Abstract
A subfamily of rhodopsin pigments was recently discovered in bacteria and proposed to function as dual-function light-driven H+/Na+ pumps, ejecting sodium ions from cells in the presence of sodium and protons in its absence. This proposal was based primarily on light-induced proton flux measurements in suspensions of Escherichia coli cells expressing the pigments. However, because E. coli cells contain numerous proteins that mediate proton fluxes, indirect effects on proton movements involving endogenous bioenergetics components could not be excluded. Therefore, an in vitro system consisting of the purified pigment in the absence of other proteins was needed to assign the putative Na+ and H+ transport definitively. We expressed IAR, an uncharacterized member from Indibacter alkaliphilus in E. coli cell suspensions, and observed similar ion fluxes as reported for KR2 from Dokdonia eikasta. We purified and reconstituted IAR into large unilamellar vesicles (LUVs), and demonstrated the proton flux criteria of light-dependent electrogenic Na+ pumping activity in vitro, namely, light-induced passive proton flux enhanced by protonophore. The proton flux was out of the LUV lumen, increasing lumenal pH. In contrast, illumination of the LUVs in a Na+-free suspension medium caused a decrease of lumenal pH, eliminated by protonophore. These results meet the criteria for electrogenic Na+ transport and electrogenic H+ transport, respectively, in the presence and absence of Na+. The direction of proton fluxes indicated that IAR was inserted inside-out into our sealed LUV system, which we confirmed by site-directed spin-label electron paramagnetic resonance spectroscopy. We further demonstrate that Na+ transport by IAR requires Na+ only on the cytoplasmic side of the protein. The in vitro LUV system proves that the dual light-driven H+/Na+ pumping function of IAR is intrinsic to the single rhodopsin protein and enables study of the transport activities without perturbation by bioenergetics ion fluxes encountered in vivo.
Introduction
Microbial rhodopsins are a widely distributed family of photoactive transmembrane proteins characterized by seven transmembrane helices and an all-trans-retinal cofactor covalently bound in a protonated Schiff base linkage to a conserved lysine residue located in the middle of the seventh helix (1,2). Photoisomerization of the retinal induces conformational changes that result in ion transport or signal transduction by different members of the family.
A microbial rhodopsin with a new ion transport function was identified in Dokdonia eikasta (formerly Krokinobacter eikastus) by Inoue et al. (3). The pH of a suspension of E. coli cells expressing the new transporter named KR2 in media containing NaCl showed light-induced alkalization, which was enhanced by CCCP and inhibited by TPP+ (3). These light-induced pH changes required the presence of Na+ and therefore strongly indicated passive proton influx into the cells caused by light-driven electrogenic Na+ transport. Moreover, light-induced acidification was observed in the cell suspensions in Na+-free media containing KCl (3). These results indicated that KR2 exhibits a dual H+/Na+ pumping activity, transporting H+ in the absence of Na+. Additional indirect evidence supporting KR2’s function as a Na+ pump are the Na+-dependent effects on flash-induced absorption changes of the pigment (3) and on light-induced helix conformational changes monitored by electron paramagnetic resonance (EPR) spin-spin dipolar coupling (4). One cannot exclude from the in vivo measurements that the proton fluxes are perturbed by the presence of the numerous proton motive force (pmf)-driven and pmf-regulated processes involving proton fluxes across the membrane in living Escherichia coli, which have been demonstrated in similar measurements in other systems to perturb and even dominate proton fluxes in response to light-driven proton ejection (5,6). Therefore a system that allows direct detection of the light-induced Na+ and H+ transport activities by this new subfamily of microbial rhodopsins is needed.
Eight additional microbial rhodopsins homologous to KR2 have been identified by genomic analysis (7,8), including NdR2 from Nonlabens (Donghaeana) dokdonensis (9), NMR2 from Nonlabens marinus (8), and GLR from Gillisia limnaea (10). From the E. coli cell suspension proton flux measurements, KR2 appeared to exhibit dual Na+/H+ pump activity whereas NMR2 and GLR were reported to exhibit only fluxes indicative of Na+ pump activity. The IAR gene was identified in the genome of Indibacter alkaliphilus (7,8) and had not been expressed and characterized before this study.
E. coli cells contain numerous proteins that mediate proton fluxes such as the Na+/H+ antiporter, voltage-gated proton channels, H+-ATPase, pmf-driven flagellar motors and transporters, and pmf-regulated processes. Influence of these components on the in vivo proton flux measurements, the most direct data implicating KR2 and its homologs as light-driven Na+ pumps, could not be excluded. Proton fluxes through these different proteins can lead to indirect effects on net proton movements making it difficult to accurately interpret results (5,6). For example, E. coli cells expressing the blue-absorbing proteorhodopsin, a proton pump, produced proton influx rather than the efflux expected from its pumping activity under illumination due to the opening of voltage-gated proton channels in the E. coli membrane that resulted from the generated and preexisting electrochemical potential across the membranes (5,6). Moreover, in Halobacterium salinarum cell suspensions, light-driven proton efflux by BR produced proton influx attributable to voltage gating of an H+-ATPase and Na+/H+ antiporter (6). Therefore, an in vitro system consisting of the purified rhodopsin in the absence of other proteins is needed to definitively assign the putative Na+ and H+ transport to the single rhodopsin protein. Accordingly, we developed an in vitro system for measurement of light-induced proton fluxes based on sealed large unilamellar vesicles (LUVs) containing purified rhodopsin proteins. LUVs have been previously used to study ion translocation activity in BR (11), halorhodopsin (12), and proteorhodopsin (13).
Two independent crystal structures of KR2 were recently reported in Gushchin et al. (14) and Kato et al. (15). Only one set of structures (4XTN) (14) showed a bound Na+, which is 23 Å from the Schiff base nitrogen. The authors concluded from its location outside of the putative ion-translocation pathway that the bound sodium ion detected is not the ion that is being transported by the protein (14).
In this report, we expressed and characterized IAR function in E. coli cells, followed by purifying and reconstituting IAR into LUVs to test the light-driven Na+ and H+ pumping activities for the first time, to our knowledge, in the absence of other proteins.
Materials and Methods
Cloning, expression, and purification of IAR
Homologs of the KR2 were identified by the protein BLAST. We chose IAR from the bacterium I. alkaliphilus, which harbors the characteristic NDQ motif (3,9). The cDNA coding for IAR was synthesized by Genewiz (South Plainfield, NJ) and subcloned between the restriction sites of NdeI and XhoI into the pET-24b(+) vector with an inherent C-terminus His6 sequence. Site-directed mutagenesis for mutants was performed with the QuikChange kit (Agilent, Santa Clara, CA) using the wild-type (WT) IAR construct as template. All the sequences were confirmed by DNA sequencing (Genewiz).
The plasmid harboring the IAR cDNA was transformed into the E. coli expression strain C43(DE3) (Lucigen, Middleton, WI), and plated on Luria Bertani (LB)-agar containing 50 μg/mL kanamycin overnight at 37°C. A single colony was used to inoculate a starter culture of 50 mL LB broth containing 50 μg/mL kanamycin and grown at 37°C with shaking at 220 rpm overnight. A 1 L LB broth culture was inoculated using 20 mL of the starter cultures and grown at 37°C in a rotary shaker at 220 rpm until the OD600 reached 0.5∼0.8. After 5 μM all-trans-retinal was added, expression of the target protein was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside, and cells were grown at 37°C and 220 rpm for 3 h. Cells expressing recombinant IAR were harvested by low-speed centrifugation.
Cells were disrupted in Buffer A (20 mM HEPES, 300 mM NaCl, 5% glycerol, pH 7.4) by a homogenizer. Cell membranes were harvested by ultracentrifugation at 40,000 rpm with a Ti45 rotor (Beckman, Brea, CA). Isolated membrane pellets were resuspended in Buffer A containing 1.5% DDM (dodecyl maltoside) from Anatrace (Maumee, OH) and incubated at 4°C overnight with gentle rotation to extract the pigment. The detergent extract was centrifuged at 50,000 rpm with a TLA-100 rotor (Beckman), and the supernatant was mixed with nickel-nitrilotriacetic acid-agarose beads (Qiagen, Valencia, CA) and incubated at 4°C for 1.5 h. After washing with 20 bed volumes of Buffer A with 20 mM imidazole, the protein was eluted with Buffer A containing 0.02% DDM and 300 mM imidazole. The pigment was exchanged into Buffer A containing 0.02% DDM to remove imidazole, and concentrated using model No. YM-10 centrifugal filters (Amicon, Billerica, MA) by centrifugation and stored at 4°C. The typical yield of protein was 5 mg/L culture. KR2 was expressed and purified in our laboratory with a similar protocol.
Proton flux measurements in E. coli
Freshly harvested E. coli from 1-L cultures expressing either IAR WT or mutants were washed with either 100 mM NaCl or 100 mM KCl three times by centrifugation and resuspended in either 100 mM NaCl or 100 mM KCl without buffering agent. The OD600 for proton flux measurements was ∼10. The light source for the illumination of the samples was a 21 V, 150 W tungsten-halogen illuminator (Cole Parmer, Vernon Hills, IL). The light was passed through a heat filter and a long-pass colored-glass filter transmitting >520 nm, and focused onto the sample in a cuvette with vigorous magnetic stirring. The pH changes were measured by a Ross glass combination micro-pH electrode and an Orion Star A211 pH meter (Thermo, Chelmsford, MA) connected to a computer.
Reconstitution of IAR into LUVs
Reconstitution of IAR into LUVs and fluorescence measurements were performed as described in Gulik-Krzywicki et al. (16) and Rigaud et al. (11,17). Briefly, 9 mg of soy PC (L-α-phosphatidylcholine) (Avanti, Alabaster, AL) and 1 mg of DOPG (1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)) (Avanti) in chloroform were mixed in a round-bottom glass tube and dried into white thin film by a gentle flow of Argon gas. The lipid film was then dried for 1 h under vacuum to remove residual chloroform. A 1 mL volume of Buffer B (either 2 mM NaPi and 100 mM NaCl, pH 7.5 or 2 mM KPi and 100 mM KCl, pH 7.5) mixed with 0.5 mM pyranine (Acros, Morris Plains, NJ) was added to resuspend the lipid film, and the mixture was incubated at room temperature for 30 min to rehydrate the lipid. The lipid suspension was passed through 0.4- and 0.2-μm polycarbonate membranes in an extruder (Avanti) to generate LUVs. For 0.5 mL of LUVs, 28 μL of 10% DDM solution was added to the lipid solution and incubated at room temperature for 1 h with occasional mixing. IAR in DDM was added to the lipid at the ratio of 1:80 (w/w) and incubated at room temperature for 1 h with gentle mixing. Bio-beads (Bio-Rad, Hercules, CA) were washed thoroughly with methanol, water, and then buffer. Bio-beads (20∼30 mg) were added to 0.5 mL of protein/lipid/DDM for 2 h at room temperature, overnight at 4°C, and then 2 h at room temperature agitated by gentle rotation to remove DDM. The IAR was reconstituted into LUVs in 20 mM HEPES buffer at pH 7.4 and 100 mM NaCl for flash photolysis measurement, without the addition of pyranine. External pyranine was removed by size-exclusion chromatography using a column with Sephadex G-50 resin (Pharmacia, Piscataway, NJ), and the LUVs were eluted with Buffer B at 4°C. The eluate was collected and the first two fractions of 1.6 mL were used for fluorescence measurements. The content of IAR (λmax 529 nm) in LUVs was assessed by ultraviolet-visible (UV-Vis) spectroscopy. Homogeneous preparation of negatively stained IAR-LUVs was confirmed by electron microscopy.
UV-Vis spectroscopy and laser flash photolysis
Absorption spectra of detergent-extracted IAR and IAR-LUV samples were collected using a Cary 4000 spectrophotometer (Varian, Palo Alto, CA) equipped with an integrating sphere. Baseline correction for scattering was performed in the software ORIGIN (OriginLab, Northampton, MA).
Flash-induced absorption changes were acquired by a Surelight I Nd-YAG laser flash (532 nm, 6 ns, 40 mJ; Continuum, Santa Clara, CA) every 5 and 20 s in 100 mM NaCl and 100 mM KCl, respectively, and recorded with a laboratory-constructed cross-beam flash spectrophotometer as described in Wang et al. (5). The signals were digitized with a Digidata 1320A using PCLAMP Ver. 8.0 software (both from Molecular Devices, Sunnyvale, CA) at the sampling rate 4 μs/point in the presence of NaCl, and 4 or 40 μs/point in the presence of KCl due to a slower photocycle. Each transient absorption change was the average of 50 or 100 traces and all traces were normalized to 1 for better comparison. Interference filters were positioned before the monochromator defining the monitored wavelength. Absorption changes monitored at 412 and 612 nm were used to measure the M and O intermediates, respectively. Rate constants were calculated by nonlinear fitting using ORIGIN.
Light-induced ion transport in IAR-LUVs monitored with fluorescence
A sample of 1.6 mL of IAR-LUVs was placed into a quartz cuvette with constant stirring and the fluorescence intensity was measured using a QuantaMaster model QM3-SS (Photon Technology International, Edison, NJ) fluorometer at 5°C. The fluorescence excitation for pyranine was set at 450 nm and emission was recorded at 500 nm. Actinic illumination of samples was performed with a halogen-tungsten light source (Ushio, Cypress, CA) equipped with a heat filter and a long-pass orange filter (530 nm < X < 700 nm) through a flexible light guide whose aperture was positioned above the cuvette. An interference filter transmitting 500 ± 20 nm was placed along the emission path to remove further stray and diffuse light arising from illumination. The excitation and emission slits were adjusted to obtain near-maximal fluorescence intensity. Ionophores including valinomycin (0.2 μM in ethanol, Sigma-Aldrich), monensin (0.1 μM in ethanol, Sigma-Aldrich), gramicidin (0.1 μM in ethanol, Sigma-Aldrich), and CCCP (0.1 μM in buffer) were used.
Site-directed spin-labeling EPR measurements
We generated a structural model of IAR based on the structure of KR2 (PDB: 3X3B) using the homology model program SWISS-MODEL (http://swissmodel.expasy.org/). Tyr12 and His277 were observed to be in the N- and C-terminal regions, respectively. The Y12C and H277C mutants of IAR were expressed, purified, and buffer-exchanged into 10 mM HEPES (pH 6.2), 100 mM NaCl, 0.02% DDM. The absorption spectra of the mutants matched that of WT IAR and E. coli cells expressing the mutants show identical Na+ and H+ pumping activities when compared to the WT pigment. Also the photocycle kinetics of the purified mutants closely matched that of WT IAR. Samples for EPR were prepared by the addition of 20-fold molar excess MTSSL (Santa Cruz Biotechnology, Dallas, TX) to each protein sample. MTSSL was dissolved in acetonitrile to 200 mM and then diluted in buffer containing 50 mM Bis-Tris (pH 6.2), 0.02% DDM (w/v), and 100 mM NaCl to a final concentration of 5 mM before addition to protein samples. The spin-labeling reaction was allowed to proceed for 16∼24 h at 4°C with gentle rotation. Excess MTSSL was removed by extensive dialysis. The labeled protein was reconstituted into LUVs in 10 mM HEPES (pH 6.2), 100 mM NaCl as described above. EPR spectra were recorded on a model No. EMX X-band spectrometer (Bruker, Billerica, MA) using a modulation amplitude of 2 G, a modulation frequency of 100 kHz, and a microwave power of 16 mW. Room temperature spectra were collected in a 50 μL capillary tube placed at the center of a 5-mm outer diameter quartz EPR tube (KIMAX-51, Gerreshimer, Queretaro, Mexico). The Heisenberg exchange reagent NiEDDA was prepared as described in Altenbach et al. (18). All data acquisition and analyses were conducted using the software WINEPR (Bruker). Measurement of the accessibility of the membrane-impermeable NiEDDA paramagnetic relaxation agent to the nitroxide spin label was performed as described in Altenbach et al. (18,19). Line broadening effects based on Heisenberg exchange caused by NiEDDA proximity to MTSSL-labeled mutants were determined by the addition of 100 mM NiEDDA to a sample of spin-labeled mutant IAR protein and the EPR spectra collected. Similar experiments were performed in the presence of 0.5% Triton X-100 to solubilize the LUVs.
Results and Discussion
UV-Vis spectra and flash photolysis of detergent-purified IAR and KR2
The sequence of IAR was identified by a protein BLAST search as a gene with 68% sequence identity with KR2. We expressed the IAR pigment in E. coli, and first characterized the absorption spectra and laser flash-induced absorption changes of the purified pigment in DDM detergent to compare with the properties of KR2 in the same conditions. The λmax of purified IAR of 529 nm (Fig. 1 A) is close to that of KR2 (523 nm) (3). No difference in λmax in DDM in NaCl versus KCl solution is observed (Fig. 1 A), as reported for KR2 (3,4).
Figure 1.
UV-Vis spectra and flash photolysis of IAR and KR2 in DDM and IAR in LUVs. (A) Absorption spectra of IAR in DDM and 100 mM Na+ (black line), DDM and 100 mM K+ (blue line), and LUVs and 100 mM Na+ (red line). Normalized flash-induced absorption changes at 412 nm (B) and 612 nm (C) in IAR in DDM and 100 mM Na+ or 100 mM K+ (solid black lines), and IAR in LUVs in 100 mM Na+ or 100 mM K+ (solid red lines). For comparison, absorption changes in KR2 in DDM and 100 mM Na+ or 100 mM K+ (dashed black lines). Results of curve fitting are located in Table 1.
IAR exhibits similar kinetics with long-lived photointermediates as in the KR2 photocycle, including the blue-shifted M and red-shifted O, monitored at 412 and 612 nm, respectively (Fig. 1, B and C; Table 1). The decay of M intermediate in IAR as in KR2 is accelerated >100-fold in NaCl compared to KCl (Fig. 1 B; Table 1). The M rise of IAR in KCl is biphasic with comparable amplitudes, and the later phase accelerates or disappears in NaCl. Also, as in KR2, the rise and decay of O in IAR in NaCl is >20 faster than in KCl (Fig. 1 C; Table 1). In NaCl, the rates of formation and decay of M and O of IAR in LUVs are very close to those in DDM; however, in KCl, the rates of M decay and O of IAR in LUVs are faster than those in DDM (Fig. 1, B and C), suggesting the conformation of Na+-bound IAR is more preserved upon DDM extraction than the conformation of K+-bound IAR. In both DDM and LUVs of IAR, a smaller amount of O is accumulated in KCl than in NaCl, as in KR2 (3). The photocycle acceleration by NaCl is one of the properties supporting the proposal that KR2 functions as a Na+ pump. Overall, in NaCl the two pigments exhibit closely similar photocycles. In KCl, the kinetics of both pigments are less similar, but the trend of more rapid photocycles in NaCl than in KCl is in good agreement with that of KR2 and GLR (3,10).
Table 1.
Kinetic constants of M (412 nm) and O (612 nm) intermediates for DDM-purified preparations of IAR and KR2 and IAR in LUVs in 100 mM NaCl or 100 mM KCl
| IAR DDM Na+ | IAR LUV Na+ | KR2 DDM Na+ | IAR DDM K+ | IAR LUV K+ | KR2 DDM K+ | |
|---|---|---|---|---|---|---|
| 412 nm | ||||||
| Rise1 | 0.033 ms | 0.025 ms | 0.039 ms | 0.033 ms (58%) | 0.246 ms (60%) | 0.037 ms (78%) |
| Rise2 | 3.09 ms (42%) | 3.10 ms (40%) | 5.38 ms (22%) | |||
| Decay1 | 0.75 ms (52%) | 0.70 ms (51%) | 0.40 ms (72%) | 294 ms | 89.10 ms | 215 ms |
| Decay2 | 6.0 ms (48%) | 3.6 ms (49%) | 1.4 ms (28%) | |||
| 612 nm | ||||||
| Rise1 | 0.75 ms (34%) | 0.58 ms (55%) | 0.70 ms | 6.80 ms (7%) | 36.40 ms | 180 ms |
| Rise2 | 2.5 ms (66%) | 2.1 ms (45%) | 90.0 ms (93%) | |||
| Decay | 17.8 ms | 15.9 ms | 8.7 ms | 470 ms | 184.3 ms | 550 ms |
Numbers in parentheses indicate the percentage of each phase in rise or decay.
Light-induced pH changes in E. coli suspensions
The key criteria used to establish light-induced pumping of a nonproton ion by pH measurements of cell suspensions are: 1) a light-induced pH increase corresponding to proton flux into the cells; 2) enhancement of the proton influx by the proton ionophore CCCP showing that the proton movement is passive; and 3) elimination of the passive proton flux by the lipid permeable cation TPP+, showing that the flux was derived from electrical hyperpolarization of the membrane. The criteria for light-induced proton pumping are 1) a light-induced outward proton flux that is 2) eliminated by CCCP.
Light-induced H+ flux measurements in E. coli expressing recombinant IAR meet criteria 1, 2, and 3 for nonproton ion pumping in NaCl solution as well as criterion 1 and 2 for proton pumping in KCl solution (i.e., no Na+ present) (Fig. 2). These light-induced pH changes are similar to those reported for KR2 in E. coli cells and implicate IAR as a Na+ pump that, like KR2, defaults to H+ pumping in the absence of Na+.
Figure 2.
Non-H+ ion pumping and H+ pumping activities of WT IAR in E. coli cell suspensions. Light-induced proton fluxes were assessed by pH changes of E. coli cell suspensions in the presence of ions and uncouplers, as indicated. Initial pH values ranged from 6.30 to 6.65.
EPR measurement of insertion orientation
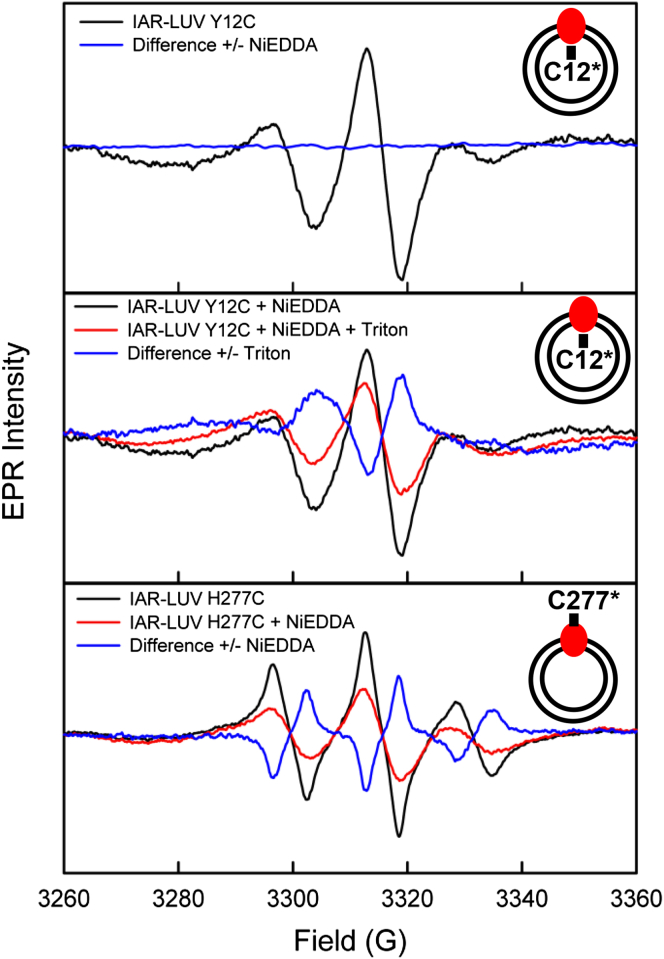
To assess the orientation of the pigment in IAR-LUVs, we used spin labeling of sites in the N- and C-terminal regions, which are on the exterior and cytoplasmic side of the membrane in cells, respectively, in all rhodopsins. We introduced single Cys residues to be used as targets for nitroxide spin labeling, followed by EPR measurements. The room-temperature continuous-wave EPR of sealed IAR-LUVs containing the spin-labeled N-terminal position Y12C and C-terminal position H277C mutants showed EPR spectra with good immobilization of the spin label (MTSSL) compared to free MTSSL. Line broadening due to Heisenberg exchange (18,19) was not detected in MTSSL-labeled Y12C-LUV upon addition of 100 mM NiEDDA, resulting in a difference spectrum that was nearly flat (Fig. 3, top panel). Lack of a NiEDDA effect suggests that the spin label is not accessible to the bulk solution consistent with its N-terminus being in the lumen of the LUVs. If this is the case, addition of 0.5% Triton X-100 solubilizes IAR-LUVs, which would thereby expose the spin label to the bulk and allow accessibility to NiEDDA. Indeed, in the presence of Triton X-100 the addition of 100 mM NiEDDA to Y12C-LUV causes considerable line broadening, a result of Heisenberg exchange between the nitroxide spin label and the exchange reagent (Fig. 3, middle panel). In spin-labeled H277C-LUVs, broadening is observed immediately after addition of NiEDDA (Fig. 3, bottom panel), indicating that the C-terminus faces the bulk. Based on the area of the difference spectrum (Fig. 3, top) compared to the absolute spectrum of IAR-LUVs, we estimate IAR is inserted with 98 ± 2% in the inside-out orientation under our experimental conditions with respect to the orientation in the E. coli inner membrane.
Figure 3.
Room temperature continuous-wave EPR of spin-labeled IAR mutants Y12C and H277C probing orientation of insertion of the pigment in LUVs. The addition of the exchange reagent NiEDDA in excess amount shows no observable effect on the line shape of the spin-labeled Y12C mutant (top). The addition of Triton X-100 exposes the labeled Y12C to NiEDDA causing a line-broadening effect (middle). The labeled H277C mutant undergoes line broadening immediately after addition of NiEDDA (bottom). Cartoon (upper right of each panel) of the spin-labeled mutant in an LUV with the orientation concluded from the data.
Light-induced ion translocation in IAR-LUVs
IAR reconstituted into LUVs with ratios of 1:80 (protein/lipid, w/w) shows a λmax at 529 nm (Fig. 1 A), indicating that the absorption spectra of IAR in detergent and LUVs are the same. We compared the kinetics of detergent-extracted IAR and IAR-LUVs in NaCl by laser flash spectroscopy. The kinetics of M and O rise are nearly the same in IAR-LUVs as in DDM-extracted IAR (Fig. 1, B and C). These data indicate that in the presence of Na+, the photocycles of IAR pigment in LUVs and in detergent micelles are very similar.
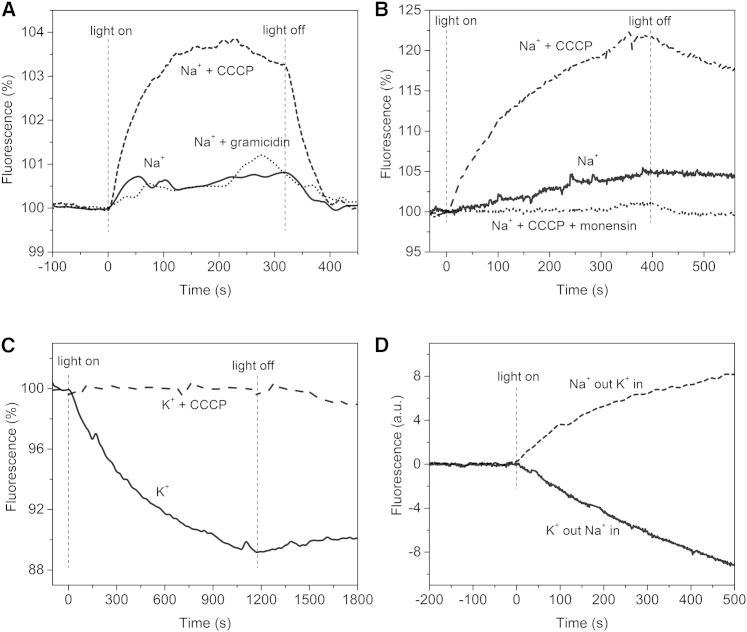
To monitor proton fluxes produced by illumination of IAR-LUVs, we trapped pyranine inside the LUVs to measure the proton concentration in the LUV lumen. Illumination in the presence of NaCl caused a fluorescence increase, indicating proton efflux from the vesicle (Fig. 4, A and B). Addition of 0.1 μM CCCP enhanced the light-induced fluorescence increase approximately fivefold, indicating that another ion was actively transported because the proton flux was passive (Fig. 4, A and B). In typical samples the fluorescence intensity increased ∼20%, which corresponds to a pH increase of ∼0.19 unit. Our interpretation is that electrogenic transport of Na+ into the vesicles produces a positive inside electrical potential across the LUV membrane, and the electrical potential drives H+ efflux through CCCP, leading to an increase of lumenal pH. When illumination is stopped, the lumenal pH decreases, as expected from equilibration of the H+ chemical gradient generated by the light (Fig. 4, A and B). The addition of the cation channel gramicidin and the cation ionophore monensin both abolished the passive proton flux (Fig. 4, A and B), indicating an uncoupler effect preventing electrogenic flux of the transported ion. The only cation present in our samples other than protons was Na+, and therefore these effects offer direct evidence that IAR transports Na+. Addition of 10 mM TPP+ to the IAR-LUV sample caused an abrupt and significant drop in fluorescence intensity in the dark, preventing clear assessment of whether it would block the proton flux as we observed in E. coli suspensions.
Figure 4.
Light-induced fluorescence changes of IAR-LUVs in NaCl or KCl. (A) Na+ pumping measurements in IAR-LUVs in the presence of NaCl only (solid line); the sample after the addition of 0.1 μM CCCP (dashed line); the sample with further addition of 0.1 μM gramicidin (dotted line). Light-induced fluorescence changes ON and OFF (vertical dashed lines). (B) Na+ pumping measurements in IAR-LUVs in NaCl only (solid line); the sample after addition of CCCP (dashed line); the sample with further addition of 0.1 μM monensin (dotted line). (C) H+ pumping measurements performed on IAR-LUVs in KCl only (solid line); the sample after the addition of 0.1 μM CCCP (dashed line). (D) IAR-LUVs with KCl outside and NaCl inside (solid line) and IAR-LUVs with NaCl outside and KCl inside in the presence of CCCP (dashed line).
Illumination of LUVs in the presence of Na+ caused alkalization of the vesicle interior, which is the opposite effect to pH changes observed in E. coli cells (Fig. 2). We conclude that the light-driven Na+ ion transport in IAR-LUVs in NaCl is from the bulk phase solution into the lumen of LUVs, opposite to the transport vectoriality in intact cells (Fig. 5). This is in good agreement with our conclusion of the inside-out orientation of the protein in the LUVs from EPR measurements (Fig. 3).
Figure 5.
Cartoon representation of light-induced Na+ flux and passive H+ flux in IAR-LUVs. IAR is preferentially oriented with its intracellular side toward the extravesicular compartment. Light-driven Na+ transport generates a membrane electrical potential positive inside. In the presence of CCCP, the electrical potential drives a proton efflux. (Green) Intravesicular pyranine.
When KCl was substituted for NaCl in the suspension solution, IAR-LUV illumination caused a fluorescence intensity change in the opposite direction, indicating inward proton flux (Fig. 4 C). Addition of CCCP abolished the fluorescence intensity change as expected from active transport of protons in the same direction as Na+ transport when Na+ is present. These data demonstrate that IAR is indeed a dual Na+/H+ pump in the absence of any other protein components.
To further characterize the transport activity in IAR-LUVs, we performed the same measurements but in the presence of different salts inside and outside of LUVs. The net proton flux with KCl in the LUV lumen and NaCl in the medium (in the presence of CCCP) is outward, as when NaCl is present on both sides of vesicles, whereas the net proton flux with NaCl in the LUV lumen and KCl in the medium is similar to that with KCl on both sides, as expected from direct inward H+ pumping (Fig. 4 D). These results confirm the inside-out orientation of the pigment in LUVs, and further imply that the transport of Na+ by IAR requires Na+ to be present only on the cytoplasmic side of the protein.
Conclusions
The in vitro LUV system proves that the dual light-driven H+/Na+ pumping function of IAR is intrinsic to the single rhodopsin protein. The in vitro system developed enables study of the transport activities without perturbation by bioenergetic ion fluxes encountered in living cells.
Our site-directed spin-labeling EPR measurements indicate that IAR was vectorially inserted almost completely with an inside-out orientation, which is in agreement with our proton flux measurements in IAR-LUVs. We also demonstrate that Na+ transport by IAR requires Na+ to be present only on the cytoplasmic side of the protein. An inside-out orientation was also found in LUVs of three other microbial rhodopsins, BR (11), HR (12), and PR (13). This orientation has been suggested to be due to the greater number of charged residues at the C-terminal than at the N-terminal region favoring more rapid hydrophobic N-terminal of BR insertion into the preformed LUVs, leading to a more stable state (20).
Photochemical reactions of rhodopsin can be studied in detergent extracts, but rhodopsin’s properties in detergent may differ from those in lipid environments (21,22) and it is not testable whether the pigment is in a native functional state in detergent micelles. Expression in E. coli allows the characterization of rhodopsin in membranes, but it is complicated by the presence of intrinsic proteins. In addition, spectroscopic measurements in E. coli are limited by the considerable light scattering caused by intact cells and complicated by the presence of other pigments such as cytochromes and flavoproteins. Rhodopsin-LUVs provide a system that can be used to characterize both the transport function and spectroscopic properties in the same state and simultaneously.
Proteoliposomes containing KR2 attached to black lipid membrane (BLM) showed photocurrents in the presence of Na+ (14). The system as presented has several disadvantages compared to the LUV system reported here: 1) The reported experiments on BLM did not resolve ion specificity or orientation of KR2. The orientation of the protein is not easily detected in BLM while various methods can be used to detect vectorial insertion in LUVs. 2) Spectroscopic measurements were not conducted and would be difficult in BLM. 3) BLMs have considerably high batch-to-batch variation, and only the values within the same experiment may be compared in Gushchin et al. (14).
In summary, we reconstituted detergent-extracted and purified IAR into LUVs, and demonstrated it as a dual-functional Na+/H+ pump using the pH-sensitive fluorescent probe pyranine to monitor light-induced lumenal pH changes. This is the first direct demonstration of Na+ transport activity in a microbial rhodopsin without other proteins sharing the membrane. The use of a sealed LUV system to study light-induced Na+ transport by this new class of microbial rhodopsins will facilitate understanding of how Na+ interacts with and moves through the protein.
Author Contributions
H.L., O.A.S., and J.L.S. designed the research; H.L., O.A.S., and G.F.Z.D. performed research and analyzed data; and H.L., G.F.Z.D. and J.L.S. wrote the article.
Acknowledgments
We thank Ah-Lim Tsai for use of his EPR instrument, Heidi Vitrac for discussions on LUV preparation and use of the fluorimeter, Brandon Goblirsch and Mikhail Bogdanov for helpful discussions, and Venkata Mallampalli for the electron microscopy measurement.
This work was supported by National Institutes of Health grant No. R01GM027750 and Endowed Chair No. AU-0009 from the Robert A. Welch Foundation.
Editor: Andreas Engel.
References
- 1.Ernst O.P., Lodowski D.T., Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spudich J.L., Sineshchekov O.A., Govorunova E.G. Mechanism divergence in microbial rhodopsins. Biochim. Biophys. Acta. 2014;1837:546–552. doi: 10.1016/j.bbabio.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue K., Ono H., Kandori H. A light-driven sodium ion pump in marine bacteria. Nat. Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 4.da Silva G.F., Goblirsch B.R., Spudich J.L. Cation-specific conformations in a dual function ion-pumping microbial rhodopsin. Biochemistry. 2015;54:3950–3959. doi: 10.1021/bi501386d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W.W., Sineshchekov O.A., Spudich J.L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 6.Helgerson S.L., Stoeckenius W. Transient proton inflows during illumination of anaerobic Halobacterium halobium cells. Arch. Biochem. Biophys. 1985;241:616–627. doi: 10.1016/0003-9861(85)90588-0. [DOI] [PubMed] [Google Scholar]
- 7.Inoue K., Kato Y., Kandori H. Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 2014;23:91–98. doi: 10.1016/j.tim.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Yoshizawa S., Kumagai Y., Kogure K. Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc. Natl. Acad. Sci. USA. 2014;111:6732–6737. doi: 10.1073/pnas.1403051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon S.K., Kim B.K., Kim J.F. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol. Evol. 2013;5:187–199. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balashov S.P., Imasheva E.S., Lanyi J.K. Light-driven Na+ pump from Gillisia limnaea: a high-affinity Na+ binding site is formed transiently in the photocycle. Biochemistry. 2014;53:7549–7561. doi: 10.1021/bi501064n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigaud J.L., Bluzat A., Buschlen S. Incorporation of bacteriorhodopsin into large unilamellar liposomes by reverse phase evaporation. Biochem. Biophys. Res. Commun. 1983;111:373–382. doi: 10.1016/0006-291x(83)90316-9. [DOI] [PubMed] [Google Scholar]
- 12.Bogomolni R.A., Taylor M.E., Stoeckenius W. Reconstitution of purified halorhodopsin. Proc. Natl. Acad. Sci. USA. 1984;81:5408–5411. doi: 10.1073/pnas.81.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dioumaev A.K., Wang J.M., Lanyi J.K. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry. 2003;42:6582–6587. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- 14.Gushchin I., Shevchenko V., Gordeliy V. Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 2015;22:390–395. doi: 10.1038/nsmb.3002. [DOI] [PubMed] [Google Scholar]
- 15.Kato H.E., Inoue K., Nureki O. Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature. 2015;521:48–53. doi: 10.1038/nature14322. [DOI] [PubMed] [Google Scholar]
- 16.Gulik-Krzywicki T., Seigneuret M., Rigaud J.L. Monomer-oligomer equilibrium of bacteriorhodopsin in reconstituted proteoliposomes. A freeze-fracture electron microscope study. J. Biol. Chem. 1987;262:15580–15588. [PubMed] [Google Scholar]
- 17.Rigaud J.L., Paternostre M.T., Bluzat A. Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry. 1988;27:2677–2688. doi: 10.1021/bi00408a007. [DOI] [PubMed] [Google Scholar]
- 18.Altenbach C., Greenhalgh D.A., Hubbell W.L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altenbach C., Froncisz W., Hubbell W.L. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys. J. 2005;89:2103–2112. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigaud J.L., Pitard B., Levy D. Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta. 1995;1231:223–246. doi: 10.1016/0005-2728(95)00091-v. [DOI] [PubMed] [Google Scholar]
- 21.Sineshchekov O.A., Govorunova E.G., Spudich J.L. Intramolecular proton transfer in channelrhodopsins. Biophys. J. 2013;104:807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duschl A., McCloskey M.A., Lanyi J.K. Functional reconstitution of halorhodopsin. Properties of halorhodopsin-containing proteoliposomes. J. Biol. Chem. 1988;263:17016–17022. [PubMed] [Google Scholar]