Abstract
Daptomycin resistance (DAPR) in Staphylococcus aureus is associated with mutations in genes that are also implicated in staphylococcal pathogenesis. Using a laboratory-derived series of DAP exposed strains, we showed a relationship between increasing DAP MIC and reduced virulence in a Galleria mellonella infection model. Point mutations in walK and rpoC led to cumulative reductions in virulence and simultaneous increases in DAP MIC. A point mutation to mprF did not impact on S.aureus virulence; however deletion of mprF led to virulence attenuation and hyper-susceptibility to DAP. To validate our findings in G. mellonella, we confirmed the attenuated virulence of select isolates from the laboratory-derived series using a murine septicaemia model. As a corollary, we showed significant virulence reductions for clinically-derived DAPR isolates compared to their isogenic, DAP-susceptible progenitors (DAPS). Intriguingly, each clinical DAPR isolate was persistent in vivo. Taken together, it appears the genetic correlates underlying daptomycin resistance in S. aureus also alter pathogenicity.
Keywords: bacterial persistence, Galleria mellonella, mprF, S. aureus, walK
Introduction
Daptomycin (DAP) is a cyclic lipopeptide antibiotic that is increasingly being used to treat Staphylococcus aureus infections.1 Unfortunately, therapeutic failures, albeit relatively uncommon, have been reported.1-3 Thus far, the mechanisms underlying daptomycin resistance (DAPR) in S. aureus have focused on point mutations in genes involved in phospholipid biosynthesis, particularly mprF,4 which codes for lysyl-phosphatidylglycerol (L-PG) synthetase, cls2, which codes for cardiolipin synthase and pgsA, which codes for CDP-diacylglycerol-glycerol-3-phosphate-3-phosphatidyltransferase.5 It is hypothesized that these mutations lead to changes in phospholipid membrane composition, which may affect membrane charge causing electro-repulsion of calcium-complexed daptomycin or may directly affect daptomycin binding.5 Interestingly, daptomycin, when complexed with calcium, appears to act similar to cationic antimicrobial peptides, and therefore genetic mutations associated with daptomycin resistance have been shown to simultaneously confer resistance to host innate immune responses.4 Other genes associated with reduced susceptibility to daptomycin may also affect S. aureus-host interactions, particularly the sensor-histidine kinase, WalK (previously YycG), which has an integral role in cell wall homeostasis but also regulates a number of genes important for virulence.6,7 The focus of this study was to assess for the first time the pathogenic consequences of DAPR in S. aureus.
Methods
Bacterial strains used in this study are shown in Table 1. For the purpose of this study, daptomycin non-susceptible isolates (defined by an MIC of DAP >1 mg/L) are referred to as DAPR. The laboratory-derived, DAP-exposed series was obtained from Friedman et al., and included a parent strain (CB1118) and 4 mutants isolated after serial in vitro DAP exposure over 20 days.8 These strains were previously genome sequenced and their cumulative mutations are shown in Table 1.8 An mprF deletion mutant (CB1118ΔmprF) was also included.9 In addition, 3 clinical S. aureus pairs were assessed, which included a daptomycin-susceptible (DAPS) parent strain with its corresponding DAPR daughter strain that developed after DAP therapy and clinical failure.5 Growth kinetic experiments were performed as described previously.10
Table 1.
Laboratory and clinically derived daptomycin-exposed Staphylococcus aureus strains used in this study
| Strain | Description | DAP MIC (mg/L) |
|---|---|---|
| Laboratory-derived isolatesa | ||
| CB1118b | Parent used for in vitro passage. | 0.5 |
| CB1618-d6 | mprF(T345A) | 2 |
| CB1618-d9 | mprF(T345A), walK(R263C) | 4 |
| CB1618-d13 | mprF(T345A), walK(R263C), rpoB(A1086V) | 4 |
| CB1618-d20 | mprF, (T345A), walK(R263C), rpoB(A1086V), rpoC(Q961K) | 16 |
| CB1118ΔmprF | mprF deletion mutant | 0.125 |
| Clinically-derived isolatesc | ||
| 1) A8819 (S) | Osteomyelitis, septic arthritis | 0.25 |
| A8817 (R) | 2 | |
| 2) A8796 (S) | Vertebral osteomyelitis | 0.5 |
| A8799 (R) | 2 | |
| 3) A9754 (S) | Endocarditis | 0.5 |
| A9757 (R) | 4 |
DAP, Daptomycin; S, Susceptible; MIC, Minimum Inhibitory Concentration; R, Resistant.
Mutations described in ref.8
Refers to previously described strain MW2.
Described in ref.5
We used Galleria mellonella as a substitute in vivo host to assess staphylococcal virulence as described previously.7,10,11 This model was chosen because G. mellonella not only have phagocytic cells in their hemolymph but they also rely on antimicrobial peptides for their immune defense, hence their utility for the study of DAPR. Briefly, bacteria were injected into the hemocoel of each caterpillar (n = 16 / strain, 1 × 105–1 × 106 CFU/larvae) using a 10 μl Hamilton syringe.10 For the mammalian experiments, 10–15 C57BL/6 or CD-1 mice per strain were injected intraperitoneally with 2–4 × 107 CFU of bacteria mixed with 6% porcine gastric mucin (Sigma Aldrich) in 500 μl, and were monitored for 7 days.12 To assess bacterial persistence, kidneys were harvested from surviving DAPR infected animals 7-days post infection, and bacterial counts were performed. Survival curves were plotted using the Kaplan-Meier method with differences calculated using log-rank tests (GraphPad Prism v 6.0). Experiments were approved by the Institutional Animal Care and Use Committee at Cubist Pharmaceuticals, Inc.. and Monash University prior to initiation of studies.
Results and discussion
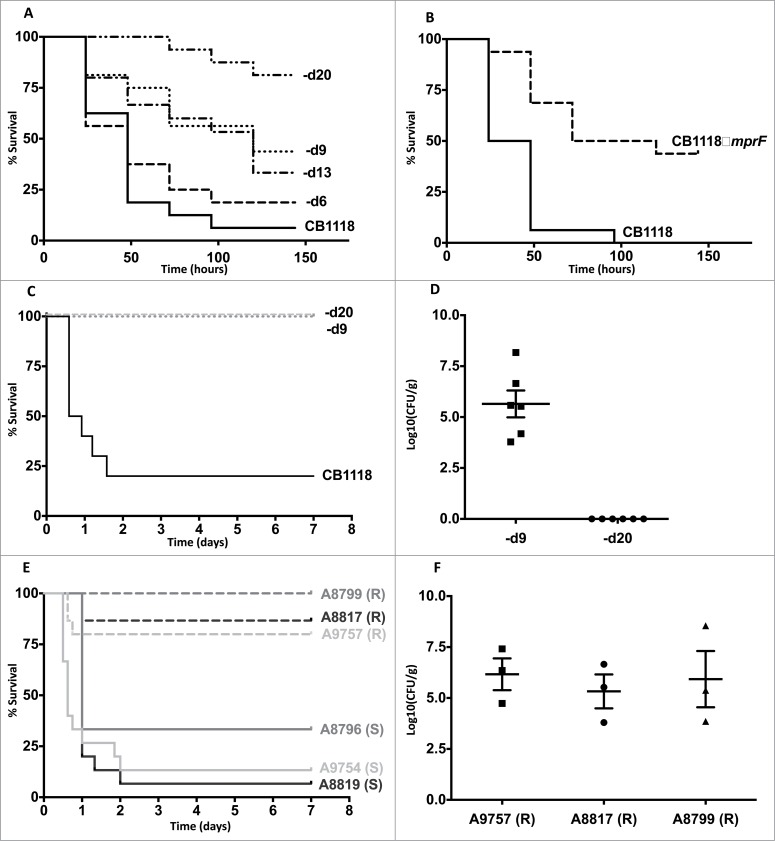
To determine the impact of DAPR on staphylococcal virulence we first infected G. mellonella with a genetically characterized in vitro derived series of S. aureus strains that had incremental increases in the MIC of DAP and a cumulative number of mutations (Table 1).8 Importantly, these strains grew similarly in vitro except for CB1618-d20, which was impaired for growth (Supplementary Fig. 1). As shown in Figure 1A, there was a significant trend of reducing virulence as the MIC of DAP increased (P < 0.05, Chi square for trend). However, when individual strains in the series were compared, significant reductions in virulence occurred only on 2 occasions. The first occurred due to a mutation (R263C) in the sensor histidine kinase known as walK (previously yycG). Working together with its cognate response regulator (WalR), this 2-component regulatory system is indispensible for cell wall homeostasis in S. aureus but has also been reported to regulate virulence.6 This isolate (CB1618-d9) had an increase in DAP MIC from 2 mg/L to 4 mg/L and it was attenuated in killing G. mellonella (P < 0.01) compared to its progenitor (CB1618-d6) (Fig. 1A). The second occurred with the final isolate of the series (CB1618-d20), which acquired a mutation in rpoC, leading to a substantial rise in MIC of daptomycin to 16 mg/L and marked virulence attenuation (Fig. 1A).
Figure 1.
Daptomycin resistance in S. aureus correlates with altered virulence and in vivo persistence. (A) G. mellonella infection of a laboratory-derived series of isolates with incremental increases in daptomycin MIC was performed (n = 16 for each strain). For clarity, CB1618-d6, CB1618-d9, CB1618-d13, CB1618-d20 are represented by d6, d9, d13 and d20 respectively. Virulence attenuation was observed for CB1618-d9 when compared to CB1618-d6 (P < 0.01) and CB1618-d20 when compared with CB1618-d13 (P < 0.01). No significant virulence attenuation was observed for CB1618-d6 (P = 0.44) and CB1618-d13 (P = 0.70) when compared to their respective progenitor strains. (B) An mprF deletion strain (CB1118ΔmprF) produced significantly less killing of G. mellonella when compared to its progenitor (P < 0.001, n = 16 for each strain). (C) CB1618-d9 and CB1618-d20 were attenuated for virulence in a murine septicaemia model (P < 0.001, n = 10 for each strain). (D) CB1618-d9 was capable of in vivo persistence as determined by bacterial densities in the kidneys of mice 7-days post infection. In contrast, bacterial burden was not observed in the kidneys of mice infected with CB1618-d20. (E) Virulence of 3 DAP-exposed clinical pairs was assessed using a murine septicaemia model. The daptomycin-resistant (R) isolates were significantly attenuated for virulence compared to their susceptible progenitors (S) (P < 0.001, n = 15 for each strain) and (F) were persistent in the kidneys of infected mice out to 7-days post-infection.
DAPR in S. aureus has most commonly been associated with ‘gain of function’ point mutations in mprF.13 This leads to greater L-PG (cationic) being produced and translocated to the outer layer of the membrane causing a reduction in the net-negative membrane charge. As a consequence, these mutations simultaneously lead to resistance to daptomycin and cationic antimicrobial peptides.14 It is not surprising then that we observed no decrease in virulence for the first isolate of the laboratory-exposed series harbouring an mprF T345A mutation (CB1618-d6) (Fig. 1A). Conversely, we would expect that deletion of mprF would create a strain not only hypersusceptible to daptomycin but also less virulent as it would be hypersusceptible to cationic antimicrobial peptides. To test this hypothesis, we assessed an mprF deletion mutant (CB1118ΔmprF) from the same wild-type strain.9 Not only was CB1118ΔmprF hypersusceptible to daptomycin (MIC 0.125 mg/L, Table 1), it was also significantly less virulent compared to its parent strain (P < 0.001, Fig. 1B). Taken together, these data further highlight MprF as an attractive drug target, as inhibition of the protein may render S. aureus simultaneously susceptible to antimicrobials and the host innate immune system.15
To validate our findings from G. mellonella and to assess the impact of DAPR on mammalian disease, we assessed the virulence of each attenuated mutant strain from the laboratory-derived series using an established murine septicaemia model.12 Consistent with that observed in G. mellonella, CB1618-d9 (DAP MIC = 4 mg/L) and CB1618-d20 (DAP MIC = 16 mg/L) were significantly attenuated for virulence when compared to their susceptible parent strain (CB1118) (P < 0.001 for each) (Fig. 1C). We were intrigued by the survival of the DAPR infected mice and wanted to assess whether the bacteria were being cleared or were persisting within the host. To test this, we assessed the bacterial burden in the kidneys of mice infected with each DAPR strain at 7-days post infection. Despite the surviving mice, the kidneys of mice infected with CB1618-d9 showed high bacterial burdens (average of 2.5 × 107 CFU/g of tissue) indicating that this strain was persistent in vivo (Fig. 1D). In contrast, no bacteria were recovered from mice infected with CB1618-d20 suggesting that mutations in this strain were associated with in vitro and in vivo fitness costs (Fig. 1D and Supp. Fig. 1A).
As a corollary to what was observed for the laboratory-derived series, we next infected mice with 3 clinical S. aureus pairs consisting of a DAPS parent strain with its corresponding DAPR daughter strain that arose after DAP therapy and clinical failure. Each of the pairs were associated with severe infections including osteomyelitis, septic arthritis and endocarditis and were confirmed to be isogenic by whole genome sequencing as described previously (Table 1).5 As shown in Figure 1E, each of the clinical DAPR strains were significantly attenuated for virulence when compared to their DAPS progenitor strain (P < 0.001). Furthermore, as shown with the laboratory derived DAPR strain (CB1618-d9), despite surviving animals out to 7 days post-infection, high bacterial loads were recovered from the kidneys of mice infected with each DAPR strain (Fig. 1F). These data lend further support that clinical DAPR S. aureus strains have the capacity to persist in vivo.
To our knowledge, this is the first study to describe an association between the development of DAPR and altered S. aureus pathogenicity. Subtle genetic changes associated with DAPR, such as a point mutation in WalK (R263C), can have significant impact on the ability of the bacteria to cause mammalian disease. Intriguingly, despite DAPR S. aureus strains causing less lethal disease, they have the capacity to persist in vivo, a finding consistent with that observed in humans.16-18 Developing a deeper understanding of the genetic determinants impacting antibiotic resistance and virulence in S. aureus will provide critical insights into novel therapeutic strategies for this aggressive human pathogen.
Supplementary Material
Conflicts of Interest
A.Y.P has been to one advisory board meeting for Ortho-McNeil-Janssen and AstraZeneca, and has received a speaker's honorarium from AstraZeneca for one presentation. G.M.E. has served on Scientific Advisory Boards for Cubist, Bayer Schering, Johnson & Johnson Pharmaceutical Research and development, Novartis, Pfizer, Shionogi, Theravance; he has had research training support from Cubist, research contracts from Novexel, Pfizer and Theravance, and speaking honoraria from Novartis. He serves on the Board of Directors of the National Foundation for Infectious Diseases. R.C.M. served as a consultant to Cubist, Forest, Merck, Novartis, Ortho, Johnston and Johnston, Pfizer, Theravance, and Wyeth. E. M received grant support and served on the advisory board for Astellas Pharma and received grant support from T2 Biosystems. L.I.M and A.R. are scientists working for Cubist Pharmaceuticals. All other authors have no conflicts of interest.
funding
D.R.C was funded by an Australian Postgraduate Award and a Monash University Postgraduate Publication Award. A.Y.P was funded by an Australian National Health and Medical Research Council R.D. Wright Biomedical Fellowship (APP1047916) and Project Grant (APP1047918). E. M was supported by National Institutes of Health grant P01 AI083214.
References
- 1. Fowler VG, Jr., Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med 2006; 355:653-65; PMID:16914701; http://dx.doi.org/ 10.1056/NEJMoa053783 [DOI] [PubMed] [Google Scholar]
- 2. Hayden MK, Rezai K, Hayes RA, Lolans K, Quinn JP, Weinstein RA. Development of Daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2005; 43:5285-7; PMID:16207998; http://dx.doi.org/ 10.1128/JCM.43.10.5285-5287.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marty FM, Yeh WW, Wennersten CB, Venkataraman L, Albano E, Alyea EP, Gold HS, Baden LR, Pillai SK. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J Clin Microbiol 2006; 44:595-7; PMID:16455920; http://dx.doi.org/ 10.1128/JCM.44.2.595-597.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 2009; 5:e1000660; PMID:19915718; http://dx.doi.org/ 10.1371/journal.ppat.1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC, Jr., Eliopoulos GM. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 2012; 7:e28316; PMID:22238576; http://dx.doi.org/ 10.1371/journal.pone.0028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 2004; 186:1175-81; PMID:14762013; http://dx.doi.org/ 10.1128/JB.186.4.1175-1181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 2011; 7:e1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 2006; 50:2137-45; PMID:16723576; http://dx.doi.org/ 10.1128/AAC.00039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rubio A, Moore J, Varoglu M, Conrad M, Chu M, Shaw W, Silverman JA. LC-MS/MS characterization of phospholipid content in daptomycin-susceptible and -resistant isolates of Staphylococcus aureus with mutations in mprF. Mol Membr Biol 2012; 29:1-8; PMID:22276671; http://dx.doi.org/ 10.3109/09687688.2011.640948 [DOI] [PubMed] [Google Scholar]
- 10. Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC, Jr., Eliopoulos GM. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis 2009; 199:532-6; PMID:19125671; http://dx.doi.org/ 10.1086/596511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 2010; 6:e1000944; PMID:20548948; http://dx.doi.org/ 10.1371/journal.ppat.1000944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silverman JA, Oliver N, Andrew T, Li T. Resistance studies with daptomycin. Antimicrob Agents Chemother 2001; 45:1799-802; PMID:11353628; http://dx.doi.org/ 10.1128/AAC.45.6.1799-1802.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang SJ, Nast CC, Mishra NN, Yeaman MR, Fey PD, Bayer AS. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob Agents Chemother 2010; 54:3079-85; PMID:20498310; http://dx.doi.org/ 10.1128/AAC.00122-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med 2001; 193:1067-76; PMID:11342591; http://dx.doi.org/ 10.1084/jem.193.9.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weidenmaier C, Kristian SA, Peschel A. Bacterial resistance to antimicrobial host defenses–an emerging target for novel antiinfective strategies? Current drug targets 2003; 4:643-9; PMID:14577655; http://dx.doi.org/ 10.2174/1389450033490731 [DOI] [PubMed] [Google Scholar]
- 16. Hayden MK, Rezai K, Hayes RA, Lolans K, Quinn JP, Weinstein RA. Development of Daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2005; 43:5285-7; PMID:16207998; http://dx.doi.org/ 10.1128/JCM.43.10.5285-5287.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marty FM, Yeh WW, Wennersten CB, Venkataraman L, Albano E, Alyea EP, Gold HS, Baden LR, Pillai SK. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J Clin Microbiol 2006; 44:595-7; PMID:16455920; http://dx.doi.org/ 10.1128/JCM.44.2.595-597.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gasch O, Camoez M, Dominguez MA, Padilla B, Pintado V, Almirante B, Martin C, Lopez-Medrano F, de Gopegui ER, Blanco JR, et al. Emergence of resistance to daptomycin in a cohort of patients with methicillin-resistant Staphylococcus aureus persistent bacteraemia treated with daptomycin. J Antimicrob Chemother 2014; 69:568-71; PMID:24107389; http://dx.doi.org/ 10.1093/jac/dkt396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.