare that they have no competing interests.
Supplementary Material
Univariate survival analysis of EpCAM/CD49f with disease-free survival (DFS) and overall survival (OS). (A-F) All the patients' tumor samples were stained with EpCAM and CD49f. The prevalence of EpCAM+/CD49f+, EpCAM+/CD49f-, and EpCAM-/CD49f- were analyzed as control, and no significant association were checked with DFS and OS.
Supplementary Table 1. Correlation of prevalence of EpCAM/CD49 with breast cancer prognosis (univariate analysis).
| No. (%) | DFS | OS | |||
|---|---|---|---|---|---|
| Log-rank | p-value | Log-rank | p-value | ||
| EpCAM-/CD49f+ (%) | 150 | 6.876 | 0.009 | 11.814 | 0.001 |
| 0 | 103 (68.7) | ||||
| >0 | 47 (31.3) | ||||
| EpCAM+/CD49f- (%) | 150 | 1.659 | 0.198 | 0.233 | 0.629 |
| 0 | 94 (62.7) | ||||
| >0 | 56 (37.3) | ||||
| EpCAM+/CD49f+ (%) | 150 | 2.761 | 0.097 | 2.754 | 0.097 |
| 0 | 110 (73.3) | ||||
| >0 | 40 (26.7) | ||||
| EpCAM-/CD49f- (%) | 150 | 0.631 | 0.427 | 0.180 | 0.671 |
| 0 | 26 (17.3) | ||||
| >0 | 124 (82.7) | ||||
EpCAM=epithelial cell adhesion molecule; DFS=disease-free survival; OS=overall survival.
Supplementary Table 2. Correlation of prevalence of EpCAM/CD49 with breast cancer prognosis (multivariate analysis).
| No. (%) | DFS | OS | |||
|---|---|---|---|---|---|
| p-value | HR (95% CI) | p-value | HR (95% CI) | ||
| EpCAM-/CD49f+ (%) | 150 | 0.010 | 0.002 | ||
| - (0) | 103 (68.7) | 1 | 1 | ||
| + ( > 0) | 47 (31.3) | 2.070 (1.188-3.608) | 3.235 (1.551-6.748) | ||
| EpCAM+/CD49f+ (%) | 150 | 0.061 | 0.042 | ||
| - (0) | 110 (73.3) | 1 | 1 | ||
| + ( > 0) | 40 (26.7) | 1.813 (0.974-3.377) | 2.250 (1.031-4.907) | ||
| EpCAM+/CD49f- (%) | 150 | 0.267 | 0.455 | ||
| - (0) | 94 (22.3) | 1 | 1 | ||
| + ( > 0) | 56 (77.7) | 1.357 (0.792-2.323) | 1.308 (0.647-2.646) | ||
| EpCAM-/CD49f- (%) | 150 | 0.302 | 0.444 | ||
| - (0) | 26 (17.3) | 1 | 1 | ||
| + ( > 0) | 124 (82.7) | 0.713 (0.376-1.351) | 0.721 (0.312-1.666) | ||
EpCAM=epithelial cell adhesion molecule; DFS=disease-free survival; OS=overall survival; HR=hazard risk; CI=confidence interval.
References
- 1.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basak SK, Veena MS, Oh S, Huang G, Srivatsan E, Huang M, et al. The malignant pleural effusion as a model to investigate intratumoral heterogeneity in lung cancer. PLoS One. 2009;4:e5884. doi: 10.1371/journal.pone.0005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boonyaratanakornkit JB, Yue L, Strachan LR, Scalapino KJ, LeBoit PE, Lu Y, et al. Selection of tumorigenic melanoma cells using ALDH. J Invest Dermatol. 2010;130:2799–2808. doi: 10.1038/jid.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Lab Invest. 2010;90:234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 11.Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012;22:187–193. doi: 10.1016/j.semcancer.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- 14.Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- 15.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 16.Honeth G, Bendahl PO, Ringnér M, Saal LH, Gruvberger-Saal SK, Lövgren K, et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stecklein SR, Jensen RA, Pal A. Genetic and epigenetic signatures of breast cancer subtypes. Front Biosci (Elite Ed) 2012;4:934–949. doi: 10.2741/E431. [DOI] [PubMed] [Google Scholar]
- 18.Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999;222:124–138. doi: 10.1177/153537029922200203. [DOI] [PubMed] [Google Scholar]
- 19.Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8:604–617. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay R, Theriault RL, Price JE. Increased levels of alpha6 integrins are associated with the metastatic phenotype of human breast cancer cells. Clin Exp Metastasis. 1999;17:325–332. doi: 10.1023/a:1006659230585. [DOI] [PubMed] [Google Scholar]
- 21.Mercurio AM, Rabinovitz I. Towards a mechanistic understanding of tumor invasion: lessons from the alpha6beta 4 integrin. Semin Cancer Biol. 2001;11:129–141. doi: 10.1006/scbi.2000.0364. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty AK, Funasaka Y, Ichihashi M, Pawelek JM. Upregulation of alpha and beta integrin subunits in metastatic macrophage-melanoma fusion hybrids. Melanoma Res. 2009;19:343–349. doi: 10.1097/CMR.0b013e32832fe121. [DOI] [PubMed] [Google Scholar]
- 23.Kwon J, Lee TS, Lee HW, Kang MC, Yoon HJ, Kim JH, et al. Integrin alpha 6: a novel therapeutic target in esophageal squamous cell carcinoma. Int J Oncol. 2013;43:1523–1530. doi: 10.3892/ijo.2013.2097. [DOI] [PubMed] [Google Scholar]
- 24.Hoogland AM, Verhoef EI, Roobol MJ, Schröder FH, Wildhagen MF, van der Kwast TH, et al. Validation of stem cell markers in clinical prostate cancer: alpha6-integrin is predictive for non-aggressive disease. Prostate. 2014;74:488–496. doi: 10.1002/pros.22768. [DOI] [PubMed] [Google Scholar]
- 25.Steglich A, Vehlow A, Eke I, Cordes N. Alpha integrin targeting for radiosensitization of three-dimensionally grown human head and neck squamous cell carcinoma cells. Cancer Lett. 2015;357:542–548. doi: 10.1016/j.canlet.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Haraguchi N, Ishii H, Mimori K, Ohta K, Uemura M, Nishimura J, et al. CD49f-positive cell population efficiently enriches colon cancer-initiating cells. Int J Oncol. 2013;43:425–430. doi: 10.3892/ijo.2013.1955. [DOI] [PubMed] [Google Scholar]
- 27.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Chaves-Pérez A, Mack B, Maetzel D, Kremling H, Eggert C, Harréus U, et al. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene. 2013;32:641–650. doi: 10.1038/onc.2012.75. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Wang Z, Brown S, Kannappan V, Tawari PE, Jiang W, et al. Liposome encapsulated Disulfiram inhibits NFkappaB pathway and targets breast cancer stem cells in vitro and in vivo. Oncotarget. 2014;5:7471–7485. doi: 10.18632/oncotarget.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Univariate survival analysis of EpCAM/CD49f with disease-free survival (DFS) and overall survival (OS). (A-F) All the patients' tumor samples were stained with EpCAM and CD49f. The prevalence of EpCAM+/CD49f+, EpCAM+/CD49f-, and EpCAM-/CD49f- were analyzed as control, and no significant association were checked with DFS and OS.
Supplementary Table 1. Correlation of prevalence of EpCAM/CD49 with breast cancer prognosis (univariate analysis).
| No. (%) | DFS | OS | |||
|---|---|---|---|---|---|
| Log-rank | p-value | Log-rank | p-value | ||
| EpCAM-/CD49f+ (%) | 150 | 6.876 | 0.009 | 11.814 | 0.001 |
| 0 | 103 (68.7) | ||||
| >0 | 47 (31.3) | ||||
| EpCAM+/CD49f- (%) | 150 | 1.659 | 0.198 | 0.233 | 0.629 |
| 0 | 94 (62.7) | ||||
| >0 | 56 (37.3) | ||||
| EpCAM+/CD49f+ (%) | 150 | 2.761 | 0.097 | 2.754 | 0.097 |
| 0 | 110 (73.3) | ||||
| >0 | 40 (26.7) | ||||
| EpCAM-/CD49f- (%) | 150 | 0.631 | 0.427 | 0.180 | 0.671 |
| 0 | 26 (17.3) | ||||
| >0 | 124 (82.7) | ||||
EpCAM=epithelial cell adhesion molecule; DFS=disease-free survival; OS=overall survival.
Supplementary Table 2. Correlation of prevalence of EpCAM/CD49 with breast cancer prognosis (multivariate analysis).
| No. (%) | DFS | OS | |||
|---|---|---|---|---|---|
| p-value | HR (95% CI) | p-value | HR (95% CI) | ||
| EpCAM-/CD49f+ (%) | 150 | 0.010 | 0.002 | ||
| - (0) | 103 (68.7) | 1 | 1 | ||
| + ( > 0) | 47 (31.3) | 2.070 (1.188-3.608) | 3.235 (1.551-6.748) | ||
| EpCAM+/CD49f+ (%) | 150 | 0.061 | 0.042 | ||
| - (0) | 110 (73.3) | 1 | 1 | ||
| + ( > 0) | 40 (26.7) | 1.813 (0.974-3.377) | 2.250 (1.031-4.907) | ||
| EpCAM+/CD49f- (%) | 150 | 0.267 | 0.455 | ||
| - (0) | 94 (22.3) | 1 | 1 | ||
| + ( > 0) | 56 (77.7) | 1.357 (0.792-2.323) | 1.308 (0.647-2.646) | ||
| EpCAM-/CD49f- (%) | 150 | 0.302 | 0.444 | ||
| - (0) | 26 (17.3) | 1 | 1 | ||
| + ( > 0) | 124 (82.7) | 0.713 (0.376-1.351) | 0.721 (0.312-1.666) | ||
EpCAM=epithelial cell adhesion molecule; DFS=disease-free survival; OS=overall survival; HR=hazard risk; CI=confidence interval.