Abstract
Bacterial invasion of the periodontal tissues has been suggested as a relevant step in the etiopathogenesis of periodontal disease. However, its exact importance remains to be defined. The present systematic review assessed the scientific evidence concerning the relationship between the quality or quantity of periodontal microbiota in periodontal tissues and development of periodontal disease. The databases Medline-PubMed, Cochrane-CENTRAL, ISI Web of Knowledge and SCOPUS were searched, up to January 2014. Studies that reported evaluation of periodontal pathogens invasion on human tissues were selected. The screening of 440 title/abstracts elected 26 papers for full-text reading. Twenty three papers were subsequently excluded because of insufficient data or a study protocol not related to the objectives of this systematic review. All included studies were case-control studies that evaluated intracellular or adherent bacteria to epithelial cells from periodontal pockets versus healthy sulci. Study protocols presented heterogeneity regarding case and control definitions and methodological approaches for microbial identification. No consistent significant differences were found related to the presence/absence or proportion of specific periopathogens across the studies, as only one study found statistically significant differences regarding the presence of A. actinomycetemcomitans (p = 0.043), T. forsythia (P < 0.001), P. intermedia (P < 0.001), C. ochracea (P < 0.001) and C. rectus (P = 0.003) in epithelial cells from periodontal pockets vs. healthy sulci. All studies reported a larger unspecific bacterial load in or on the epithelial cells taken from a diseased site compared to a healthy sulcus. The current available data is of low to moderate quality and inconsistent mainly due to study design, poor reporting and methodological diversity. As so, there is insufficient evidence to support or exclude the invasion by periodontal pathogens as a key step in the etiopathogenesis of periodontal disease. Further research is needed.
Keywords: bacteria, gingival epithelial cells, intracellular invasion, periodontal disease, tissue invasion
Introduction
Over the past 50 years the role of invasion of bacteria in the complex pathogenic process of periodontal disease has undergone cycles of acceptance and rejection. Since Listgarten,1 and its first electron microscopy images of spirochetal infiltration in gingival tissues from patients with necrotizing ulcerative gingivitis, the invasiveness of oral bacteria emerged as a potentially important mechanism that mediates initiation and progression of periodontal disease.
It is not difficult to conjecture that the presence of microorganisms within gingival tissues would not augur good things to the periodontium. Tissue invasion allows direct discharge of destructive bacterial products2-4 and promotes the release of lysosomal contents from neutrophils in the periodontal tissues.5,6 Besides that, invasive bacteria seem to have mechanisms to evade host defenses. To be sheltered from the humoral immune surveillance invasive bacteria penetrate and remain within the epithelial cells, in a nutritious environment, where they can replicate and spread to neighboring cells.7 Additionally, in vitro studies have shown that Porphyromonas gingivalis impedes transepithelial migration of neutrophils and prevents epithelial cells from secreting IL-8 in response to bacterial challenge.8,9 Several studies have also suggested that periodontal bacteria actively suppress cell-mediated immunity and this, presumably, contributes to periodontal lesion development.10 Standard periodontal treatment is also undermined as intracellular bacteria are less likely to be physically removed by scaling and root planning 11 and are more resistant to antibiotics.12
Although there is a theoretical basis for a role of invasion in the etiopathogenesis of periodontal disease and in vitro studies that support that bacteria possess the molecular machinery to perform invasion and deceive host defenses,7 few in vivo studies were conducted,13–16 letting the idea in abeyance throughout the years.
Findings of putative periodontal pathogens in healthy sulci,17 the impossibility to discriminate periodontal disease by microbiologic analysis,18 the inability to categorically refute the possibility that bacteria were not artificially introduced into the tissues during collection or processing, in some studies,19 and evidence of internalized putative periodontal pathogens in epithelial buccal cells of healthy individuals20,21 helped to sustain this doubt.
As the comprehensive knowledge of the whole dynamic of the periodontal interface is crucial for improving diagnostics and setting effective and rational treatments and the exact importance of bacteria invasiveness in the etiopathogenesis of periodontal disease remains unclear there is a need for a systematic assessment of the literature on this topic. The aim of the present systematic review was to assess the existing scientific literature to ascertain the relationship between the quality or quantity of periodontal microbiota in periodontal tissues and periodontal disease.
Results
Search and selection results
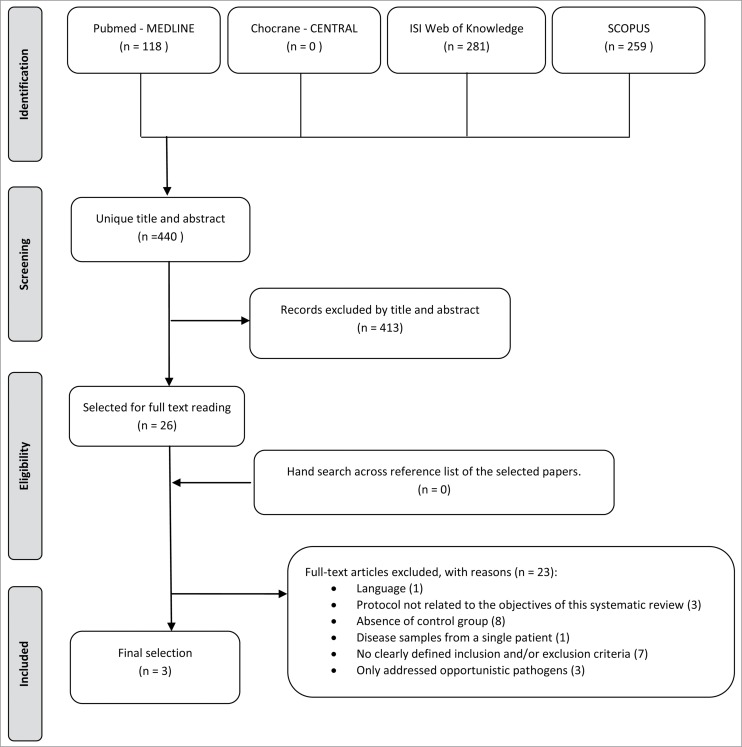
The search resulted in 440 unique papers, in which titles and abstracts were screened (for details see Fig. 1). Of these, 26 met the eligibility criteria14,22-46 and were selected for full text reading. One study was in Japanese and was excluded.36 Twenty-two studies were subsequently excluded for specific reasons: 3 studies34,35,37 had a study protocol not related to the objectives of this systematic review, 8 studies14,25-27,30,39,40,42 had absence of control group (non-diseased samples), one study38 only presented disease samples from a single patient, 7 studies28,29,31-33,41,44 hadn't clearly defined inclusion and/or exclusion criteria, and finally 3 studies43,45,46 were excluded because they only addressed opportunistic pathogens. Additional hand searching of the reference lists of the selected papers didn't retrieve any additional studies. As such, only 3 papers were included in the present systematic review.
Figure 1.
Flowchart of literature search and study selection.
General trial characteristics and heterogeneity
The three included studies were case-control studies with considerable heterogeneity. In all studies subjects with systemic diseases and antibiotic intake in the previous 6 months were excluded. Two studies (#2, #3) also excluded pregnant women. In two studies (#2, #3) diseased sites were defined as having probing pocket depth (PPD) and clinical attachment lost (CAL) ≥ 4 mm and healthy sites as having PPD ≤3 mm and CAL < 4 mm while in the other study (#1) diseased sites were defined as having PPD>5, bleeding on probing (BOP) and suppuration and healthy sites as having PPD < 5 with no BOP or suppuration. No information about CAL thresholds was provided in this last study. One study (#2) provided smoking habits information of the participants while in the other 2 (#1, #3) this information was not reported. Two studies (#1, #3) had separate case and control groups of participants. In one study (#2) each participant provided samples from healthy sites and diseased sites.
Two studies used DNA-DNA checkerboard (#1, #2) to assess the presence of intracellular or adherent bacteria contrasting with study #3 where fluorescence in situ hybridization (FISH) was used. All studies applied probes for the detection of Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Tannerella forsythia and Treponema denticola. Study #1 additionally aimed to detect Prevotella intermedia, Prevotella nigrescens, Capnocytophaga ochracea, Fusobacterium nucleatum subsp vicentii, Campylobacter rectus, Veillonella parvula, Streptococcus sanguis, Streptococcus oralis, Streptococcus intermedius and Peptostreptococcus micros. In addition to all these, study #2 also aimed to detect Actinomyces naeslundii, Actinomyces viscosus, Actinomyces odontolyticus, Actinomyces israelii, Actinomyces gerencseriae, Capnocytophaga sputigena, Capnocytophaga gingivalis, Campylobacter showae, Eubacterium nodatum, Eikenella corrodens, Fusobacterium periodonticum, Fusobacterium nucleatum subsp. polymorphum, Fusobacterium nucleatum subsp. nucleatum, Gemella morbillorum, Leptotrichia buccalis, Neisseria mucosa, Propionibacterium acnes, Streptococcus anginosus, Streptococcus constellatus, Streptococcus gordonii, Streptococcus mitis and Selenomonas noxia. For global bacteria counts study #3 used DNA probe EUB338. This probe is universal for eubacteria. The studies #1 and #3 inferred global bacteria counts by the available data but didn´t use any specific method. Detailed information regarding the studies characteristics is presented in Table 1.
Table 1.
Characteristics of included studies
| A. Subject characteristics |
|||||||
|---|---|---|---|---|---|---|---|
| # | Study design | n | Population | Male/Female%(age ± SD ) | Smokers (%) | ||
| 1 | Dibart et al. (1998) | Case-control | Cases | 24 | Patients from Center for Clinical Research in Periodontal Disease | Unknown | Unknown |
| Controls | 27 | Periodontal healthy dental students | |||||
| 2 | Colombo et al. (2006) | Case-control | Cases | 120 | Patients with moderate to severe CP from Federal University of Rio de Janeiro | 43/57% (46.3 ± 1.4) | Non-smokers (51%) Past smokers (27%) Smokers (22%) |
| Controls | 92 | ||||||
| 3 | Colombo et al. (2007) | Case- Control | Cases | 175 | Patients with CP from Federal University of Rio de Janeiro | 57/43% (44.2 ± 1.4) | Unknown |
| |
|
|
Controls |
68 |
Periodontal healthy subjects |
21/79 % (29.9 ± 2.3) |
|
| |
|
|
B. Intervention characteristics |
||||
| # |
Sample |
|
Sample site characteristics |
Intervention |
Microorganisms |
Microbiological differences Bacteria (disease vs. health, p) |
|
| 1 | Epithelial cells from periodontal pockets | Cases | PPD = 6.5 | DNA-DNA checkerboard | P. gingivalis, A. actinomycetemcomitans, T. forsythia,T. denticola, P. intermedia, P. nigrescens, C. ochracea, F. nucleatum subsp vicentii, C. rectus, V. parvula, S. sanguis, S. oralis, S. intermedius and P. micros | Aa. (16.7% vs. 0%, p = 0.043) T f. (75% vs. 11.1%, p < 0.001) P i. (54.2% vs. 7.4%, p < 0.001) Co. (37.5% vs. 0%, p < 0.001) Cr. (29.2% vs. 0%, p = 0.003) |
|
| Controls | PPD = 2.5 | ||||||
| 2 | Epithelial cells from periodontal pockets | Cases | PPD = 5.8 ± 0.3 CAL = 6.8 ± 0.3 | DNA-DNA checkerboard | P. gingivalis, A. actinomycetemcomitans, T. forsythia,T. denticola, P. intermedia, P. nigrescens, C. ochracea, F. nucleatum subsp vicentii, C. rectus, V. parvula, S. sanguis, S. oralis, S. intermedius, P. micros A. naeslundii, A. viscosus, A. odontolyticus, A. israelii, A. gerencseriae, C. sputigena, C. gingivalis, C. showae, E. nodatum, E. corrodens, F. periodonticum, F. nucleatum subsp. polymorphum, F. nucleatum subsp. nucleatum, G. morbillorum, L. buccalis, N. mucosa, P. acnes, S. anginosus, S. constellatus, S. gordonii, S. mitis, and S. noxia, | No statistically significant differences regarding specific bacteria | |
| Controls | PPD = 1.8 ± 0.1 CAL = 2.3 ± 0.1 | ||||||
| 3 | Epithelial cells from periodontal pockets, gingival crevice and buccal mucosa | Cases | PPD = 5.8 ± 0.3 CAL = 6.8 ± 0.3 PI = 86 ± 11 BOP = 70 ± 18 |
Fluorescence in situ hybridization laser-scanning confocal microscopy | P. gingivalis, A. actinomycetemcomitans, T. forsythia and T. denticola. | No statistically significant differences regarding specific bacteria GBC (35.8% vs. 0%, p < 0.05). |
|
| Controls | PPD = 1.8 ± 0.1 CAL = 2.3 ± 0.1 PI = 27 ± 15 no BOP | ||||||
Note: PPD = probing pocket depth (mm); CAL = clinical attachment loss (mm); PI= plaque index (%); BOP = bleeding on probing; Aa. = A. actinomycetemcomitans; Tf. = T. Forsythia; Pi. = P. intermedia; Co. = C. ochracea; Cr.= C. rectus; GBC= global bacterial counts in or on epithelial cells (>100 ).
Quality analysis
Methodological quality scores were given according to predetermined criteria (Table 2). The highest level of evidence, with low risk of confounding or bias and a moderate probability that the relationship is causal, was attributed to Study #3.
Table 2.
Quality assessment of included studies
| # | Appropriate and clearly focused question | Defined Exclusion Criteria | Case Selection | Control Selection | Representati-veness of cases | Comparability of cases and controls | Ascertainment of Exposure | Blindness | Statistical Analysis | Funding | Overall assessment |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Dibart et al. (1998) | 2 | 2 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 2 | − |
| 2 Colombo et al. (2006) | 2 | 2 | 2 | 1 | 1 | 2 | 1 | 0 | 1 | 2 | − |
| 3 Colombo et al. (2007) | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 0 | 1 | 2 | + |
Note: 0 = not addressed, 1 = poorly addressed, 2 = adequately addressed.
++ All or most of the criteria have been fulfilled. Where they have not been fulfilled the conclusions of the study or review are thought very unlikely to alter; + some of the criteria have been fulfilled. Those criteria that have not been fulfilled or not adequately described are thought unlikely to alter the conclusions; − some of the criteria have been fulfilled. Those criteria that have not been fulfilled or not adequately described are thought likely to alter the conclusions.
Outcome measurements
Primary outcome - Study #1 found statistically significant differences (Fisher´s exact test) regarding the presence of A. actinomycetemcomitans (16.7% vs. 0%, P = 0.043), T. forsythia (75% vs. 11,1%, P < 0.001), P. intermedia (54.2% vs. 7.4%, P < 0.001), C. ochracea (37.5% vs. 0%, P < 0.001) and C. rectus (29.2% vs. 0%, p = 0.003) in epithelial cells from periodontal pockets versus healthy sulci. Studies #2 and #3 did not found statistically significant differences regarding specific bacteria.
Secondary outcome - All 3 studies reported that there were quantitatively more unspecific bacteria in or on the epithelial cells taken from a diseased site when compared with a healthy sulcus, however objective data is only shown in study #3. This study presented a statistically significant higher percentage (chi-square test) of epithelial cells with more than 100 bacteria (>100) in periodontal pockets of subjects with periodontitis compared with epithelial cells from healthy sulci of periodontally healthy subjects (35.8% vs. 0%, P < 0.05). No statistically significant differences were found for cells harboring 1–20 and 21–100 bacteria. Interestingly, there were also no differences in the percentage of epithelial cells with 1–20, 21–100 or >100 bacteria from periodontal pockets and healthy sulci of periodontitis subjects.
Evidence profile
For quality rating of evidence a several aspects were taken into consideration. The case-control design (observational) of all included studies, the low number of studies, the imprecise or sparse data, the uncertainty about directness and the high risk of bias suggest that results are subject to limitations and therefore must be interpreted with caution (very low quality rating).
Discussion
This systematic review attempted to gather the available evidence on the in vivo presence of periodontal pathogens in periodontal tissues and its relationship with periodontal disease.
Summary of main results
All selected studies describe the presence of intracellular and/or adherent bacteria to gingival epithelial cells, in vivo, either in periodontal pockets or in healthy sulci. These observations not only corroborate the in vitro findings of this bacterial ability,47-52 but also sustain that invasion of periodontal tissues occurs even in the presence of a hostile and apparently competent (potential cases with systemic diseases were excluded) immune system. They also substantiate that bacterial invasion, per se, is a common event both in health and in disease as suggested by several authors.16,20,21,53
Study #1 found significantly more A. actinomycetemcomitans, T. forsythia, P. intermedia, C. ochracea and C. rectus intracellularly or adhered to epithelial cells from periodontal pockets when compared to healthy sulci. However, this observation was not supported by the other 2 case-control studies. In fact, none of these 2 latter studies found a statistically significant difference in any of the putative periodontal pathogens when comparing health and disease.
The lack of consistency in the findings could result from different methodological approaches regarding microbial identification and case and control definitions. Sudies #1 and #2 used the DNA-DNA checkerboard technique. In this technique a positive reaction was recorded only when a signal was greater or equal to (104) bacterial cells of the searched specie. This means that only substantial differences in the bacterial load were detected. Study #1 hypothetically potentiated differences by defining cases as having suppuration besides PPD >5 and BOP. A much higher polymorphonuclear neutrophil (PMN) and microbial load in suppurative pockets when compared with healthy sulci54 could be a possible explanation for the significant differences found, with such a small sample. Study #2 hypothetically attenuated possible existing differences by comparing periodontal pockets and healthy sulci from the same subjects with periodontitis. It has been recognized that healthy sulcus from subjects with periodontal disease harbor greater number of pathogens when compared with a subject with a healthy periodontium.55,56 Study #3 used the FISH technique. In this technique a sample was considered positive when at least one epithelial cell in each microscopic field observed presented fluorescent bacteria from the hybridized species-specific probe. Bacterial numbers were estimated by direct counting. Unlike DNA-DNA checkerboard, FISH allows smaller differences to be detected, however as clusters of bacteria are not dispersed the real number of bacteria could be underestimated. Despite all this, all studies seemed to be unanimous in considering that there were quantitatively more bacteria in or on the epithelial cells taken from a diseased site than from a healthy sulcus.
The absence of a significantly higher amount of the ‘traditional’ periopathogens and the apparent increase of the ‘commensal’ flora in disease could also be interpreted with reference to the concept of keystone periopathogen described by Hajishengallis.57 Accordingly to Hajishengallis, keystone pathogens, specifically P. gingivalis, seemed to be able to remodel a symbiotic community into a dysbiotic state by imparing innate immunity in ways that enhanced uncontrolled growth of other species, while being a quantitatively minor constituent of the periodontal microbiota.
Limitations
The studies included in this review are all non-randomized case-control studies. Although having inherent limitations and less level of significance, systematic reviews of observational studies of etiology are especially important. This type of studies are often limited in size and so, only by the simultaneous examination of data from similar studies we can achieve insight into real and spurious associations.58
Concerning, however, the scope of this review, the reduced number and the limitations of the included studies preclude a high or even moderate evidence of association between invasion of the periodontium by periodontal pathogens and periodontal disease. The current available data is of low to moderate quality and inconsistent mainly due to study design, poor reporting and methodological diversity.
Conclusion
There is insufficient evidence to support or exclude the invasion by periodontal pathogens as a key step in the etiopathogenesis of periodontal disease. This review highlights the need of further studies on this topic. Future research should rely on large sample studies, with clear case and control definitions ensuring representativeness of cases and comparability of cases and controls. It´s also important to ascertain exposure to invasive periodontal pathogens with maximum detail (e.g. strains, intracellular/intercellular, synergisms) using a blinded operator.
Methods
This systematic review was conducted in accordance with the guidelines of Cochrane Collaboration59 and Transparent Reporting of Systematic Reviews and Meta-Analyses [PRISMA statement].60 The focused question was given in the following: What is the relationship between the quality or quantity of putative periodontal pathogens in periodontal tissues and periodontal disease?
Eligibility criteria
All studies that reported evaluation of periodontal pathogens invasion on human tissues were considered eligible. To be as inclusive as possible, no restrictions were applied with regard to the year of publication or to language. Exclusion decision of non-English papers was delayed until the next step. Reviews and case reports were excluded.
Search strategy
Four Internet sources of evidence were used in search of appropriate papers that fulfilled the purpose of the study: the National Library of Medicine, Washington, DC (MEDLINE-PubMed), the Cochrane Central Register of Controlled Trials (CENTRAL), ISI Web of Knowledge (Thomson Reuters) and SCOPUS (Elsevier). The databases were searched up to January 22, 2014. A structured search strategy was developed to include any published paper that investigated the invasion of periodontal pathogens in patients with periodontal disease. The following strategy was used in the search: (((periodontitis OR “periodontal disease” OR “periodontal pocket”) AND (identification OR detection OR localization) AND bacteria*) AND invas* OR intracellular OR tissue* OR “epithelial cells”)). Key words were combined with Boolean operators and the asterisk symbol (*) was used as truncation. The search strategy was customized according to the database being used.
Screening and selection
Firstly, 2 independent reviewers (LM and MP) screened titles and abstracts for eligible papers. Secondly, the full texts of those papers were obtained and screened in relation to the study purpose, protocol and reported data. An additional hand search across the reference list of the selected studies was made in an attempt to find additional papers that could meet the eligibility criteria.
Data extraction and analysis
Data from the papers that met the selection criteria were processed for analysis. Information regarding year of publication, study design, population, inclusion and exclusion criteria, case and control definitions, characteristics of participants, type of intervention, ascertainment of exposure, cell sample, microorganisms found and its location and statistical analysis were collected by LM e MP using data extraction sheets.
Heterogeneity of trials
The heterogeneity across the trials was detailed according to the following factors: population characteristics, study design, case and control definition, used methodology and type of pathogens found.
Quality assessment
The methodological quality of the selected studies was independently scored by 2 reviewers (LM and MP). Any disagreement was resolved after additional discussion. As all eligible papers were non-randomized studies (case-control studies) methodological quality was assessed combining several proposed criteria of the STROBE statement,61 the Newcastle – Ottawa quality assessment scale,62 the Downs and Black checklist for non-randomized studies63 and SIGN50 guidelines64 as suggested by the Quality Assessment Tools Project Report elaborated by the Canadian Agency for Drugs and Technologies in Health.65 In short, a study was classified as high quality with a very low risk of confounding or bias and a high probability that the relationship is causal (++) when all of the following criteria were adequately addressed: appropriate and clearly focused question, defined exclusion criteria, case selection, control selection, representativeness of cases, comparability of cases and controls, ascertainment of exposure, blindness, funding and statistical analysis. When some of the criteria have not been fulfilled or not adequately described but this fact is thought unlikely to alter the conclusions the study was classified as well conducted study with low risk of confounding or bias and a moderate probability that the relationship is causal (+). When some of the criteria have not been fulfilled or not adequately described and this fact is thought likely to alter the conclusions the study was classified as a study with a high risk of confounding or bias and a significant risk that the relatioship is not causal (−).
Outcome variables
Primary outcome measures of interest were intracellular or adherent (to cell /periodontal tissues) mean value or percentage of known pathogens. Secondary outcomes of interest were global counts of intracellular or adherent (to cell /periodontal tissues) unspecific bacteria.
Evidence Profile
The Grading of Recommendations Assessment, Development and Evaluation (GRADE) system66 as proposed by the GRADE working group was used for grading the collective evidence emerging from this review. Two reviewers (LM and MP) rated the quality of the evidence for outcomes across studies. Any disagreement was resolved after additional discussion.
Acknowledgements
The authors report no conflicts of interest related to this systematic review. This investigation was self founded by the authors and their institutions.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Listgarten MA. Electron microscopic observations on the bacterial flora of acute necrotizing ulcerative gingivitis. J Periodontol 1965; 36:328-39; PMID:14326701; http://dx.doi.org/ 10.1902/jop.1965.36.4.328 [DOI] [PubMed] [Google Scholar]
- 2. Lamont RJ, Jenkinson HF. Life below the gum line. Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 1998; 62: 1244-63; PMID:9841671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Socransky SS, Haffajee AD. Microbial mechanisms in the pathogenesis of desctructive periodontal disease: a critical assessment. J Periodontol Res 1991; 26 (3 pt 2):195-212; http://dx.doi.org/ 10.1111/j.1600-0765.1991.tb01646.x [DOI] [PubMed] [Google Scholar]
- 4. Curtis MA, Kuramitsu HK, Lantz M, Macrina FL, Nakayama K, Potempa J, Reynolds EC, Aduse-Opoku J. Molecular genetics and nomenclature of protéases of Porphyromonas gingivalis. J Periodontol Res 1999; 34(8):464-72; PMID:NOT_FOUND; http://dx.doi.org/ 10.1111/j.1600-0765.1999.tb02282.x [DOI] [PubMed] [Google Scholar]
- 5. Van Dyke TE. Neutrophil receptor modulation in the pathogenesis of periodontal diseases. J Dent Res 1984; 63(3):452-4; PMID:6583245; http://dx.doi.org/ 10.1177/00220345840630031701 [DOI] [PubMed] [Google Scholar]
- 6. Ding Y, Haapasalo M, Kerosuo E, Lounatmaa K, Kotiranta A, Sorsa T. Release and activation of human neutrophil matrix metallo and serine proteinases during phagocytosis of fusobacterium nucleatum, Porphyromonas gingivalis, and Treponema denticola. J Clin Periodontol 1997; 24(4):237-48; PMID:9144046; http://dx.doi.org/ 10.1111/j.1600-051X.1997.tb01837.x [DOI] [PubMed] [Google Scholar]
- 7. Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000. 2010; 52(1):68-83; PMID:20017796; http://dx.doi.org/ 10.1111/j.1600-0757.2009.00323.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Darveu RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun 1998; 66 (4):1660-5; PMID:9529095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Madianos PN, Papapanou PN, Sandros J. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect Immun 1997; 65(10):3983-90; PMID:9316996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gemmell E., Yamazaki K., Seymour G.J.. The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontol 2000. 2007; 43: 14-40; PMID:17214833; http://dx.doi.org/ 10.1111/j.1600-0757.2006.00173.x [DOI] [PubMed] [Google Scholar]
- 11. Johnson JD, Chen R, Lenton PA, Zhang G, Hinrichs JE, Rudney JD. Persistence of extracrevicular bacterial reservoirs after treatment of aggressive periodontitis. J Periodontol 2008; 79(12):2305-12; PMID:19053921; http://dx.doi.org/ 10.1902/jop.2008.080254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eick S, Pfister W. Efficacy of antibiotics against periodontopathogenic bacteria within epithelial cells: an in vitro study. J Periodontol 2004; 75(10):1327-34; PMID:15562909; http://dx.doi.org/ 10.1902/jop.2004.75.10.1327 [DOI] [PubMed] [Google Scholar]
- 13. Vitkov L, Krautgartner WD, Hannig M. Bacterial internalization in periodontitis. Oral Microbiol Immunol 2005; 20(5):317-21; PMID:16101968; http://dx.doi.org/ 10.1111/j.1399-302X.2005.00233.x [DOI] [PubMed] [Google Scholar]
- 14. Manor A, Lebendiger M, Shiffer A, Tovel H. Bacterial invasion of periodontal tissues in advanced periodontitis in humans. J Periodontol 1984; 55(10):567-73; PMID:6593450; http://dx.doi.org/ 10.1902/jop.1984.55.10.567 [DOI] [PubMed] [Google Scholar]
- 15. Delacourt-Debruyne EM, Boutigny HR, Hildebrand HF. Features of severe periodontal disease in a teenager with Chédiak-Higashi syndrome. J Periodontol 2000; 71(5):816-24; PMID:10872965; http://dx.doi.org/ 10.1902/jop.2000.71.5.816 [DOI] [PubMed] [Google Scholar]
- 16. Noiri Y, Ozaki K, Nakae H, Matsuo T, Ebisu S. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J Periodont Res 1997;32(7):598-607; PMID:9401932; http://dx.doi.org/ 10.1111/j.1600-0765.1997.tb00937.x [DOI] [PubMed] [Google Scholar]
- 17. Van Winkelhof AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol 2002; 29(10):1023-8; PMID:12472995; http://dx.doi.org/ 10.1034/j.1600-051X.2002.291107.x [DOI] [PubMed] [Google Scholar]
- 18. Mombelli A, Casagni F, Madianos PN. Can presence or absence of periodontal pathogens distinguish between subjects with chronic and aggressive periodontitis? A systematic review. J Clin Periodontol 2002; 29(suppl 3):10-21; discussion 37-8; PMID:12787203; http://dx.doi.org/ 10.1034/j.1600-051X.29.s3.1.x [DOI] [PubMed] [Google Scholar]
- 19. Lamont RJ, Yilmaz O. In or out: the invasiveness of oral bacteria. Periodontol 2000. 2002; 30:61-69; PMID:12236896; http://dx.doi.org/ 10.1034/j.1600-0757.2002.03006.x [DOI] [PubMed] [Google Scholar]
- 20. Rudney JD, Chen R, Zhang D. Streptococci dominate the diverse flora within buccal cells. J Dent Res 2005; 84(12):1165-71; PMID:16304448; http://dx.doi.org/ 10.1177/154405910508401214 [DOI] [PubMed] [Google Scholar]
- 21. Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res 2005; 84(1):59-63; PMID:15615877; http://dx.doi.org/ 10.1177/154405910508400110 [DOI] [PubMed] [Google Scholar]
- 22. Dibart S, Skobe Z, Snapp KR, Socransky SS, Smith CM, Kent R. Identification of bacterial species on or in crevicular epithelial cells from healthy and periodontally diseased patients using DNA-DNA hybridization. Oral Microbiol Immunol 1998; 13(1):30-35; PMID:9573819; http://dx.doi.org/ 10.1111/j.1399-302X.1998.tb00747.x [DOI] [PubMed] [Google Scholar]
- 23. Colombo AV, da Silva CM, Haffajee A, Colombo APV. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol 2006; 55(pt 5):609-15; PMID:16585650; http://dx.doi.org/ 10.1099/jmm.0.46417-0 [DOI] [PubMed] [Google Scholar]
- 24. Colombo AV, da Silva CM, Haffajee A, Colombo APV. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodont Res 2007; 42(3):236-43; PMID:17451543; http://dx.doi.org/ 10.1111/j.1600-0765.2006.00938.x [DOI] [PubMed] [Google Scholar]
- 25. Saglie FR, Carranza FA, Jr, Newman MG, Cheng L, Lewin KJ. Identification of tissue-invading bacteria in human periodontal disease. J Periodontal Res 1982; 17(5):452-5; PMID:6218269; http://dx.doi.org/ 10.1111/j.1600-0765.1982.tb02024.x [DOI] [PubMed] [Google Scholar]
- 26. Saglie R, Newman MG, Carranza FA, Jr, Pattison GL. Bacterial invasion of gingival in advanced periodontitis in humans. J Periodontol 1982; 53(4):217-22; PMID:6951991; http://dx.doi.org/ 10.1902/jop.1982.53.4.217 [DOI] [PubMed] [Google Scholar]
- 27. Allenspach-Petrzilka GE, Guggenheim B. Bacterial invasion of the periodontium: an important factor in the pathogenesis of periodontitis? J Clin Periodontol 1983; 10(6):609-17; PMID:6581176; http://dx.doi.org/ 10.1111/j.1600-051X.1983.tb01299.x [DOI] [PubMed] [Google Scholar]
- 28. Saglie FR, Carranza FA, Jr, Newman MG. The presence of bacteria within the oral epithelium in periodontal disease. I. A scanning and transmission electron microscopic study. J Periodontol 1985; 56(10):618-24; PMID:3863912; http://dx.doi.org/ 10.1902/jop.1985.56.10.618 [DOI] [PubMed] [Google Scholar]
- 29. Saglie FR, Smith CT, Newman MG, Carranza FA, Jr, Pertuiset JH, Cheng L, Auil E, Nisengard RJ. The presence of bacteria in the oral epithelium in periodontal disease. II. Immunohistochemical identification of bacteria. J Periodontol 1986; 57(8):492-500; PMID:2427680; http://dx.doi.org/ 10.1902/jop.1986.57.8.492 [DOI] [PubMed] [Google Scholar]
- 30. Christersson LA, Wikesjö UM, Albini B, Zambón JJ, Genco RJ. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. II. Correlation between immunofluorescence and culture techniques. J Periodontol 1987; 58(8):540-5; PMID:3305857; http://dx.doi.org/ 10.1902/jop.1987.58.8.540 [DOI] [PubMed] [Google Scholar]
- 31. Saglie FR, Pertuiset JH, Smith CT, Nestor MG, Carranza FA, Jr, Newman MG, Rezende MT, Nisengard R. The presence of bacteria in the oral epithelium in periodontal disease. III. Correlation with Langerhans cells. J Periodontol 1987; 58(6):417-22; PMID:2439676; http://dx.doi.org/ 10.1902/jop.1987.58.6.417 [DOI] [PubMed] [Google Scholar]
- 32. Saglie FR, Marfany A, Camargo P. Intragingival occurrence of actinobacillus actinomycetemcomitans and bacteroides gingivalis in active destructive periodontal lesions. J Periodontol 1988; 59(4):259-65; PMID:3290428; http://dx.doi.org/ 10.1902/jop.1988.59.4.259 [DOI] [PubMed] [Google Scholar]
- 33. Saglie FR, Pertuiset J, Rezende MT, Nestor M, Marfany A, Cheng J. In situ correlative immune-identification of mononuclear infiltrates and invasive bacteria in diseased gingival. J Periodontol 1988; 59(10):688-96; PMID:2460612; http://dx.doi.org/ 10.1902/jop.1988.59.10.688 [DOI] [PubMed] [Google Scholar]
- 34. Tonetti MS, Cortellini D, Lang NP. In situ detection of apoptosis at sites of chronic bacterially induced inflammation in human gingival. Infect Immun 1998; 66(11):5190-5; PMID:9784521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hillmann G, Dogan S, Geurtsen W. Histopathological investigation of gingival tissue from patients with rapidly progressive periodontitis. J Periodontol 1998; 69(2):195-208; PMID:9526920; http://dx.doi.org/ 10.1902/jop.1998.69.2.195 [DOI] [PubMed] [Google Scholar]
- 36. Ishizuka M. Identification of periodontopathic bacteria DNA from periodontitis-involved gingival tissue using a PCR method. Kokubyo Gakkai Zasshi 2002; 69(1):15-26; PMID:11968834; http://dx.doi.org/ 10.5357/koubyou.69.15 [DOI] [PubMed] [Google Scholar]
- 37. Rautemaa R, Järvensivu A, Kari K, Wahlgren J, DeCarlo A, Richardson M, Sorsa T. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis 2004; 10(5):298-305; PMID:15315648; http://dx.doi.org/ 10.1111/j.1601-0825.2004.01021.x [DOI] [PubMed] [Google Scholar]
- 38. Noiri Y, Li L, Yoshimura F, Ebisu S. Localization of Porphyromonas gingivalis-carrying fimbriae in situ in human periodontal pockets. J Dent Res 2004; 83(12):941-5; PMID:15557402; http://dx.doi.org/ 10.1177/154405910408301210 [DOI] [PubMed] [Google Scholar]
- 39. Thiha K, Takeuchi Y, Umeda M, Huang Y, Ohnishi M, Ishikawa I. Identification periodontopathic bacteria in gingival tissue of Japanese periodontitis patients. Oral Microbiol Immunol 2007; 22(3):201-7; PMID:17488447; http://dx.doi.org/ 10.1111/j.1399-302X.2007.00354.x [DOI] [PubMed] [Google Scholar]
- 40. Silva N, Dutzan N, Hernandez M, Dezerega A, Rivera O, Aquillon JC, Aravena O, Lastres P, Pozo P, Vernal R, Gamonal J. Characterization of progressive periodontal lesions in chronic periodontitis patients: levels of chemokines, cytokines, matrix metalloproteinase-13, periodontal pathogens and inflammatory cells. J Clin Periodontol 2008; 35(3):206-14; PMID:18269660; http://dx.doi.org/ 10.1111/j.1600-051X.2007.01190.x [DOI] [PubMed] [Google Scholar]
- 41. Kim YC, Ko Y, Hong SD, Kim KY, Lee YH, Chae C, Choi Y. Presence of Porphyromonas gingivalis and plasma cell dominance in gingival tissues with periodontitis. Oral Dis 2010; 16(4) 375-81; PMID:20233323; http://dx.doi.org/ 10.1111/j.1601-0825.2009.01649.x [DOI] [PubMed] [Google Scholar]
- 42. Guyodo H, Meuric V, Le Pottier L, Martin B, Pers JO, Bonnaure-Mallet M. Colocalization of Porphyromonas gingivalis with CD4+ T cells in periodontal disease. FEMS Immunol Med Microbiol 2012; 64(2):175-83; PMID:22066676; http://dx.doi.org/ 10.1111/j.1574-695X.2011.00877.x [DOI] [PubMed] [Google Scholar]
- 43. Colombo AV, Barbosa GM, Higashi D, di Micheli G, Rodrigues PH, Simionato MR. Quantitative detection of Staphylococcus aureus, Enterococcus faecalis and Pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol 2013; 62(Pt 10):1592-600; PMID:23800598; http://dx.doi.org/ 10.1099/jmm.0.055830-0 [DOI] [PubMed] [Google Scholar]
- 44. Riviere GR, Weisz KS, Simomson LG, Lukhart SA. Pathogen-related spirochetes identified within gingival tissue from patients with acute necrotizing ulcerative gingivitis. Infect Immun 1991; 59(8):2653-57; PMID:1855985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kubar A, Saygun I, Özdemir A, Yapar M, Slot J. Real-time polymerase chain reaction quantification of human cytomegalovirus and Epstein-Barr virus in periodontal pockets and the adjacent gingiva of periodontitis lesions. J Periodontal Res 2005; 40(2):97-104; PMID:15733143; http://dx.doi.org/ 10.1111/j.1600-0765.2005.00770.x [DOI] [PubMed] [Google Scholar]
- 46. Reed SG, Lopatin DE, Foxman B, Burt BA. Oral Chlamydia trachomatis in patients with established periodontitis. Clin Oral investig 2000; 4 (4):226-32; PMID:11218493; http://dx.doi.org/ 10.1007/s007840000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol 1999; 1(3):215-23; PMID:11207554; http://dx.doi.org/ 10.1046/j.1462-5822.1999.00022.x [DOI] [PubMed] [Google Scholar]
- 48. Fives-Taylor P, Meyer D, Mintz K. Characteristics of Actinobacillus actinomycetemcomitans invasion of ans adhesion to cultured epithelial cells. Adv Dent Res 1995; 9(1):55-62; PMID:7669215; http://dx.doi.org/ 10.1177/08959374950090011001 [DOI] [PubMed] [Google Scholar]
- 49. Dorn BR, Leung KL, Progulske-Fox A. Invasion of human oral epithelial cells by Prevotella intermedia. Infect Immun 1998; 66(12):6054-7; PMID:9826397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mooney A, Byrne C, Clyne M, Johnson-Henry K, Sherman P, Bourke B. Invasion of human epithelial cells by Campylobacter upsaliensis. Cell Microbiol 2003; 5(11):835-47; PMID:14531898; http://dx.doi.org/ 10.1046/j.1462-5822.2003.00325.x [DOI] [PubMed] [Google Scholar]
- 51. Dickinson BC, Moffatt CE, Hagerty D. Interaction of oral bacteria with gingival epithelial cell multilayers. Mol Oral Microbiol 2011; 26(3):210-220; PMID:21545698; http://dx.doi.org/ 10.1111/j.2041-1014.2011.00609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saito A, Inagaki S, Ishihara K. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb Pathog 2009; 47(6):329-33; PMID:19818393; http://dx.doi.org/ 10.1016/j.micpath.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 53. Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 2003; 30(7):644-54; PMID:12834503; http://dx.doi.org/ 10.1034/j.1600-051X.2003.00376.x [DOI] [PubMed] [Google Scholar]
- 54. Herrera D, Roldán S, González I, Sanz M. The periodontal abscess (I). Clinical and microbiological findings. J Clin Periodontol 2000; 27(6):387-94; PMID:10883867; http://dx.doi.org/ 10.1034/j.1600-051x.2000.027006387.x [DOI] [PubMed] [Google Scholar]
- 55. Riviere GR, Smith KS, Tzagaroulaki E, Kay SL, Zhu X, DeRouen TA, Adams DF. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. J Periodontol 1996; 67(2):109-115; PMID:8667130; http://dx.doi.org/ 10.1902/jop.1996.67.2.109 [DOI] [PubMed] [Google Scholar]
- 56. Sakamoto M, Huang Y, Ohnishi M, Umeda M, Ishikawa I, Benno Y. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J Med Microbiol 2004; 53(pt 6):563-71; PMID:15150339; http://dx.doi.org/ 10.1099/jmm.0.45576-0 [DOI] [PubMed] [Google Scholar]
- 57. Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 2012; 27(6):409-419; PMID:23134607; http://dx.doi.org/ 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dickersin K. Systematic reviews in epidemiology: why we are so far behind? Inter J Epidemiol 2002; 31(1):6-12; PMID:11914282; http://dx.doi.org/ 10.1093/ije/31.1.6 [DOI] [PubMed] [Google Scholar]
- 59. Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions. The Cochrane Collaboration, 2011; v.5.0.2 Available at http://www.cochrane-handbook.org. Accessed 1 Dec. 2013. [Google Scholar]
- 60. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA G. Preferred reporting items for systematic reviews and meta analyses: the PRISMA statement. Open Med 2009; 3(3):123-30. [PMC free article] [PubMed] [Google Scholar]
- 61. von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP, STROBE initiative The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med 2007; 4(10):1623-7; http://dx.doi.org/ 10.1371/journal.pmed.0040296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomized studies in meta-analysis. Available at http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 1 Dec 2012 (2011) [Google Scholar]
- 63. Downs SH, Black N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. J Epidemiol Community Health 1998; 52(6):377-84; PMID:9764259; http://dx.doi.org/ 10.1136/jech.52.6.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scottish Intercollegiate Guidelines Network . SIGN 50: A guideline developer's handbook. Annex C. 2008; Available: http://www.sign.ac.uk/guidelines/fulltext/50/ [Google Scholar]
- 65. Bai A, Shukla VK, Bak G, Wells G. Quality assessment tools project report. Ottawa: Canadian Agency for Drugs and Technologies in Health; 2012; Available: http://www.cadth.ca/en/products/methods-and-guidelines/publication/3458 [Google Scholar]
- 66. Atkins D, Best D, Briss PA, Eccles M, Falck-Ytter Y, Flottorp S, Guyatt GH, Harbour RT, Haugh MC, Henry D, et al. Grading quality of evidence and strength of recommendations. BMJ 2004; 328(7454):1490; PMID:15205295; http://dx.doi.org/ 10.1136/bmj.328.7454.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.