Abstract
Efforts to differentiate bovine spongiform encephalopathy (BSE) from scrapie in prion infected sheep have resulted in effective methods to decide about the absence of BSE. In rare instances uncertainties remain due to assumptions that BSE, classical scrapie and CH1641–a rare scrapie variant–could occur as mixtures. In field samples including those from fallen stock, triplex Western blotting analyses of variations in the molecular properties of the proteinase K resistant part of the disease‑associated form of prion protein (PrPres) represents a powerful tool for quick discrimination purposes. In this study we examined 7 deviant ovine field cases of scrapie for some typical molecular aspects of PrPres found in CH1641‑scrapie, classical scrapie and BSE. One case was most close to scrapie with respect to molecular mass of its non-glycosylated fraction and N-terminally located 12B2‑epitope content. Two cases were unlike classical scrapie but too weak to differentiate between BSE or CH1641. The other 4 cases appeared intermediate between scrapie and CH1641 with a reduced molecular mass and 12B2‑epitope content, together with the characteristic presence of a second PrPres population. The existence of these 2 PrPres populations was further confirmed through deglycosylation by PNGaseF. The findings indicate that discriminatory diagnosis between classical scrapie, CH1641 and BSE can remain inconclusive with current biochemical methods. Whether such intermediate cases represent mixtures of TSE strains should be further investigated e.g. in bioassays with rodent lines that are varying in their susceptibility or other techniques suitable for strain typing.
Keywords: CH1641, prion, sheep, typing, Western blot
Abbreviations
- AVG
average
- BSE
bovine spongiform encephalopathy
- IHC
immunohistochemistry
- ic.
intracerebrally
- ip.
intraperitoneally
- PK
proteinase K
- PrPC
prion protein in cellular form
- PrPres
proteinase K resistant fragment of PrPSc
- PrPSc
prion protein in TSE associated form
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TE
tissue equivalents
- triplex-WB
triplex Western blotting method
- TSE
transmissible spongiform encephalopathy
- VC
variation coefficient
Introduction
Transmissible spongiform encephalopathy's (TSEs) are infectious neurodegenerative diseases with long incubation times and are diagnostically best characterized by depositions of prion protein (PrP) mainly in the central nervous system. In healthy conditions this protein is present as a cell membrane bound component (PrPC) while in disease it is present as a refolded and aggregated product that is named PrPSc ‑ Sc referring to scrapie ‑, the archetypal prion disease condition occurring naturally in sheep and goats. Because of the peculiar nature of the infectious agent, composed only of protein, the term prion diseases has been generally used as an alternative term for TSEs after the introduction of the concept of the agent as prions.1
In the 1980s, the emergence of an epidemic prion disease in cattle known as bovine spongiform encephalopathy (BSE) or mad cow disease has revealed the epizootic and zoonotic risks of TSEs. BSE not only affected at least 2 hundred thousand cattle mostly in the UK, but also has affected about 2 hundred people,2-4 cats,5 zoo-animals,6 and 2 goats.7-9 In humans the rise and decline of the disease known as variant Creutzfeldt-Jakob disease (vCJD) was epidemiologically linked to the BSE outbreak. Moreover, vCJD has strain properties similar to BSE both by typing in mice and by analyzing the molecular properties of its PrPSc. The potential source of BSE is unknown but most probably has been a ruminant source either from a small ruminants scrapie variant or from a sporadic case of bovine TSE. The event for it to happen so recently was most likely an insufficiently safe process of rendering prion contaminated animal proteins into meat and bone meal.10
The recognition and discrimination of different TSEs is an essential task. Diagnosis targeted on the N-terminal proteolytic processing of PrPSc is being an effective tool in discriminating TSE strains either without the use of protease digestion as in immunohistochemistry (IHC)11-13 or with additional digestion using proteinase K as performed in Western blotting, bead-based multiplex assays and ELISA assays.14-18 Of these techniques, Western blotting is able to both recognize PrP and provide essential information on different molecular PrPSc properties adding to the phenotypical palette of parameters for TSE strain typing. In this way, several types have been discovered in ruminants. In cattle 3 rather well recognized BSE variants have been defined being C‑type, H‑type and L‑type BSE.19-21 Variant distinct TSE forms have been discriminated in sheep and goat by immunohistochemistry in tissue sections or Western blotting on PK digested brain homogenates such as classical scrapie, CH1641 scrapie, Nor98/atypical scrapie, and (experimental) BSE.8,12-16,22-30 However, at least in small ruminants there are potentially several phenotypes present in the same host which pose basic questions about the nature of the agent, the source of the BSE epidemic and how to recognize the presence of BSE.31-36
Recently we introduced 2 unique and powerful multiplex TSE differentiation non-enzymatic assays using fluorescence detection: one bead‑based assay for TSE‑typing in sheep18,37 and one probing technique for Western blots.38 These 2 techniques were effective to differentiate in brain homogenates between classical scrapie, CH1641, BSE, and Nor98/atypical scrapie in one test using 3 antibodies in a single mixture. The blotting technique, named triplex Western blotting (triplex-WB), is rather unique since the same protein (in casu PrP) is simultaneously detected by 3 different primary PrP‑specific antibodies on the same blotting membrane. The method eliminates potential errors in conventional Western blotting due to small migrational differences between gels as well as to variations in sample application when comparing different antibodies. Interestingly, the fluorescence detection method also is comparably sensitive to enzyme enhancement techniques and relatively quick with a low hands-on time.
In this study 7 French ovine field TSE cases were investigated by the triplex-WB technique.38 These cases were previously detected in France as deviating in conventional Western blots from the usual migration pattern of PrPres of classical scrapie, and were categorized as CH1641-like in our bead-based methodology.37 By using a set of samples as reference for classical scrapie, CH1641 and BSE, the new triplex–WB data support the idea that especially in some of those deviant field cases variable levels of a second PrPres population (also having a different N-terminal susceptibility to proteolytic cleavage) occur. The implications of the data indicate that biochemical diagnosis in brain homogenates alone is not always straightforward and that therefore other methods such as rodent bioassays for small ruminant scrapie typing remain essential.
Results
The triplex-WB analyses on 7 deviant French TSE cases in this study (F1-F7, Table 1, Fig. 1) were intended to probe different structural properties of PrPres by using antibodies 12B2, L42, and SAF84 that are specific for different PrPres regions (Fig. 2). As references were used: ovine brain stem samples from sheep with confirmed classical scrapie (natural cases N1-N32 and experimental J1 distributed over 6 different PrP genotypes), experimental CH1641 (C1-C9 with genotypes ARQ/ARQ or AHQ/AHQ), and experimental BSE (B1-B4), respectively. Also one CH1641 infected goat case was included (C-gt). Where appropriate, bovine C–type (NL86) and H-type BSE (AU8) were used as additional controls.
Table 1.
Brain stem tissue detailsa
| caseb | TSE typec | statusd | routee | species | PrPf genotype | sourceg |
|---|---|---|---|---|---|---|
| F1/06-0287 | unusual, study cases | f.s. | natural | sheep | ARQ/ARQ | ANSES (FR) |
| F2/06-0412 | f.s. | natural | ||||
| F3/PS048 | cl. | natural | VRQ/VRQ | INRA (FR) | ||
| F4/PS129 | cl. | natural | ||||
| F5/PS1650 | cl. | natural | ||||
| F6/PS227 | cl. | natural | ||||
| F7/PS303 | cl. | natural | ||||
| N1/02203685-0369 | classical scrapie | sl. | natural | sheep | ARH/VRQ | CVI (NL) |
| N2/96785103 | f.s. | natural | ||||
| N3/601677-2861 | cl. | natural | ARR/VRQ | |||
| N4/1957877 | sl. | natural | ||||
| N5/R07545 | f.s. | natural | ||||
| N6/R09123-10 | f.s. | natural | ||||
| N7/547189-4333 | cl. | natural | ||||
| N8/612945-3921 | cl. | natural | VRQ/VRQ | |||
| N9/623789-5488 | cl. | natural | ||||
| N10/12042 | sl. | natural | ||||
| N11/38161 | sl. | natural | ||||
| N12/378512-296 | f.s. | natural | ||||
| N13/2082428-0444 | f.s. | natural | ||||
| N14/0959773-0472 | f.s. | natural | ||||
| N15/358927-1063 | f.s. | natural | ||||
| N16/0238243-0180 | sl. | natural | ARQ/ARQ | |||
| N17/390246-0732 | sl. | natural | ||||
| N18/735007-0083 | sl. | natural | ||||
| N19/2173083-1999 | f.s. | natural | ||||
| N20/0060019-2171 | f.s. | natural | ||||
| N21/R07900-6 | f.s. | natural | ||||
| N22/36782-0763 | f.s. | natural | ARQ/ARH | |||
| N23/1660717-6641 | f.s. | natural | ||||
| N24/621553-4035 | cl. | natural | ARQ/VRQ | |||
| N25/R04768 | f.s. | natural | ||||
| N26/R05096 | f.s. | natural | ||||
| N27/123547-117 | f.s. | natural | ||||
| N28/1256389-1156 | f.s. | natural | ||||
| N29/R06925-11 | f.s. | natural | ||||
| N30/465377-073 | sl. | natural | ||||
| N31/2200994-0068 | f.s. | natural | ||||
| N32/R07589-8 | f.s. | natural | ||||
| J1/#73 | cl. | exptl | ARQ/ARQ | NIAH (USA) | ||
| C1/#56 | CH1641 | cl. | exptl | sheep | ARQ/ARQ | NIAH (USA) |
| C2/PG1271/05 | cl. | exptl | AHQ/AHQ | AHVLA (UK) | ||
| C3/PG1275/05 | cl. | exptl | ||||
| C4/PG1276/05 | cl. | exptl | ||||
| C5/PG1283/05 | cl. | exptl | ||||
| C6/PG0851/05 | cl. | exptl | ||||
| C7/PG1284/05 | cl. | exptl | ||||
| C8/8015470-6867 | cl. | exptl | ARQ/ARQ | CVI (UK) | ||
| C9/8015471-8173 | cl. | exptl | ||||
| Cgt/8018337-1587 | cl. | exptl | goat | wt | ||
| B1/6019555-8163 | BSE 1st pass. | cl. | exptl | sheep | wt | CVI (NL) |
| B2/6021075-8167 | cl. | exptl | ||||
| B3/6019325-8024 | BSE 2nd pass. | cl. | exptl | |||
| B4/6020305-8028 | cl. | exptl | ||||
| NL86 | boBSE -C | sl. | natural | cow | wt | CVI (NL) |
| AU8 | boBSE-H | f.s. | natural | cow | wt | AGES (Austria) |
Notes. aThe seven unusual TSE cases F1-F7 are the study cases which were diagnosed as CH1641-like in the bead-based assay37. bCase numbers: first two or three characters in the numbers preceding the slash are referring to F, French study case; NL, classical scrapie from the Netherlands; J, Japan; C, CH1641-scrapie; B, BSE in sheep; NL86 and AU08, respectively bovine C-type BSE from The Netherlands and H-type BSE from Austria. Numbers after the slash are institutional source numbers. cTSE-type: cases B1-B2 and B3-B4 were derived respectively after 1st and 2nd passage in ARQ/ARQ sheep. dStatus at collection: f.s., fallen stock; cl., clinical; sl. healthy at slaughter detected by active monitoring. eRoute: exptl, experimental (C1-C7 ic.; C8, C9 and C-gt: ic.ip.). fGenotype: PrP genotype of sheep is usually described for both PrP alleles for the three codons 136 (A = alanine or V = valine), 154 (R = arginine or H = histidine), and 171 (Q = glutamine, R = arginine, or H = histidine), where the most common one is ARQ/ARQ (wild type); wt, wild type PrP genotype. gSource: supplying laboratory with between parentheses the original country source where the original isolate originated from.
Figure 1.
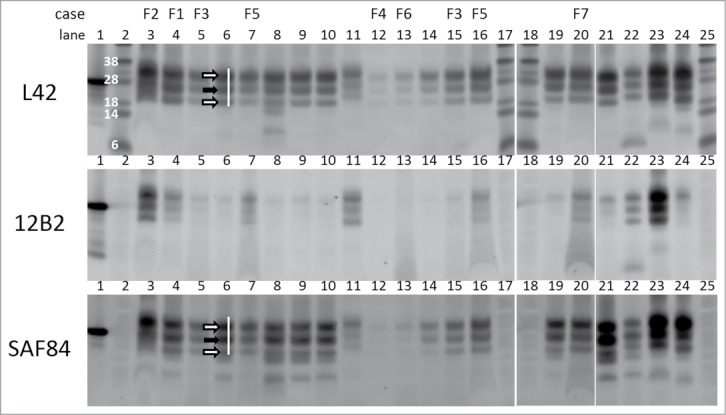
Triplex-WB analysis picture of study cases together with some reference samples. Selection of samples analyzed include all study cases numbers F1–F7. All lanes contain ovine samples except lanes 10 and 22, respectively caprine CH1641 Cgt and bovine BSE-H. Lanes 6, 8, 9, 14, 19, and 21: ovine CH1641 samples C8, C9, C1, C7, C3, and C6; lane 11 experimental scrapie J1; lanes 23 and 24 respectively ovine classical scrapie N18 and BSE B4 (details of case codes in Table 1). In the triplex‑WBs a mix of the 3 antibodies L42, 12B2 and SAF84 was applied. As an example for the glycoprofile estimation, the white bar shows the typical triple band (3 arrows) area in the L42 and SAF84 channels, thus neglecting the 2 lower bands that are visible in CH1641 samples. The migration position of mono‑glycosylated PrPres fraction (at ± 24 kDa) is indicated with a black arrow, the di-glycosylated and non-glycosylated fractions (white arrows) migrate respectively above and below the 24kDa band in this triplet region. Marker proteins used are: rec‑ovPrP in lane 1; molecular mass markers in lanes 2, 17, 18 and 25 with in lane 2 the molecular masses in kDa. TE applied per lane: 0.5 mg in lanes 3, 21–24, 0.8 mg in lane 7, 1 mg in lanes 4–6, 8–16, 19–20. See Methods section for antibody details and fluorescence detection.
Figure 2.
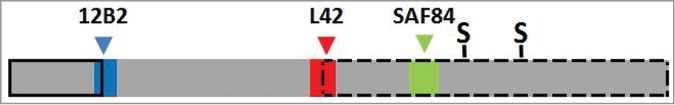
Schematic bar diagram of PrPres to illustrate the relative position of the 3 epitopes used in this study. The bar length reflects the length of a PrP monomer in classical scrapie PrPres (PrP residues ± 76–234). The epitope position of antibodies 12B2, L42 and SAF84 are reflected by the colored boxes with respectively blue, red and green (color choices agree with wave length of fluorescence emission signals, see Methods section). The PrP location of antibodies 12B2, L42 and SAF84 corresponds respectively with that of group A, B and C antibodies as described by us previously.35 The location of the 2 glycosylgroups are indicated by S. The solid black lined box indicates the removal by proteinase K of N-terminal PrP region in BSE and CH1641, which in CH1641 is even a few residues further down stream than in BSE. The broken black lined box indicates the length of the second PrPres population (PrPres#2) as occurring in CH1641, where the 12B2 epitope is absent and the L42 epitope incomplete, thus only allowing full binding by SAF84. Sugar moieties are variably present in both PrPres populations of CH1641, and in PrPres from classical scrapie and BSE.
Protease susceptibility estimations of PrPres N-terminus
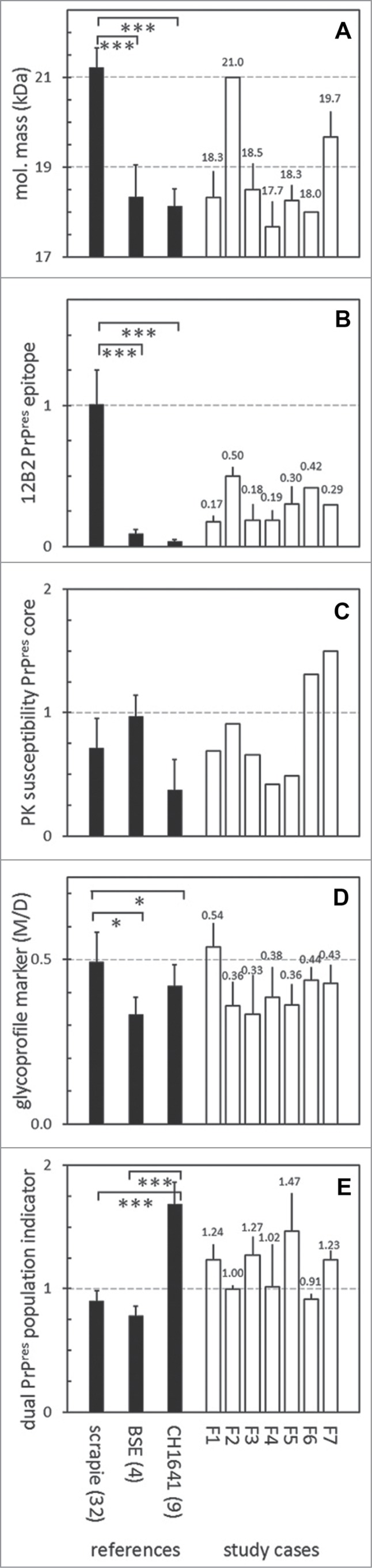
Molecular mass of the non-glycosylated PrPres moiety was probed in each sample with antibody L42. The variations in kDa values per individual scrapie sample were low, varying per sample (3 measurements per sample) with VCs between 1–8%. The molecular mass values in classical scrapie samples averaged at 21 kDa (average ± SD 21.2 ± 0.4 kDa, range 20.3-22.0), ovine CH1641 at 18.1 kDa (18.1 ± 0.9, range 17.7–19.0) and for ovine BSE at 18.3 kDa (18.3 ± 0.7, range 17.7-19.3) (Fig. 2A). These molecular mass values in the BSE and CH1641 cases differed significantly from those of the scrapie cases (P < 0.001). The molecular mass values in the study cases F1 and F3–F7 varied considerably between 17.7–19.7 kDa which is in the range of CH1641 and BSE references, while case F2 migrated as a 21.0 kDa protein, comparable to classical scrapie (Fig. 3A).
Figure 3.
Western blot results transformed into values to express 5 different PrPres properties. In the 5 panels are expressed: (A) molecular mass of non-glycosylated PrP as observed with mAb L42 (see Fig. 1); (B) N-terminal epitope content expressed as 12B2/L42 ratio; (C) PK resistance as a ratio of L42 signals at pH 8 and pH 6.5; (D) glycoprofile expressed as L42 signal ratio M/D, where M and D are the respective mono-glycosylated and di-glycosylated fractions of the 3 PrPres bands (see Fig. 1); (E) marker for the presence of a dual PrPres population as estimated by SAF84/L42 ratio of the 24 kDa fraction of total PrPres triplet (see Fig. 1). Black bars represent averages ± SD of the reference samples indicated on the horizontal axis with the number of different samples per group between parentheses. The open bars represent data of the 7 study samples F1–F7 with averages ± SD for 2–7 measurements per sample, except for panel c where single measurements have been carried out; for these cases bar-values are specified above the bars.
Next, the 12B2 epitope content relative to core epitope L42 was tested as a method for measuring the resistance of the PrPres amino-terminus to PK processing under neutral pH conditions. The 12B2/L42 epitope ratios per individual scrapie sample varied with an average VC of 8% (range 1–24%). These ratios were similar between BSE and CH1641 isolates with values of respectively 0.04 ± 0.01 (range 0.02–0.05) and 0.09 ± 0.03 (range 0.06–0.12), and highly significantly higher in natural scrapie with values of 1.00 ± 0.25 (range 0.57–1.75, P < 0.001) (Fig. 3B). In the 7 F-cases, this parameter varied strongly with 12B2/L42 ratios ranging from 0.10–0.50, case F2 having the highest value. These 12B2/L42 values were clearly between on the one hand those of the natural scrapie and on the other hand of both CH1641 and BSE isolates.
Though the 2 parameters -molecular mass and relative 12B2 epitope content- are both related to cleavage of the PrPres N–terminus by PK, there was no obvious correlation between the 2 different measurements for these 7 F–samples (not shown). This discrepancy in these specific examples was highly probably due to variation in the amount of available ovPrP 93–97 12B2–epitope over the 3 triplet bands. The estimation of the relative 12B2 epitope content becomes critical if protease cleavage occurs around amino acid tryptophan 9337,39 where presence or absence of this single amino acid will have an all or none effect on antibody 12B2 binding.40 Nevertheless, especially sample F2 is most likely a classical scrapie case with a sufficiently high amount of 21 kDa non-glycosyl band to yield a 12B2/L42 ratio of 0.5.
Protease susceptibility estimations of the PrPres core
A method to test the protease susceptibility of the PrPres core has previously been successfully used to discriminate between the bovine TSEs where C-BSE was found to be rather resistant compared to BSE type H.21 The method uses 2 PK digestion conditions–mild at pH 6.5 and stringent at pH8, at respective PK concentrations of 50 and 500 μg,ml−1– and a core specific antibody such as L42 as detecting antibody. The extent of resistance can be expressed as the ratio between L42 signal estimated at pH8 and pH6.5. Application to the ovine samples yielded susceptibility values for classical scrapie PrPres ranging between 0.26–1.25 (0.72 ± 0.25), CH1641 between 0.11–0.89 (0.37 ± 0.25), and BSE as the most resistant between 0.82–1.16 (0.97 ± 0.17) (Fig. 3C). None of these differences however were statistically significant. The 7 F-cases varied between 0.42–1.50, like the variable behavior of the classical scrapie and CH1641 samples. Remarkably, 2 cases–F6 and F7–exhibited the highest values encountered in all sheep isolates of this study, being 1.31 and 1.50.
As a side note, it is a remarkable novel observation that the PrPres‑core PK resistance of the BSE samples is tending to be higher than that of classical scrapie, while the PrPres N-terminal region of BSE is highly PK susceptible under mild conditions as was mentioned above for the 12B2/L42 ratios. The latter parameter nowadays being of critical discriminatory value in the diagnosis between BSE/CH1641 and classical scrapie.22,24
Glycosylation patterns
Glycosylation profiles of PrPres are usually presented as the monoglycosylated (M) and diglycosylated (D) fraction in the total triple band area, using a B-group antibody for detection i.c. L42. We employed the M/D ratio of these 2 glycosylated moieties for typing. The M/D ratio of BSE PrPres mono- and di-glycosyl fractions in the scrapie samples (0.49 ± 0.09) differed statistically (P < 0.05) from that of the set of CH1641 (0.42 ± 0.06) and BSE samples (0.33 ± 0.05) (Fig. 3D). None of the 7 study cases were outstanding in this respect, case F1 having an M/D ratio (0.54); the other cases F2-F7 ranged between 0.33–0.44.
Thus, consistent with other reports there is a tendency that ovine BSE cases contain a diglycosyl PrPres fraction around 65% (range 61-68%) of total PrPres which is higher than in scrapie cases (average 56%, range 47–67%), at the expense of mainly the mono‑glycosylated moiety. The CH1641 cases contained a diglycosyl PrPres level similar to that in BSE (average 64%, range 62-70%), but mainly at the expense of the non‑glycosylated moiety. This non‑glycosylated PrPres fraction, was highest in scrapie cases (average 17%, range 11–23%), followed by BSE (average 14%, range 11–16%) and CH1641 (average 9%, range 7–11%).
Investigating presence of a PrPres#2 population
A feature of TSEs like CH1641 in sheep and H-type BSE in cattle is the presence of 2 populations of PrPres: population PrPres#1 exists as a triple band with molecular masses in the usual range between 17–32 kDa, and reactive with group B antibody L42 as well as group C antibody SAF84, while PrPres#2 also contains a triple band composition of lower molecular mass in the 10–25 kDa range but only reactive with the group C antibody SAF84. Evidence for the existence of the #2 population can be deduced from the ratio of SAF84/L42 signal in the fraction of mono-glycosylated PrPres (or 24kDa) region within the triple band region of the L42 reactive bands38. In the sheep derived samples all experimental CH1641 samples exhibited such SAF84/L42 ratios between 1.4-2.0, and 5 of the 7 field samples under study–F1, F3, F4, F5 and F7 – yielded ratios of 1.2 or higher, indicative for presence of a certain amount of PrPres#2 population (Fig. 3E). Both the 3two classical scrapie reference samples, the experimental classical scrapie J1 and the 4 experimental BSE cases appeared to contain only one PrPres population based on ratio values ranging between 0.7-1.1. The variation in the SAF84/L42 ratios at the 24kDa band for these samples were low at an average VC of 10 ± 6%, indicative for a reliable way of estimating this dual population parameter.
To find more support for this dual population property, deglycosylations were carried out with PNGaseF. As expected for CH1641 (and H-type BSE), this treatment yielded 2 deglycosylated PrPres bands, of which band 1 migrated at the usual 17-21 kDa position, and band 2 at the CH1641 specific position 10-13 kDa (Fig. 4A). All CH1641 samples yielded a dual PrPres population result by the presence of band2 (band2/band1 ratios of > 0.2). In the classical scrapie samples investigated band 2 was hardly or not present, with values between 0.05 and 0.15 while reference cases N7 and N22 (and B1 as well) had values close 0.2. Four study cases–F1, F2, F5 and F7–yielded values >0.20, but not F3 while cases F4 and F6 were too weak for analysis. Finally, when the SAF84/L42 ratio of all samples analyzed for both parameters was plotted against band2/band 1 ratio for linear regression analysis, it appeared that there was a positive correlation between these 2 variables (R2 = 0.74), though study samples F2 and F3 did poorly fit this correlation (Fig. 4B).
Figure 4.
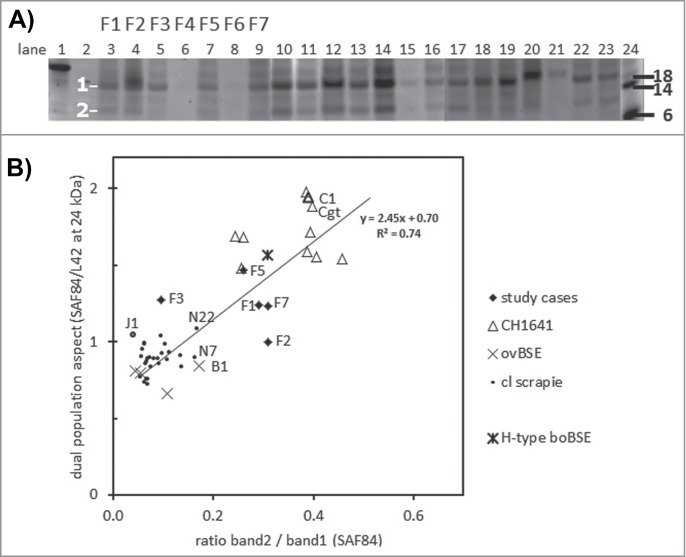
Dual PrPres population composition analysis with SAF84 antibody. (A) Triplex-WB of an selection of the samples analyzed, containing 7 study cases (lanes 3–9) and a set of CH1641 cases (lanes 10–17, 22–23), ovine BSE (lanes 18–19), and ovine scrapie (lanes 20-21). The PrPres samples were deglycosylated using PNGaseF. The position of deglycosylated bands 1 and 2 at respectively at 10–12 kDa and 18–21 kDa is indicated. For case details see Table 1. All lanes presented were scanned at 532 nm excitation wavelength, except lane 24 at 633 nm. Molecular mass references: lane 1 rec-ovPrP, lanes 2 and 24 blue molecular mass markers that yield a strong signal at excitation wavelength 633 nm (in kDa). TE applied per lane: 0.25 mg in lanes 4, 18–20, and 0.5 mg in lanes 3, 5–17, 21–23. (B) Dotplot of the 2 methods of dual population estimation. Vertical axis, the ratio between SAF84/L42 fraction of the 24kDa band in the PrPres triplet (see Fig. 1). Horizontal axis, the band 2 versus band 1 ratio estimated in blots of the 54 cases as exemplified in panel a. For all samples analyzed a linear regression correlation curve is presented together with the mathematical curve data and regression value R2.
Combining the observations
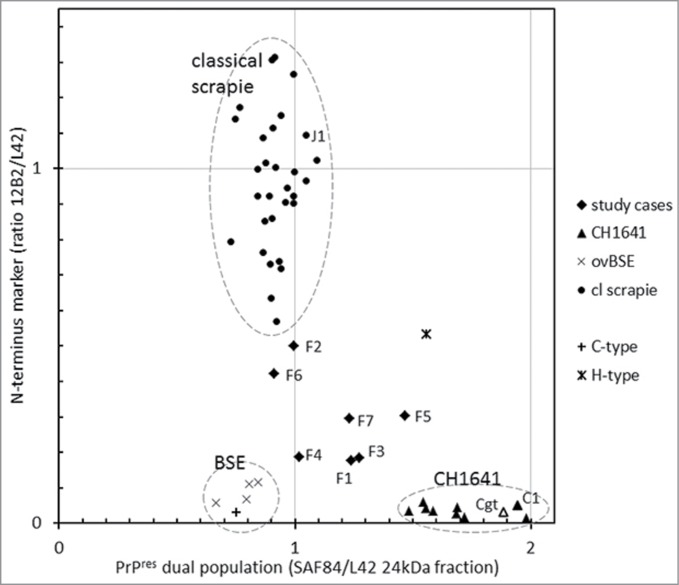
To classify the TSE type in the 7 French field sheep cases for their PrPres features by Western blotting, we used confirmed cases of classical scrapie cases and experimentally generated infected BSE– and CH1641–brain tissues from sheep as reference. Within these reference groups, the 3 most crucial and discriminating molecular PrPres parameters were the molecular mass of the non‑glycosylated moiety (kDa), the relative presence of the 12B2 N–terminus epitope (12B2/L42 signal ratio in the triple band area), and the occurrence of a second population (indicated by the SAF84/L42 signal ratio of the 24 kDa band fraction). The 7 study samples exhibited some CH1641 property (cases F1-F3, F5, and F7), or similarities with classical scrapie (F6) or BSE (F4), the latter 2 both weak cases for which a band 2 estimation was not possible; however neither of these 2 did fit completely a diagnosis for classical scrapie, CH1641 or BSE (Fig. 5). Other references included: an experimentally generated goat CH1641 sample (Cgt) which behaved similar to the experimental sheep CH1641 cases C2-C8 (inoculum originally sourced from the same UK source); experimentally generated sheep scrapie (J1) and CH1641 (C1) of which both were subpassaged in sheep from the same USA classical scrapie infected animal36 and which behaved similarly respectively to Dutch natural scrapie and experimentally generated UK sourced CH1641; and bovine C- and H-type BSE samples included to compare with ovine BSE and CH1641 (Fig. 5). Furthermore, there were no statistically significant differences for any of the 5 parameters discussed above between the 6 different sheep genotypes with classical scrapie. Bovine and ovine BSE were very comparable in all parameters estimated. Bovine H-type BSE and ovine CH1641 did show presence of a dual PrPres population but they differed in the relative content of the 12B2 N‑terminus epitope being amply present in H-type, but close to zero in CH1641.
Figure 5.
Two features for BSE and CH1641 recognition in TSE-infected sheep plotted for individual samples. The sample numbers for the 7 French samples and for some other samples (goat CH1641 case Cgt, NIAH experimental scrapie case J1, NIAH experimental CH1641 case C1). By surrounding data points by the dashed circles, it is clear that the 7 study cases exhibit an intermediate behavior between scrapie, BSE and CH1641.
Discussion
The molecular analyses by triplex-Western blotting in this study of 7 ovine field TSE cases from France confirmed their separate status with respect to the features that usually allow a diagnosis of classical scrapie, BSE and CH1641 in sheep. Whether these isolates represent separate scrapie strains or mixtures of different TSE strains remained undetermined, but either possibility might well be the case. Nevertheless, the data obtained here point to TSE heterogeneity, the quasi‑species nature of sheep prions or the existence of different strains in field TSEs of sheep. The strongest features for type recognition in Western blots appear to be 3 parameters: the molecular mass of the non–glycosylated PrPres moiety, the estimation of the amounts of (ovine) PrP epitope 93–97 (12B2) in relation to the core 148–153 epitope (L42), and the presence of a second PrPres population (PrPres#2) that is typical for CH1641 and well recognized only by group C antibodies (e.g., SAF84) reactive with an epitope C–terminal of the ovine PrP amino acid position 154. The 2 other parameters–PrPres glycoprofile and PK susceptibility of the PrPres core estimated by using a group B antibody–were too variable within the reference TSE types to allow a firm recognition.
The results of the Western blotting technique, even though not fulfilling the expectation to clearly recognize these 7 sheep TSE cases as classical scrapie, BSE or CH1641, do confirm the power of the 2 discriminative parameters defined in our previous study: N-terminal processing of PrPres by PK and presence of 2 PrPres populations.38 The blotting system allowed to deduce these 2 aspects with a selected triplex antibody usage applied on the same protein in one single blot leading to low interassay variations. Variations in proteolytic processing of the PrPSc N–terminus can be expressed as molecular mass (kDa) or the ratio of the immuno‑signals between e.g. 12B2 and L42, while the presence of 2 PrPres populations is evident by the SAF84/L42 ratio in the 24 kDa fraction of the PrPres triple band region38. Glycoprofile estimation of BSE cases estimated with a group B antibody (like L42, 6H4, Sha31) is another aspect characterized by a high diglycosyl fraction22,24 which indeed was the case varying between 60–70% of total PrPres, while that in the natural scrapie cases varied between 47–67% and CH1641 between 62–70%. However, this overlapping result in diglycosyl PrPres concentration between the 3 sheep TSE types makes it an indicative but inadequate marker for a distinct typing diagnosis.
The existence of a separate TSE status of the study cases does not seem genotype or tissue quality dependent since one of the 2 fallen stock ARQ/ARQ samples (F1) and several of the VRQ/VRQ freshly collected samples (F3, F4, F5, F7) could be categorized as cases with a dual PrPres population. Moreover, extra N-terminal PrPres processing by autolysis or microbial breakdown would have shown up in many more instances of European monitoring or in the fallen stock classical scrapie references of this study. Also, PrPres properties might be influenced by the brain region chosen for analysis.41-43 A difference in outcome of a sheep was suggested where a scrapie as well as a BSE diagnosis had been obtained by using different techniques in probably different laboratories in respectively the caudal medulla and the brain stem.34 The chance that our observations on PrPres properties might be dependent on the region in the brain cannot be ruled out, but seems negligible since in all cases homogenized brain stem material was used.
In a previous study using a bead-based multiplex immuno-assay for TSE type discrimination,37 we diagnosed all 7 French cases as CH1641‑like, while in the current triplex-WB analysis a CH1641‑like diagnosis was applicable to cases F1, F3, F5, and F7, maybe to F2 but not to weak cases F4 and F6. In that study the binding of the group C antibody 94B4 was essential for capture to a bead of both the PrPres#1 and PrPres#2 population, while antibody Sha31 – a group B antibody very similar in epitope specificity to L42 by epitope mapping–was used as a reporter for bead-captured PrPres. However, it should be realized that the 2 methods have really different principles. The triplex-WB yields a more detailed result due to the fact that binding of all 3 antibodies is observed on one single molecule while in the bead-based assay only one antibody can bind per PrP molecule. Therefore, the bead‑based multiplex assay remains a remarkable test system with unique structural resolution, different from Western blots and ELISAs.
The molecular properties of the 7 cases with their PrPres properties intermediate between BSE, classical scrapie and CH1641 pose a basic question about the nature of prions: represent these cases separate strains, or mixtures of different strains, or are prions appearing as quasi-species where the fittest dominates under a certain biological condition44? These are questions that would need further investigations. The existence of mixtures of scrapie types in sheep has been discussed since the time that genetic susceptibility for TSEs has been described using CH1641 and SSBP/1 as experimental challenge material.25,29,31,32,45,46 Furthermore, for 4 French ovine CH1641-like cases with a low P4 (with nearly similar PrP binding location as 12B240) epitope content in PrPres it was shown in transgenic TgovPrP4 mice expressing ovine ARQ PrP that indeed a CH1641 feature was transmitted characterized by the presence of the 2 PrPres populations PrPres#1 and PrPres#2 characteristic for experimental CH1641.26 However, the presence of a typical classical scrapie feature was not shown in these mice, for which reasons the presence of a mixture of classical scrapie and CH1641 remains uncertain. In our study, the 7 cases variably exhibited a 12B2 epitope content intermediate between that found in classical scrapie and reference ovine BSE cases; 4 of these 7 (F1, F3, F5 and F7) exhibited evidence for the presence of a PrPres#2 population as in CH1641‑like isolates (Fig. 5), while 2 cases (F2, F6) were close to classical scrapie and one (F4) most close to BSE. However cases F4 and F6 were rather low in PrPres content limiting the reliability of the detection and hampering an enzymatic deglycosylation analysis with PNGaseF. Low levels of PrPres#2 in naturally TSE infected sheep brain samples seem anyway to be sometimes present in sheep with classical scrapie26 and this also might be the case in some of our sheep reference samples. Nevertheless Western blot analyses only are not sufficient to understand the nature of sheep prions, but maybe these cases could serve as interesting targets for further research.
The existence of deviant field TSE cases in sheep as found in this study and others based on PrPres properties16,26 call for further investigation. Bioassays including limited titration experiments in rodents and transgenic mice, or even other techniques such as in vitro conversion studies or permissive TSE‑susceptible cell systems are necessary to get additional confirmation of strain typical features. This will not only be required for TSEs in sheep but also in goats which are also susceptible to BSE and different scrapie forms as found in sheep.
Materials and Methods
Materials
To serve molecular TSE-typing 3 antigenic PrP regions A, B and C have been defined recognizing respective epitope regions A, B and C which are located from N- to C-terminus in the respective ovine PrP sequence positions 60–100, 101–154 and 154–234 (Fig. 2).35 Purified monoclonal PrP-specific antibodies used (with their antigenic group specificity, Ig class type, and source) were: 12B2 (group A, IgG1, CVI-WageningenUR), L42 (group B, IgG2a, R-Biopharm), and SAF84 (group C, IgG2b, SpiBio, Bertin Pharma).40,47-49 The respective epitope specificities of 12B2, L42 and SAF84 were 93WGQGG97, 148YEDRYY153, and 166YRPVDQY172 (sheep PrP numbering, glutamine on position 17150 as mapped by Pepscan analysis using solid phase synthetic peptides based on the ovine PrP sequence.51 Mouse IgG class-specific detection kits containing isotype-specific goat anti-mouse-Fc Fab-Alexa Fluor 488, 555, and 647 conjugates were purchased as Zenon® labels (Life Technologies). Proteinase K was from Merck (30 U/mg; 124568; Darmstadt, Germany). Purified recombinant ovine PrPARQ was a kind gift from Human Rezaei, (INRA, Jouy‑en‑Josas). Molecular mass markers kit See Blue was from Life Technologies.
Tissues
Fifty 6 different brain stem samples have been investigated, nearly all from sheep except one goat sample and 2 bovine BSE cases. These tissues are collectively detailed in Table 1 including the supplying institute and the country from where the tissue was originating from. Also PrP genotypes of the individual sheep samples are explained in Table 1.
Seven deviant scrapie cases from different flocks (F1-F7) were investigated in this study because of unusual behavior in our bead-based assays37: F1 and F2, both of PrP genotype ARQ/ARQ, were collected from fallen stock monitoring, and F3-F5 (all VRQ/VRQ) from clinical surveillance.
As references in this study, CH1641 samples were obtained from 5 institutes to cover the places world wide where CH1641 materials have been discovered or generated. Three different sheep TSE types were used: 3two natural classical scrapie cases (N1-N32) from active and passive surveillance and varying in PrP genotype, 8 CH1641 experimentally infected samples from sheep (C2-C9) and one goat (Cgt) challenged with CH1641/3 which is a UK derived brain pool from 5 Sippa/pa sheep (presumed PrP genotype ARQ/ARQ) and kindly provided by Roslin Institute, (University of Edinburgh, UK),32 4 experimentally BSE infected samples after 1st (B1, B2) or 2nd (B3, B4) passage. In addition included were brain stems from 2 different Cheviot sheep i.c. infected with brain from either experimental classical scrapie case #2314 (J1) or experimental CH1641 case #294 (C1); the original scrapie isolate used at 1st passage in both sheep #2314 and #294 was a classical scrapie isolate form the USA.36 Furthermore, 2 bovine cases of different BSE type have also been used, one of which was a case of C-type (NL86) and the other of H‑type BSE (AU8).52 Confirmed TSE-negative brain stem tissues from sheep served as negative controls. All cases were confirmed by immuno-histochemistry and/or Western blotting.
PrPres preparation and triplex Western (triplex-WB) blotting analyses
Preparation of 10 per cent tissue homogenates in lysis buffer, conditions for digestion with proteinase K (PK) to prepare PrPres from PrPSc, precipitation of PrPres with 1-propanol, sodium dodecyl sulfate PAGE (SDS-PAGE), subsequent triplex-WB and fluorimetric detection were as described38 with the following details. Blots were developed with a mixture of the antibodies 12B2, L42, and SAF84 at respective concentrations of 0.1, 0.1 and 0.5 μg IgG ml−1, and buffers used were freshly prepared and filtered through a 0.45 μm filter. 12B2, SAF84 and L42 detection was carried out with the respective Alexa Fluor labels 488, 555, 647 (blue, green and red in Fig. 2). Blots were dried in a vacuum desiccator for 1 hour at ambient temperature. Molecular masses of triplet PrPres bands detected with L42 were calculated using GelPro software (Gel-Pro analyzer; Media Cybernetics) and with InvitroGen SeeBlue® prestained molecular mass markers (Life Sciences) as applied in 2 lanes per gel; these standards have a high fluorescence yield in the 488 nm channel. In all runs rec-ovPrPARQ was included as a correction reference for signal level differences between emission light intensities in the 3 different Typhoon Trio laser light channels. As a further correction to quantify the relative N-terminal 93–97 epitope presence in 12B2/L42 ratio estimations, the average 12B2/L42 value for all scrapie samples in an experiment (usually n = 32) was considered to be 1,39 therefore each individual 12B2x/L42x outcome was multiplied with 1/(12B2scr/L42scr) where the 12B2scr/L42scr ratio is representing the average 12B2/L42 value of all 3two classical scrapie cases in the experiment. Glycoprofiles were estimated as the percentual fraction of di-, mono- and non-glycosylated PrPres in the triple band area.38 For estimating the presence of a second population of PrPres, the L42 and SAF84 signals in the monoglycosylated PrPres band (migrating at 24-25 kDa) expressed as fraction of the total PrPres-triplet signal were used to calculate the SAF84/L42 ratio; a value >1.2 points to the presence of a significant amount of PrPres#2 population that is typical for CH1641-like cases.
Differential PK digestions to test protease susceptibility of PrPres core
Determination of the resistance of the PrPres core to PK was performed as described21 by comparing samples digested at pH 6.5 and 8 and respective enzyme concentrations 50 μg ml−1 500 μg ml−1 PK. Triplex-WB was used with antibody L42 as PrPres core region detecting antibody. The relative breakdown of the PrPres core was expressed as the pH8/pH6.5 signal ratio between each set of lanes probed in the triple band region, where values between zero and one related to respectively high and low PK susceptibility.
Deglycosylation of PrPres by treatment with PNGase F
Deglycosylation of PrPres preparations with peptide N-glycosidase F (PNGase F; New England Biolabs) was performed as described previously.40
Statistical analyses
Interassay variation for a sample value was expressed as variation coefficient (VC) being the percentage of standard deviation (SD) of the average value (AVG) of a group of values found for that sample, thus VC = (SD/AVG)×100%. One-way analyses of variance were carried out to establish whether variations between groups of data were larger than expected; if so, subsequent differences between pairs of groups were considered significant if the probability (P) of a non–difference was <0.05 in multiple-comparisons tests, according to the Student-Newman-Keuls test. The software used for these calculations was with Prism 5.02 software from Graph-Pad Software.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Jim Foster from Roslin institutes (UEDIN, Edinburgh, UK) for supply of CH1641 challenge material used for generating the different experimental CH1641 tissues at CVI and AHVLA. Dr. Jacques Grassi (CEA, Saclay, France) and Dr. Martin Groschup (FLI, Riems-Greifswald, Germany) provided kindly an initial batch of respectively antibodies SAF84 and L42. We thank Dr. Human Rezaei (INRA, Jouy-en-Jozas, France) for kind gift of rec-ovine PrPARQ.
Funding
This work was largely supported by the Dutch Ministry of Economic Affairs (grant WOT-01-002-01). Part of this work was also possible thanks to Department for Environment, Food and Rural Affairs (Defra, UK; project SE1700/CSA 7335), European Commission funded STREP project GoatBSE, FOOD-CT-2006-36353 and EMIDA ERA-NET project GOAT-TSE-FREE.
References
- 1. Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136-44; PMID:6801762; http://dx.doi.org/ 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 2. Wells GA, Scott AC, Johnson CT, Gunning RF, Hancock RD, Jeffrey M, Dawson M, Bradley R. A novel progressive spongiform encephalopathy in cattle. Vet Rec 1987; 121:419-20; PMID:3424605; http://dx.doi.org/ 10.1136/vr.121.18.419 [DOI] [PubMed] [Google Scholar]
- 3. Will RG, Ironside JW, Zeidler M, Cousens SN, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith PG. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet 1996; 347:921-5; PMID:8598754; http://dx.doi.org/ 10.1016/S0140-6736(96)91412-9 [DOI] [PubMed] [Google Scholar]
- 4. Bruce ME, Boyle A, Cousens S, McConnell I, Foster J, Goldmann W, Fraser H. Strain characterization of natural sheep scrapie and comparison with BSE. J Gen Virol 2002; 83:695-704; PMID:11842264 [DOI] [PubMed] [Google Scholar]
- 5. Wyatt JM, Pearson GR, Smerdon TN, Gruffydd-Jones TJ, Wells GA, Wilesmith JW. Naturally occurring scrapie-like spongiform encephalopathy in five domestic cats. Vet Rec 1991; 129:233-6; PMID:1957458; http://dx.doi.org/ 10.1136/vr.129.11.233 [DOI] [PubMed] [Google Scholar]
- 6. Pattison J. The emergence of bovine spongiform encephalopathy and related diseases. Emerg Infect Dis 1998; 4:390-4; PMID:9716952; http://dx.doi.org/ 10.3201/eid0403.980311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eloit M, Adjou K, Coulpier M, Fontaine JJ, Hamel R, Lilin T, Messiaen S, Andreoletti O, Baron T, Bencsik A, et al. BSE agent signatures in a goat. Vet Rec 2005; 156:523-4; PMID:15833975; http://dx.doi.org/ 10.1136/vr.156.16.523-b [DOI] [PubMed] [Google Scholar]
- 8. Jeffrey M, Martin S, Gonzalez L, Foster J, Langeveld JP, van Zijderveld FG, Grassi J, Hunter N. Immunohistochemical features of PrP(d) accumulation in natural and experimental goat transmissible spongiform encephalopathies. J Comp Pathol 2006; 134:171-81; PMID:16542672; http://dx.doi.org/ 10.1016/j.jcpa.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 9. Spiropoulos J, Lockey R, Sallis RE, Terry LA, Thorne L, Holder TM, Beck KE, Simmons MM. Isolation of prion with BSE properties from farmed goat. Emer Infect Dis 2011; 17:2253-61; PMID:22172149; http://dx.doi.org/ 10.3201/eid1712.110333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilesmith JW, Ryan JB, Atkinson MJ. Bovine spongiform encephalopathy: epidemiological studies on the origin. Vet Rec 1991; 128:199-203; PMID:1823120; http://dx.doi.org/ 10.1136/vr.128.9.199 [DOI] [PubMed] [Google Scholar]
- 11. Jeffrey M, Martin S, Gonzalez L, Ryder SJ, Bellworthy SJ, Jackman R. Differential diagnosis of infections with the bovine spongiform encephalopathy (BSE) and scrapie agents in sheep. J Comp Pathol 2001; 125:271-84; PMID:11798244; http://dx.doi.org/ 10.1053/jcpa.2001.0499 [DOI] [PubMed] [Google Scholar]
- 12. Thuring CM, van Keulen LJ, Langeveld JP, Vromans ME, van Zijderveld FG, Sweeney T. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J Comp Pathol 2005; 132:59-69; PMID:15629480; http://dx.doi.org/ 10.1016/j.jcpa.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 13. Jeffrey M, Gonzalez L, Chong A, Foster J, Goldmann W, Hunter N, Martin S. Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: immunochemical similarities can be resolved by immunohistochemistry. J Comp Pathol 2006; 134:17-29; PMID:16324707; http://dx.doi.org/ 10.1016/j.jcpa.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 14. Baron TG, Madec JY, Calavas D, Richard Y, Barillet F. Comparison of French natural scrapie isolates with bovine spongiform encephalopathy and experimental scrapie infected sheep. Neurosci Lett 2000; 284:175-8; PMID:10773427; http://dx.doi.org/ 10.1016/S0304-3940(00)01047-8 [DOI] [PubMed] [Google Scholar]
- 15. Hill AF, Sidle KC, Joiner S, Keyes P, Martin TC, Dawson M, Collinge J. Molecular screening of sheep for bovine spongiform encephalopathy. Neurosci Lett 1998; 255:159-62; PMID:9832197; http://dx.doi.org/ 10.1016/S0304-3940(98)00736-8 [DOI] [PubMed] [Google Scholar]
- 16. Hope J, Wood SC, Birkett CR, Chong A, Bruce ME, Cairns D, Collinge J. Molecular analysis of ovine prion protein identifies similarities between BSE and an experimental isolate of natural scrapie, CH1641. J Gen Virol 1999; 80 (Pt 1):1-4; PMID:9934675 [DOI] [PubMed] [Google Scholar]
- 17. Simon S, Nugier J, Morel N, Boutal H, Creminon C, Benestad SL, Andréoletti O, Lantier F, Bilheude JM, Feyssaguet M, et al. Rapid typing of transmissible spongiform encephalopathy strains with differential ELISA. Emer Infect Dis 2008; 14:608-16; PMID:18394279; http://dx.doi.org/ 10.3201/eid1404.071134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang Y, Thorne J, Whatling K, Jacobs JG, Langeveld J, Sauer MJ. A single step multiplex immunofluorometric assay for differential diagnosis of BSE and scrapie. J Immunol Methods 2010; 356:29-38; PMID:20214905; http://dx.doi.org/ 10.1016/j.jim.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 19. Biacabe AG, Laplanche JL, Ryder S, Baron T. Distinct molecular phenotypes in bovine prion diseases. EMBO Rep 2004; 5:110-5; PMID:14710195; http://dx.doi.org/ 10.1038/sj.embor.7400054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M. Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A 2004; 101:3065-70; PMID:14970340; http://dx.doi.org/ 10.1073/pnas.0305777101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobs JG, Langeveld JP, Biacabe AG, Acutis PL, Polak MP, Gavier-Widen D, Buschmann A, Caramelli M, Casalone C, Mazza M, et al. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. J Clin Microbiol 2007; 45:1821-9; PMID:17442800; http://dx.doi.org/ 10.1128/JCM.00160-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stack MJ, Chaplin MJ, Clark J. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol 2002; 104:279-86; PMID:12172914 [DOI] [PubMed] [Google Scholar]
- 23. Nonno R, Esposito E, Vaccari G, Conte M, Marcon S, Di Bari M, Ligios C, Di Guardo G, Agrimi U. Molecular analysis of cases of Italian sheep scrapie and comparison with cases of bovine spongiform encephalopathy (BSE) and experimental BSE in sheep. J Clin Microbiol 2003; 41:4127-33; PMID:12958236; http://dx.doi.org/ 10.1128/JCM.41.9.4127-4133.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thuring CM, Erkens JH, Jacobs JG, Bossers A, Van Keulen LJ, Garssen GJ, Van Zijderveld FG, Ryder SJ, Groschup MH, Sweeney T, et al. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J Clin Microbiol 2004; 42:972-80; PMID:15004040; http://dx.doi.org/ 10.1128/JCM.42.3.972-980.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baron T, Biacabe AG. Molecular behaviors of "CH1641-like" sheep scrapie isolates in ovine transgenic mice (TgOvPrP4). J Virol 2007; 81:7230-7; PMID:17442721; http://dx.doi.org/ 10.1128/JVI.02475-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baron T, Bencsik A, Vulin J, Biacabe AG, Morignat E, Verchere J, Betemps D. A C-terminal protease-resistant prion fragment distinguishes ovine "CH1641-like" scrapie from bovine classical and L-Type BSE in ovine transgenic mice. PLoS Pathog 2008; 4:e1000137; PMID:18769714; http://dx.doi.org/ 10.1371/journal.ppat.1000137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benestad SL, Arsac JN, Goldmann W, Noremark M. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet Res 2008; 39:19; PMID:18187032; http://dx.doi.org/ 10.1051/vetres:2007056 [DOI] [PubMed] [Google Scholar]
- 28. Vulin J, Biacabe AG, Cazeau G, Calavas D, Baron T. Molecular typing of protease-resistant prion protein in transmissible spongiform encephalopathies of small ruminants, France, 2002-2009. Emerg Infect Dis 2011; 17:55-63; PMID:21192855; http://dx.doi.org/ 10.3201/eid1701.100891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vulin J, Beck KE, Bencsik A, Lakhdar L, Spiropoulos J, Baron T. Selection of distinct strain phenotypes in mice infected by ovine natural scrapie isolates similar to CH1641 experimental scrapie. J Neuropathol Exp Neurol 2012; 71:140-7; PMID:22249459; http://dx.doi.org/ 10.1097/NEN.0b013e3182439519 [DOI] [PubMed] [Google Scholar]
- 30. Wemheuer WM, Benestad SL, Wrede A, Wemheuer WE, Brenig B, Bratberg B, Bratberg B, Schulz-Schaeffer WJ. PrPSc spreading patterns in the brain of sheep linked to different prion types. Vet Res 2011; 42:32; PMID:21324114; http://dx.doi.org/ 10.1186/1297-9716-42-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimberlin RH, Walker CA. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol 1978; 39:487-96; PMID:96212; http://dx.doi.org/ 10.1099/0022-1317-39-3-487 [DOI] [PubMed] [Google Scholar]
- 32. Foster JD, Dickinson AG. The unusual properties of CH1641, a sheep-passaged isolate of scrapie. Vet Rec 1988; 123:5-8; PMID:3140468; http://dx.doi.org/ 10.1136/vr.123.1.5 [DOI] [PubMed] [Google Scholar]
- 33. Lezmi S, Martin S, Simon S, Comoy E, Bencsik A, Deslys JP, Grassi J, Jeffrey M, Baron T. Comparative molecular analysis of the abnormal prion protein in field scrapie cases and experimental bovine spongiform encephalopathy in sheep by use of Western blotting and immunohistochemical methods. J Virol 2004; 78:3654-62; PMID:15016886; http://dx.doi.org/ 10.1128/JVI.78.7.3654-3662.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stack M, Jeffrey M, Gubbins S, Grimmer S, Gonzalez L, Martin S, Chaplin M, Webb P, Simmons M, Spencer Y, et al. Monitoring for bovine spongiform encephalopathy in sheep in Great Britain, 1998-2004. J Gen Virol 2006; 87:2099-107; PMID:16760414; http://dx.doi.org/ 10.1099/vir.0.81254-0 [DOI] [PubMed] [Google Scholar]
- 35. Biacabe AG, Jacobs JG, Bencsik A, Langeveld JP, Baron TG. H-type bovine spongiform encephalopathy: complex molecular features and similarities with human prion diseases. Prion 2007; 1:61-8; PMID:19164888; http://dx.doi.org/ 10.4161/pri.1.1.3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yokoyama T, Masujin K, Schmerr MJ, Shu Y, Okada H, Iwamaru Y, Imamura M, Matsuura Y, Murayama Y, Mohri S. Intraspecies prion transmission results in selection of sheep scrapie strains. PloS one 2010; 5:e15450; PMID:21103326; http://dx.doi.org/ 10.1371/journal.pone.0015450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang Y, Gielbert A, Jacobs JG, Baron T, Andreoletti O, Yokoyama T, Langeveld JP, Sauer MJ. All major prion types recognised by a multiplex immunofluorometric assay for disease screening and confirmation in sheep. J Immunol Methods 2012; 380:30-9; PMID:22498749; http://dx.doi.org/ 10.1016/j.jim.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 38. Jacobs JG, Sauer M, van Keulen LJ, Tang Y, Bossers A, Langeveld JP. Differentiation of ruminant transmissible spongiform encephalopathy isolate types, including bovine spongiform encephalopathy and CH1641 scrapie. J Gen Virol 2011; 92:222-32; PMID:20943889; http://dx.doi.org/ 10.1099/vir.0.026153-0 [DOI] [PubMed] [Google Scholar]
- 39. Gielbert A, Davis LA, Sayers AR, Hope J, Gill AC, Sauer MJ. High-resolution differentiation of transmissible spongiform encephalopathy strains by quantitative N-terminal amino acid profiling (N-TAAP) of PK-digested abnormal prion protein. J Mass Spectrom 2009; 44:384-96; PMID:19053160; http://dx.doi.org/ 10.1002/jms.1516 [DOI] [PubMed] [Google Scholar]
- 40. Langeveld JP, Jacobs JG, Erkens JH, Bossers A, van Zijderveld FG, van Keulen LJ. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet Res 2006; 2:19; PMID:16764717; http://dx.doi.org/ 10.1186/1746-6148-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Somerville RA, Hamilton S, Fernie K. Transmissible spongiform encephalopathy strain, PrP genotype and brain region all affect the degree of glycosylation of PrPSc. J Gen Virol 2005; 86:241-6; PMID:15604453; http://dx.doi.org/ 10.1099/vir.0.80251-0 [DOI] [PubMed] [Google Scholar]
- 42. Yull HM, Ritchie DL, Langeveld JP, van Zijderveld FG, Bruce ME, Ironside JW, Head MW. Detection of type 1 prion protein in variant Creutzfeldt-Jakob disease. Am J Pathol 2006; 168:151-7; PMID:16400018; http://dx.doi.org/ 10.2353/ajpath.2006.050766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parchi P, Strammiello R, Notari S, Giese A, Langeveld JP, Ladogana A, Zerr I, Roncaroli F, Cras P, Ghetti B, et al. Incidence and spectrum of sporadic Creutzfeldt-Jakob disease variants with mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol 2009; 118:659-71; PMID:19718500; http://dx.doi.org/ 10.1007/s00401-009-0585-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weissmann C, Li J, Mahal SP, Browning S. Prions on the move. EMBO Rep 2011; 12:1109-17; PMID:21997298; http://dx.doi.org/ 10.1038/embor.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dickinson AG, Stamp JT, Renwick CC, Rennie JC. Some factors controlling the incidence of scrapie in Cheviot sheep injected with a Cheviot-passaged scrapie agent. J Comp Pathol 1968; 78:313-21; PMID:4970193; http://dx.doi.org/ 10.1016/0021-9975(68)90007-8 [DOI] [PubMed] [Google Scholar]
- 46. Thackray AM, Hopkins L, Lockey R, Spiropoulos J, Bujdoso R. Emergence of multiple prion strains from single isolates of ovine scrapie. J Gen Virol 2011; 92:1482-91; PMID:21270287; http://dx.doi.org/ 10.1099/vir.0.028886-0 [DOI] [PubMed] [Google Scholar]
- 47. Harmeyer S, Pfaff E, Groschup MH. Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J Gen Virol 1998; 79(Pt 4):937-45 [DOI] [PubMed] [Google Scholar]
- 48. Demart S, Fournier JG, Creminon C, Frobert Y, Lamoury F, Marce D, Lasmézas C, Dormont D, Grassi J, Deslys JP. New insight into abnormal prion protein using monoclonal antibodies. Biochem Biophys Res Commun 1999; 265:652-7; PMID:10600476; http://dx.doi.org/ 10.1006/bbrc.1999.1730 [DOI] [PubMed] [Google Scholar]
- 49. Jacobs JG, Bossers A, Rezaei H, van Keulen LJ, McCutcheon S, Sklaviadis T, Lantier I, Berthon P, Lantier F, van Zijderveld FG, et al. Proteinase K-resistant material in ARR/VRQ sheep brain affected with classical scrapie is composed mainly of VRQ prion protein. J Virol 2011; 85:12537-46; PMID:21917981; http://dx.doi.org/ 10.1128/JVI.00448-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goldmann W, Hunter N, Foster JD, Salbaum JM, Beyreuther K, Hope J. Two alleles of a neural protein gene linked to scrapie in sheep. Proc Natl Acad Sci U S A 1990; 87:2476-80; PMID:1969635; http://dx.doi.org/ 10.1073/pnas.87.7.2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Slootstra JW, Puijk WC, Ligtvoet GJ, Langeveld JP, Meloen RH. Structural aspects of antibody-antigen interaction revealed through small random peptide libraries. Mol Divers 1996; 1:87-96; PMID:9237197; http://dx.doi.org/ 10.1007/BF01721323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Langeveld JP, Erkens JH, Rammel I, Jacobs JG, Davidse A, van Zijderveld FG, Bossers A, Schildorfer H. Four independent molecular prion protein parameters for discriminating new cases of C, L, and h bovine spongiform encephalopathy in cattle. J Clin Microb 2011; 49:3026-8; PMID:21677067; http://dx.doi.org/ 10.1128/JCM.01102-11 [DOI] [PMC free article] [PubMed] [Google Scholar]