Abstract
In eukaryotes, the origin recognition complex (ORC) heterohexamer preferentially binds replication origins to trigger initiation of DNA replication. Crystallographic studies using eubacterial and archaeal ORC orthologs suggested that eukaryotic ORC may bind to origin DNA via putative winged-helix DNA-binding domains and AAA+ ATPase domains. However, the mechanisms how eukaryotic ORC recognizes origin DNA remain elusive. Here, we show in budding yeast that Lys-362 and Arg-367 residues of the largest subunit (Orc1), both outside the aforementioned domains, are crucial for specific binding of ORC to origin DNA. These basic residues, which reside in a putative disordered domain, were dispensable for interaction with ATP and non-specific DNA sequences, suggesting a specific role in recognition. Consistent with this, both residues were required for origin binding of Orc1 in vivo. A truncated Orc1 polypeptide containing these residues solely recognizes ARS sequence with low affinity and Arg-367 residue stimulates sequence specific binding mode of the polypeptide. Lys-362 and Arg-367 residues of Orc1 are highly conserved among eukaryotic ORCs, but not in eubacterial and archaeal orthologs, suggesting a eukaryote-specific mechanism underlying recognition of replication origins by ORC.
Formation of higher-order nucleoprotein complexes at chromosomal replication origin(s) is a prerequisite for once-per-cell-cycle DNA replication in all three domains of life1,2,3,4. In eukaryotes, the earliest step of DNA replication is ATP-dependent association of the origin recognition complex (ORC), which consists of Orc1/2/3/4/5/6, with chromosomal replication origins. In yeast, these replication origins are called autonomous replicating sequences (ARSs). In G1 phase, ORC binding to replication origins triggers recruitment of Cdc6, Cdt1, and the MCM2–7 heterohexamer onto double-stranded (ds) DNA to form the ORC–Cdc6–Cdt1–MCM2–7 (OCCM) complex5. This step is followed by ordered dissociation and association processes, which allows formation of the ORC–Cdc6–MCM2–7 (OCM) intermediate and recruitment of another MCM2–7 hexamer to yield a salt-stable, head-to-head MCM2–7 double hexamer. The resultant double hexamer is the substrate for formation of the Cdc45–MCM2–7–GINS (CMG) complex at the G1–S transition. The CMG complex, which is an active helicase in vitro, is thought to serve in S phase as the replicative helicase component of the replisome for daughter-strand synthesis.
Basic structures of all of the ORC subunits except for Orc6 are suggested to be highly conserved among eukaryotes. Orc1/2/3/4/5 contain a motif related to the AAA+ (ATPase associated with various cellular activities) family in the middle region of each primary sequence, as well as one or two putative DNA-binding winged-helix (WH) motifs in the C-terminal region. A typical AAA+ fold is a globular structure with 5 parallel β-strands that make up a β-sheet, which is encircled by helical structures. A typical WH fold is a compact α/β structure bearing twisted antiparallel β strands and the two characteristic loops (a.k.a. ‘wings’) at the C-terminus of the fold6. Orc1, the largest subunit of ORC, has an extension at the N-terminus not found in other ORC subunits. The N-terminal half of the extension contains a bromo-adjacent homology (BAH) domain that has affinity for transcription-related proteins7. The BAH domain in Saccharomyces cerevisiae (Sc) Orc1 is a globular structure in the crystal, consisting of a conserved BAH core rich in β-strands and a non-conserved helical sub-domain8. The BAH domain is dispensable for cell viability in S. cerevisiae, but stabilizes the association between ORC and a subset of ARSs in vivo9. Mechanism of this stabilization process might not be simple, as another group shows that ScORC-ARS binding is stabilized by nucleosome in vitro and that the BAH domain is dispensable for this process10. So far, more than 400 ARSs throughout the genome have been experimentally confirmed11. Comprehensive mutagenesis of ARS plasmids has revealed two major functional elements of these sequences: the essential A element containing the ARS consensus sequence (ACS) and the stimulatory B elements12.
ScORC binds specifically to ARSs in vitro in a competitor DNA-dependent manner13,14. By contrast, in Schizosaccharomyces pombe (Sp), Drosophila (Dm), and human (Hs), ORC-origin DNA binding is less sequence-specific, with a preference for AT-rich or negatively supercoiled DNA15,16,17,18. In light of this difference, ScORC represents a unique model for analyzing the crucial mechanisms that support molecular recognition of ARSs.
ORC orthologs in eubacteria and archaea have properties similar to those of eukaryotic ORC. In Escherichia coli, DnaA binds to the unique chromosomal replication origin oriC to recruit DnaB helicase4. Like Orc1, DnaA has a protein–protein interaction domain at the N-terminus, an AAA+ domain in the middle region, and a helix-turn-helix (HTH) DNA-binding domain (a simplified WH) at the C-terminus. An unstructured linker connects the N-terminal and AAA+ domains. The HTH domain directly binds to DNA19. In archaea, Sulfolobus solfataricus has three ORC orthologs, two of which form a heterodimer on one of three replication origins. By contrast, Aeropyrum pernix has only one ORC ortholog. In crystal structures of both archaeal proteins, the WH and AAA+ domains contact origin DNA20,21; the WH domain determines affinity for origin DNA, whereas the AAA+ domain contributes to origin specificity22.
In contrast to the situation in eubacteria and archaea, the common mechanisms by which eukaryotic ORC recognizes replication origins remain unknown. Requirements of AAA+ and WH domains for eukaryotic ORC binding to ARS have not been reported. Mechanism in BAH domain-dependent stimulation of ScORC-ARS binding in vivo remains elusive. Indeed, the mechanisms experimentally verified to date have emphasized diversity rather than similarity among species. In Drosophila, the least conserved subunit of ORC, Orc6, is essential for DNA binding, whereas ScORC lacking Orc6 can bind to ARS and recruit Cdc623,24,25. Hs Orc6 has little affinity for Orc1/2/3/4/5, yielding a substoichiometric hexamer in vitro26. In S. pombe, the predominant DNA-binding domain of ORC is in the N-terminal extension in Orc4, which is not conserved in other eukaryotes17,18,27. Recently, we performed single-particle analysis of the Cdc6–ORC–ARS complex in S. cerevisiae, on the basis of which we proposed that ARS tracks along the interior surface of the complex; this model is consistent with previous studies using different approaches, such as in vitro subunit mapping by crosslinking and observations of DNA bending by ScORC24,28,29. Given that eubacterial DnaA and archaeal ORC bind origin DNA(s) via their HTH and WH domains, respectively, with a contribution from the AAA+ domain in the case of archaeal ORC, some researchers have inferred that eukaryotic ORC may also bind origin DNA via WH and AAA+ domains3,7,26. So far, none of these hypotheses have been experimentally validated using eukaryotic mutant proteins.
In this study, we predicted a domain of ScORC involved in ARS binding, based on the structural model described above. Mutational analysis of this domain revealed that two residues play crucial roles in the specific interaction between the ARS and ORC hexamer. Contrary to the hypotheses described above, the identified residues are located in a basic residue-rich region outside the WH and AAA+ domains. Because these residues are highly conserved among eukaryotes, we propose a general mechanism in which eukaryotic ORC recognizes replication origins via the basic patch in Orc1, which we refer to as the eukaryotic origin sensor (EOS).
Results
Prediction of ORC residues that interact with the essential element of ARS1
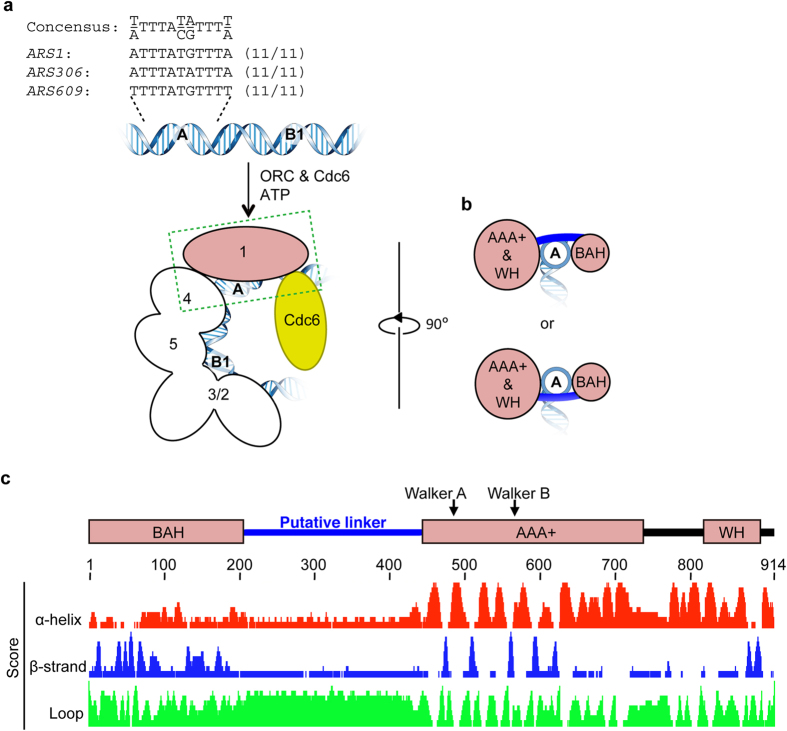
To identify the subunits, domains, and residues of ORC essential for ARS binding, we first examined the structural model of the ORC–Cdc6–ARS1 DNA complex we reported previously5,28. The AAA+ and WH domains of Orc1 are located on the same surface of the elongated, crescent-like, two-lobed shape, whereas the BAH domain is located on the other side of the structure (Fig. 1a,b). ARS1 is located in the center of ORC, tracking along the interior surface; the A element of ARS1 is close to the Orc1 subunit, consistent with the results of an in vitro mapping experiment in which purified ORC and ARS1 were incubated in the presence of a crosslinker24. We noticed that the putative linker domain of Orc1, not mentioned in the original reports, should be located between the BAH and AAA+ domains, and that the A element of ARS1 would thus be near the linker domain as well as the AAA+ and WH domains (Fig. 1b,c).
Figure 1. Structural models of ScOrc1.
(a) Cartoons summarizing the ORC binding regions in typical ARSs and the structural model of the ORC-Cdc6-ARS1 complex5,28. ACSs in ARS1, ARS306, and ARS609 are shown on top. Orc1 and Orc2/3/4/5 are shown in pink and white, respectively. Possible locations of the A and B1 elements are indicated. For clarity, Orc6 and the WH domains of Orc2/3/4/5 are omitted. (b) Side view of Orc1 complexed with DNA (as indicated by a dotted rectangle in panel (a) rotated 90° around the vertical axis from panel (a). Possible loci of a linker region that connects BAH and AAA+ domains are indicated as blue lines. (c) Domain structure and prediction of secondary structure of ScOrc1. Representative domains and motifs are indicated along with the predicted score of each residue. Walker A and B indicate the ATP-binding motifs. See text for BAH, AAA+ and WH.
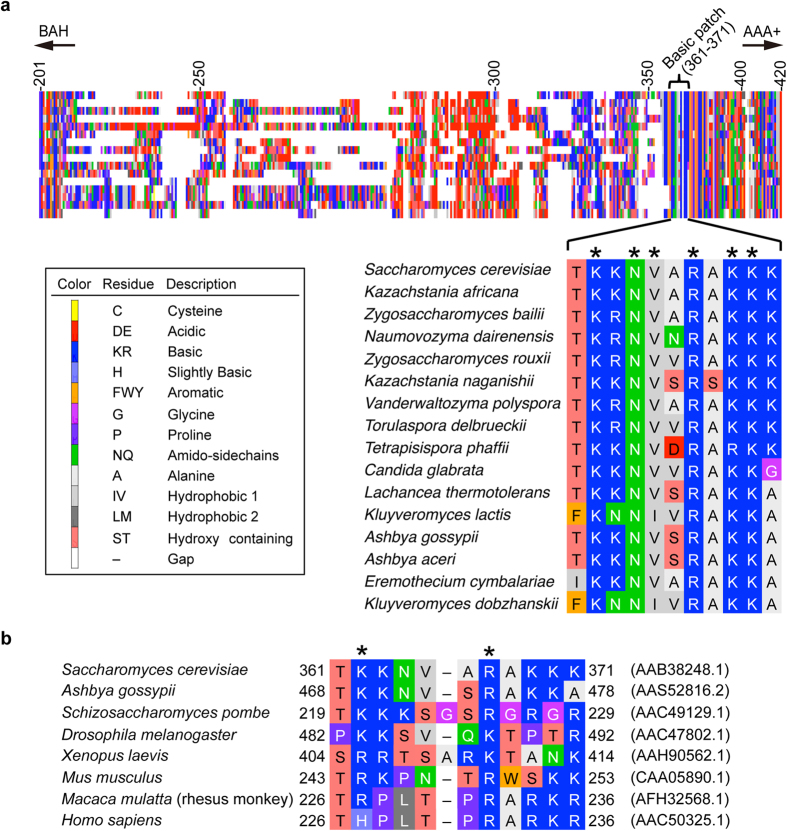
If this hypothesis is true, the linker domain may contain residues that could contact ARS1. To test this idea, we performed an in silico screen to identify conserved residues in the linker domain. BLAST search and multiple alignment analysis unveiled an 11 amino acid “patch” (ScOrc1361–371) conserved in fungi that is rich in basic residues (i.e., the EOS; Fig. 2a and Supplementary Data S1). A similar basic patch is also present in a wide range of eukaryotic cells including fungi, metazoans, and human, but not in representative eubacterial and archaeal orthologs (Fig. 2b and Supplementary Fig. S1). Because basic residues are positively charged at physiological pH, and DNA is negatively charged, the EOS is likely to contain DNA-binding residues.
Figure 2. In silico analyses of Orc1.
(a) Multiple alignment of a BLAST search with ScOrc1201–420 is shown on top using a colored rectangular pixel for each residue. A magnified alignment of the basic patch is shown below. The residues assigned to each color are indicated by the legend at left *, conserved residues. Full details of the alignment are shown in Supplementary Data S1. (b) Multiple alignment of the basic patch of eukaryotic Orc1 homologs, including representative model species. NCBI accession numbers are indicated on the right. Residues are colored as in panel (a). Histidine, though less basic than lysine or arginine, can play a crucial role for DNA binding at physiological pH19. *, Lys-362 and Arg-367 residues in ScOrc1 and equivalents in homologs.
Orc1 residues Lys-362 and Arg-367 have a crucial role for ARS-binding in vivo
To determine whether any of the residues in the EOS play a critical role in vivo, we performed a plasmid-shuffle assay using an orc1 disruptant pre-complemented with a URA3 plasmid bearing ORC1 as a tester strain. If the URA3 plasmid was swapped for a LEU2 plasmid bearing ORC1, it still complemented the orc1 disruption, but if it was swapped for a plasmid bearing orc1 K485T, an ATP-binding Walker A mutant, complementation was abolished (Fig. 3a), consistent with previous studies30,31. Notably, alanine-scanning experiments of the EOS using this assay revealed that orc1 K362A and orc1 R367A also failed to complement: these mutations resulted in slight residual activity and almost no activity, respectively (Fig. 3a). Both residues are highly conserved from yeast to human (Fig. 2b).
Figure 3. In vivo activities of orc1 mutants.
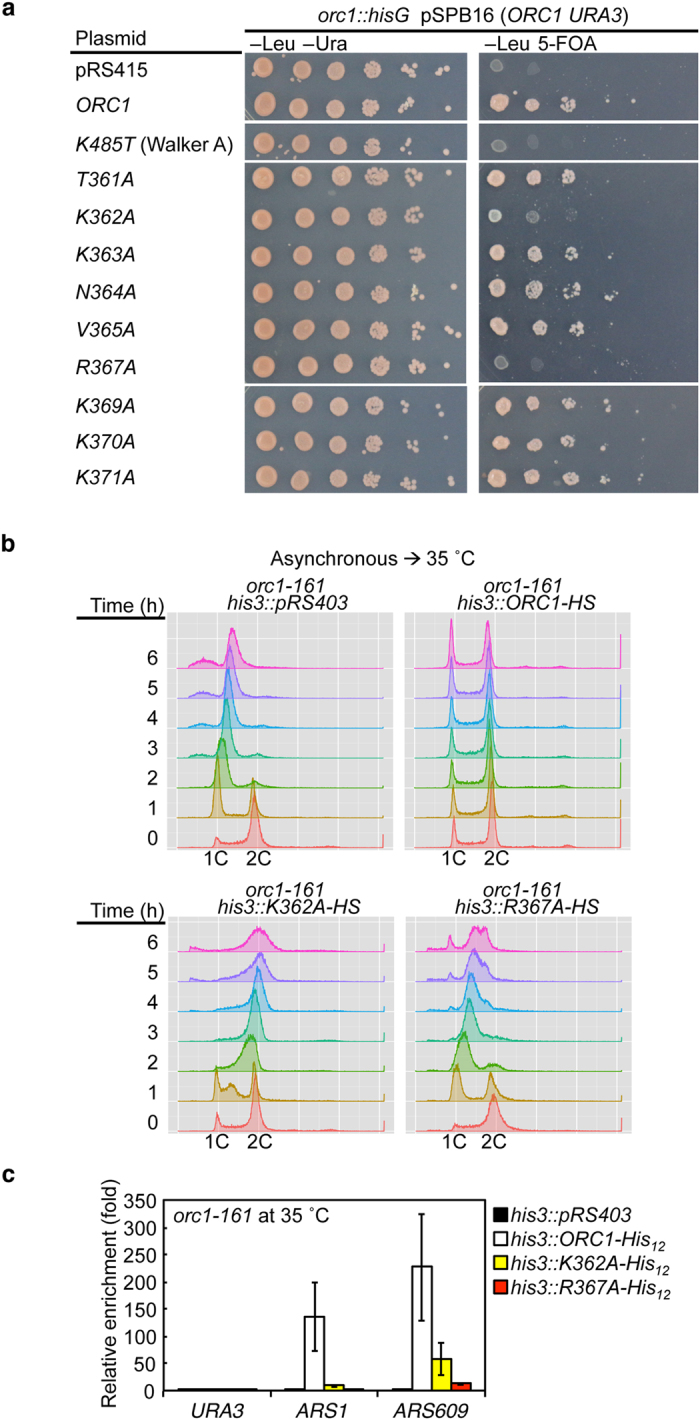
(a) Plasmid-shuffle assay. The tester strain YB838 (orc1::hisG pSPB16) was transformed with derivatives of pSPB15 bearing the indicated orc1 alleles and incubated at 30 °C. Serially diluted cells were spotted on the indicated selective media and further incubated at 30 °C. (b) Flow cytometry analysis. YHK26 (orc1-161 his3::pRS403), YHK27 (orc1-161 his3::ORC1-HS), YHK28 (orc1-161 his3::orc1 K362A-HS), and YHK29 (orc1-161 his3::orc1 R367A-HS) were grown at 23 °C to early log phase and then further incubated at 35 °C. Samples were taken at the indicated time points after the temperature shift. (c) ChAP assay. Cells of YHK26 (orc1-161 his3::pRS403), YHK33 (orc1-161 his3::ORC1-His12), YHK34 (orc1-161 his3::orc1 K362A-His12), and YHK35 (orc1-161 orc1 R367A-His12) were grown at 23 °C and further incubated at 35 °C. The amounts of indicated chromosomal loci crosslinked to wild-type or mutant Orc1-His12 were quantified and shown as relative enrichment compared with those using YHK26 (n = 3; mean ± SE).
Given that ORC is essential for the cell cycle, we predicted that the orc1 K362A and orc1 R367A strains would be defective in cell-cycle progression. To test this, we performed a cell-cycle execution point analysis of the orc1 K362A and orc1 R367A mutants. Because orc1 K362A and orc1 R367A strains obtained by plasmid shuffling did not grow (Fig. 3a), we constructed conditionally lethal variants by integrating His–Strep II (HS)-tagged orc1 K362A or orc1 R367A into the his3 locus of a temperature-sensitive orc1-161 strain. Cells expressing only the orc1-161 allele grow at the permissive temperature of 23 °C, but fail to grow at 30–37 °C due to a reduction in Orc1 protein level32,33,34. Addition of HS-tag sequences to endogenous ORC1 did not significantly affect doubling time or cell-cycle progression (Supplementary Fig. S2). orc1-161/ORC1-HS cells grew normally at 35°C, but orc1-161/orc1 K362A-HS and orc1-161/orc1 R367A-HS cells both exhibited severe cell-cycle delays after shift to the non-permissive temperature (Fig. 3b). Cell cycle of most of those cells was arrested in G2/M phase at the non-permissive temperature, which is consistent with the fact that loss of ORC1 function activates the DNA damage and spindle assembly checkpoint pathways33. Cell-cycle execution in orc1-161/K362A-HS cells was faster than that in orc1-161/orc1 R367A-HS cells. This is consistent with the results of the plasmid shuffle assay (Fig. 3a). ORC hexamers containing Orc1 K362A might have slight residual activity in initiation (see below). Hereafter, ORC hexamers containing Orc1 K362A or Orc1 R367A are referred to as ORC-1K362A and ORC-1R367A, respectively,
Based on those results, a plausible idea is that ORC-1K362A and ORC-1R367A have less or no affinity for ARS. To test this possibility, we first established a chromatin affinity purification (ChAP) assay in which His12-tagged Orc1 (Orc1-His12) is crosslinked with DNA and is recovered using a pull-down method under a denatured condition. The previously published Orc1 ChIP assay under a native condition underestimates Orc1-ARS binding in G1 phase due to steric inhibition of immunoprecipitation by ORC-bound replication proteins34. Integration of the ORC1-His12 gene into the his3 locus of the orc1-161 strain suppressed temperature sensitive phenotype (Supplementary Fig. S3), indicating that Orc1-His12 is active in vivo. Using this strain, association of Orc1-His12 with ARS1 and ARS609, a representative early and late origin, respectively, was observed in a His12-tag-dependent manner (Fig. 3c). In contrast, association of Orc1-His12 with URA3, a representative non-origin locus, was only minimal (Fig. 3c), as shown by Orc1 ChIP34.
Next, similar assays were performed using strains bearing mutant orc1-His12. Orc1 K362A-His12 and Orc1 R367A-His12 were impaired in binding to ARS1 and ARS609 although Orc1 K362A-His12 retained slight residual activity (Fig. 3c). These results are well consistent with the data shown above (Fig. 3a,b). Binding of these mutant Orc1-His12 to URA3 was kept to only a minimum (Fig. 3c), suggesting that it is unlikely that DNA replication promiscuously initiates from non-ARS regions which mutant ORC binds less sequence-specifically although the possibility of firing at unidentified non-ARS regions is not completely denied. A likely idea is that replication process on ARS is impaired in these mutant cells. Taken together, we conclude that Orc1 Lys-362 and Arg-367 residues have a crucial role for ARS binding of Orc1 in vivo.
Rapid purification of ORC using a mammalian overexpression system
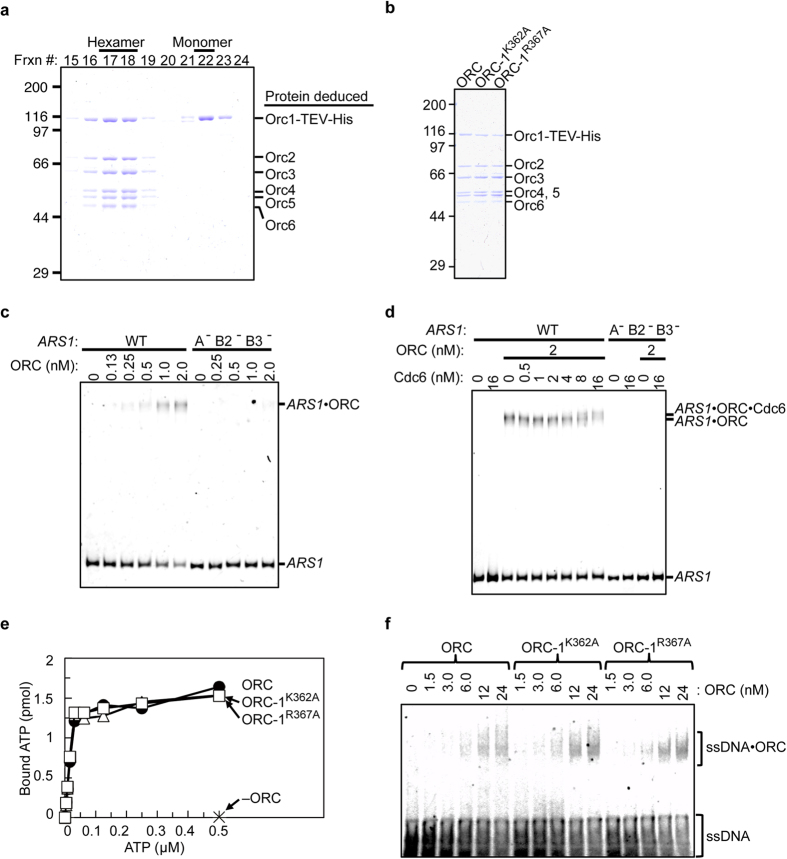
Previously published protocols for purification of recombinant ORC for EM studies and quantitative biochemistry involve co-overexpression of ORC components5,28,35,36. For preparation of mutant ORC, these methods are time-consuming due to the necessity of combining baculoviruses expressing multiple ORC subunits and the number of traditional fractionation steps that must be performed without affinity tags. To accelerate mutational analyses of ORC in vitro, we developed a novel method for overexpression and purification of ORC in mammalian cells. In this system, ORC-overproducing plasmids, once mutagenized and sequenced, can be directly subjected to efficient co-transfection into mammalian cells37, thereby eliminating the need to construct and amplify baculoviruses. In addition, the use of an affinity column enabled omission of several purification steps. Using our new method, we obtained 0.72 mg of wild-type ORC from 400 ml of medium with a purity of ~90%, comparable to the yield from insect cells (Fig. 4a,b).
Figure 4. In vitro activities of ScORC purified from a mammalian overexpression system.
(a) Gel-filtration analysis of affinity-purified ScOrc1-TEV-His co-overexpressed with ScOrc2/3/4/5/6 in 293T cells. Fractions were analyzed by 9% SDS-PAGE, followed by Coomassie staining. Gel positions of the ORC subunits are indicated. (b) Purified wild-type ORC, ORC-1K362A, and ORC-1R367A (1.2 pmol each) were analyzed by 9% SDS-PAGE, followed by Coomassie staining. (c) EMSA of Cy5-labeled DNA with wild-type ORC. DNA fragments contained wild-type (WT) or mutant (A− B2− B3−) ARS1. Protein concentrations are indicated. (d) EMSA of Cy5-labeled DNA, as in panel c, in the presence of wild-type ORC and Cdc6. (e) Affinities for ATP. Wild-type ORC or mutants (2.4 pmol each) were incubated with the indicated amount of [α-32P]ATP, and bound nucleotide was quantified by filter binding assay. (f) EMSA of Cy5-labeled ssDNA with wild-type ORC or mutants.
To determine whether this ORC preparation could be used for downstream applications (e.g., for biochemical assays), we analyzed the affinity of the purified ORC for ARS1 DNA by electrophoretic mobility shift assay (EMSA) using a fluorescent ARS1 fragment and non-labeled competitor DNA. The results revealed that our purified ORC retained sequence-specific affinity for ARS1 (Fig. 4c). Furthermore, when Cdc6 was added to this assay, the ORC–Cdc6 interaction was detectable as a Cdc6-dependent supershift of the ORC–ARS1 complex (Fig. 4d). These results are consistent with previous studies using ORC purified from baculovirus-transfected cells38, indicating that our novel ORC preparation is as useful for downstream applications as conventionally obtained material.
Next, we independently purified ORC-1K362A and ORC-1R367A. The purity of the mutant ORCs was comparable to that of the wild-type ORC (Fig. 4b).
Affinities of ORC-1K362A and ORC-1R367A for ATP and single-stranded (ss) DNA
To determine whether K362A and R367A mutations affect overall ORC activities, we first analyzed nucleotide binding by a nitrocellulose membrane-retention assay. The affinities of ORC-1K362A and ORC-1R367A for ATP were similar to that of wild-type ORC (Fig. 4e and Table 1).
Table 1. Affinities of wild-type and mutant ORC for ATP.
| Kd (nM) | Stoichiometry | |
|---|---|---|
| ORC | 31 | 0.78 |
| ORC-1K362A | 35 | 0.82 |
| ORC-1R367A | 32 | 0.81 |
Kd and stoichiometry were estimated from Fig. 4e using Scatchard plots.
ORC binds non-specific ssDNA in vitro in an ATP-independent manner39. We tested the mutant ORCs for this affinity by EMSA using 39-mer fluorescent ssDNA and competitor dsDNA. Wild-type ORC, ORC-1K362A, and ORC-1R367A all bound to ssDNA in an ORC concentration-dependent manner (Fig. 4f). These results suggest that Orc1 Lys-362 and Arg-367 residues are not required for ssDNA binding.
ORC-1K362A and ORC-1R367A are defective in ARS binding
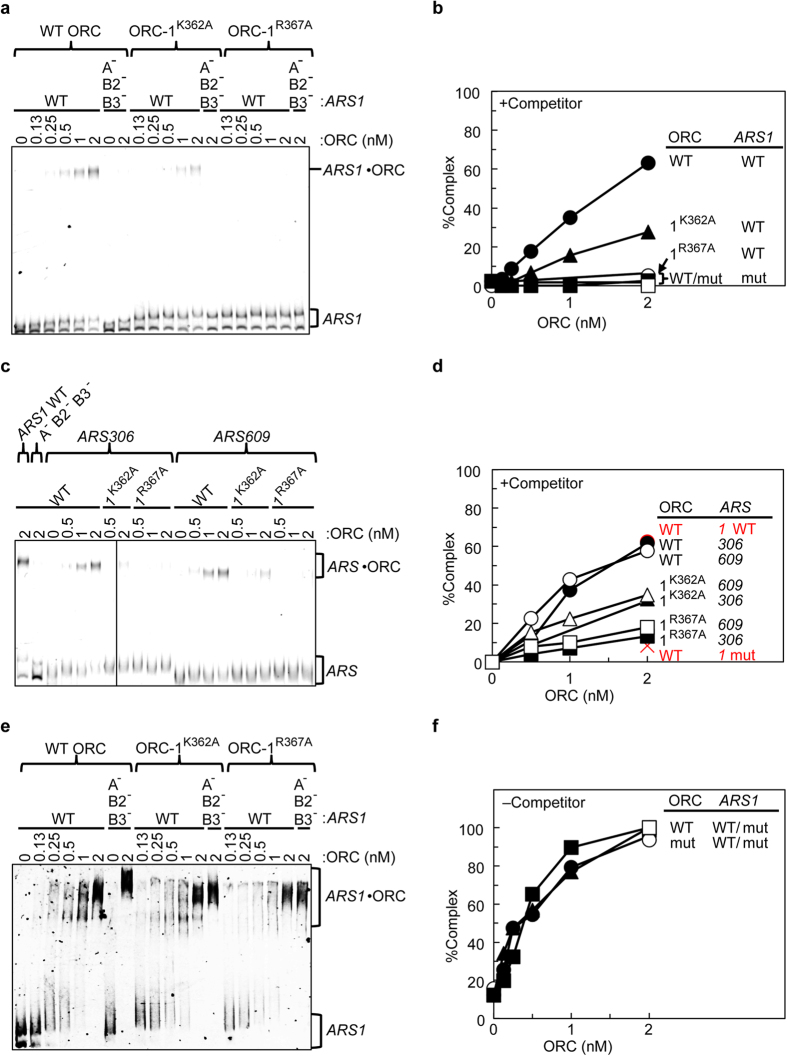
Next, we examined the ARS-binding activities of the mutant ORCs by EMSA using Cy5-labeled ARS1 and unlabeled competitor DNA. Both ORC-1K362A and ORC-1R367A had a lower affinity for ARS1 than wild-type ORC: binding of K362A was low but detectable, whereas the binding of R367A was almost undetectable (Fig. 5a,b). Similar results were obtained using ARS306 and ARS609, which are representative early and late origins, respectively (Fig. 5c,d), suggesting that the dependency of ARS binding upon Orc1 Lys-362 and Arg-367 is general, rather than an ARS1-specific phenomenon. The relative residual activities of Orc1 K362A and Orc1 R367A were consistent with those observed in vivo (Fig. 3), suggesting that the in vivo phenotypes of these mutants can be essentially explained by the defects in ARS-binding activity observed in vitro.
Figure 5. Affinities of ScORC for dsDNAs.
(a) EMSA of Cy5-labeled wild-type (WT) or mutant (A− B2− B3−) ARS1 DNA with wild-type or mutant ORC in the presence of competitor DNA. (b) Quantified results from panel (a) are shown. (c) EMSA of Cy5-labeled ARS306 or ARS609 DNA with wild-type or mutant ORC in the presence of competitor DNA. Wild-type (WT) or mutant (A− B2− B3− or mut) ARS1 was also used as controls. (d) Quantified results from panel c are shown. (e,f) EMSA of Cy5-labeled DNA in the absence of competitor DNA. DNAs were the same as in panels (a,b).
Next, we examined the non-specific DNA binding activity of ORC. As noted above, omission of competitor DNA eliminates the sequence specificity of ORC-DNA binding13. When EMSA was performed in the absence of competitor DNA, ORC-1K362A and ORC-1R367A both retained significant affinity for mutant ARS1 DNA (Fig. 5e,f). This result indicates that the binding mechanisms of ORC with non-specific and ARS DNA are mediated by at least partially independent mechanisms in terms of their requirement for the Orc1 Lys-362 and Arg-367 residues (see below).
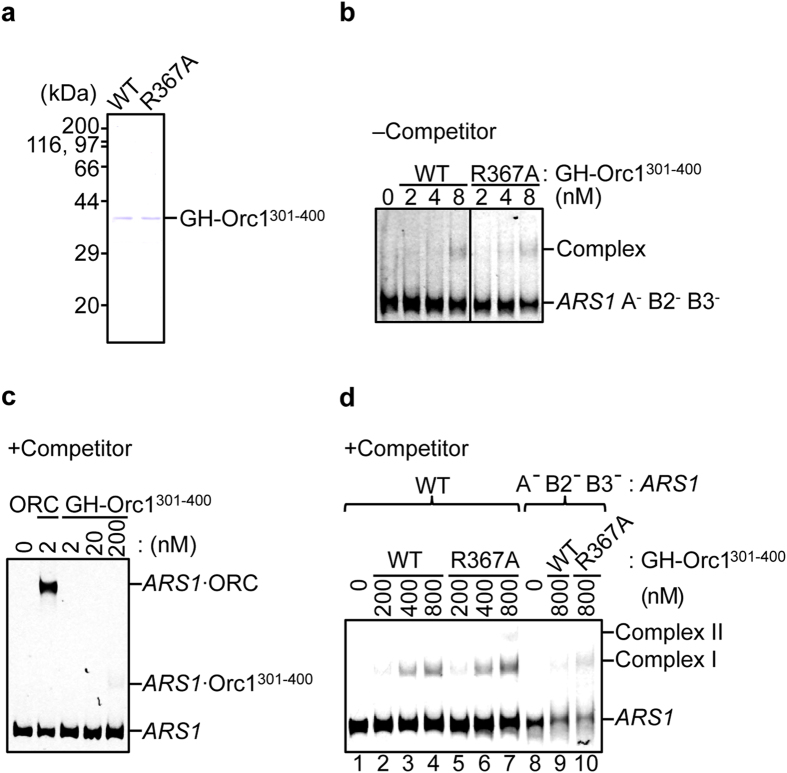
Role for Orc1301–400 in recognition of ARS
To investigate if Orc1 binds to ARS DNA via EOS in vitro, Orc1301–400 polypeptides with or without R367A substitution were purified as a GST-His (GH) tagged form up to near homogeneity (Fig. 6a). Orc1301–400 bears a part of the linker domain including EOS, but the AAA+ and WH domains are excluded (Fig. 1c). In the absence of competitor DNA, both wild-type and mutant GH-Orc1301–400 exhibited affinity for mutant ARS1 DNA (Fig. 6b), indicating that Orc1301–400 basically has EOS-independent non-specific DNA binding activity. In the presence of competitor DNA, wild-type GH-Orc1301–400 retained affinity for wild-type ARS1 DNA, but its affinity was at least 102-fold lower than that of ORC hexamer (Fig. 6c). This is consistent with the fact that ORC subcomplexes lacking one of Orc1/2/3/4/5 subunits are severely impaired in binding to ARS24.
Figure 6. Affinities of Orc1301–400 for dsDNAs.
(a) Wild-type and mutant GH-tagged Orc1301–400 (WT and R367A; 0.5 μg each) were analyzed by 12% SDS-PAGE, followed by Coomasie Brilliant Blue staining. (b) EMSA of Cy5-labeled mutant (A− B2− B3−) ARS1 DNA with wild-type or mutant GH-Orc1301–400 in the absence of competitor DNA. (c) EMSA of Cy5-labeled wild-type ARS1 DNA with ORC hexamer or GH-Orc1301–400 in the presence of competitor DNA. (d) EMSA of Cy5-labeled wild-type (WT) or mutant (A− B2− B3−) ARS1 DNA with wild-type or mutant GH-Orc1301–400 in the presence of competitor DNA.
Intriguingly, competitor DNA effectively reduced binding of wild-type GH-Orc1301–400 to mutant ARS1 and wild-type ARS sequence-dependent binding was evidently observed (Fig. 6d, lanes 4 and 9). These results suggest that Orc1301–400 contains ARS1-specific binding activity even though its affinity is low. When similar assay was performed using GH-Orc1301–400 bearing R367A, interaction with wild-type ARS1 DNA was slightly enhanced compared with wild-type GH-Orc1301–400 and even a supershift was observed (Fig. 6d, lanes 2–4 vs. lanes 5–7). Competitor DNA did not inhibit binding of GH-Orc1301–400 R367A to mutant ARS1 DNA as effectively as it did binding of wild-type GH-Orc1301–400 (Fig. 6d, lane 10). These results can be explained by a possibility that R367A mutation in GH-Orc1301–400 reduced sequence specificity of the polypeptide binding and stimulated abnormal binding modes of the polypeptide with DNA. In ORC hexamers, if Arg367-dependent recognition of ARS1 is impaired, abnormal binding modes with DNA might be conformationally inhibited by other domains of Orc1 or another ORC subunit(s) (see Discussion). Taken together, these results suggest that Orc1301–400 solely recognizes ARS sequence with low affinity and that Arg-367 residue stimulates sequence specific binding mode of Orc1301–400.
Discussion
In this study, we used a combination of in silico screening, yeast genetics, and biochemical analyses using mutant ORC proteins to identify Orc1 residues in the EOS that are essential for specific ORC hexamer–ARS binding. The EOS is located in a putative intrinsically disordered region rich in basic residues, between the BAH and the AAA+ domains of Orc1, which had not been previously proposed to be a DNA-binding domain. The critical residues identified in the EOS are highly conserved among eukaryotes, but not in the eubacterial and archaeal orthologs (Fig. 2; Supplementary Data S1; Supplementary Fig. S1). To our knowledge, this is the first evidence of a conserved structure–function relationship relevant to origin DNA binding by eukaryotic ORC. Intrinsically disordered basic regions in certain proteins facilitate or modulate selectivity by engaging in specific interactions with DNA40, which is consistent with our results (Figs 3c and 5). Taken together, we suggest that the EOS directly involves in effective and specific recruitment of ORCs onto ARSs. This mechanism might underlie the previous in vivo observation suggesting that the number of the total ORC molecules in a cell and the number of ORC-bound ARSs in a G1-phase cell are similar41,42: EOS might effectively support recruitment of ORC molecules to ARSs. Moreover, although the above model was constructed on the basis of findings in S. cerevisiae, it is consistent with a previous subcellular localization study in human cells showing that deletion of a region in HsOrc1 including both the nuclear localization signal and the entire EOS-homologous motif abolishes colocalization of HsOrc1 foci with DAPI43. We do not exclude a possibility that another region as well as EOS is important in regulation of specific localization of ORC.
It is unlikely, however, that the EOS is sufficient for ORC hexamer to successfully bind an ARS, as affinity of Orc1301–400 for ARS1 is at least 102-fold weaker than that of ORC hexamer (Fig. 6c). We suggest that EOS-flanking region in Orc1301–400 and other domain(s), as well as EOS, play mutually supportive roles to achieve high-affinity ARS binding, as with the WH and AAA+ domains of archaeal ORC (but not eubacterial DnaA)22. This is consistent with our finding that ORC-1K362A, ORC-1R367A, and GH-Orc1301–400 R367A retain non-specific DNA binding in the absence of competitor DNA (Figs 5e,f and 6b). Indeed, the ARS region bound to ORC (i.e., from A to B1) is longer than the EOS. In the archaeal ORC ortholog Orc1-1, mutations in an origin-binding site of AAA+ domain impair origin sequence specificity but only slightly reduce affinity for origin DNA22, which is likely similar to our observation that GH-Orc1301–400 bearing R367A exhibited non-specific binding activity even in the presence of competitor DNA (Fig. 6d). Thus, we suggest a model in which EOS directly recognizes the A element of ARS (Fig. 7), although we do not exclude an alternative model in which EOS might regulate direct interaction between somewhere in Orc1301–400 and the A element.
Figure 7. Model for EOS-mediated origin recognition in eukaryotic ORC.
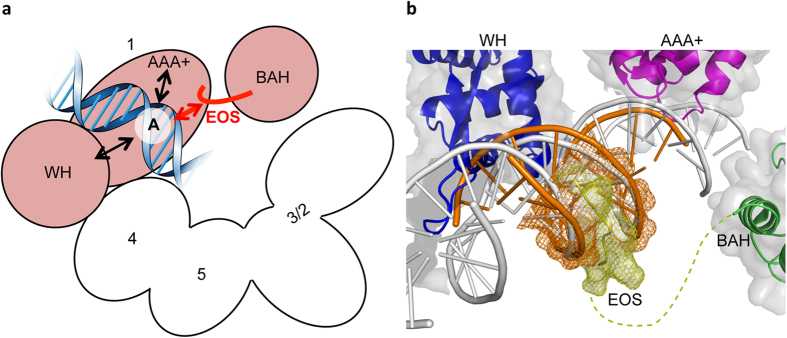
(a) A cartoon of ORC illustrating recognition of the A element via EOS and other domains in a mutually supportive manner. Orc1 and Orc2/3/4/5 are shown in pink and white, respectively. For clarity, Orc6 and the WH domains of Orc2/3/4/5 are omitted. (b) An atomic model predicts putative location of EOS. EOS (yellow) complexed with ACS DNA (orange) was computer-modeled and superimposed on the crystal structures of S. solfataricus Orc1-3 bound to DNA (PDB ID: 2QBY)21 and ScOrc1 BAH domain (green; PDB ID: 1M4Z)8. The AAA+ and WH domains and DNA in the crystal structure are shown in magenta, blue, and gray, respectively. See Discussion for details.
Taken together, we suggest the following model for recognition of replication origins and initiation thereon. First, ORC scans DNA non-specifically in an EOS-independent manner. When ORC reaches an ARS, unstructured EOS recognizes the A element via weak but specific affinity, followed by stabilization of the binding by other structured domains such as WH and AAA+ (Fig. 7a). This stabilization assumes certain steric configuration by EOS-mediated specific DNA recognition. In this case, EOS is likely to contact DNA from the opposite side of the WH and AAA+ domains: a minor groove might be involved in this contact (Fig. 7b). The Orc1 Arg-367 residue makes a greater contribution to this process than Lys-362. This recognition might stimulate conformational changes in the overall ORC structure, particularly Orc1. In the ORC–ARS intermediate, the conformationally altered ORC transiently bends the DNA to pull in the B1 element and facilitate its recognition, presumably via Orc2, 3, or 5 (Fig. 1a). This step, which confirms that the bound DNA region lies within an ARS, may be enhanced by additional conformational changes that occur upon ORC–Cdc6 binding. The resultant ORC–Cdc6–ARS complex ensures ARS-specific recruitment of the Cdt1–MCM complex. This model is consistent with the observation that ORC-1K362A and ORC-1R367A retain non-specific DNA-binding activity (Fig. 5e,f), as well as the results of a recent single-molecule study showing that ORC can slide along non-specific DNA sequences to reach an ARS region44. Also, this model explains a possible mechanism underlying the observation that GH-Orc1301–400 bearing R367A binds DNA non-specifically, but ORC-1R367A does not bind to ARS. The different degree of contribution by Lys-362 and Arg-367 residues in the model are consistent with the differences we observed in the residual activities of ORC-1K362A and ORC-1R367A, both in vitro in the ARS binding assay (Fig. 5a–d) and in vivo in the plasmid-shuffle, temperature-shift/flow cytometry, and ChAP assays (Fig. 3). Furthermore, this model agrees with the results of previous in vivo comprehensive mutagenesis studies using ARS1 and ARS307, in vitro footprinting using A− and B1− mutants, and our structural model12,14,28,42. Notably, our model explains why the ACS is more highly conserved throughout the genome than the B1 element, but still not sufficient for ORC binding42,45. Also, this model is consistent with previously reported models in which ORC bends the ARS24,28,29, analogous to bending of DNA by the intrinsically disordered C-terminus of histone H146.
Origin sequences in eukaryotes are divergent compared with those in eubacteria and archaea15,16,47, but it remains unclear why the mechanisms of origin recognition by eukaryotic ORC could have diversified despite the fact that WH and AAA+ domains in ORC are highly conserved. Because EOS-homologous motifs contain slight deviations, except for the two essential residues in ScOrc1 (Fig. 2b), we hypothesize that eukaryotic ORC evolved its origin recognition strategy via mutation of the EOS and/or flanking disordered region. This is consistent with the reported robustness of disordered polypeptides with respect to mutation and the role of disordered regions in defining DNA sequence-specific contacts48,49. This hypothesis may provide a key to solving species-specific mechanisms of ORC in other organisms.
Another noteworthy finding of this study is that the EOS-homologous motif in Hs Orc1 is conserved even though a G-rich RNA/ssDNA-binding domain is present immediately upstream of the AAA+ domain, a region flanking the EOS-homologous motif (Fig. 2b)50. Replication origins in human cells associate with a G quadruplex (G4)-forming consensus motif 51; thus there is an intriguing coincidence between the location and function of the G-rich RNA/ssDNA-binding domain. The EOS-homologous motif and the G-rich RNA/ssDNA-binding domain in HsOrc1 may have distinct, specific roles in recognition of origin DNA.
A similar idea could be extended to the possible roles of the EOS-homologous motifs in S. pombe, Drosophila, and human, in which the unique N-terminal extension of SpOrc4 and the transcription factor-homologous domain in DmOrc6 and HsOrc6, respectively, are solely responsible for DNA binding (Fig. 2b)17,23,27,52. We suggest that the EOS-homologous motifs of Orc1 in these species might play a distinct role in DNA binding and formation of active nucleoprotein complexes even when SpOrc4, DmOrc6, and HsOrc6 are bound to origin DNA. Consistent with this idea, SpORC binds to DNA in at least two steps in vitro, of which only the first step involves the extension of the SpOrc4 subunit53. In S. cerevisiae, Orc6 is dispensable for ORC–ARS binding and contains a long insertion not found in other eukaryotes7,24.
It has been reported that the BAH domain in metazoan Orc1 regulates ORC chromatin association, cell cycle progression, and development via interaction with a histone H4K20me254. However, core histone-mediated regulation of ORC-DNA binding may be diversified among species. For example, ScOrc1 and SpOrc1 do not bind to H4K20me254. In S. cerevisiae, stabilization of ORC-ARS binding by nucleosomes in vitro is independent of the BAH domain of Orc110 even though the BAH domain in ScOrc1 stabilizes in vivo association of ORC to a subset of ARSs9. As this apparent complexity is of future interest, further dissection of ORC functional structures will pave the way to understand detailed mechanisms in ORC-nucleosome interactions.
During preparation of this manuscript, the crystal structure of a DNA-free, truncated form of DmORC was reported55. The reported structure is fully consistent with our model. The overall structures of ScORC and DmORC share a similar architecture28 and DmOrc1 also bears an EOS-homologous motif (Fig. 2b), which was eliminated in order to achieve crystallization.
Methods
Yeast techniques
Yeast media and cell synchronization were as described56. For plasmid-shuffle assay, medium was supplemented with 1 g/L 5-fluoroorotic acid (FOA). For flow cytometry, cells were stained with SYTOX Green (Life Technologies) and analyzed on a FACSCalibur using the CellQuest Pro software (BD Biosciences) as described previously56.
Plasmids and yeast strains
Yeast pRS vectors and pSPB15 (ORC1 ARS CEN LEU2) were gifts from Dr. Bruce Stillman. The mammalian expression vector version 3–537 was a gift from Dr. Hisao Masai. Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent) and verified by sequencing. pHK142 (GH-ORC1901–1200) and pHK143 (GH-orc1901–1200 R367A) were constructed by Gibson assembly using pGEX-6P-1 (GE Healthcare), followed by site-directed mutagenesis. Yeast strains are listed in Supplementary Table S1.
Computational analysis
The secondary structure of ScOrc1 was predicted using PROFphd57. Multiple alignment of ScOrc1201–420 was generated using COBALT58 with a semi-automated pipeline: the raw data of a BLAST search were used as the initial data set, and the duplicated or truncated sequences in the alignment were removed manually. Structure of ScOrc1362–367 complexed with an 11-bp ACS DNA was modeled using MacroModel (Schrödinger). Graphical representations of the 3D model and the flow cytometry data were generated in PyMOL and R/Bioconductor using the ggplot2 package, respectively.
Chromatin affinity precipitation (ChAP)
ChAP experiments were carried out as described34,56,59 with some modifications. Briefly, 25-ml culture of cells was incubated at 23 °C until A600 reached 0.5, followed by further incubation at 35 °C for 2 h. Cells were crosslinked, spheroplasted, and lyzed. Chromatin-containing insoluble materials were pelleted, re-suspended, and sonicated to yield a mean DNA size of 500 bp. A cleared lysate was prepared, mixed with 2.5 volumes of buffer B (20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 8 M urea, 500 mM NaCl, 2 mM PMSF, and 10 mM imidazole), and subjected to a pull down method using Dynabeads His-tag (Invitrogen). The eluate with buffer E (50 mM Tris-HCl [pH 7.5], 500 mM imidazole, 1% SDS, 10 mM EDTA, 10 mM DTT, and 500 mM NaCl) was treated with 20 μg RNase A for 15 min at 37 °C and decrosslinked by incubation at 65 °C for 6 h. DNA was column-purified, subjected to qPCR, and quantified using the second derivative maximum method: the primers used were HK297 (GCTCCCTAGCTACTGGAGAATA) and HK298 (CTCTTCCACCCATGTCTCTTTG) for URA3, HK299 (TGTTTGTGCACTTGCCT) and HK300 (CATTGCGGTGAAATGGTAAA) for ARS1, and HK313 (TTTGTGAATTTAGCCAATTCCG) and HK314 (GCCATTGAATTATGCCGAGAG) for ARS609, respectively. As a control, 4% of the input sample was similarly processed.
Proteins
ScCdc6 was overexpressed in E. coli and purified as described5,28,38. Overexpression and purification of ScORC will be described in detail elsewhere. Briefly, derivatives of the version 3–5 vector37 carrying ORC1-TEV-His, ORC2, ORC3, ORC4, ORC5, and ORC6 were transiently co-transfected to 293T cells by the PEI method37 ORC hexamer was affinity-purified and further fractionated using Superdex 200. GH-Orc1301–400 was overexpressed in E. coli BL21(DE3) bearing pCodonPlus (Agilent) and pHK142 or pHK143, and a cleared lysate was prepared. GH-Orc1301–400 was isolated by a pull-down method using Ni-Sepharose, and eluted by step gradient of imidazole. Composition of buffers was the same as that used for purification of ORC hexamer.
ATP-binding assay
ATP bound to ORC was determined as described31,60 with some modifications. Briefly, ORC (2.4 pmol) and [α-32P]ATP were incubated for 5 min at 30 °C in 50 μl of buffer K (45 mM HEPES–KOH [pH 7.6], 4.5 mM magnesium acetate, 140 mM KCl, 9% [v/v] glycerol). The reaction mixture was filtered through a nitrocellulose membrane (Merck Millipore) pre-equilibrated with buffer K. The membrane was washed with buffer K and dried. Radioactivity retained on the membrane was quantified using a liquid scintillation counter.
Electrophoretic mobility shift assay (EMSA)
EMSA with ds or ssDNA was carried out as described38,39 with minor modifications. A 290 bp segment containing wild-type ARS1 or a mutant (A− B2− B3−) were PCR amplified as described38. Fragments carrying ARS306 or ARS609 were similarly amplified using primers HK278 (CCCACATGTAAGCTTAACTTCTTCGTGAGGAAGGAAAGTG) and HK279 (CCGACGTCAGATCTTAATTTATCTCATGAAGTAATGATAC) for ARS306, and HK280 (CCCACATGTCAATTTAGTATATTACTGTATATCTAGTTC) and HK281 (CCGACGTCGTTAAAAACAGAAAAGTAAAAATTCCGATCTTG) for ARS609. For ssDNA binding, DL11 (GTTACCATGGCATCGAGTTCTTCAACAAGACTACAATGG)39 was used. These DNAs were labeled with Cy5-ddUTP using terminal transferase. ORC and Cdc6 were incubated with labeled DNA (1.6 nM as a fragment for Cy5-ARS; 3 nM for Cy5-DL11) and 300 ng of GC-rich competitor38 for 10 min at room temperature in 5 μl of binding buffer (25 mM HEPES–KOH [pH 7.6], 100 mM potassium glutamate, 5 mM magnesium acetate, 5 mM calcium chloride, 5 mM DTT, 5% [v/v] glycerol, 0.1% [v/v] Triton X-100, 2 mg/ml BSA, and 1 mM ATP). DNA was separated by 4% 29:1 polyacrylamide gel electrophoresis, and fluorescent signals were detected in an ImageQuant LAS4010 imager (GE Healthcare).
Additional Information
How to cite this article: Kawakami, H. et al. Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues. Sci. Rep. 5, 14929; doi: 10.1038/srep14929 (2015).
Supplementary Material
Acknowledgments
We thank Dr. Bruce Stillman, Dr. Steve Bell, Dr. Hisao Masai, and the National Bio-Resource Project (NBRP) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan for strains and plasmids; Dr. Takashi Hishida and Dr. Kenji Keyamura for advice on yeast gene manipulation; and the Kyushu University Research Support Center for shared resources. This work was supported in part by JSPS KAKENHI (26291004, 24870021, and 15K18504), Platform for Drug Discovery, Informatics, and Structural Life Science from MEXT, Japan, the Takeda Science Foundation, and the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development.
Footnotes
Author Contributions H.K. performed the computational analysis and most of the experiments. E.O. and T.T. contributed to the experiments with mammalian cells. S.K. constructed and characterized an ORC1-HS strain. H.K. and T.K. conceived this study and wrote the manuscript.
References
- Yardimci H. & Walter J. C. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol 21, 20–5 (2014). [DOI] [PubMed] [Google Scholar]
- O’Donnell M., Langston L. & Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 5, a010108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt K. E. & Berger J. M. A structural framework for replication origin opening by AAA+ initiation factors. Curr Opin Struct Biol 23, 144–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H. & Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem Cell Biol 88, 49–62 (2010). [DOI] [PubMed] [Google Scholar]
- Sun J. et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol 20, 944–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harami G. M., Gyimesi M. & Kovács M. From keys to bulldozers: expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem Sci 38, 364–71 (2013). [DOI] [PubMed] [Google Scholar]
- Duncker B. P., Chesnokov I. N. & McConkey B. J. The origin recognition complex protein family. Genome Biol 10, 214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hayashi M. K., Merkel O., Stillman B. & Xu R.-M. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J 21, 4600–11 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P. et al. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev 24, 1418–33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizume K., Yagura M. & Araki H. Concerted interaction between origin recognition complex (ORC), nucleosomes and replication origin DNA ensures stable ORC-origin binding. Genes Cells 18, 764–79 (2013). [DOI] [PubMed] [Google Scholar]
- Siow C. C., Nieduszynska S. R., Müller C. A. & Nieduszynski C. A. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res 40, D682–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y. & Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255, 817–23 (1992). [DOI] [PubMed] [Google Scholar]
- Remus D. et al. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H., Marahrens Y. & Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol Cell Biol 14, 7643–51 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D., Beall E. L. & Botchan M. R. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 23, 897–907 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S. et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17, 1894–908 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Moon K. Y., Jiang Y. & Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc Natl Acad Sci USA 98, 13589–94 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R. Y. & Kelly T. J. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA 96, 2656–61 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa N. et al. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res 31, 2077–86 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M., Schuwirth B. S., Westcott S. L. & Wigley D. B. Structural basis of DNA replication origin recognition by an ORC protein. Science 317, 1213–6 (2007). [DOI] [PubMed] [Google Scholar]
- Dueber E. L. C., Corn J. E., Bell S. D. & Berger J. M. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317, 1210–3 (2007). [DOI] [PubMed] [Google Scholar]
- Dueber E. C., Costa A., Corn J. E., Bell S. D. & Berger J. M. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res 39, 3621–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M., Huijbregts R. P. H. & Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol 27, 3143–53 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. G. & Bell S. P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol 17, 7159–68 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 105, 10326–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Stillman B. The origin recognition complex: a biochemical and structural view. Subcell Biochem 62, 37–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D. & DePamphilis M. L. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol 21, 8095–103 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. et al. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure 20, 534–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain P. D., Bowers J. L., Lee D. G., Bell S. P. & Griffith J. D. Mapping subunit location on the Saccharomyces cerevisiae origin recognition complex free and bound to DNA using a novel nanoscale biopointer. J Biol Chem 279, 36354–62 (2004). [DOI] [PubMed] [Google Scholar]
- Klemm R. D. & Bell S. P. ATP bound to the origin recognition complex is important for preRC formation. Proc Natl Acad Sci USA 98, 8361–7 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R. D., Austin R. J. & Bell S. P. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 88, 493–502 (1997). [DOI] [PubMed] [Google Scholar]
- Vader G. et al. Protection of repetitive DNA borders from self-induced meiotic instability. Nature 477, 115–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Bell S. P. & Aparicio O. M. Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes Cells 11, 557–73 (2006). [DOI] [PubMed] [Google Scholar]
- Aparicio O. M., Weinstein D. M. & Bell S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69 (1997). [DOI] [PubMed] [Google Scholar]
- Samel S. A. et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev 28, 1653–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Mitchell J., Leber J., Kobayashi R. & Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83, 563–8 (1995). [DOI] [PubMed] [Google Scholar]
- Uno S., You Z. & Masai H. Purification of replication factors using insect and mammalian cell expression systems. Methods 57, 214–21 (2012). [DOI] [PubMed] [Google Scholar]
- Speck C., Chen Z., Li H. & Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 12, 965–71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. G., Makhov A. M., Klemm R. D., Griffith J. D. & Bell S. P. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. EMBO J 19, 4774–82 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth-Petróczy A., Simon I., Fuxreiter M. & Levy Y. Disordered tails of homeodomains facilitate DNA recognition by providing a trade-off between folding and specific binding. J Am Chem Soc 131, 15084–5 (2009). [DOI] [PubMed] [Google Scholar]
- Belsky J. A., MacAlpine H. K., Lubelsky Y., Hartemink A. J. & MacAlpine D. M. Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes Dev 29, 212–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Cocker J. H., Harwood J. & Diffley J. F. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J 14, 2631–41 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidonnici M. R. et al. Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. J Cell Sci 117, 5221–31 (2004). [DOI] [PubMed] [Google Scholar]
- Duzdevich D. et al. The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. Mol Cell 58, 483–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Aparicio J. G., Aparicio O. M. & Tavaré S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7, 276 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterino T. L., Fang H. & Hayes J. J. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol Cell Biol 31, 2341–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C. & Méchali M. DNA replication origins. Cold Spring Harb Perspect Biol 5, a010116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A. N.-S., Bradley P., Cernadas R. A., Bogdanove A. J. & Stoddard B. L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellay J. et al. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol 12, R14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshina S. et al. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem 288, 30161–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E. et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19, 837–44 (2012). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci USA 108, 7373–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchens C. R. et al. Multiple mechanisms contribute to Schizosaccharomyces pombe origin recognition complex-DNA interactions. J Biol Chem 283, 30216–24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A. J. et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484, 115–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F., Botchan M. R. & Berger J. M. Crystal structure of the eukaryotic origin recognition complex. Nature 519, 321–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y.-J. & Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24, 101–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachdav G. et al. PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 42, W337–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos J. S. & Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–9 (2007). [DOI] [PubMed] [Google Scholar]
- Kasho K., Fujimitsu K., Matoba T., Oshima T. & Katayama T. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res 42, 13134–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H., Keyamura K. & Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem 280, 27420–30 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.