abstract
Even though prion (encoded by the PRNP gene) diseases like bovine spongiform encephalopathy (BSE) are fatal neurodegenerative diseases in cattle, their study via gene deletion has been limited due to the absence of cell lines or mutant models. In this study, we aim to develop an immortalized fibroblast cell line in which genome-engineering technology can be readily applied to create gene-modified clones for studies. To this end, this study is designed to 1) investigate the induction of primary fibroblasts to immortalization by introducing Bmi-1 and hTert genes; 2) investigate the disruption of the PRNP in those cells; and 3) evaluate the gene expression and embryonic development using knockout (KO) cell lines. Primary cells from a male neonate were immortalized with Bmi-1and hTert. Immortalized cells were cultured for more than 180 days without any changes in their doubling time and morphology. Furthermore, to knockout the PRNP gene, plasmids that encode transcription activator-like effector nuclease (TALEN) pairs were transfected into the cells, and transfected single cells were propagated. Mutated clonal cell lines were confirmed by T7 endonuclease I assay and sequencing. Four knockout cell lines were used for somatic cell nuclear transfer (SCNT), and the resulting embryos were developed to the blastocyst stage. The genes (CSNK2A1, FAM64A, MPG and PRND) were affected after PRNP disruption in immortalized cells. In conclusion, we established immortalized cattle fibroblasts using Bmi-1 and hTert genes, and used TALENs to knockout the PRNP gene in these immortalized cells. The efficient PRNP KO is expected to be a useful technology to develop our understanding of in vitro prion protein functions in cattle.
Keywords: cattle embryos, immortalization, knockout, PRNP, TALEN
Introduction
Prion diseases, a group of neurodegenerative disorders affecting humans and animals (e.g. bovine spongiform encephalopathy (BSE)) still lack an effective treatment. If the function of the cellular prion protein (PrPC) in bovine cell lines were studied, knowledge of its function could be enhanced by regulating the expression of the PRNP in cell lines.1 Thus, to understand the molecular pathways of specific genes in vitro or in vivo, the most common methods currently used are to induce the inhibition or overexpression of the gene in Knockout (KO)/Knockin (KI) cell lines or animals. One of the easiest ways to approach these basic biomedical studies is to introduce DNA or RNA constructions designed for aimed research into the suitable cell lines that are isolated from target organs or tissues via primary culture, and then to analyze the expression of the target genes or other related ones.2 Because of this, primary cells have been widely used in the area of biological and biotechnological science for a long time. However, it is hard to get the results with replicative experiments, because primary cells are capable of a limited number of cell divisions in in vitro culture and then enter a state where they can no longer grow.3 If the primary cells arrive at replicative senescence about halfway through the experiments, another trial to isolate the same cells should be carried out, and there can sometimes be significant unintentional variation between preparations.
To overcome this replicative senescence in primary cells, several genes (e.g., simian virus 40 large T antigen (SV40T), cMyc or shp53, Bmi-1 and human telomerase reverse transcriptase (hTert)) have been introduced into the cells, resulting in their immortalization. 4 Many types of cell lines have been developed in human or mice species in previous studies, and there have not been many reports about the generation of immortalized cell lines in livestock, such as goat,5 sheep,6 and pig.7 In terms of the molecular understanding of specific or zoonotic diseases, a need for the establishment of in vitro cell lines in livestock has been raised. Particularly in bovine species, only a few studies about the induction and characterization of immortalization have been reported.8-10 Therefore, in this study, immortalized bovine cell lines were established with the aim of releasing the basic or translating understanding in molecular pathways after gene regulation treatments such as overexpression, knock-down, or KO/KI. These cells are believed to be more convenient than primary cells.
Among the gene-editing approaches, KO is one of the most useful methods for revealing the related molecular mechanisms or generating animal models. However, conventional homologous recombination (HR) is a very difficult and time-consuming process, and it produces KO cells or animals with low efficiency. Recently, genome-editing technologies such as Zinc Finger Nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs), and Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 have emerged as powerful methods to understand gene function via KO cell lines or animals with high efficiency and easy DNA preparation.11-16
Accordingly, the purpose of this study is to establish and characterize the bovine immortalized fibroblasts for disrupting specific genes using TALENs. To verify the KO efficiency of TALENs in immortalized cells, the PRNP gene, which encodes the prion protein of which conformational changes are potentially related to the incidence of BSE1, was selected among several candidates in cattle. Next, the PRNP KO immortalized cells were cloned via somatic cell nuclear transfer (SCNT) to study the feasibility of embryonic development.
Materials and Methods
Ethics Statement
In this study, we did not use live animals. Ovaries needed for embryo culture were obtained from a local slaughterhouse. The embryos were not transferred to live animals. A biopsy punch was used to obtain a small piece of ear skin tissue. The primary cells from the tissue, which is published in our study17 were used for this study.
Primary Culture
Ear skin tissues taken from a male neonate calf using biopsy punch were washed several times in PBS supplemented with antibiotics, chopped into tiny pieces using a surgical blade, and incubated in collagenase type I overnight at 38°C, in 5% CO2 humidified air. The tissues were placed in culture dish containing cell culture medium (Dulbecco Modified Eagle Medium (DMEM) (Gibco, Carlsbad, California, USA) supplemented with 15% FBS (Gibco), 1% Penicillin/streptomycin (P/S) (Gibco), 1% Non-essential amino acids (NEAA) (Gibco), and 100 mM β-Mercaptoethanol (2-ME) (Sigma-aldrich). Expanded primary cells were maintained in a cell culture medium at 38°C, in 5% CO2 humidified air, and then frozen to −196°C for further use.
Transfection for Immortalization
Based on previous reports,4,18,19 2 immortalization genes, Bmi-1 and hTert, were selected. Bmi-1 (from addgene, # 12240) was linked to an RFP marker with T2A sequences by PCR amplification, and inserted into to the piggybac transposon expression vector (PB-CA, addgene plasmid # 20960). hTert (from addgene, # 12245) was cloned into the plasmid DNA (CMV-DsReds containing neomycin resistance gene, Clontech catalog No. 632420) (see Fig. S1). After the transfection of PB-CA-Bmi-1-T2A-RFP and transposase (named pCy43 provided by Sanger Institute, Hinxton, UK) using nucleofection (Neon, Invitrogen), the RFP positive cells were mechanically isolated and sub-cultured. CMV-hTert DNA strands were also subsequently transfected into those cell lines, and then the hTert-introduced cells were selected under the treatment of 800 mg/ml of neomycin for 10 days.
Cell Characterization
Doubling time
At every 2–5 passages, control cells and the 2 kinds of immortalized cells (Bmi-1 and Bmi-1+hTert) were plated at a density of 4 × 104 cells in 12-well plates. Then, each of the 3 wells was trypsinized, and the total cell numbers were counted manually using a hemocytometer. The population doubling time was calculated using an online doubling time calculator (http://www.doubling-time.com/compute.php).20
Single cell colony formation and colony staining
At both the early and late passages of each cell line, hundred cells after trypsinization were seeded into 100 mm cell culture dishes (BD Falcon) with the cell culture medium. To evaluate the colony-forming competence of single cells, colony formation assay was performed, as described below (clonogenic assay of cells in vitro). After two weeks of seeding, each culture dish was rinsed with PBS, and 3 ml of a mixture of 6% glutaraldehyde and 0.5% crystal violet was added. The dishes were incubated at room temperature for 1 hour, the staining solution was carefully removed, and the colonies were rinsed with tap water. Among the stained colonies, only the colonies with a cell number over 10,000 were counted for viable status (Fig. S2). To expand a single colony, the colony was mechanically isolated and sequentially sub-cultured into 48-, 12- and 6-well plates. Then, half of the cells from the 6-well plates were used for nuclear transfer, and the other cells were subjected to mutation analysis.
Karyotyping
To perform karyotyping, cultured cells were treated as follows. First, 0.2 μg/ml of colcemid (Gibco) solution in a culture medium was added to the culture plate. Then, the cells were incubated at 37°C for 4 h. After incubation, cells were collected in 15 ml tubes and centrifuged at 1000 rpm for 10 min. After the medium had been carefully aspirated, 5 ml of hypotonic solution (0.075M KCl) was added and allowed to stand at 37°C for 10 min. Then, 500 μl of Carnoy's fixative solution (Methanol:Acetic acid 3:1) was added and mixed by inverting the tube, followed by centrifugation at 1000 rpm for 10 min. The hypotonic solution was aspirated carefully and 3 ml of Carnoy's fixative was added and well mixed. After more than 20 min, the mixture was centrifuged at 1000 rpm for 10 min. The supernatant fixative solution was carefully aspirated to achieve approximately 2:1 volume to pellets ratio. The pellets were spread on a prepared glass slide, which was then baked at 60°C for 30 min. The slide was treated with 50% H2O2 for 3 min, and then baked again at 60°C for 30 min. Finally, the slide was stained with the Giemsa stain-GTG banding method. Chromosome imaging was accomplished with the ChIPS-Karyo (Chromosome Image Processing System) (GenDix, Inc., Seoul, Korea).
Gene expression
Total RNAs were extracted to analyze the gene expression in the several types of tissues, cells, and embryos by the easy-spin Total RNA Extraction Kit (iNtRON). Total RNAs were added to Maxime RT Premix kit (oligo (dT) primer; iNtRON) for the synthesis of complementary DNAs (cDNAs), and then the synthetized cDNAs were used as RT-PCR templates. Primer information is described in Table S1. All of the PCR reactions were performed under the same condition. (94°C for 2 min, 35 cycles of 94°C for 20 sec/60°C for 10 sec/72°C for 30 sec, and 72°C for 5 min). The gene expression level of p53 was also measured with real-time PCR (StepOnePlus Real-Time PCR System, Applied Biosystems) and PCR amplicons of the PRNP and PRND mRNA transcripts were sequenced to confirm the amplified products (Fig. S6).
Telomerase activity test
The quantification and characterization of telomerase activity were evaluated by the telomeric repeat amplification protocol (TRAP). For this test, a TeloTAGGG Telomerase PCR ELISAPLUS kit (Roche, Basel, Switzerland) was used, following the manufacturer's indications. Relative telomerase activities (RTA) within different samples in an experiment were obtained using the following formula: RTA=[(AS-AS0)/AS,IS]/[(ATS8-ATS8,0)/ATS3,IS]×100 (AS: Absorbance of the sample. AS,0: Absorbance of the heat-treated sample. AS, IS: Absorbance of the internal standard (IS) of the sample. ATS8: Absorbance of the control template. ATS8,0: Absorbance of the lysis buffer. ATS8,IS: Absorbance of the IS of the control template).
Disruption of PRNP Gene and Cell Line
TALENs and CRISPR/Cas9 nucleases for cow PRNP gene exon 3 (Genbank #AJ298878) were designed and synthesized as described.21,22 To validate the activities of synthesized endonucleases, an equal amount of expression vectors for the left and right TALENs (for TALEN) or sgRNA were transferred into immortalized bovine cell lines via Neon (Life technologies) or Nucleofector 4D (Lonza), and T7E1 mismatch-specific nuclease assay was performed as described previously.23
To increase the proportion of PRNP KO cells in whole transfected pools, PRNP TALENs were transferred together with reporter vectors that consisted of a MACS surrogate reporter and the synthetic target sites of PRNP TALEN into immortalized bovine cells and subjected to a KO cell enrichment process24-26 using the MACS system (Miltenyl biotec). The sorted cell population with H2KK cell surface protein whose expression was induced by the co-expression of PRNP TALENs was subjected to limiting dilution in a 96-well plate and a monoclonal cell population was selected and propagated. Genomic DNA from each isolated clone was analyzed by a T7E1 assay and dideoxy sequencing on the PRNP target locus to identify and characterize the PRNP KO alleles.
SCNT and Embryo Culture
The transfer of a donor cell (primary, immortalized, or PRNP KO-immortalized cells) into an enucleated oocyte was carried out as previously described.27 Reconstructed embryos were electrically fused, activated for 4 min with ionomycin followed by culturing for 4 h in 6-DMAP. Cloned embryos were cultured in 25 µL microdrops of a chemically defined medium overlaid with mineral oil for 7–8 days at 38°C in an atmosphere of 5% O2, 5% CO2, and 95% N2. The chemically defined medium was prepared as described in previous reports.28 Cleaved and blastocysts stage embryos were observed at 24 and 192 h of culture, respectively.
Results
Characteristics of Immortalized Fibroblasts
After serial transfection with Bmi-1 and hTert, the cell morphology and doubling time were observed (Fig. 1A and B). Whereas control cells were senescent around 80 days after culture initiation, Bmi-1 and Bmi-1+hTert-transfected cells were populated over 120 days without reduction of proliferative competence. As Bmi-1-immortalized cells were culturing after long-term passage, their doubling time was increased (from about 20 hr to 40 hr) and they gradually became senescent about 130 days after in vitro culturing. However, the doubling time of Bmi-1+hTert-immortalized cells was not changed (around 20 hrs), in spite of the high number of culture passages (greater than 180 days, Fig. 1B).
FIGURE 1.
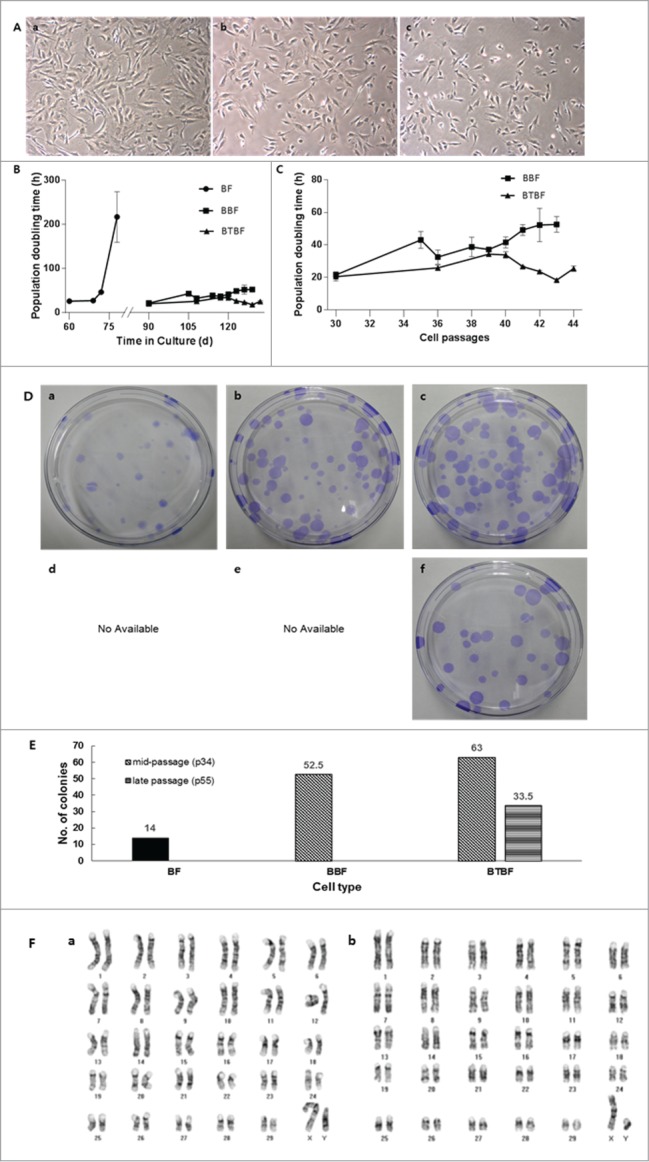
(See previous page).Analysis of immortalized bovine fibroblasts. (A) Bright-field pictures of in vitro cultured bovine fibroblasts. BFs at passage 7 were used as a control (a), and BBFs (b) and BTBFs (c) represented non-morphological changes at passage 42 compared with the early passage of BFs. (B) The population doubling time of immortalized bovine fibroblasts. Senescent cells did not show any cellular growth after 80 days of primary culture, but immortalized cells were populated continuously after the BFs' senescence. (C) The doubling time was only increased in BBFs as their passage increased, and they arrived at cellular senescence after passage 43 (b). (D) Clonogenic assay of each cell line; single colonies of BFs, BBFs, and BTBFs were stained using crystal blue, and the number of colonies in each group were counted (E). (F) Karyogram of BTBFs at passage 52. BTBFs have normal karyotype (b) like BFs (a). (BFs: Primary cultured bovine fibroblasts, BBFs: Bmi-1-introduced bovine fibroblasts, BTBFs, Bmi-1+hTert-introduced bovine fibroblasts).
Single Cell Forming Assays
To evaluate the proliferative activity of immortalized cells, the number of single cell–derived colonies was counted (Fig. 1D). Additionally, the representative growing appearance of a single cell–derived colony was captured by a time-lapse image recorder (Juli Br., H/S system, Seoul Korea; Supplementary Movie 1). In control cells (at 7 passages), 14 colonies had grown up on average. Bmi-1 and Bmi-1+hTert-immortalized cells at 34 passages formed 52 and 63, respectively. Additionally, Bmi-1+hTert-immortalized cells at 52 passages formed 33 colonies (Fig. 1D and E).
Karyotyping
Chromosome preparation in the early and late passage of Bmi-1+hTert-immortalized cells showed a normal number of karyoplasts (2n = 60, XY), and there were no abnormalities compared with the control group (Fig. 1F).
Embryonic Development
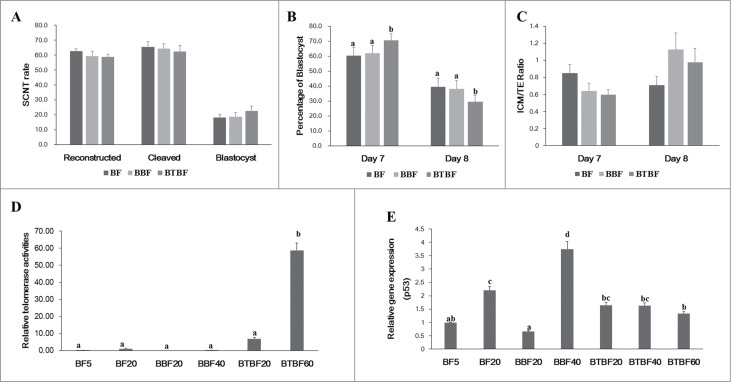
Cloned embryonic development from control, Bmi-1-, and Bmi-1+hTert-immortalized cells were evaluated by cleavage (65.5 ± 3.3, 64.4 ± 3.2, and 62.4 ± 4.0, respectively) and blastocyst formation (18.1 ± 2.1, 18.9 ± 2.5, and 22.7 ± 3.2, respectively) (Fig. 2A). There were no significant differences in cleavage, total blastocyst formation (Day 7 + Day 8) (Fig. 2A and B). However, at a specific time (Day 7) upon observing blastocyst formation, a significantly higher number had developed in the Bmi-1+hTert-immortalized group (70.6 ± 4.3 vs 60.4 ± 5.5 and 62.0 ± 5.2) (Fig. 2B). In cell number ratio (Inner cell mass/Trophectoderm), there was no significant difference between experimental groups (Day 7: 0.85 ± 0.1, 0.64 ± 0.09, 0.6 ± 0.06, Day 8: 0.71 ± 0.1, 1.13 ± 0.19, 0.98 ± 0.16) (Fig. 2C).
FIGURE 2.
Embryonic development, telomerase activity, and gene expression. (A) Development rate of cloned embryos of BFs, BBFs, and BTBFs. Reconstructed embryos' fusion, cleavage, and blastocyst formation rates were estimated one hour after electric fusion, on day 2, and on day 7, respectively. (B) Among the blastocysts in each group, bastocyst formation ratio was differently observed on Day 7 and Day 8. (C) The cell number of the blastocysts in each group was counted after differential staining, and the ICM/TE ratio was calculated. (D) Telomerase activities were measured in control cell lines (passage 5 (BF5), passage 20 (BF20), BBF lines (passage 20 (BBF20), passage 40 (BBF40)), BBBF lines (passage 20 (BTBF20), passage 40 (BTBF 60)). In the BTBF60, telomerase activity was detected in the highest level. (E) p53 gene expression by quantitative RT-PCR was measured in various cell passages of BFs, BBFs, and BTBFs. (BFs: Primary cultured bovine fibroblasts, BBFs: Bmi-1-introduced bovine fibroblasts, BTBFs, Bmi-1+hTert-introduced bovine fibroblasts).
Telomerase Activity
The RTAs of control cells, Bmi-1- and Bmi-1+hTert-immortalized cells (early/late passages) were 0.14 ± 0.09/ 1.05 ± 0.27, 0.00 ± 0.02/ 0.24 ± 0.03, and 6.94 ± 0.71/ 58.87 ± 4.18, respectively. Telomerase activity was significantly increased in Bmi-1+hTert-immortalized cells compared to both the control cells and Bmi-1-immortalized cells (Fig. 2D).
p53 Gene Expression
The expression of one of the tumor suppressor genes, p53, was analyzed. In the late passage of control cells and Bmi-1-derived cells, the expression levels were increased, while the expression was not changed in the early and late passages in the Bmi-1+hTert-immortalized cells (Fig. 2E).
PRNP KO in Immortalized Bovine Fibroblasts Using TALENs and CRISPR/Cas9
To compare primary and immortalized bovine fibroblasts in the genome engineering process, we designed a TALEN pair targeting the bovine PRNP gene (Fig. 3A) and isolated PRNP KO monoclonal cell clones from each fibroblast cell population. While PRNP TALEN induced mutations in both primary and immortalized fibroblasts, the frequency of mutant PRNP alleles induced by TALEN expression was higher in immortalized fibroblasts (66%), likely due to the more efficient DNA delivery (Fig. 3C). Additionally, while monoclonal cell colonies with PRNP gene mutations were successfully isolated from both cell lines after the transient expression of PRNP TALENs, only mutant cell populations from immortalized fibroblasts were successfully expanded to form established independent cell lines. Importantly, 19 of the 66 PRNP mutant clonal cell populations from the immortalized fibroblasts had mutations on both PRNP alleles, showing the efficient engineering of the PRNP gene using TALEN. Mutations on the PRNP gene were mostly small insertion and deletion on the TALEN spacer, but a large deletion of >100 bp was also observed (Fig. 3D). Six clones had frameshift mutations in all PRNP alleles representing PRNP KO cell lines.
FIGURE 3.
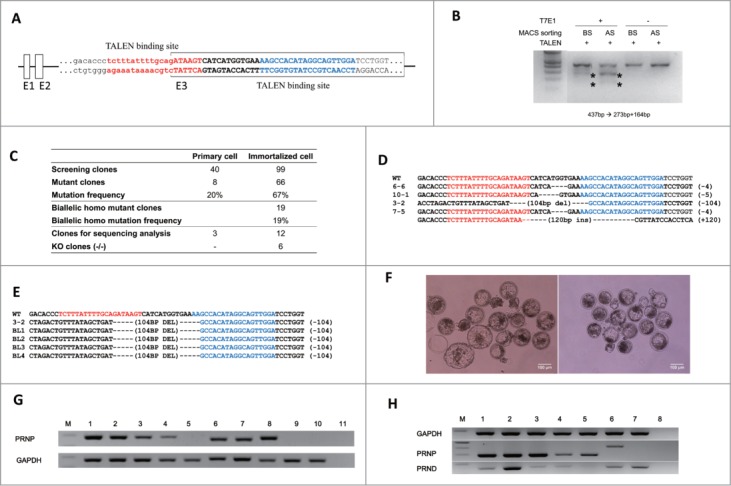
Knockout of the PRNP gene in immortalized bovine fibroblast cell lines. (A) TALEN target site on exon 3 of bovine PRNP gene. Recognition sequences for left TALEN (red) and right TALEN (blue) are flanking a 12-bp spacer (bold). (B) Enrichment of PRNP-modified cells via a surrogate reporter-mediated MACS assessed by T7E1 assay. The cleavage products by T7E1 due to the TALEN-induced mutation on the PRNP gene are depicted with asterisks. (C) Summary of the PRNP knockout results in primary and immortalized bovine fibroblasts. (D) The genotype of the PRNP mutation in 4 representative knockout clones. Note that identical mutations were introduced to both alleles in selected clones, except for 7–5. (E) Mutant allele analysis of cloned blastocysts derived from a knockout immortalized cell line. (F) Representative pictures of cloned blastocysts from immortalized cells (left) and knockout cells (right). (G) Expression of PRNP mRNA in knockout blastocysts (M: DNA ladder, 1: Spinal cord, 2: Cumulus cells, 3: Primary cells, 4: Immortal cells, 5: KO immortal cells, 6: in vitro fertilized blastocysts, 7 and 8: Blastocysts from immortalized cells, 9 and 10: Blastocysts from knockout immortalized cells, 11: Negative controls). (H) PRNP and PRND expression by RT-PCR in PRNP KO cell lines. (M: DNA ladder, 1: Spinal cord, 2: Testis, 3: Primary cells, 4: Immortal cells, 5: KO cells 6–6, 6: KO cells 7–5, 7: KO cells 3–2, 8: Negative controls).
While TALEN successfully supported gene KO in immortalized bovine fibroblasts, we also tested if CRISPR/Cas9-based engineered nucleases, which recently emerged as a robust genome engineering technology, can also be used to establish KO cell lines. Indeed, when CRISPR/Cas9 nuclease for bovine PRNP was expressed in immortalized bovine cell lines, cells with mutations on a PRNP gene were induced efficiently. Additionally, a clonal PRNP mutant cell population was readily established with high efficiency (Figs. S9 and S10).
PRNP Expression in Cloned Blastocysts
Four PRNP KO cell lines were subjected to SCNT tests to determine whether these cell lines could support the establishment of gene KO-cloned blastocysts (Fig. 3F). The average cleavage and blastocyst formation rates were 81.3% and 20.6% (Table S2). To prove PRNP deletion functionally, based on the previous report in which blastocysts expressed PRNP,32 we analyzed PRNP expression in blastocysts from in vitro fertilized, immortalized cells including KO-cells or spinal cords as positive controls for expression (Fig. 3G). While blastocysts from in vitro fertilized or control donor cells expressed as expected, PRNP mRNA expression in blastocysts from the KO-cell line was not detected (Fig. 3G). Furthermore, they showed the same mutation pattern as donor cells (Fig 3E).
PRND Expression
Before PRND expression analysis in PRNP-KO cell lines, PRNP and PRND expression screening was conducted in various bovine tissues (spinal cord, heart, liver, lung, spleen, kidney, testis, and ovary) to obtain control data (Fig. S5). For each gene expression, PRNP and PRND, the strongest expressing tissues, spinal cord and testis, were used as controls, respectively. Based on the results of PRND expression in the PRNP KO cell lines, 3 kinds of established KO cell lines (small and large deletion, and insertion) were analyzed. The PRNP and PRND mRNA transcripts exhibited different expression patterns (Fig. 3H). In the 6-6 KO cell line (small deletion), PRNP expression was detected, although 4bp deletions were confirmed in this cell clone, and PRND was not expressed. In the case of cell line 7-5, RT-PCR amplicons were shifted approximately 100bp up (120 inserts sequenced), and PRND was also detected in this KO cell line. Only the KO cell line 3-2 did not express the PRNP mRNA transcripts, whereas PRND was expressed.
Discussion
Numerous cell lines in human or mice along with developing gene-regulation technologies have been provided to find out the molecular and biological functions of genes. However, a few studies in livestock, particularly cattle, were not only reported on generating cell lines, but also gene KO in culture cells.8-10,29 Here, we demonstrated the feasibility of a TALEN-based approach for efficient gene KO in bovine immortalized ear skin fibroblast cells, and then those cells in enucleated oocytes were reprogrammed to develop up to pre-implantation-stage embryos. The targeted PRNP gene mutation in the mutation frequency was sufficient to find bi-allelic KO after a single TALEN treatment and without any selection process.
After the release of the bovine genome, a better understanding of this species has emerged. In livestock, genomic understanding has long been considered important, along with concerns about genetic diseases and zoonosis pathways for diseases such as BSE (prion diseases). BSE is induced by the conformational transition of the cellular prion protein (PRNP gene) and its accumulation in the brain. One of the easiest ways to elucidate the genomic events in in vitro studies is to use cattle cell lines or embryos. Through this study, we found that ear skin fibroblasts from a male neonate could be changed to immortalized status using 2 gene transfections (Bmi-1 and hTert). Even though single gene of Bmi-1 or hTert previously induced immortalization in cells, we used a combination of these genes for our cell immortalization studies.4,18,19 As a result, while Bmi-1-derived cells have grown up into 40 passages, Bmi-1+hTert-derived cells proliferated into more than 60 passages. Moreover, those immortalized cells can not only colonize into a single population cell line without any replicative senescence or chromosomal changes (see Fig. 1 and Supplementary Movie 1), but also be reprogrammed into cloned embryonic development with no detrimental effects. It indicated that a single mutated cell line could be isolated without any selective gene transfection such as antibiotic resistance by using an immortalized cell line.
As immortalized cells were employed in SCNT, neither their embryonic development nor cell numbers were changed (Fig. 2). In addition, even though Bmi-1+hTert-derived immortalized cells with long-term culture were used for SCNT, their developmental competence was not decreased. This phenomenon can be explained in our analysis as follows. First is telomerase activity. In a previous study, telomerase activity was found to be related to the reprogramming of donor cells for SCNT.30 Thus, we suggest that the increase of telomerase activity in Bmi-1 + hTert-derived immortalized cells supports the maintenance of developmental competence in SCNT embryos. Second is that p53 expression in Bmi-1+hTert-derived immortalized cells was not changed, while its expression was increased in the other cells (control and Bmi-1 derived cells). P53 expression was related to SCNT embryonic development in a previous study.31 One interesting finding in this study is blastocyst formation speed. When we observed the blastocyst formation on Day 7, a higher number of blastocysts was found in Bmi-1+hTert-derived immortalized cells. While previous studies did not report the embryonic development speed, it was investigated in our study. However, a more advanced understanding of the molecular approach may be required to examine this issue in detail.
The PRNP gene is well known as a potential reason for prion diseases such as BSE in cattle and Creutzfeldt–Jakob disease (CJD) in humans. Even though BSE is a severe zoonotic disease, our understanding of the role or function of PRNP in bovine cells or embryos has been limited to date. Most studies on prion issues have examined in mouse cell lines or KO mice. However, because mice are not a host model for prion diseases, its application and understanding is limited in practice. Thus, one good example of PRNP function in cattle (host model) is that prion-lacking models were generated using serial homologous recombination transfection and SCNT, as in a previous report.29 In that study,29 it is time consuming and complicated to produce bi-allelic knockout cattle. However, recent genome editing technologies such as ZFNs, TALENs, and CRISPR/Cas9 can disrupt the gene efficiently with time- and cost-saving works. Here, PRNP-TALEN mutated the target region in the control and immortalized cells. However, while control cells (primary cells with limited proliferative potency) showed low frequency mutations in the target region, immortalized cells with high proliferative and single cell population potency showed high efficiency after the enrichment sorting system.24 Finally, a 66% bi-allelic mutation frequency was detected in immortalized cells (Fig. 3C). Even more, isolated single cell populations can be grown and propagated into a large enough number of cells to use somatic cell nuclear transfer. Another genome-editing tool, CRISPR/Cas9, was very effective for PRNP gene deletion after double transfection with similar mutation efficiency (Fig. S10) in immortalized cells. Despite double nucleofections, proliferative potency was not affected in immortalized cells. Collectively, these data suggest that an immortalized bovine cell line can be a highly useful cell resource in which efficient targeted gene editing with engineered nucleases can be achieved.
Four representative KO cell lines among those from the TALEN approach showed comparable embryonic development competence compared with the ones from the non-mutated immortalized cells in terms of blastocyst formation. This suggests that PRNP-KO had not functionally affected the embryonic development at the in vitro level.
Because the study of the transcript abundances of bovine PRNP interacting genes is necessary for finding out the unknown function of prion proteins, its knockdown using RNA interference has been studied,33,34 without analyzing interacting genes. However, in previous studies on other species, using PRNP knockdown in murine neuroblastoma cell lines 35 or microarrays from human PRNP cell lines,36,37 the expression of several interacting genes (cell proliferative and PRNP-related) was analyzed and identified. In this study, to prove that these were related to gene expression in the bovine species, the affected genes were chosen and analyzed using bovine PRNP-KO cells. After primer design and PCR amplification of the same genes in the bovine genome, 3 genes (CSNK2A1, FAM64A, and MPG) were investigated in terms of their expression on primary, immortalized, and KO-immortalized cells. Previously, these genes were up regulated in murine PRNP knockdown cells, and their expression after KO in our study was inconsistently up-regulated (Fig. S8). Rather, their expression was down regulated via an immortalization (Bmi-1and hTert) process. Additionally, FAM64A and MPG expression after PRNP-KO was different from the mutation pattern.
For further analysis in the PRNP-KO cell lines, we investigated the PRND expression in KO cells. In a previous mice study, PRND was expressed after disrupting the PRNP gene. To prove the results in bovine cells, we first analyzed the PRND expression level in several tissues by RT-PCR and indicating that in testis tissue, its expression was dominantly observed in the same line with a previous report.38 The sequence of amplified RT-PCR was the same as with the database information (Fig. S5). Interestingly, while PRND in the primary and control immortalized cells was not detectable, its expression was upregulated in PRNP KO cells lines. However, PRND expression was dependent on the PRNP mutation pattern (large deletion and insertion in our study). This result is the same as that found in PRNP KO mice studies.39 In PRNP KO mice studies, PRND expression affected the animals to show ataxia and Purkinje cell loss.39 Because PRND up-regulation after PRNP disruption had not been investigated in the previous PRNP KO cattle,29 for the first time, we reported the expression of PRND in PRNP KO cell lines. If we were to perform SCNT using PRNP KO cells with PRND expression for generating KO cattle using TALENs, it might cause ataxia or Purkinje cell loss in the KO cattle phenotype, as in PRNP KO mice. As a future study, before generating PRNP KO cattle using TALEN, the relation of PRNP and PRND expression could be examined to elucidate the function of PRNP in cattle.
Bovine skin fibroblasts were immortalized using Bmi-1 and hTert with the competence of single cell–derived colony formation, and the immortalized cells were found to have comparable embryonic development via SCNT. Furthermore, we demonstrated that the PRNP gene in this cell line could be efficiently disrupted by genome editing tools such as TALEN and CRISPR/Cas9. These results represent the first bovine PRNP KO cell line model and opened a powerful opportunity to provide the in vitro molecular pathways of the PRNP functions of the bovine cell model or prion diseases (BSE or CJD).
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
FUNDING
This study was financially supported by the BK21 PLUS Program for Creative Veterinary Science Research, IPET (# 109023-05-5-CG000, 111078-03-3-CG000), Biogreen (PJ0090962014), the National Research Foundation of Korea (J.-S.K., 2012-0001225), and IBS (IBS-R021-D1).
SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed on the publisher's website.
REFERENCES
- 1.Prusiner SB. Prion diseases and the BSE crisis. Science 1997; 278:245-51; PMID:9323196; http://dx.doi.org/ 10.1126/science.278.5336.245 [DOI] [PubMed] [Google Scholar]
- 2.Büssow K. Stable mammalian producer cell lines for structual biology. Curr Opin Struct Biol 2015; 32:81-90; PMID:25804355; http://dx.doi.org/ 10.1016/j.sbi.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 3.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8:729-40; PMID:17667954; http://dx.doi.org/ 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Escudero V, Garcia-Gomez A, Gargini R, Martin-Bermejo MJ, Langa E, de Yebenes JG, Delicado A, Avila J, Moreno-Flores MT, Lim F. Prevention of senescence progression in reversibly immortalized human ensheathing glia permits their survival after deimmortalization. Mol Ther: The J Am Soc Gene Ther 2010; 18:394-403; PMID:19935779; http://dx.doi.org/ 10.1038/mt.2009.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He YL, Wu YH, He XN, Liu FJ, He XY, Zhang Y. An immortalized goat mammary epithelial cell line induced with human telomerase reverse transcriptase (hTERT) gene transfer. Theriogenology 2009; 71:1417-24; PMID:19303628; http://dx.doi.org/ 10.1016/j.theriogenology.2009.01.012 [DOI] [PubMed] [Google Scholar]
- 6.Cui W, Wylie D, Aslam S, Dinnyes A, King T, Wilmut I, Clark AJ. Telomerase-immortalized sheep fibroblasts can be reprogrammed by nuclear transfer to undergo early development. Biol Reproduct 2003; 69:15-21; PMID:12606403; http://dx.doi.org/ 10.1095/biolreprod.102.013250 [DOI] [PubMed] [Google Scholar]
- 7.Oh HY, Jin X, Kim JG, Oh MJ, Pian X, Kim JM, Yoon MS, Son CI, Lee YS, Hong KC. Characteristics of primary and immortalized fibroblast cells derived from the miniature and domestic pigs. BMC Cell Biol 2007; 8:20; PMID:17543094; http://dx.doi.org/ 10.1186/1471-2121-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donai K, Kiyono T, Eitsuka T, Guo Y, Kuroda K, Sone H, Isogai E, Fukuda T. Bovine and porcine fibroblasts can be immortalized with intact karyotype by the expression of mutant cyclin dependent kinase 4, cyclin D, and telomerase. J Biotechnol 2014; 176C:50-7; PMID:24589663; http://dx.doi.org/ 10.1016/j.jbiotec.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 9.Veitonmaki N, Fuxe J, Hultdin M, Roos G, Pettersson RF, Cao Y. Immortalization of bovine capillary endothelial cells by hTERT alone involves inactivation of endogenous p16INK4A/pRb. FASEB 2003; 17:764-6; PMID:12586745 [DOI] [PubMed] [Google Scholar]
- 10.Saito M, Handa K, Kiyono T, Hattori S, Yokoi T, Tsubakimoto T, Harada H, Noguchi T, Toyoda M, Sato S, et al.. Immortalization of cementoblast progenitor cells with Bmi-1 and TERT. J Bone Min Res 2005; 20:50-7; PMID:15619669; http://dx.doi.org/ 10.1359/JBMR.041006 [DOI] [PubMed] [Google Scholar]
- 11.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 2012; 109:17382-7; PMID:23027955; http://dx.doi.org/ 10.1073/pnas.1211446109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, et al.. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Gen Res 2014; 24:125–31; PMID:24253447; http://dx.doi.org/21502977 10.1101/gr.163394.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, Zhao Y, Fan N, Song J, Tian J, et al.. Generation of PPARgamma mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res 2011; 21:979-82; PMID:21502977; http://dx.doi.org/ 10.1038/cr.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S, Luo J, Song Z, Ding F, Dai Y, Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res 2011; 21:1638-40; PMID:21912434; http://dx.doi.org/ 10.1038/cr.2011.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet 2014; 15:321-34; PMID:24690881; http://dx.doi.org/ 10.1038/nrg3686 [DOI] [PubMed] [Google Scholar]
- 16.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31:230-2; PMID:23360966; http://dx.doi.org/ 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 17.Jang G, Jeon HY, Ko KH, Park HJ, Kang SK, Lee BC, Hwang WS. Developmental competence and gene expression in preimplantation bovine embryos derived from somatic cell nuclear transfer using different donor cells. Zygote 2005; 13:187-95; PMID:16261763; http://dx.doi.org/ 10.1017/S0967199405003217 [DOI] [PubMed] [Google Scholar]
- 18.Tatrai P, Szepesi A, Matula Z, Szigeti A, Buchan G, Madi A, Uher F, Német K. Combined introduction of Bmi-1 and hTERT immortalizes human adipose tissue-derived stromal cells with low risk of transformation. Biochem Biophys Res Commun 2012; 422:28-35; PMID:22554522; http://dx.doi.org/ 10.1016/j.bbrc.2012.04.088 [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Soda Y, Takahashi K, Bai Y, Mitsuru A, Igura K, Satoh H, Yamaguchi S, Tani K, Tojo A, et al.. Successful immortalization of mesenchymal progenitor cells derived from human placenta and the differentiation abilities of immortalized cells. Biochem Biophys Res Commun 2006; 351:853-9; PMID:17094946; http://dx.doi.org/ 10.1016/j.bbrc.2006.10.125 [DOI] [PubMed] [Google Scholar]
- 20.Widera D, Zander C, Heidbreder M, Kasperek Y, Noll T, Seitz O, Saldamli B, Sudhoff H, Sader R, Kaltschmidt C, et al.. Adult palatum as a novel source of neural crest-related stem cells. Stem cells 2009; 27:1899-910; PMID:19544446; http://dx.doi.org/ 10.1002/stem.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al.. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 2013; 31:251-8; PMID:23417094; http://dx.doi.org/ 10.1038/nbt.2517 [DOI] [PubMed] [Google Scholar]
- 22.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Gen Res 2014; 24:132-41; PMID:24253446; http://dx.doi.org/ 10.1101/gr.162339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Gen Res 2009; 19:1279-88; PMID:19470664; http://dx.doi.org/ 10.1101/gr.089417.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Um E, Cho SR, Jung C, Kim H, Kim JS. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 2011; 8:941-3; PMID:21983922; http://dx.doi.org/ 10.1038/nmeth.1733 [DOI] [PubMed] [Google Scholar]
- 25.Kim YH, Ramakrishna S, Kim H, Kim JS. Enrichment of cells with TALEN-induced mutations using surrogate reporters. Methods 2014; 69:108–17; PMID:24780521; http://dx.doi.org/10.1016/j.ymeth.2014.04.01223441197 [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Kim MS, Wee G, Lee CI, Kim H, Kim JS. Magnetic separation and antibiotics selection enable enrichment of cells with ZFN/TALEN-induced mutations. PloS One 2013; 8:e56476; PMID:23441197; http://dx.doi.org/ 10.1371/journal.pone.0056476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saadeldin IM, Choi W, Roibas Da Torre B, Kim B, Lee B, Jang G. Embryonic development and implantation related gene expression of oocyte reconstructed with bovine trophoblast cells. J Reproduct Dev 2012; 58:425-31; PMID:22522228; http://dx.doi.org/ 10.1262/jrd.11-112H [DOI] [PubMed] [Google Scholar]
- 28.Saadeldin IM, Kim B, Lee B, Jang G. Effect of different culture media on the temporal gene expression in the bovine developing embryos. Theriogenology 2011; 75:995-1004; PMID:21220156; http://dx.doi.org/ 10.1016/j.theriogenology.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 29.Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, Sathiyaseelan J, Wu H, Matsushita H, Koster J, et al.. Production of cattle lacking prion protein. Nat Biotechnol 2007; 25:132-8; PMID:17195841; http://dx.doi.org/ 10.1038/nbt1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betts D, Bordignon V, Hill J, Winger Q, Westhusin M, Smith L, King W. Reprogramming of telomerase activity and rebuilding of telomerase length in cloned cattle. Proc Natl Acad Sci USA 2001;98: 1077-82; PMID:11158597; http://dx.doi.org/ 10.1073/pnas.98.3.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma PJ, Zhang H, LI R, Wang YS, Zhang Y, Hua S. P53-Mediated Repression of the Reprogramming in Cloned Bovine Embryos Through Direct Interaction with HDAC1 and Indirect Interaction with DNMT3A. Reprod Domesti Anim. 2015;50:400-9; PMID:25753134; http://dx.doi.org/ 10.1111/rda.12502 [DOI] [PubMed] [Google Scholar]
- 32.Peralta OA, Huckle WR, Eyestone WH. Developmental expression of the cellular prion protein (PrP(C) ) in bovine embryos. Mol Reproduct Dev 2012; 79:488-98; PMID:22674901; http://dx.doi.org/ 10.1002/mrd.22057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutou S, Kunishi M, Kudo T, Wongsrikeao P, Miyagishi M, Otoi T. Knockdown of the bovine prion gene PRNP by RNA interference (RNAi) technology. BMC Biotechnol 2007; 7:44; PMID:17655742; http://dx.doi.org/ 10.1186/1472-6750-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Lv X, Zhang K, Lin T, Liu X, Yuan J, Dai Y, Li N. Knockdown of the prion gene expression by RNA interference in bovine fibroblast cells. Mol Biol Rep 2010; 37:3193-8; PMID:19821149; http://dx.doi.org/ 10.1007/s11033-009-9900-0 [DOI] [PubMed] [Google Scholar]
- 35.Kang SG, Roh YM, Lau A, Westaway D, McKenzie D, Aiken J, Kim YS, Yoo HS. Establishment and characterization of Prnp knockdown neuroblastoma cells using dual microRNA-mediated RNA interference. Prion 2011; 5:93-102; PMID:21494092; http://dx.doi.org/ 10.4161/pri.5.2.15621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satoh J, Yamamura T. Gene expression profile following stable expression of the cellular prion protein. Cell Mol Neurobiol 2004; 24:793-814; PMID:15672681; http://dx.doi.org/ 10.1007/s10571-004-6920-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satoh J, Obayashi S, Misawa T, Sumiyoshi K, Oosumi K, Tabunoki H. Protein microarray analysis identifies human cellular prion protein interactors. Neuropathol Applied Neurobiol 2009; 35:16-35; PMID:18482256; http://dx.doi.org/ 10.1111/j.1365-2990.2008.00947.x [DOI] [PubMed] [Google Scholar]
- 38.Moore RC, Lee IY, Silverman GL, Harrison PM, Strome R, Heinrich C, Karunaratne A, Pasternak SH, Chishti MA, Liang Y, et al.. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J Mol Biol 1999; 292:797-817; PMID:10525406; http://dx.doi.org/ 10.1006/jmbi.1999.3108 [DOI] [PubMed] [Google Scholar]
- 39.Weissmann C, Flechsig E. PrP knock-out and PrP transgenic mice in prion research. Br Med Bull 2003; 66:43-60; PMID:14522848; http://dx.doi.org/ 10.1093/bmb/66.1.43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.