Abstract
Mammalian KIF3AC is classified as a heterotrimeric kinesin-2 that is best known for organelle transport in neurons, yet in vitro studies to characterize its single molecule behavior are lacking. The results presented show that a KIF3AC motor that includes the native helix α7 sequence for coiled-coil formation is highly processive with run lengths of ∼1.23 μm and matching those exhibited by conventional kinesin-1. This result was unexpected because KIF3AC exhibits the canonical kinesin-2 neck-linker sequence that has been reported to be responsible for shorter run lengths observed for another heterotrimeric kinesin-2, KIF3AB. However, KIF3AB with its native neck linker and helix α7 is also highly processive with run lengths of ∼1.62 μm and exceeding those of KIF3AC and kinesin-1. Loop L11, a component of the microtubule-motor interface and implicated in activating ADP release upon microtubule collision, is significantly extended in KIF3C as compared with other kinesins. A KIF3AC encoding a truncation in KIF3C loop L11 (KIF3ACΔL11) exhibited longer run lengths at ∼1.55 μm than wild-type KIF3AC and were more similar to KIF3AB run lengths, suggesting that L11 also contributes to tuning motor processivity. The steady-state ATPase results show that shortening L11 does not alter kcat, consistent with the observation that single molecule velocities are not affected by this truncation. However, shortening loop L11 of KIF3C significantly increases the microtubule affinity of KIF3ACΔL11, revealing another structural and mechanistic property that can modulate processivity. The results presented provide new, to our knowledge, insights to understand structure-function relationships governing processivity and a better understanding of the potential of KIF3AC for long-distance transport in neurons.
Introduction
The kinesin-2 subfamily of molecular motors associates with microtubules and undergoes a characteristic ATPase cycle for intracellular transport (1–5). Mammalian motors classified as kinesin-2s include homodimeric KIF17 and heterotrimeric KIF3AC and KIF3AB. In multiple species heterodimeric kinesin-2 motors have been shown to associate with a nonmotor, adaptor polypeptide designated KAP to form a heterotrimer (4,5). The motor domains of KIF3AB and KIF3AC are intriguing motors because they exist as dimers from three different gene products: KIF3A, KIF3B, and KIF3C. Kinesin-2s are ubiquitously expressed and are known in particular for their long-distance transport roles (6–26). Mammalian KIF17 as well as its Caenorhabditis elegans homolog OSM-3 have been characterized as fast and highly processive motors like kinesin-1, functioning in multiple cell types including cilia and neurons (4,5,12,27,28). In contrast, KIF3AB has been reported to be a much slower transporter and not as processive as kinesin-1 or KIF17, yet KIF3AB has been reported to be very sensitive to force (7,20).
Although there has been significant interest in KIF3AB to understand its role in intraflagellar transport, KIF3AC has not been as well studied. In part, this is because the model organisms like Drosophila and C. elegans do not have a true KIF3C ortholog. Furthermore, KIF3AC like kinesin-1 transports organelles in neurons, therefore, a direct comparison of the motor properties of KIF3AC to those of kinesin-1 may reveal new insights to better understand why there are so many different kinesins expressed in neurons and the specific capabilities of each (3,11,12). KIF3AB and KIF3AC also share similarities in protein sequence and structure, therefore, this analysis may also provide a better understanding of the key principles for force generation shared by heterotrimeric kinesin-2s in general relative to homodimeric KIF17 and other N-terminal processive kinesins.
To pursue these studies the KIF3AC and KIF3AB heterodimers were designed to include the N-terminal native sequence of each motor domain, neck linker, and native helix α7, followed by a dimerization motif to stabilize the native coiled coil. A single molecule approach was used to determine whether KIF3AC was processive as assumed and how similar its motor properties were to kinesin-1 and KIF3AB.
Materials and Methods
M. musculus KIF3 motor proteins
M. musculus KIF3A and KIF3B cDNA were generous gifts from William O. Hancock (Pennsylvania State University, University Park, PA). The M. musculus KIF3C cDNA was purchased from Open Biosystems (GE Life Sciences, Lafayette, CO). The KIF3A (M1-L374), KIF3B (M1-L369), and KIF3C (M1-L396) constructs were amplified and introduced into a modified pET24d (Novagen, Madison, WI) plasmid that encodes a C-terminal tobacco etch virus (TEV) protease-cleavable His8 tag (TTSENLYFQGASHHHHHHHH). However, the KIF3A construct for heterodimerization was introduced into a modified pET31b plasmid (Novagen) that encodes a C-terminal TEV protease-cleavable StrepII tag (TTSENLYFQGASNWSHPQFEK). The KIF3CΔL11 polypeptide contains a P258A substitution and deletion of the C-specific extension in loop L11 (N259–S284). All constructs include the native N-terminal motor domain sequence, neck linker, and α7 helix, which are C-terminally fused to an in-register segment of the dimerization motif from EB1 (DFYFGKLRNIELICQENEGENDPVLQRIVDILYATDE). See Fig. 1, which illustrates the construct design for the expression of the KIF3 polypeptides with the amino acid residue sequences reported in Fig. S1 in the Supporting Material. The resulting constructs were sequence-verified over the entire open reading frame insert. Note that the KIF3AB heterodimer used in this report was similarly designed with its dimerization stabilized through a synthetic heterodimerization domain (SHD) (29). A KIF3AC-SHD motor was also characterized using the SHD dimerization motif (Figs. S1 and S2).
Figure 1.
Construct design for analysis of KIF3 motors. (A) Constructs of KIF3A (M1-L374), KIF3B (M1-L369), and KIF3C (M1-L396) encode each native N-terminal motor domain, neck linker, and helix α7 C-terminally fused in-register to a segment of the dimerization motif of EB1, followed by a cleavable TEV site, and the C-terminal purification tags (Strep tag (S) or His8 tag (H)). Loop L11 is illustrated for KIF3A, KIF3B, and KIF3C with the KIF3C-specific L11 extension illustrated in red. (B) Sequence alignment of the KIF3 neck-linker and helix α7 sequences in comparison with other kinesins. C-terminal fusion sequences that were used for dimerization and purification of KIF3 motors are also shown. (C) Table listing details of all constructs used for experiments reported here. See Materials and Methods and Supporting Material for specific sequences of the heterodimeric and homodimeric motors and their characterization.
The KIF3 motors were expressed in Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Strategene, La Jolla, CA) with heterodimers resulting from cotransformation of two plasmids. See the Supporting Material for detailed expression and purification procedures. After purification homodimeric or heterodimeric state was confirmed by analytical gel filtration and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (see Fig. 2). Before each experiment, the purified motors were clarified for 5 min at 4°C by high-speed centrifugation (Beckman Coulter Optima TLX Ultracentrifuge, TLA-100 rotor, 313,000 × g) followed by concentration determination using IgG as a protein standard and the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA).
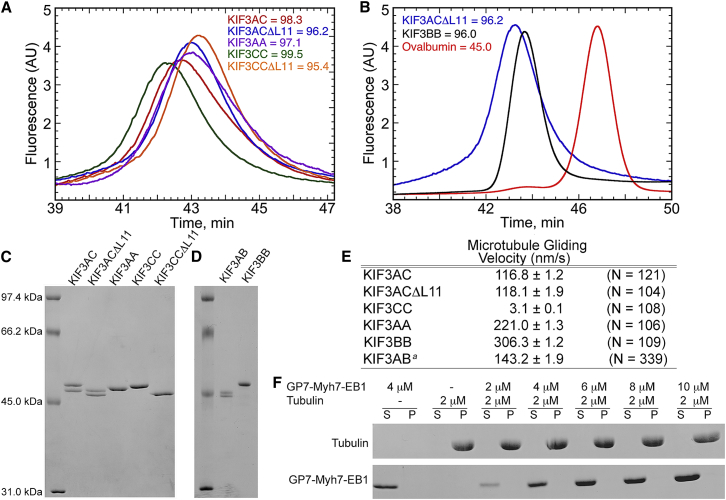
Figure 2.
Characterization of purified KIF3 motors. (A) Analytical gel filtration elution profiles of KIF3 motors plotted as intrinsic fluorescence versus time suggest stable dimers for each motor. Estimated relative molecular mass (Mr) based on amino acid sequence are provided for each dimer (see also Fig. 1 C). (B) Additional analytical gel filtration profiles of KIF3BB and monomeric ovalbumin, Mr = 45.0 kDa compared with KIF3ACΔL11. (C) Purified dimers analyzed by 10% acrylamide/2 M urea SDS-PAGE confirm purity of proteins and that heterodimers were at a 1:1 ratio. Molecular mass markers are presented to the left side of the gel with associated Mr. Estimated Mr of each dimer based on amino acid sequence: KIF3AC = 98.3; KIF3ACΔL11 = 96.2; KIF3CC = 99.5; KIF3CCΔL11 = 95.4; and KIF3AA = 97.1. (D) Purified KIF3AB and KIF3BB separated by 12% acrylamide/2 M urea SDS-PAGE. Estimated Mr based on amino acid sequence: KIF3AB = 92.0; KIF3BB = 96.0. (E) Table of microtubule gliding rates of KIF3 dimers (mean ± SE) obtained from Gaussian fits. Data were collected from three replicate experiments. aSee (29). (F) Equilibrium binding analysis of MYH7-EB1 dimer (0–10 μM) incubated with 2 μM tubulin polymer. Supernatant (S) and pellet (P) samples were analyzed by 12% acrylamide/2 M urea SDS-PAGE gels stained with Coomassie Blue R-250.
Quantum dot-motor attachment
Streptavidin-coated quantum dots (Qdot 525-Streptavidin conjugate, Life Technologies, Carlsbad, CA) were diluted to 200 nM and preincubated in a 1:1 ratio with biotinylated-Penta-His antibody (Qiagen, Valencia, CA) for 60 min at room temperature in PME80 buffer (80 mM PIPES, pH 6.9 with KOH, 5 mM MgCl2, and 1 mM EGTA). His8-tagged motors were then added to the Qdot-antibody complex at 20 nM dimer, which resulted in a 1:10 ratio of dimer/Qdots (20 nM dimer, 200 nM Qdot-antibody complex) that were then incubated for 60 min at 4°C. Based on a Poisson distribution this procedure results in a working stock in which 9% of the Qdots were estimated to have one motor bound and 0.5% with ≥2 motors bound. To determine if the 1:10 ratio of KIF3 motors/Qdots was sufficient for single molecule conditions, experiments using KIF3AC were also performed with dimer/Qdots at a 1:20 ratio (10 nM KIF3AC, 200 nM Qdot-antibody complex). For both 1:10 and 1:20 ratio experiments, the Qdot complexes were introduced into the reaction chamber to yield a final concentration of 2 nM KIF3AC (10× dilution of 1:10 ratio in Activity Buffer; 5× dilution of 1:20 ratio in Activity Buffer; see below). The results show that the KIF3AC velocity and run length data were comparable for 1:10 and 1:20 KIF3AC/Qdot ratios (Fig. S2).
Microtubule preparation
The long microtubule tracks (13–30 μm) used for this report were polymerized from lyophilized X-rhodamine tubulin (Cytoskeleton, Denver, CO). The tubulin was first resuspended in PME80 buffer containing 10% glycerol followed by incubation on ice with 1 mM MgGTP for 5 min. The tubulin was then centrifuged at 16,000 × g for 10 min at 4°C (Galaxy 16D Micro 1816 centrifuge, VWR, Bridgeport, NJ). The supernatant containing soluble tubulin was transferred to a new tube, and microtubule polymerization initiated at 37°C for 20 min. The microtubules were then stabilized with paclitaxel (33 μM final concentration) and incubated for an additional 10 min. This procedure yields a final microtubule stock at 30 μM tubulin polymer.
Processivity assay
Perfusion chambers were formed from a silanized coverslip mounted on a glass slide separated by strips of double-sided tape to generate a 10-μl flow cell (30). The chamber was first incubated with 0.4% rat anti-α-tubulin antibody (ABD Serotec, Raleigh, NC) for 5 min followed by a surface blocking step using 5% Pluronic F-127 (Sigma, St. Louis, MO) for 5 min. Diluted microtubules (1:300 in PME80 supplemented with paclitaxel to 22 μM) were then introduced into the chamber and incubated for 10 min. Unbound microtubules were removed by flowing into the chamber PME80 supplemented with 10 mM DTT and 20 μM paclitaxel. The working stock of Qdot-motor complexes (1:10, 20 nM dimer/200 nM Qdot-antibody complex; or 1:20, 10 nM dimer/200 nM Qdot-antibody complex) was diluted in Activity Buffer (PME80, 0.5% Pluronic F-127, 30 μM paclitaxel, 125 μg/ml bovine serum albumin, 50 μM DTT, 25 mM glucose, 0.2 mg/ml glucose oxidase, 175 μg/ml catalase, 0.3 mg/ml creatine phosphokinase, 2 mM phosphocreatine with varying MgATP) resulting in a final concentration of 2 nM Qdot-bound dimer in the perfusion chamber.
The human kinesin-1 KIF5B K560 protein was coupled to Qdots via the C-terminal His8 tag as described for the KIF3 motors. For the Qdot-K560 complexes, 1 mM adenosine 5′-(β, γ-imido)triphosphate (AMP-PNP) was added to generate the microtubule•K560-Qdot complex. Therefore, the perfusion chamber setup was adjusted such that immediately following the 10 min microtubule incubation, the Qdot-K560 complexes were introduced in PME80 supplemented with 10 mM DTT, 20 μM paclitaxel, and 1 mM AMP-PNP and incubated in the chamber for 5 min. This step also served to wash out unbound microtubules. Qdot-K560 motors were then activated by introduction of Activity Buffer containing 1.5 mM MgATP. A similar technique was used for the KIF3BB experiments such that the motors were initially bound to the microtubule tracks in the perfusion chamber through a 5 min incubation in the presence of 0.5 mM AMP-PNP followed by perfusion of the Activity Buffer containing 1 mM MgATP.
Total internal reflection fluorescence microscopy and image acquisition
Immediately upon addition of Activity Buffer, chambers were imaged by total internal reflection fluorescence (TIRF) microscopy using a Zeiss Inverted Axio Observer Z1 MOT fluorescence microscope with the 100X oil 1.46 N.A. Plan-Apochromat objective (Carl Zeiss Microscopy, Jena, Germany). Digital images were collected through a Hamamatsu EM-CCD digital camera using the AxioVision 4.8.2 software package. This setup yielded 512 × 512 pixel images with 0.16 μm/pixel in both x and y planes. The Qdot complexes were tracked by imaging at 488 nm (5% laser power) every ∼0.6 s for 5 min using 100 ms exposure. Due to the slow velocity of KIF3CC and KIF3CCΔL11, videos were acquired over 30 min with 5 s intervals. Reference images of the X-rhodamine microtubule tracks were taken at 564 nm (2% laser power) with 300 ms exposure both before and after motility. Qdot videos were then overlaid with the microtubule image using National Institutes of Health (NIH, Bethesda, MD) ImageJ software.
Data analysis
Analysis of Qdot processivity was conducted using the MultipleKymograph plugin for ImageJ (J. Rietdorf and A. Seitz, European Molecular Biology Laboratory, Heidelberg, Germany). Velocities were plotted as histograms, and a Gaussian function was applied to determine the mean velocity ± SE. Run lengths were plotted as histograms using a single exponential decay equation fit to the data to calculate mean run length:
where A is the maximum amplitude and l is the mean run length reported ± SE, with masking of the first bin due to the resolution limit of the TIRF microscope (<0.25 μm). Statistical analyses for run lengths were performed using the Comparing Means (t-test assuming different variances) algorithm of StatPlus software using an α-reliability level of 5%.
To most accurately represent the maximum run length potential of the motors, Qdot complexes that traveled the entire microtubule length and paused at the end, began or ended a run outside the timescale of the experiment, or detached from the end of the microtubule were excluded from the analysis. Both long and short runs were obtained from the same microtubule tracks to avoid data bias. Pausing and stalling occurred for all motors tested but represented only a minor fraction (2–6%) of each data set and showed no apparent correlation between pause frequency or duration versus MgATP concentration or L11 length. The data that included pausing events were eliminated from further analysis. Furthermore, there was no evidence of torque exerted by the KIF3 motors tested in these experiments, which is consistent with the report from Brunnbauer et al. (31), which reported no evidence of torque generation by M. musculus KIF3AB.
Microtubule cosedimentation of the EB1 motif
To examine the microtubule binding properties of the segment of EB1 used for designing the KIF3 constructs reported here, an equivalently designed segment of M. musculus β-cardiac myosin (MYH7 rod; K1783-T1854) was developed (32). The dimerized myosin rod segment contained both an N-terminally fused segment of GP7 scaffolding protein with linker sequence (ASMPLKPEEHEDILNKLLDPELAQSERTEALQQLRVNYGSFVSEYNDLTKS) (33) and a C-terminally fused segment of EB1, matching that used for the KIF3 proteins, including linker residues in-register to the heptad repeat of the rod sequence (EDLEKERDFYFGKLRNIELICQENEGENDPVLQRIVDILYATDE).
For cosedimentation analysis, a 200-μl reaction mix included 0–10 μM GP7-MYH7-EB1 dimer, 100 mM KCl, 2 μM tubulin polymer, and 40 μM paclitaxel in ATPase buffer. Reactions were incubated for 30 min at 25°C followed by centrifugation at 25°C (Beckman Optima TLX Ultracentrifuge with TLA-100 rotor, 313,000 × g, 30 min). For each reaction, the top 100 μl of supernatant was collected for an SDS-PAGE sample with the remaining supernatant discarded. Pellets were gently washed with buffer, resuspended in 100 μl ATPase buffer containing 5 mM CaCl2, and incubated at 4°C for 10 min. After transferring the resuspended pellet to another tube, the reaction tubes were washed with an additional 100 μl buffer, which was then added to the resuspended pellet sample. Laemmli 5× sample buffer was added to both the supernatant (100 μl + 25 μl sample buffer) and the pellet (200 μl + 50 μl sample buffer) samples. Each aliquot was then denatured at 100°C for 3 min. Aliquots of equal volume from the resulting samples were subjected to SDS-PAGE on a 12% acrylamide/2 M urea gel followed by staining with Coomassie Brilliant Blue R-250.
Results and Discussion
Previously, we reported the construct design and mechanistic analysis of a murine KIF3AB heterodimer that included the respective native motor domains, neck region, and native helix α7 for coiled-coil formation followed by a SHD that stabilized the KIF3AB heterodimeric state (29,34,35). For the study reported here a KIF3AC-SHD heterodimer was similarly designed and the motor characterized, but another strategy for dimerization was also pursued using a portion of the homodimeric coiled coil and four-helix bundle motif of EB1 (Figs. 1 and Fig. S1) (33,36). To purify the heterodimeric and homodimeric KIF3 motors (Supporting Materials), selection was based on the C-terminal affinity tags (Strep tag or His8 tag) (29). The results in Fig. 2 show that both the heterodimeric and homodimeric KIF3 motors purified as stable dimers, and each promoted robust microtubule gliding.
The EB1 dimerization motif was reported previously not to interact with microtubules (37,38). To test this hypothesis directly, microtubule cosedimentation experiments were performed with a fragment of human cardiac β-myosin dimerized by the EB1 motif and designated GP7-Myh7-EB1. Fig. 2 F shows that indeed GP7-Myh7-EB1 remained in the supernatant and did not partition to the pellet with the microtubules even at GP7-Myh7-EB1 concentrations at fivefold excess of the microtubule concentration. In addition, single molecule experiments were also pursued with KIF3AC-SHD (Fig. S2, C and D), for comparison to the results for KIF3AC-EB1 (Fig. 3 A). These data show that the velocity and run length for KIF3AC-SHD and KIF3AC-EB1 are comparable (Fig. S2 E), therefore confirming that the EB1 dimerization motif used to stabilize the native helical coiled coil is not interacting with the microtubule lattice to enhance the velocity or run length of the KIF3AC single molecules.
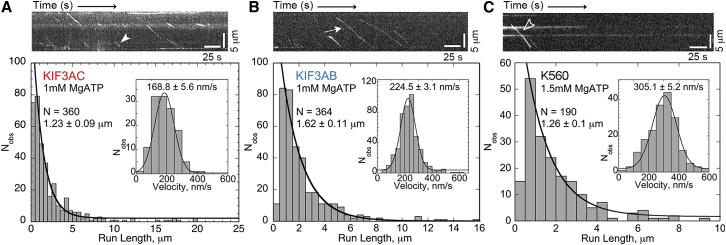
Figure 3.
KIF3AC and KIF3AB are as processive as conventional kinesin-1. (A) KIF3AC run length and velocity (inset) data at 1 mM MgATP including a kymograph of a 13.3 μm-long microtubule. Highlighted trace (solid arrowhead) illustrates a 1.44 μm run at 160 nm/s (scale bars: 25 s along x axis; 5 μm along y axis) See also Movie S2. (B) KIF3AB run length and velocity (inset) data at 1 mM MgATP including a kymograph of a 13.0 μm-long microtubule. Highlighted trace (arrow) represents an 11.0 μm run at 182 nm/s. See also Movie S3. (C) Histograms of data collected for human kinesin-1 K560 run length and velocity (inset) at 1.5 mM MgATP including a kymograph of a 13.1 μm-long microtubule. Highlighted trace (outlined arrowhead) illustrates an 8.48 μm run at 294 nm/s. See also Movie S1. To see this figure in color, go online.
The processivity of KIF3AC matches that of conventional kinesin-1 in single molecule experiments
In single molecule quantum dot (Qdot) experiments, KIF3AC moved at a rate of 169 nm/s with a characteristic run length of 1.23 μm in 1 mM MgATP (Fig. 3 A; Movie S2). Because KIF3AC transports organelles in neurons much like kinesin-1, comparative experiments were pursued using the well characterized truncated motor human kinesin-1 K560 (39–42). The results show that K560 exhibits a similar run length of 1.26 μm and velocity of 305 nm/s (see Fig. 3 C; Movie S1). These values are within the range of previously reported single molecule results for human K560 with velocity reported at 260–400 nm/s and run length results at 1.3–2 μm (39–42). The single molecule behavior of KIF3AB was also examined (29). KIF3AB was observed to travel at an average rate of 224 nm/s (Fig. 3 B; Movie S3), mirroring the steady-state ATPase kcat previously reported (29). Furthermore, KIF3AB exhibited a characteristic run length of 1.62 μm in 1 mM MgATP (Fig. 3 B).
The results presented in Fig. 3 were surprising because kinesin-1 and kinesin-2 KIF3AB have been extensively studied and have been reported to have very different run lengths attributed to the length and charge of the neck linker peptide (41,43–45). However, the data presented clearly show that KIF3AC, another heterodimeric kinesin-2 with the canonical 17-residue neck linker, can run equivalently long distances as kinesin-1 K560. Moreover, KIF3AB moves on average longer distances than either K560 or KIF3AC. These results imply that processivity is being tuned by mechanisms in addition to the well-studied role of the neck linker and are consistent with a recent report on KIF3AB by Andreasson et al. (7).
Loop L11 tunes the processivity of KIF3AC
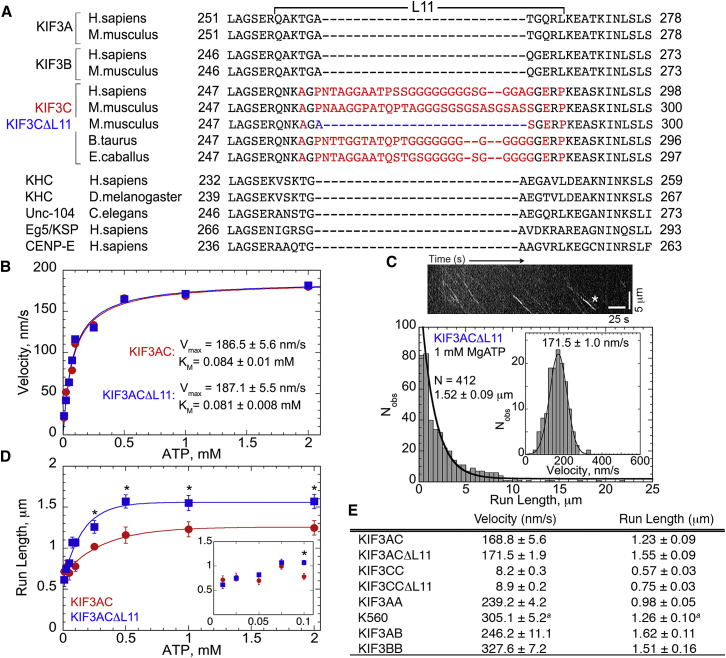
KIF3C contains a prominent 25-residue insert enriched in glycines and serines in surface loop L11 that constitutes a major difference between KIF3C and other kinesins (Fig. 4 A) (12,18,46). Loop L11 is a component of the microtubule-motor interface that has been implicated in activating ADP release upon microtubule collision (47–56). Therefore, KIF3AC provided the opportunity to explore the impact of loop L11 on KIF3AC motor behavior in the single molecule studies. An analysis of heterodimeric KIF3AC encoding a truncation in KIF3C-L11 (KIF3CΔL11) was pursued. This construct when expressed shortens the extended L11 sequence of KIF3C back to the length seen in other kinesins with the specific design of this polypeptide emulating the L11 region of KIF3B (Fig. 4 A).
Figure 4.
Altering loop L11 modifies KIF3AC run length. (A) Species-specific alignment of loop L11 sequences between KIF3A, B, and C in comparison with other processive kinesins. Red sequence represents extended KIF3C-specific residues with blue font showing the residue changes for KIF3CΔL11. (B) Mean velocities of KIF3AC and KIF3ACΔL11 from Qdot experiments plotted as a function of increasing MgATP concentration with the Michaelis-Menten equation fit (mean ± SE). See also Movies S2 and S4. (C) KIF3ACΔL11 run length and velocity (inset) data at 1 mM MgATP including a kymograph of a 13.4 μm-long microtubule. Highlighted trace (∗) represents a 3.2 μm run at 173 nm/s (scale bars: 25 s along x axis; 5 μm along y axis). See also Movie S4. (D) Mean run length (±SE) of KIF3AC and KIF3ACΔL11 calculated from single exponential decay fits compiled and plotted as a function of MgATP concentration from 0.01–2 mM. At low MgATP (0.01–0.075 mM, inset), KIF3AC and KIF3ACΔL11 behave similarly. The difference in run length becomes statistically significant (∗p < 0.001) from 0.1 to 2 mM MgATP with KIF3ACΔL11 exhibiting significantly longer runs. (E) Table of compiled Qdot data for KIF3 dimers and human kinesin-1 K560 (mean ± SE) at 1 mM MgATP or a1.5 mM MgATP.
The single molecule experiments were conducted over a range of MgATP concentrations for both the wild-type KIF3AC heterodimer as well as KIF3ACΔL11 that carries a truncation in loop L11. Both exhibited an ATP-concentration-dependence for the velocity of stepping as well as the run length (see Fig. 4, B and D). The results from the ATP-concentration dependence revealed that shortening loop L11 did not alter KIF3AC velocity. KIF3AC and KIF3ACΔL11 reached a Vmax of 187 nm/s (Fig. 4 B; Movies S2 and S4). In contrast, the mean run length of KIF3ACΔL11 increased to 1.52 μm at 1 mM MgATP (Fig. 4 C) and approached the mean run length of KIF3AB at 1.62 μm at 1 mM MgATP. Fig. 4 D shows that both wild-type KIF3AC and KIF3ACΔL11 exhibit an ATP-concentration-dependent increase in run length with a statistically significant difference existing between the two motors at high ATP concentrations (0.1–2 mM; ∗, p < 0.001).
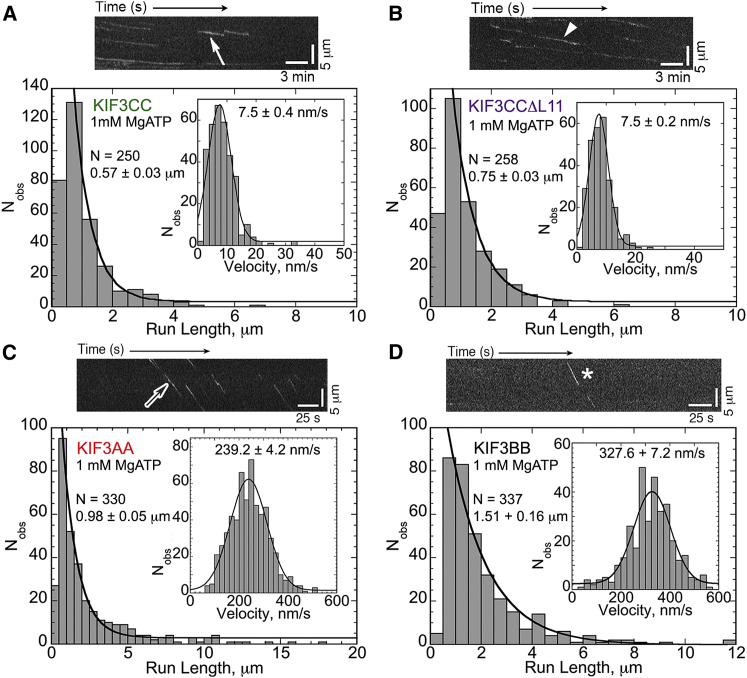
The effect of shortening L11 was further examined using homodimers of KIF3CC and KIF3CCΔL11 (Fig. 5; Movies S5 and S6). Both of these motors moved exceedingly slowly at ∼8 nm/s, yet both did move processively along microtubule tracks. KIF3CC moved processively with a mean run length of 0.57 μm, but note that homodimeric KIF3CCΔL11 exhibited a longer mean run length of 0.75 μm than KIF3CC at 0.57 μm. This is a highly significant difference (p < 0.00001) and mirrors the trend seen when comparing heterodimeric KIF3AC with either KIF3ACΔL11 or KIF3AB, whereby shortening L11 increased the run length. For comparison engineered homodimeric KIF3AA and homodimeric KIF3BB (Fig. 5; Movie S7 and S8) were also analyzed at 1 mM MgATP. KIF3AA exhibited a characteristic run length of 0.98 μm and velocity of 239 nm/s, whereas the mean run length of KIF3BB was 1.51 μm and velocity of 328 nm/s (Figs. 4 E and 5). These results suggest that the KIF3B contribution is to enhance the run length of KIF3A to account for the behavior of the KIF3AB heterodimer whose run length is 1.62 μm and is not significantly different from the run length of KIF3BB.
Figure 5.
Single molecule analyses of homodimers illustrate differences in KIF3 motors. (A–D) Single molecule run length and velocity (insets) histograms with representative kymographs for homodimeric KIF3 motors (mean ± SE). See also Fig. 4E. (A) KIF3CC data with a highlighted trace (solid arrow) representing a 1.28 μm run at 6 nm/s (scale bars: 3 min along x axis; 5 μm along y axis). See also Movie S5. (B) KIF3CCΔL11 data with a highlighted trace (arrowhead) representing a 2.4 μm run at 6 nm/s (scale bars: 3 min along x axis; 5 μm along y axis). See also Movie S6. (C) KIF3AA data, with a highlighted trace (open arrow) representing a 6.1 μm run at 288 nm/s (scale bars: 25 s along x axis; 5 μm along y axis). See also Movie S7. (D) KIF3BB data with a highlighted trace (∗) representing a 5.8 μm run at 409 nm/s (scale bars: 25 s along x axis; 5 μm along y axis). For KIF3BB the microtubule-motor complex was preformed with 0.5 mM MgAMP-PNP before activation by 1 mM MgATP. See also Movie S8. To see this figure in color, go online.
Shortening L11 increases microtubule affinity
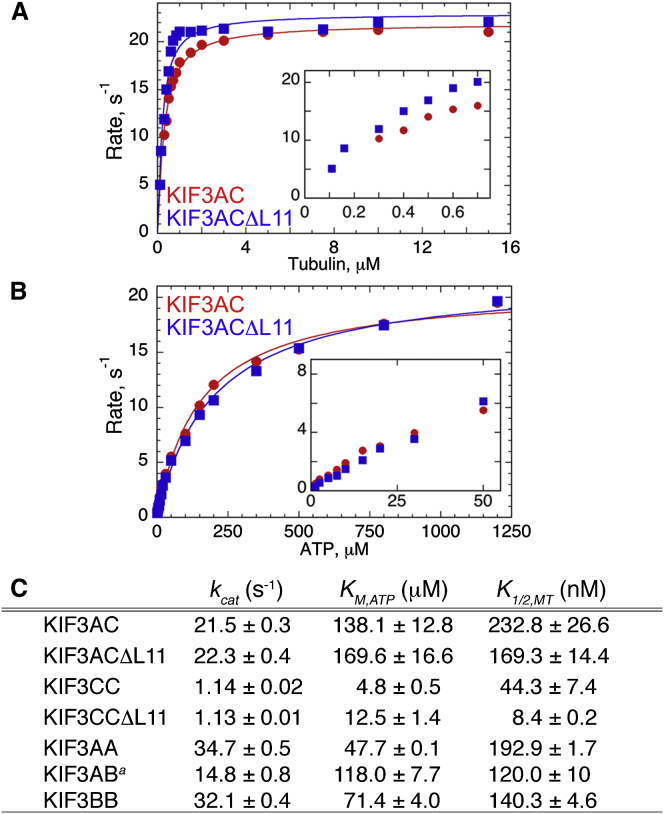
The single molecule results show that there was no difference in the single molecule velocity of KIF3AC and KIF3ACΔL11, yet the run length for KIF3ACΔL11 increased significantly. To test the hypothesis that the difference in run length was modulated through microtubule affinity, steady-state ATPase experiments were pursued. The results in Fig. 6 show that the kcat for KIF3AC and KIF3ACΔL11 were comparable at ∼22 s−1, yet the apparent microtubule affinity based on the K1/2,MT was tighter for KIF3ACΔL11 at 169 nM than for KIF3AC at 233 nM. The steady-state ATPase analysis for the homodimers, KIF3CC and KIF3CCΔL11, also show a difference in the K1/2,MT with KIF3CC at 44 nM and KIF3CCΔL11 at 8 nM. Furthermore, the steady-state ATPase results for KIF3AB and KIF3BB also support the premise that a shorter L11 results in an increase in the apparent microtubule affinity in these dimers (see Fig. 6 C).
Figure 6.
Shortening loop L11 increases apparent microtubule affinity. (A–C) Steady-state ATPase kinetics of KIF3AC and KIF3ACΔL11 as a function of microtubule (A) or MgATP (B) concentration. (A) Final concentrations: 0.1 μM KIF3AC or KIF3ACΔL11 active sites, 2 mM Mg[α-32P]ATP, 40 μM paclitaxel, and 0.1–15 μM tubulin polymer. The quadratic fit provided the kcat and K1/2,MT for each motor. (Inset) Low concentrations of tubulin polymer (0–0.8 μM) illustrate the difference in apparent microtubule affinity of KIF3AC vs. KIF3ACΔL11. (B) Final concentrations: 0.1 μM KIF3AC or KIF3ACΔL11 active sites, 20 μM tubulin polymer, 40 μM paclitaxel, and 1.5–1200 μM Mg[α-32P]ATP. The Michaelis-Menten fit provided the kcat and KM,ATP for each motor. (C) Table of the steady-state ATPase parameters reported as mean ± SE.
Additionally, the comparison of the steady-state ATPase kcat for KIF3CC in comparison to KIF3CCΔL11 is also consistent with the single molecule velocity results where there was not a statistical difference in the velocity data, and both homodimers promoted very slow yet processive movement on microtubules at ∼8 nm/s (Figs. 4 E and 5). The kcat for KIF3CC and KIF3CCΔL11 were similar at 1.1 s−1 (Fig. 6 C). A single molecule velocity of ∼8 nm/s (Fig. 5) with each 8-nm step tightly coupled to one ATP turnover predicts an ATP turnover rate of ∼1 s−1. Therefore, the steady-state ATPase kinetics are consistent with the single molecule results and provide additional mechanistic insights.
Do the properties of KIF3CC support its role as a homodimeric motor in vivo?
The observation that KIF3CC can support slow processive movement at ∼8 nm/s (Figs. 4 E and 5 A) posits that if this motor were to exist as a physiological subpopulation it could function as a slow, processive cargo transporter or as a microtubule tether that promotes microtubule dynamics as proposed previously (9,10,12,16). However, Huang and Banker (12) reported that homodimers of KIF3C failed to accumulate in either dendrites or axons of hippocampal neurons suggesting that homodimeric KIF3CC may not be an organelle transporter. Gumy, et al. (10) proposed that KIF3C is an injury-specific kinesin that functions to regulate microtubule-dynamics for axon regeneration at growth cones. Although the results reported here for homodimeric KIF3CC reveal its single molecule capability, these studies cannot provide direct evidence to conclude that homodimeric KIF3CC functions in vivo as a transporter or regulator of microtubule dynamics. These intriguing hypotheses await further experimentation.
The sequence and structure of helix α7 influences motor properties
This study highlights the importance of the native helix α7 for processive potential. Whereas the KIF3AC and KIF3AB constructs reported here encode the full native motor domain, neck linker, and helix α7 sequence for initiation of the coiled coil, earlier studies of kinesin-2 used an engineered KIF3A motor specifically built to compare the effect of differences in the motor and neck linker regions of kinesin-2 to other processive kinesins (43,44). This was accomplished by fusing the motor head and neck linker of KIF3A directly to the helix α7 and proximal coiled-coil region of Drosophila kinesin-1, thereby eliminating the sequence of the native helix α7. Although the results did provide evidence of the influence of neck linker properties on motor processivity, the engineered design made the assumption that all kinesins exhibit the same coiled-coil structure for helix α7, even though the sequence similarity between these motifs in different kinesin families is exceedingly low (Fig. 1 B). The structure of dimeric rat kinesin-1 (3KIN (57),) shows that helix α7 starts at Ala-339, which is equivalent to Ala-345 in Drosophila kinesin-1 where the sequence is conserved between rat and Drosophila. However, the coiled-coil prediction with programs such as Marcoil (58) does not indicate a strong coiled-coil propensity for this residue, suggesting the difficulty of predicting the exact start of the coiled coil. Examination of the coiled-coil propensity for KIF3 helix α7 suggests that it is not entirely clear where helix α7 starts. Regardless of where the helix starts, the KIF3AB and KIF3AC results reported here emphasize the importance of the native neck linker peptide in conjunction with the native helix α7 for defining the extent of KIF3AB and KIF3AC processivity.
The impact of helix α7 on kinesin stepping behavior has been contested with early studies suggesting that the extended orientation of the neck linkers with the leading head neck linker undocked and the trailing head neck linker docked onto the catalytic core does not provide sufficient distance for the two-head-bound state and that there must be some degree of melting in the initial native neck coil to accommodate the two-head-bound state (Fig. 7, Intermediate E5) (59–61). Computational modeling and high-resolution force spectroscopy by atomic force microscopy support the argument that there is no unwinding of helix α7 or the initial coiled coil (62–64). Others however have indicated that flexibility can be encoded into this region by the specific amino acid sequence and define the critical properties for processivity (39–41,65,66). Although the studies reported here do not directly evaluate these hypotheses, they clearly indicate that the combination of the native helix α7 in the context of the 17-residue neck linkers of kinesin-2 KIF3A, KIF3B, and KIF3C is responsible for the dramatic increase in the run lengths observed for KIF3AC and KIF3AB. Furthermore, because the sequence of helix α7 of KIF3B and KIF3C are identical, the results suggest that the shorter run length of KIF3AC is due to the extended loop L11 in KIF3C.
Figure 7.
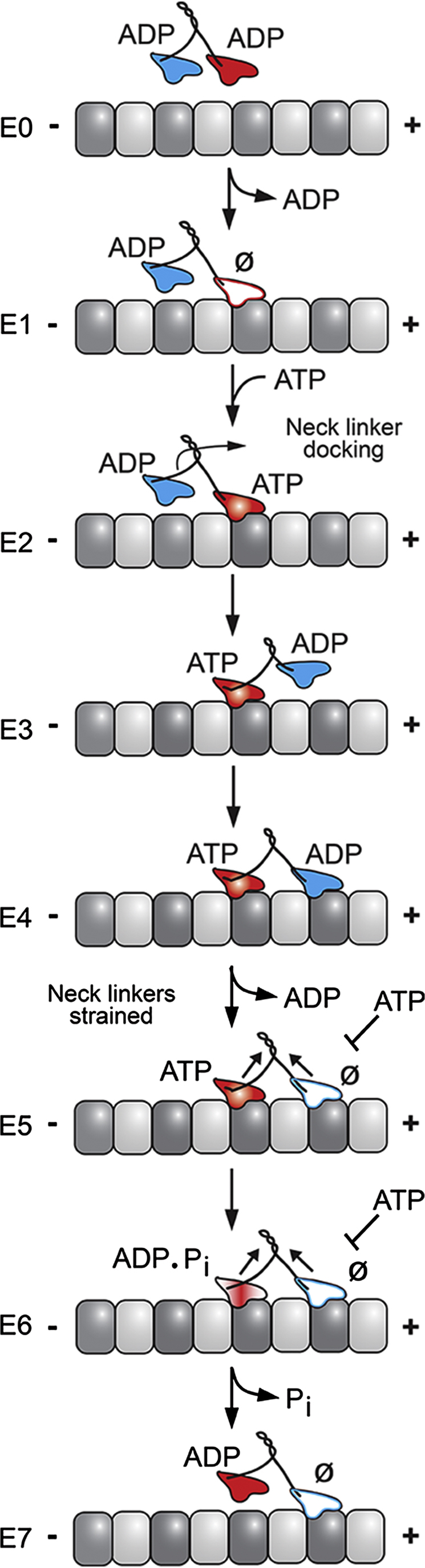
Kinesin-2 stepping model illustrates key transitions that impact processivity. A processive run is initiated upon microtubule collision followed by rapid ADP release to form the E1 intermediate in which the leading head is microtubule-bound and nucleotide-free (Ø), whereas the trailing head is detached with ADP tightly bound. ATP binding to the leading head induces a series of structural transitions including neck linker docking and positioning of the trailing ADP-head 16-nm ahead to its new microtubule-binding site (E2–E4). ADP release (E4–E5) results in the E5 two-head bound state, thereby generating intermolecular strain, which inhibits ATP binding at the leading head. ATP hydrolysis at the trailing head followed by phosphate (Pi) release generates the ADP state, which is weakly bound to the MT (E6–E7). Detachment of the trailing head as the ADP intermediate relieves the intermolecular strain (E7), and the next processive step continues as ATP binds to the leading head.
Tuning of kinesin-2 KIF3AC processive stepping
The major difference in the processivity of kinesin-1 and kinesin-2 has been reported to be the difference in their response to force, where kinesin-2 KIF3AB is much more likely to detach at submaximal forces (7,20). The kinesin-2 stepping model in Fig. 7 highlights two transitions that could result in termination of a processive run that may be the points in the cycle most sensitive to hindering loads. The first occurs between E2 to E4 during the process of neck linker docking to the motor domain. The efficiency of this transition is crucial as it allows for the ADP-bound motor to find its subsequent binding site along the microtubule track before the ATP-bound head undergoes ATP hydrolysis and loses its tight binding. A processive run could end prematurely as a result of inefficient neck linker docking, which could be a likely scenario if the neck linker were engineered to be longer or shorter or different than what is naturally encoded in the native sequence (39,41,43,44,66–68).
The series of events that occurs in the kinesin motor domain upon microtubule binding (Fig. 7, E0–E1 and E4–E5) has been found by structural studies to be linked to changes in L11. The C-terminus of L11 becomes partially ordered upon microtubule collision which results in elongation of the switch II helix (α4) by ∼2.5 turns. The ordered residues of L11 interact with the α-tubulin subunit causing a rearrangement of the switch II residues for nucleophilic attack in the nucleotide binding pocket (48,53–55) and affecting release of the bound ADP. The efficiency of this collision-induced ADP release would be at least partially dependent upon loop L11 and would be expected to be variable between different motors dependent upon the length and sequence of this flexible surface loop. The engineered KIF3C motor with a shortened L11 resulted in a KIF3AC heterodimer with enhanced processivity, which the data reveal was due in part to an increase in apparent microtubule binding affinity. Therefore, the results for KIF3AC and KIF3ACΔL11 suggest that the extended L11 in KIF3C is decreasing the run length of KIF3AC relative to KIF3AB and doing so through tuning the microtubule affinity.
The second transition that would determine the fate of a processive run occurs when the dimer is in a strained two-head-bound configuration between E4 to E6 (Fig. 7). The degree of strain between the two heads is critical for maintaining the asynchrony of the two ATPase cycles, putting the nucleotide-free head in an orientation less favorable for accepting ATP until the trailing head hydrolyzes its bound ATP and releases phosphate. If the nucleotide-free leading head in this strained configuration were able to bind ATP, even rarely, those instances would result in the loss of asynchrony such that both heads would enter a microtubule weakly-bound ADP state. The result would be premature release of the dimer from its processive run.
Impact of KIF3A on KIF3C during the processive run
Because of the significant difference in the motor properties of KIF3AA and KIF3CC revealed by the results presented, questions remain about the behavior of the individual motor heads once KIF3AC enters a processive run. For example, do KIF3A and KIF3C become more similar in their stepping mechanochemistry because of intermolecular communication that maintains the ATPase cycle of each head out-of-phase with other? The data show (Fig. 4 E) that the velocity of KIF3AC is somewhat slower than KIF3AA but KIF3AC is more processive than KIF3AA or KIF3CC. Furthermore, we do not yet know whether the response of KIF3AC to force is more similar to KIF3AB or kinesin-1. For example, is there an intrinsic bias maintained within KIF3AC such that either the KIF3A or KIF3C head is more sensitive to hindering loads and more likely to end the processive run, which would imply potential differences in their gating mechanisms? These hypotheses warrant further investigation and may provide new insights to understand the unique contributions of KIF3AC for intracellular transport in neurons.
Perhaps most noteworthy is that the results presented here show that kinesin-2, when examined with the 17-residue neck linker in conjunction with the native helix α7 for entry into its coiled-coil domain, exhibits extensive run lengths and velocities comparable to conventional kinesin-1. These results argue against a view that all coiled coils are equal or simply provide an inert architecture for the dimerization of the motor domains into a functional unit. Rather, these characterizations of KIF3AC and KIF3AB that include the native neck linker-helix α7 motif highlight the importance of this domain for the processive properties of kinesin motors. Furthermore, to our knowledge, the novel finding reported here that both KIF3AC and KIF3AB can traverse long distances over the microtubule track reveal that these motors are adept and well-tuned for their specialized roles as cargo transporters.
Author Contributions
S.G.-L., K.C.R., I.R., and S.P.G. designed research; S.G.-L., K.C.R., K.C.T., and B.M.B performed research; and S.G.-L., K.C.R., B.M.B., I.R., and S.P.G. analyzed data and wrote the article.
Acknowledgments
We thank Jennifer L. Ross and Leslie Conway (University of Massachusetts, Amherst) for assistance in the experimental design and analysis of the single molecule experiments, and Megan Valentine (University of California, Santa Barbara) for thoughtful discussions. We also acknowledge the members of the Susan P. Gilbert lab and Ivan Rayment lab for thoughtful discussions and input on these experiments and analyses.
This work was supported by National Institutes of Health grants R37-GM54141 (to S.P.G.) and R01-GM086351 (to I.R.). K.C.R. was supported by NIH training grant T32-GM07215.
Editor: Ram Dixit.
Footnotes
Stephanie Guzik-Lendrum and Katherine C. Rank contributed equally to this work.
Supporting Materials and Methods, two figures, and eight movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00809-7.
Contributor Information
Ivan Rayment, Email: ivan_rayment@biochem.wisc.edu.
Susan P. Gilbert, Email: sgilbert@rpi.edu.
Supporting Material
Average velocity 305.1 nm/s, mean run length 1.26 μm (see also Fig. 3 C). Video playback speed at 20× real-time.
Average velocity 168.8 nm/s, mean run length 1.23 μm (see also Fig. 3 A). Video playback speed at 20× real-time.
Average velocity 224.5 nm/s, mean run length 1.62 μm (see also Fig. 3 B). Video playback speed at 20× real-time.
Average velocity 171.5 nm/s, mean run length 1.52 μm (see also Fig. 4 C). Video playback speed at 20× real-time.
Average velocity of 7.5 nm/s, mean run length 0.57 μm (see also Fig. 5 A). Timeframe of video is 30 min, with playback speed at 360× real-time.
Average velocity 7.5 nm/s, mean run length 0.75 μm (see also Fig. 5 B). Timeframe of video is 30 min, with playback speed at 360× real-time.
Average velocity 239.2 nm/s, mean run length 0.98 μm (see also Fig. 5 C). Video playback speed at 20× real-time.
Average velocity 327.6 nm/s, mean run length 1.51 μm (see also Fig. 5 D). Video playback speed at 20× real-time.
References
- 1.Marx A., Hoenger A., Mandelkow E. Structures of kinesin motor proteins. Cell Motil. Cytoskeleton. 2009;66:958–966. doi: 10.1002/cm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirokawa N., Noda Y., Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 3.Hirokawa N., Tanaka Y. Kinesin superfamily proteins (KIFs): various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 2015;334:16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Scholey J.M. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu. Rev. Cell Dev. Biol. 2013;29:443–469. doi: 10.1146/annurev-cellbio-101512-122335. [DOI] [PubMed] [Google Scholar]
- 5.Verhey K.J., Kaul N., Soppina V. Kinesin assembly and movement in cells. Annu. Rev. Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- 6.Aizawa H., Sekine Y., Hirokawa N. Kinesin family in murine central nervous system. J. Cell Biol. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreasson J.O., Shastry S., Block S.M. The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 2015;25:1166–1175. doi: 10.1016/j.cub.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter B.S., Barry R.L., Allen B.L. The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J. Cell Sci. 2015;128:1034–1050. doi: 10.1242/jcs.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidovic L., Jaglin X.H., Khandjian E.W. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 2007;16:3047–3058. doi: 10.1093/hmg/ddm263. [DOI] [PubMed] [Google Scholar]
- 10.Gumy L.F., Chew D.J., Fawcett J.W. The kinesin-2 family member KIF3C regulates microtubule dynamics and is required for axon growth and regeneration. J. Neurosci. 2013;33:11329–11345. doi: 10.1523/JNEUROSCI.5221-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirokawa N., Niwa S., Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 12.Huang C.F., Banker G. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 2012;13:549–564. doi: 10.1111/j.1600-0854.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo S., Sato-Yoshitake R., Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marszalek J.R., Liu X., Goldstein L.S. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 15.Morris R.L., Scholey J.M. Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J. Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muresan V., Abramson T., Schnapp B.J. KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol. Biol. Cell. 1998;9:637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navone F., Consalez G.G., Cabibbo A. Expression of KIF3C kinesin during neural development and in vitro neuronal differentiation. J. Neurochem. 2001;77:741–753. doi: 10.1046/j.1471-4159.2001.00277.x. [DOI] [PubMed] [Google Scholar]
- 18.Sardella M., Navone F., Cabibbo A. KIF3C, a novel member of the kinesin superfamily: sequence, expression, and mapping to human chromosome 2 at 2p23. Genomics. 1998;47:405–408. doi: 10.1006/geno.1997.5123. [DOI] [PubMed] [Google Scholar]
- 19.Scholey J.M. Kinesin-2 motors transport IFT-particles, dyneins and tubulin subunits to the tips of Caenorhabditis elegans sensory cilia: relevance to vision research? Vision Res. 2012;75:44–52. doi: 10.1016/j.visres.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder H.W., 3rd, Hendricks A.G., Holzbaur E.L. Force-dependent detachment of kinesin-2 biases track switching at cytoskeletal filament intersections. Biophys. J. 2012;103:48–58. doi: 10.1016/j.bpj.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenova I., Ikeda K., Rodionov V. Regulation of microtubule-based transport by MAP4. Mol. Biol. Cell. 2014;25:3119–3132. doi: 10.1091/mbc.E14-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman M.A., Kaech S., Banker G. Expression of kinesin superfamily genes in cultured hippocampal neurons. Cytoskeleton. 2010;67:784–795. doi: 10.1002/cm.20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki H., Nakata T., Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z., Goldstein L.S. Characterization of the KIF3C neural kinesin-like motor from mouse. Mol. Biol. Cell. 1998;9:249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z., Roberts E.A., Goldstein L.S. Functional analysis of mouse kinesin motor Kif3C. Mol. Cell Biol. 2001;21:5306–5311. doi: 10.1128/MCB.21.16.5306-5311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C., Omori Y., Malicki J. Kinesin-2 family in vertebrate ciliogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:2388–2393. doi: 10.1073/pnas.1116035109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond J.W., Blasius T.L., Verhey K.J. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirajuddin M., Rice L.M., Vale R.D. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albracht C.D., Rank K.C., Gilbert S.P. Kinesin-2 KIF3AB Exhibits Novel ATPase Characteristics. J. Biol. Chem. 2014;289:27836–27848. doi: 10.1074/jbc.M114.583914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixit R., Ross J.L. Studying plus-end tracking at single molecule resolution using TIRF microscopy. Methods Cell Biol. 2010;95:543–554. doi: 10.1016/S0091-679X(10)95027-9. [DOI] [PubMed] [Google Scholar]
- 31.Brunnbauer M., Dombi R., Okten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol. Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Taylor K.C., Buvoli M., Rayment I. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc. Natl. Acad. Sci. USA. 2015;112:E3806–3815. doi: 10.1073/pnas.1505813112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frye J., Klenchin V.A., Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition. Biochemistry. 2010;49:4908–4920. doi: 10.1021/bi100349a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindhout D.A., Litowski J.R., Sykes B.D. NMR solution structure of a highly stable de novo heterodimeric coiled-coil. Biopolymers. 2004;75:367–375. doi: 10.1002/bip.20150. [DOI] [PubMed] [Google Scholar]
- 35.Rank K.C., Chen C.J., Rayment I. Kar3Vik1, a member of the Kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern. J. Cell Biol. 2012;197:957–970. doi: 10.1083/jcb.201201132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen I., Veprintsev D., Steinmetz M.O. End binding proteins are obligatory dimers. PLoS One. 2013;8:e74448. doi: 10.1371/journal.pone.0074448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bu W., Su L.K. Characterization of functional domains of human EB1 family proteins. J. Biol. Chem. 2003;278:49721–49731. doi: 10.1074/jbc.M306194200. [DOI] [PubMed] [Google Scholar]
- 38.Komaki S., Abe T., Hashimoto T. Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J. Cell Sci. 2010;123:451–459. doi: 10.1242/jcs.062703. [DOI] [PubMed] [Google Scholar]
- 39.Romberg L., Pierce D.W., Vale R.D. Role of the kinesin neck region in processive microtubule-based motility. J. Cell Biol. 1998;140:1407–1416. doi: 10.1083/jcb.140.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorn K.S., Ubersax J.A., Vale R.D. Engineering the processive run length of the kinesin motor. J. Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildiz A., Tomishige M., Vale R.D. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conway L., Wood D., Ross J.L. Motor transport of self-assembled cargos in crowded environments. Proc. Natl. Acad. Sci. USA. 2012;109:20814–20819. doi: 10.1073/pnas.1209304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shastry S., Hancock W.O. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shastry S., Hancock W.O. Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoeprich G.J., Thompson A.R., Berger C.L. Kinesin’s neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 2014;106:1691–1700. doi: 10.1016/j.bpj.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rank K.C., Rayment I. Functional asymmetry in kinesin and dynein dimers. Biol. Cell. 2013;105:1–13. doi: 10.1111/boc.201200044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kull F.J., Sablin E.P., Vale R.D. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y.H., Marx A., Mandelkow E. Structure of a fast kinesin: implications for ATPase mechanism and interactions with microtubules. EMBO J. 2001;20:6213–6225. doi: 10.1093/emboj/20.22.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sindelar C.V., Budny M.J., Cooke R. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat. Struct. Biol. 2002;9:844–848. doi: 10.1038/nsb852. [DOI] [PubMed] [Google Scholar]
- 50.Marx A., Muller J., Mandelkow E. The structure of microtubule motor proteins. Adv. Protein Chem. 2005;71:299–344. doi: 10.1016/S0065-3233(04)71008-6. [DOI] [PubMed] [Google Scholar]
- 51.Sindelar C.V., Downing K.H. The beginning of kinesin’s force-generating cycle visualized at 9-A resolution. J. Cell Biol. 2007;177:377–385. doi: 10.1083/jcb.200612090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parke C.L., Wojcik E.J., Worthylake D.K. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J. Biol. Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gigant B., Wang W., Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- 54.Shang Z., Zhou K., Sindelar C.V. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. eLife. 2014;3:e04686. doi: 10.7554/eLife.04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao L., Wang W., Gigant B. The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat. Commun. 2014;5:5364. doi: 10.1038/ncomms6364. [DOI] [PubMed] [Google Scholar]
- 56.Morikawa M., Yajima H., Hirokawa N. X-ray and Cryo-EM structures reveal mutual conformational changes of Kinesin and GTP-state microtubules upon binding. EMBO J. 2015;34:1270–1286. doi: 10.15252/embj.201490588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozielski F., Sack S., Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 58.Delorenzi M., Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 59.Tripet B., Vale R.D., Hodges R.S. Demonstration of coiled-coil interactions within the kinesin neck region using synthetic peptides. Implications for motor activity. J. Biol. Chem. 1997;272:8946–8956. doi: 10.1074/jbc.272.14.8946. [DOI] [PubMed] [Google Scholar]
- 60.Thormahlen M., Marx A., Mandelkow E. The coiled-coil helix in the neck of kinesin. J. Struct. Biol. 1998;122:30–41. doi: 10.1006/jsbi.1998.3986. [DOI] [PubMed] [Google Scholar]
- 61.Hoenger A., Sack S., Mandelkow E. Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with x-ray structure and implications for motility. J. Cell Biol. 1998;141:419–430. doi: 10.1083/jcb.141.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyeon C., Onuchic J.N. Mechanical control of the directional stepping dynamics of the kinesin motor. Proc. Natl. Acad. Sci. USA. 2007;104:17382–17387. doi: 10.1073/pnas.0708828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyeon C., Onuchic J.N. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc. Natl. Acad. Sci. USA. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bornschlogl T., Woehlke G., Rief M. Single molecule mechanics of the kinesin neck. Proc. Natl. Acad. Sci. USA. 2009;106:6992–6997. doi: 10.1073/pnas.0812620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomishige M., Vale R.D. Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J. Cell Biol. 2000;151:1081–1092. doi: 10.1083/jcb.151.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toprak E., Yildiz A., Selvin P.R. Why kinesin is so processive. Proc. Natl. Acad. Sci. USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clancy B.E., Behnke-Parks W.M., Block S.M. A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duselder A., Thiede C., Lakamper S. Neck-linker length dependence of processive Kinesin-5 motility. J. Mol. Biol. 2012;423:159–168. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average velocity 305.1 nm/s, mean run length 1.26 μm (see also Fig. 3 C). Video playback speed at 20× real-time.
Average velocity 168.8 nm/s, mean run length 1.23 μm (see also Fig. 3 A). Video playback speed at 20× real-time.
Average velocity 224.5 nm/s, mean run length 1.62 μm (see also Fig. 3 B). Video playback speed at 20× real-time.
Average velocity 171.5 nm/s, mean run length 1.52 μm (see also Fig. 4 C). Video playback speed at 20× real-time.
Average velocity of 7.5 nm/s, mean run length 0.57 μm (see also Fig. 5 A). Timeframe of video is 30 min, with playback speed at 360× real-time.
Average velocity 7.5 nm/s, mean run length 0.75 μm (see also Fig. 5 B). Timeframe of video is 30 min, with playback speed at 360× real-time.
Average velocity 239.2 nm/s, mean run length 0.98 μm (see also Fig. 5 C). Video playback speed at 20× real-time.
Average velocity 327.6 nm/s, mean run length 1.51 μm (see also Fig. 5 D). Video playback speed at 20× real-time.