Abstract
Aspergillus fumigatus is the most common mold involved in human infections. However, the number of non-fumigatus species able to cause disease is continuously increasing. Among them, Aspergillus lentulus is reported in hematological and cystic fibrosis patients and in those treated with corticosteroids. A. lentulus differs from A. fumigatus in some clinically relevant aspects such as virulence and antifungal susceptibility, showing high MICs to most antifungals. Previous studies proved that A. lentulus was pathogenic in immunocompromised mice, although the course of the infection was delayed compared to A. fumigatus. These differences could explain why A. lentulus is mostly found in mixed infections with A. fumigatus challenging the diagnosis and treatment. We used the alternative model host Galleria mellonella to compare virulence, host interaction, fungal burden and antifungal response when larvae were infected with A. fumigatus or A. lentulus alone, and with a mixture of both species. A. lentulus was pathogenic in G. mellonella but infected larvae did not respond to therapeutic doses of voriconazole. We were able to simultaneously detect A. fumigatus and A. lentulus by a multiplex Nested Real Time PCR (MN-PCR). Comparative analysis of larvae histological sections showed melanization of both species but presented a different pattern of immune response by haemocytes. Analysis of fungal burden and histology showed that A. lentulus survived in the G. mellonella despite the antifungal treatment in single and mixed infections. We conclude that the simultaneous presence of antifungal susceptible and resistant Aspergillus species would likely complicate the management of these infections.
Keywords: Aspergillus spp, cyp51A, Galleria mellonella infection model, mixed infection, one-step single tube real time PCR
Introduction
The incidence of invasive fungal diseases (IFD) together with the number of fungal species pathogenic for humans has increased in the past decade.1-3 Aspergillus fumigatus continues to be the most common fungi isolated from Aspergillus related mycosis. However, the epidemiology of these infections is changing with new emerging cryptic species becoming isolated and occasionally causing disseminated infections in immunocompromised hosts.3,4 The risk factors associated with these cryptic species are still undefined but it is known that they show remarkable high MICs to multiple antifungal agents.5,6 The reasons for this change in fungal populations, as well as the emergence of drug resistance, are still poorly understood.
Aspergillus lentulus, one of these sibling species belonging to the section Fumigati, can be considered as an uncommon fungal pathogen for humans. However, it has been increasingly reported in severely immunocompromised patients such as hematological and cystic fibrosis patients, those suffering from chronic obstructive pulmonary disease (COPD) or other populations receiving high doses of corticosteroids.7-10 Invasive aspergillosis caused by A. lentulus is associated with high mortality and clinical isolates of this species show high MICs to azoles, echinocandins and amphotericin B.6-9,11 Resistance to voriconazol (VRC) is especially worrisome considering that this antifungal is the primary drug used to treat invasive aspergillosis. A. lentulus cannot be distinguished by classical morphological methods from other Fumigati section species including A. fumigatus. To solve this problem, molecular methods based on amplification and sequencing of the β-tubulin, rodlet or calmodulin genes are currently used in order to differentiate them at species level. Colonies of A. lentulus appear to be less sporulated and are white colored compared to A. fumigatus. Pathogenicity of A. lentulus has been proven in immunocompromised mice although the course of infection was delayed compared to A. fumigatus when the same intranasally administered inocula size was used.12 This difference in the developmental fungal growth in vivo might explain why A. lentulus is frequently isolated in mixed infections with A. fumigatus or other Aspergillus spp.7,9 The simultaneous co-infection by multiple fungal species could represent a possible threat as A. lentulus is often misidentified and shows resistance to most antifungals in clinical use. In addition, population dynamics on a fungal mixed infection and species interaction in the host and in the environment remains entirely unexplored.
To date, murine models are considered to be the gold standard to study fungal pathogenesis and analyze efficacy of antifungal treatment. However, economic and logistical factors, together with ethical considerations, are limiting the use of mammals in infection experiments. To overcome these limitations, an alternative approach using different invertebrate infection models has been developed, including amoebae, Caenorhabditis elegans, Drosophila melanogaster, and larvae of the wax moth Galleria mellonella.13 All of them are inexpensive, ethically approved and do not require specialized facilities for maintenance. Compared to other invertebrates, G. mellonella offers several advantages that include: the size of the larvae which facilitates easy handling; and the possibility of injecting a defined inoculum and drug concentration. Furthermore, in contrast to other invertebrate hosts, G. mellonella larvae can be maintained at temperatures up to 37°C, equivalent to the temperature in mammalian hosts.14,15
In this study we first optimized a one-step diagnostic tool based on multiplex nested PCR (MN-PCR) for simultaneous detection of A. fumigatus and A. lentulus that allowed fungal quantification from tissue samples. We then established a host model of mixed infection with A. fumigatus and A. lentulus in the alternative host G. mellonella. The model was used to evaluate differences in virulence, fungal burden, pathogenesis and antifungal drug response in presence of Aspergillus species mixtures in vivo.
Material and Methods
Strains, media and growth conditions
For the standardization and validation of the one step multiplex nested-PCR (MN-PCR) a total of 20 clinical strains for each species (20 A. fumigatus and 20 A. lentulus) were employed. All strains belonged to the Mycology Reference Laboratory collection, except the strain AF1416 that was kindly donated by Dr. J.A. Calera. The specificity of the technique was assessed with other fungal species with clinical origin: Aspergillus flavus (CM-2669), Aspergillus terreus (CM-2013), Fusarium verticillioides (CM-2975), Fusarium oxysporum (CM-2914), Scedosporium prolificans (CM-1627), Scedosporium apiospermum (CM-3169), Cryptococcus neoformans (CL-2132), Rhizopus oryzae (CM-3020), Rhizomucor variabilis (CM-2437), Candida albicans (ATCC64551) and Candida glabrata (CL-5533). In addition, human and murine genomic DNAs (Promega, Madrid, Spain) were included.
A. fumigatus (akuBKU80),17 CM-237,18 AF1416 and A. lentulus (CM-1290) strains were selected for our infection model and further analysis.
Fungal strains were routinely grown at 37°C on different media: GYEP (2% glucose, 0.3% yeast extract, 1% peptone), Sabouraud (2% glucose, 1% mycopeptone) and potato dextrose agar. Minimal media (MM) containing 5 mM ammonium tartrate, 1% (w/v) glucose, and 2% (v/v) salt solution was used for phenotypic testing.19
Radial growth was measured for 5 d by point inoculation of 5 × 104 spores of A. fumigatus and A. lentulus on MM, at 37°C. Aspergillus species susceptibility profiles to voriconazol6 was confirmed by E-test according to the manufacturer's instructions (TecLaim S.A).
DNA extraction
Genomic DNA from Aspergillus spp and other fungal strains were isolated using a rapid extraction procedure.20 Two μL of extracted DNA were used for each PCR reaction. DNA extraction from lung tissue samples used to validate the PCR assay, was performed using the QiAmp DNA Kit (Qiagen, Izasa, Madrid, Spain) following manufacturer recommendations. Elution was performed in 50 μL of elution buffer. DNA from G. mellonella was isolated from 0.85 % saline homogenized larvae as follows: DNA from 25 mg of homogenized larvae was isolated using the QiAmp DNA Kit for tissues, following manufacturer's instructions and eluting in 50 μL. All samples were stored at –20°C and they were allowed to thaw at room temperature before testing.
Multiplex Nested-PCR assay
Primers and Probes design
Primers and molecular beacon probes were designed on the basis of the nucleotide sequence of the cyp51A gene from 10 strains of each A. fumigatus and A. lentulus. The Beacon Designer 7.0 software (Premier Biosoft, Palo Alto, CA) was employed to design primers and probes for the MN-PCR assay. Selected primers and probes were subjected to a blast search into the GenBank sequence database (http://www.ncbi.nih.gov/Genbank/) to avoid cross-homology with other organisms. The primers for the first PCR were designed with a primer-annealing temperature higher than the primers for the second PCR segment (Table 1).21 The first PCR segment at that temperature did not allow the primers for the second PCR to anneal to the template. The outer primers were degenerated to hybridize with all species belonging to the Fumigati section to be used in future assays. The Molecular Beacon probes were designed based on the species specific cyp51A intron sequences (Supplementary Fig. 1) and were labeled with 5′ 6-carboxifluorescein (FAM) for A. fumigatus and 4,7,2′, 4′, 5′, 7′-hexacloro-6-carboxyfluorescein (HEX) for A. lentulus (Sigma-Genosys, Spain). Both were labeled with Black Hole Quencher1 (BHQ1) in the 3′ end as quencher.
Table 1.
Primers and Probes Sequences designed for the Multiplex Nested Real Time PCR assay.
| Primer Name | Sequence (5’-3’)a |
|---|---|
| OliNestCyp51A1 (Forward. First PCR) | CTYGCTCAATGTYGTTTATCAAYTATTCTTYCGGC |
| OliNestCyp51A2 (Reverse. First PCR) | CGCMGACTGAGTYAAGCCGTACTTGATGAACTTC |
| OliCyp51int-3 (Forward, second PCR) | CAGAACCGCCAATGGTCT |
| OliCyp51int-2 (Reverse, second PCR) | GACRTCCTTGAGCTTGCC |
| Cyp51int-AF (A.fumigatus probe) | FAM-CGCGATCAGATTGTAGTTTGACATTCATTCCTGGGGATCGCG-BHQ1 |
| Cyp51int-AL (A. lentulus probe) | HEX-CGCGATCAAGCTAGCGTCTGACATTTATCCCGGGATCGCG-BHQ1 |
Underlining regions indicates the PCR target sequence
PCR conditions and standardization in vitro
PCR reactions were undertaken in a LC480 unit (Roche Diagnostics, Madrid, Spain). The SensiMix (dT) kit 2x (Bioline, Ecogen, Spain) was used following manufacturer's instructions. The PCR was performed in a total volume of 20 μL. The PCR mixture contained 0,05 μM each of the first-PCR primers, 0,5 μM each of the second-PCR primers, 0,4 μM of both probes, 3mM MgCl2 and a 2 μL aliquot of DNA from the extracted sample. Cycling conditions included a first step for pre-incubation (activation of the enzyme) and denaturation of the template DNA at 95°C during 10 minutes. The first PCR consisted on only 10 cycles which included 25s at 95°C, 30s at 58°C and 5s at 72°C. The first PCR was immediately followed by the second PCR: 40 cycles which included 25s at 95°C, 30s at 52°C and 5s at 72°C. Quantification standards were run in conjunction which each set of samples as well as negative controls.
Standard curves were constructed with PCR results from 5 repetitions of different dilutions of A. fumigatus (CM-237) and A. lentulus (CM-1290) genomic DNA. Dilutions ranged from 20 ng to 2 fg DNA/20 uL of reaction. A regression line (Supplementary Fig. 2) was constructed by plotting the Log10 concentration of DNA vs. its corresponding threshold cycle value (Ct). If this curve exhibited a linear regression coefficient value of >0.980, the standard curve was then used to determine the efficiency of the PCR, sensitivity and reproducibility of the assay. In order to evaluate the specificity, 2 ng of DNA/20 μl from the species mentioned before (see Control strains) as well as human and mouse genomic DNA were included in the PCR assay.
Data were analyzed using LC480 software (Roche Diagnostics, Madrid, Spain) and a color compensation experiment was created to avoid signal overlapping between both fluorophores. Each PCR run contained both positive and negative controls consisting of water and different concentrations of genomic DNA from A. fumigatus or A. lentulus.
Murine infections
Murine models for lung sample collection were performed according to the models previously described.22,23 Murine infections were performed in compliance with Real Decreto 223/1988 for the protection of experimental animals and the Project License CBA PA_02_2013. The experiments were carried out with 8 mice per group. Leukopenic and corticosteroid treated mice were infected by intranasal instillation of 105 spores/mice conidia in 30 μl of saline solution. Survival was monitored over a 14 day period, cages were checked twice daily and those mice that developed signs of severe respiratory distress and reduction in body weight were sacrificed. Control groups (not infected and not immunosuppressed, and not infected but immunosuppressed) had no symptoms of infection and were sacrificed at the end of the experiment.
Whole lungs were collected and homogenized in saline. An aliquot of this homogenate was plated onto Sabouraud plus chloramphenicol and gentamicin for fungal infection validation. The rest of the homogenate was stored at −20°C until DNA extraction was performed.
Galleria mellonella survival assay
Wax-moth larvae were infected with A. fumigatus strain akuBKU80 or A. lentulus CM-1290, or both in the mixed infection experiments. Wax moth larvae killing assays were carried out as described previously.24,25 Briefly, groups of 10 larvae (0.3–0.5 g, Alcotan S.L., Valencia, Spain) were inoculated with 10 μL of a 107 conidia/ml suspension in PBS into the haemocoel, so the final inoculum in each group was 105 conidia per larva. Mixed infected larvae were inoculated with A. fumigatus and A. lentulus (ratio 1:1) to a final concentration of 105 conidia per larvae.
Within two hours of the infection, 5 μl of voriconazole (VRC, Pfizer SA) antifungal solution (VRC: 400 μg/ml) (2 μg/larva equivalent to 10 mg/kg therapeutic dose of VRC) was injected into a different proleg using the same technique. After that, caterpillars were incubated at 37˚C for 10 d. Mortality, defined by lack of movement in response to stimulation and melanization of the cuticle, was recorded daily. Control groups were included: untouched, pierced, PBS and antifungal toxicity. Each experiment was performed in duplicate at least 3 times and the results were reported as mean values. To simplify some of the Figures, PBS control, toxicity and untouched groups were omitted.
Histopathological analysis of infected G. mellonella
Larva histology was performed as described before.26 Three larvae per group were longitudinally sectioned and fixed for 24 h in 4% buffered formalin and dehydrated with increasing concentrations of ethanol (70%, 80%, 90%, 96% and 100%). The samples were then treated with xylene and paraffin embedded. Tissue sections of 5 microns were stained with periodic acid Schiff (PAS) and sections examined with a Leica DMI 3000B microscope.
Fungal burden quantification in G. mellonella
In order to assess fungal burden as a quantitative criterion for virulence and antifungal response, the MN-PCR assay described before was performed on total genomic DNA extracted from homogenized G. mellonella at different time points (2, 4 and 6 d post infection). DNA was extracted as previously described. Fungal burden was expressed as ng of fungal DNA / gram of larvae.
Statistical analysis
Statistical analyses were performed with GraphPad Prism, version 5 Project (GraphPad Software, San Diego, CA). The statistical significance of variances between fungal burdens was calculated by using a nonparametric Mann-Whitney t test. A p value <0.01 was considered significant.
Kaplan-Meier survival curves were analyzed by using a log-rank (Mantel-Cox) test for significance. A p value <0.01 was considered significant.
Nucleotide sequence accession numbers
The full nucleotide sequences of the cyp51A genes used in this work appear in the GenBank nucleotide sequence database under Accession Ns. GU479991 for A. lentulus cyp51A and Accession Ns. AF338659 was the reference sequence used for A. fumigatus.
Results
Detection of A. fumigatus and A. lentulus by one step Multiplex Nested Real Time PCR
As accurate species identification could influence the infection outcome, a one-step diagnostic tool based on a multiplex nested PCR (MN-PCR) was optimized for differential detection of A. fumigatus and A. lentulus. Two sets of primers were designed for the first and second round of amplification including 2 degenerated outer primers, 2 specific inner primers and 2 Molecular Beacon probes labeled with FAM and HEX specific for A. fumigatus and A. lentulus, respectively. The design was based in a region of the cyp51A gene that contained a common intron of 71 bp. This region allowed the design of common primers for all the Aspergilli of the section Fumigati and specific probes for each species within the group (Fig. S1).
The detection limit of the designed PCR was calculated at 200 fg of DNA in 20 μl of PCR reaction mixture for both A. fumigatus and A. lentulus. The efficiency for both PCR amplifications was higher than 86% with coefficients (r2) of 0.99 (Fig. S2). The averages of coefficients of variation for all DNA concentrations were 3.4% for A. fumigatus and 2.7% for A. lentulus. Once the PCR conditions were optimized, the developed MN-PCR assay was validated on 40 clinical strains of A. fumigatus and A. lentulus with a specificity of 100%. Sequencing of all PCR amplified fragments confirmed the specificity. No signal was detected when 2 ng of DNA from other fungi (different species of Aspergillus, Fusarium, Candida, mucorales etc…) and genomic DNA from human and mouse were also tested. In addition, no cross-reaction was observed between A. fumigatus and A. lentulus.
In order to validate the MN-PCR assay in vivo, first we used an experimental model of pulmonary aspergillosis using cyclophosphamide treated mice.22 Mice were infected with A. fumigatus (CM-237) or A. lentulus (CM-1290) and the infection was confirmed with positive cultures from lung tissues. A total of 8 mice inoculated with A. fumigatus had proven lung infection and positive PCR results from lung tissue. Similarly, PCR was positive in 13 of 14 lung samples (93%) from mice with proven infection with A. lentulus. The average of DNA concentration detected per μL in samples was 1.5 × 104 fg.
A second type of cortisone immunosuppressed murine model, closer to the cystic fibrosis patients, was performed.23 In this model the PCR assay was able to detect 100% of samples from mice with proven infection infected with A. fumigatus (10/10) or A. lentulus (10/10).
All together, the MN-PCR technique was positive in 41 of 42 samples (98%) and was able to distinguish both species in the 2 immunosuppressed mice models.
Differential pathogenesis of A. lentulus and A. fumigatus in G. mellonella
Prior to any assay in vivo, the growth rate and the antifungal susceptibility profile of A. fumigatus versus A. lentulus strains were compared in vitro. There were no differences in terms of colony size and radial growth between both species. As shown before,6 A. lentulus was resistant to VRC (MIC of 4 μg/L) while A. fumigatus was susceptible (MIC 0.5 μg/L) (Fig. S3). The virulence in the alternative host Galleria mellonella was determined by infecting the larva with an inoculum of each Aspergillus spp (1.5 × 105 cfu/larva). When A. fumigatus was used, 100% of infected larvae died between the 3rd and 4th day post infection. However, virulence of A. lentulus was slower and only 80% of larvae had died by day 10 post infection (Fig. 1A). The survival pattern of infection mimics that observed using the same inocula of Aspergillus spp in immunocompromised mice12 (Fig. S4).
Figure 1.
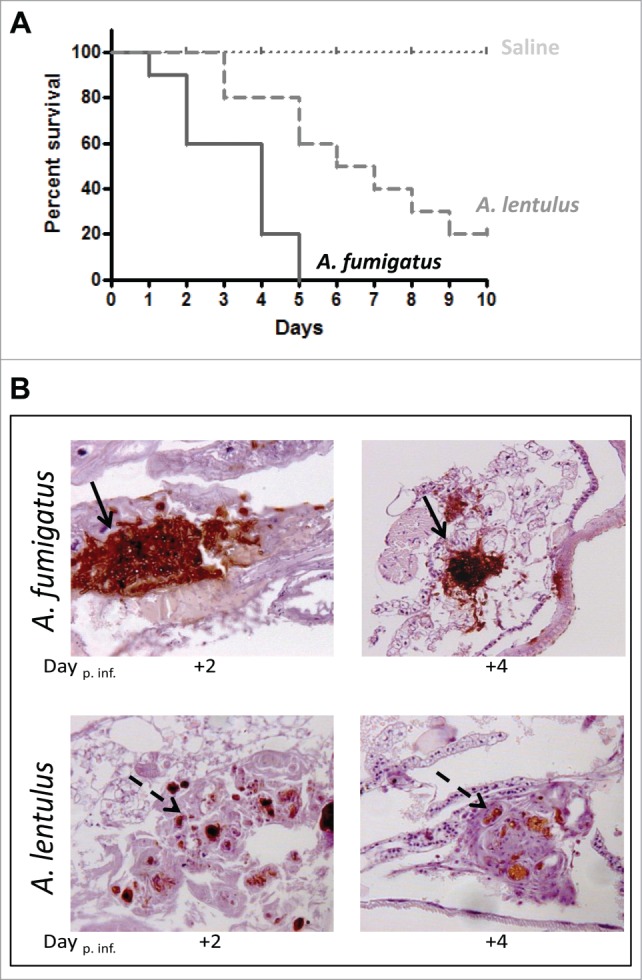
(A) Survival rate of G. mellonella infected with A. fumigatus (CM-237) or A. lentulus (CM-1290) (105 spores/larvae). Control group (dotted line) represent survival of larvae inoculated with saline only. (B) Histopathology of G. mellonella infected with A. fumigatus and A. lentulus, at different times points of the infection (Day +2 and +4). 20 x magnification, PAS stained. Differences between the A. fumigatus and A. lentulus infection pattern: larvae showed branched invasive hyphal growth of A. fumigatus (steady arrows) while A. lentulus was encapsulated by immune cells (dash arrows).
In order to explore the effect of Aspergillus infection on G. mellonella physiology, the infected larvae were fixed and paraffin-embedded sections were stained with PAS and evaluated for histological changes at different time points post infection (Fig. 1B). Histological analysis revealed differences between the A. fumigatus and A. lentulus infection pattern. Larvae showed branched invasive hyphal growth of A. fumigatus while A. lentulus was encapsulated by immune cells. Hemocytes with clearly visible nodule melanization were observed in nodules attached to organ structures and the fungus (Fig. 1B).
Voriconazole efficacy in an in vivo model of infection with Aspergillus spp using G. mellonella
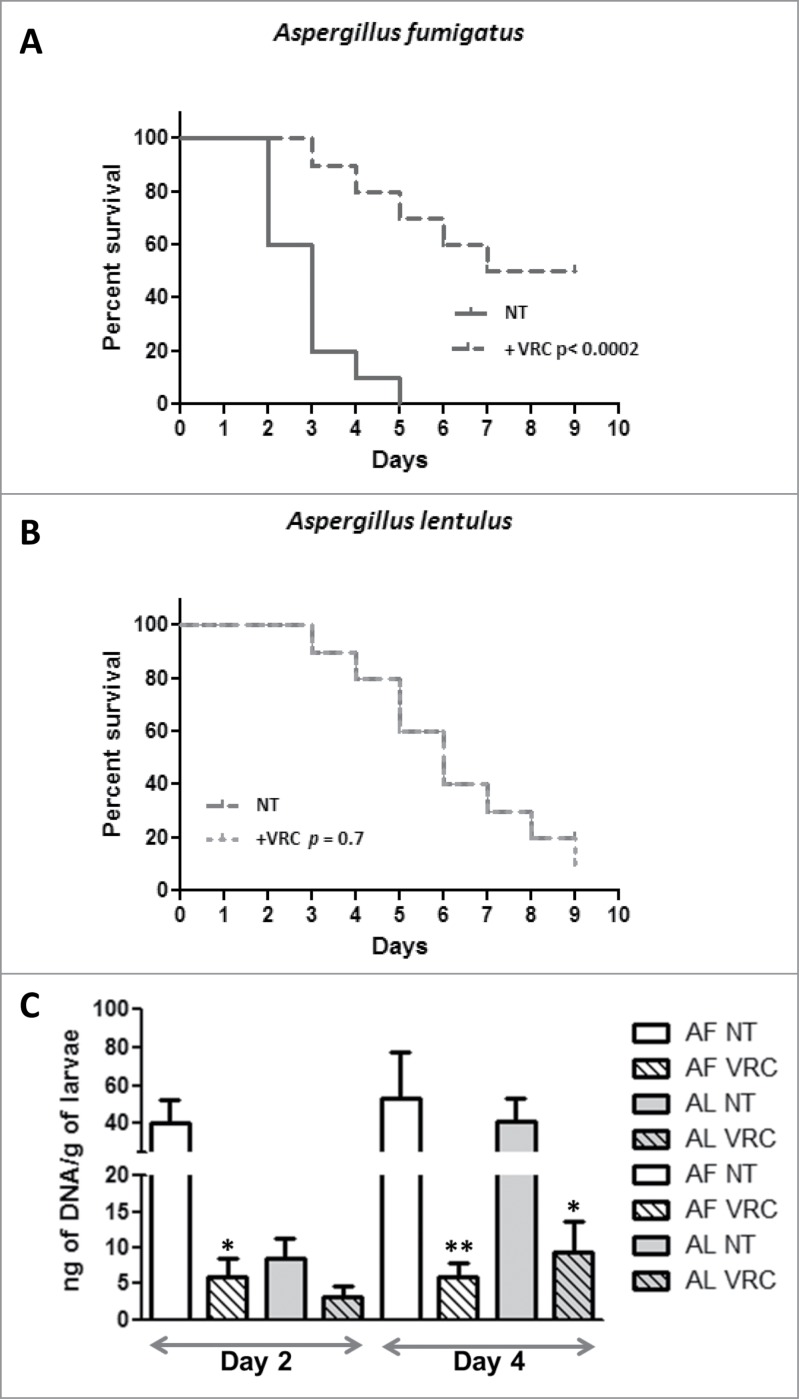
The G. mellonella alternative model of infection was used to compare the efficacy of antifungal agents to treat A. fumigatus or A. lentulus infected larva. Groups of 20 larvae were infected with 1.5 × 105 spores per larvae and treated groups were inoculated with therapeutic doses of VRC (4 μg/larva equivalent to 10 mg/Kg/day) within 2 hours of infection. Virulence was reported as survival rate against time (Fig. 2A).
Figure 2.
(A) Killing rate comparison of G. mellonella infected with A. fumigatus with or without VRC treatment. (B) Killing rate comparison of G. mellonella infected with A. lentulus with or without VRC treatment. (C) Fungal burden determination in G. mellonella infected with A. fumigatus (AF) or A. lentulus (AL), untreated (NT) or treated with VRC. Statistical significance by unpaired t test of VRC treated vs. untreated data for each time point is marked as (*P < 0.01 or **P < 0.002).
The results showed that at therapeutic doses of VRC, larvae infected with azole susceptible A. fumigatus (akuBKU80) strain significantly improved survival in relation to the untreated group (P < 0.0002). In contrast, when G. mellonella were infected with the azole resistant A. lentulus strain (CM-1290), the percentage of survival showed no significant differences between this group and the respective untreated group (p = 0 .7) (Fig. 2B).
To test VRC efficacy of A. fumigatus or A. lentulus in vivo, we used the developed MN-PCR to examine the fungal burden of G. mellonella infected with each Aspergillus strains. Time points were set at 2 and 4 d post infection. The analysis identified a significant decrease (P < 0.01) in the fungal burden retrieved from VRC treated larvae infected with A. fumigatus compared to A. lentulus infected larvae, and in comparison with untreated animals (Fig. 2C) at both time points.
Histopathology analysis showed that when the larvae were infected with A. fumigatus and treated with VRC, the fungus was almost cleared with tissues appearing healthy. In contrast, A. lentulus CM-1290 strain persisted in the tissues even after 6 d of infection (Fig. 3).
Figure 3.
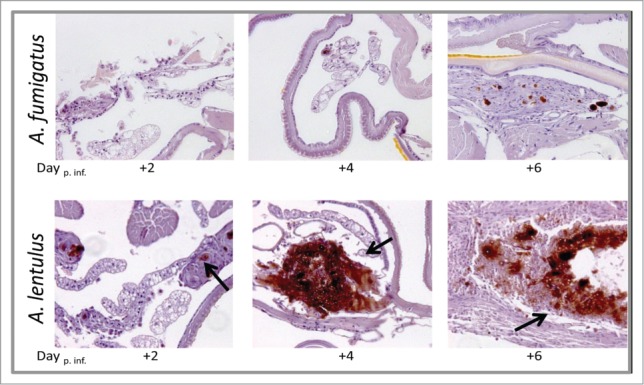
Histopathology of G. mellonella infected with A. fumigatus and A. lentulus and treated with VRC, at different time points of the infection (Day +2, +4 and +6). 20 x magnification, PAS stained. Upper panel: when the larvae were infected with A. fumigatus and treated with VRC, the fungus was almost cleared with tissues appearing healthy. Lower panel: A. lentulus CM-1290 strain persisted in the tissues with an increase of fungal burden after 4 and 6 d post-infection (black arrows).
Mixed infection model
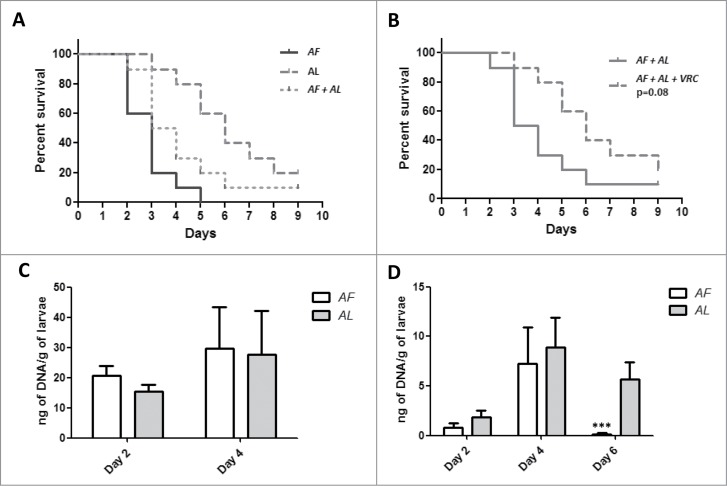
A model of A. fumigatus and A. lentulus mixed fungal infection (1:1) in G. mellonella was used to evaluate the influence of intrinsically resistant species on the antifungal treatment outcome. Using the previously described model, larvae were infected with an equal amount of spores of both species (5 × 104 spores per larvae of each Aspergillus species). VRC response (4 μg/larva equivalent to 10 mg/Kg) of the G. mellonella larvae infected with the mixture of A. fumigatus and A. lentulus was monitored by larvae survival rate and fungal burden using MN-PCR.
When the larvae were infected with a combination of both species, survival of infected larvae was intermediate to those of each single species infection respectively (Fig. 4A). In addition, Aspergillus spp mixed infected G. mellonella did not respond to therapeutic doses of VRC (p = 0 .08) (Fig. 4B).
Figure 4.
(A) Survival rate of G. mellonella infected with a mixture of A. fumigatus (AF):A. lentulus (AL) (1:1) compared to single infections. (B) Comparative analysis of VRC treated and untreated larvae (This data is the same that was used for Fig. 4A). (C) Fungal burden determination in G. mellonella infected with a mixed inocula of both A. fumigatus and A. lentulus. (D) Fungal burden determination and VRC treatment efficacy in a mixed fungal infection. Statistical significance by unpaired t test of A. fumigatus versus A. lentulus at 6 d is marked as (***P <0.003).
Analysis of fungal burden showed no differences of fungal DNA per larvae between 2 and 4 d for both Aspergillus species (Fig. 4C). When VRC was administered fungal burden dropped for A. fumigatus at both, 2 and 4 d post infection, compared to the untreated group (P < 0.01)( (Fig. 4C and D). In the case of A. lentulus this decrease was statistically significant at 2 d (p = 0 .002) but not at 4 d (p = 0 .35) (Fig. 4C and D). The fungal burden was maintained at 6 d with no differences when it was compared to fungal burden of VRC treated larvae at 4 d post infection (p = 0 .44)(Fig. 4D).
Discussion
Mixed infections are expected to occur in nature where interaction between co-infecting microorganisms will depend on the host environment. The clinical relevance and the epidemiology of fungal mixed infections is relatively unknown but recent reports highlight the fact that these might be underestimated.27 Mixed infections that include more than one fungal species are reported in superficial tissues, such as in onychomycosis infections, as well as in immunocompromised hosts.28 Remarkably high death rates (75%) among bone marrow transplant patients infected with Aspergillus spp and among patients infected with Aspergillus spp. and Candida spp (100%) have been reported.29 Also, a report of a mixed infection of the lung with C. albicans and A. fumigatus in a patient on prolonged steroid therapy was recently published.30 Several case reports have described invasive aspergillosis caused by mixed infection with A. flavus and A. fumigatus31 or by cryptic Aspergillus species.32 These studies are concluding that mixed infections are very complex and they could challenge current fungal diagnosis. Furthermore, they are often associated to lack of response to antifungal treatment and to bad outcome, and normally linked to high mortality rates. Among mixed infections cryptic species are especially important because fungal identification at species level is not straightforward. Additional methods are required, such as molecular identification, and this involve a longer time for diagnosis. In addition, the antifungal profile of Aspergillus species complex frequently shows high MICs to multiple antifungals.5,6
The goal of our study was to develop and validate in tissue samples a PCR based assay able to distinguish between A. fumigatus (azole susceptible) and A. lentulus (azole resistant) as well as to standardize an alternative mixed infection model in order to correlate in vitro susceptibility profiles with antifungal drug response.
Aspergillus fumigatus accounts for most of the infections produced by filamentous fungi and it is usually susceptible to azole drugs, while A. lentulus, a species belonging to section Fumigati, has high MICs to multiple antifungals including VRC and AmB.6 A correct and early diagnosis leading to a prompt antifungal therapy is one essential step to improve clinical outcome. In this sense, molecular techniques have been proven to be very useful for the diagnosis of fungal infections, and most of them are based on the detection of the Internal transcriber Spacer (ITS) regions of the rDNA.33 However, ITS sequences cannot be used to differentiate species within the A. fumigatus complex.34 This is a great disadvantage as, until now, only single locus genes have been able to discriminate between those species. Among them, the more usual targets have been β-tubuline, calmodulin, and rodlet A genes.6,35 The use of single copy genes shows better discriminatory power but have the inconvenience of lack of sensitivity when clinical samples such as blood or BALs are used. In order to improve this, we developed a multiplex quantitative one-step nested PCR (MN-PCR) targeting the intron sequences of the azole target, 14-α sterol demethylase (cyp51A).
The developed MN-PCR is useful for the correct identification of A. fumigatus and A. lentulus. Moreover, it allows discrimination of A. fumigatus and closely related species and it can be implemented for the correct identification of other Aspergillus species in the section Fumigati in clinical specimens (under way in our laboratory). Also, the assay has been validated in cultured strains and in lung samples from 2 experimental mice models of invasive aspergillosis with excellent sensitivity (98%) and specificity (100%).
The G. mellonella model was used to explore Aspergillus spp fungal virulence and the in vivo correlation with in vitro antifungal susceptibility data. Results showed a high correlation in terms of survival rate between the murine and the wax moth model of aspergillosis12 (Supplementary Fig. 4). Similarly to the murine model, A. lentulus was pathogenic in G. mellonella, although survival of infected larvae was prolonged compared to larvae infected with A. fumigatus. Those differences in the virulence pattern would suggest a more rapid or invasive growth characteristic of A. fumigatus in vivo.
Also, comparative analysis of tissue sections of G. mellonella infected with each Aspergillus spp showed a different pattern. Larvae melanization occurred with both species at all time points, but recruitment of haemocytes differed. Branched invasive hyphal growth of A. fumigatus was observed in untreated larvae while the pattern of infection with A. lentulus was encapsulated hypha by immune cells producing nodules. This data suggests that A. fumigatus and A. lentulus might use different strategies to respond to host defenses. While A. fumigatus escapes from haemocytes, germinates and quickly develops in the host, A. lentulus is encapsulated by immune cells. No statistical differences were found in terms of haemocyte density between the 2 species in any of the time points, although it seems that there is a delay of the immune response by haemocytes against A. lentulus compared to A. fumigatus (data not shown). Changes in haemocyte density following microbial challenge have been previously observed36 correlating high haemocyte concentration with longer survival rates.
Galleria mellonella presents certain benefits in comparison with other non-mammalian models of infection.14 Previous studies have documented that G. mellonella is a valid model to evaluate the microbial virulence and/or the efficacy of different antimicrobial agents in infections caused by different bacteria37 and fungi, such as C. neoformans,38,39 Candida spp,40,41 Histoplasma spp.42 and A. fumigatus.25 In addition, the G. mellonella model appears to be a promising approach to evaluate fitness of drug-resistant strains.43
Since VRC is the first line antifungal used to treat aspergillosis, we also analyzed the in vivo response of G. mellonella infected with A. fumigatus or A. lentulus and treated with this antifungal. G. mellonella was a useful model in the evaluation of the efficacy of VRC at concentrations equivalent to the therapeutic doses used in humans. The results correlated very well with the antifungal susceptibility phenotype shown in vitro. While VRC was capable of improving the survival of larvae infected with azole susceptible A. fumigatus, this drug was ineffective for larvae infected with azole resistant strains (A. lentulus).
To further investigate the effect of antifungals during infection, we analyzed fungal burden of Aspergillus infected larvae by the MN-PCR described before. MN-PCR showed progressive increases in the Aspergillus burden during the time of infection at 2 and 4 d post infection, although it was not significant, when larvae were not treated with VRC. Antifungal treatment significantly reduced A. fumigatus DNA per gram of larvae at 2 d of infection. Reduction of fungal burden at 4 d of infection was observed for larvae infected with A. fumigatus and A. lentulus although that decrease was more prominent for A. fumigatus than for A. lentulus (P < 0.002 and P < 0.01 respectively) (Fig. 2C). These results suggest that isolates with higher VRC MICs might show a poorer response to antifungal treatment in vivo.7
Following the same steps, we used the model of G. mellonella to evaluate the efficacy of VRC in the mixed infection model. A similar approach to evaluate a mixed infection of Rhizopus oryzae and A. fumigatus using corticosteroid-immunosuppressed mice has been used to explore if the exposure to antifungals could provoke a selection pressure for breakthrough mucormycosis during infection.44 Results of survival analysis and fungal burden of mice infected with both species showed that VRC pre-exposure in vitro promotes breakthrough infection in vivo with R. oryzae and increases mortality due to R. oryzae.44
In our model, the results show that at therapeutic doses VRC did not significantly improved larval survival when they were co-infected with equal amounts of A. fumigatus and A. lentulus. In addition, quantitative analysis of fungal burden at 6 d post infection demonstrated A. lentulus selection by VRC in the mixed infection. This suggests that in a mixed infection with A. fumigatus and A. lentulus the treatment with VRC may facilitate invasive growth of A. lentulus. The eventual domination of A. lentulus over A. fumigatus is consistent with concepts of antimicrobial selection, where suppression of more susceptible populations presages the emergence of resistant pathogens with reduced fitness cost.45 Similarly, with malaria it has been proposed that when sensitive and resistant parasites co-infect the same host individuals, drug use would further increase the relative fitness of drug-resistant clones by removing drug-sensitive competitors.46
It may seem that there is a discrepancy between the decrease in fungal burden and the lack of response in survival of G. mellonella infected with A. lentulus in the single and the mixed infection. It has been described that A. lentulus VRC MICs range from 4 to 6 mg/L.6 Therefore, the reduction of fungal burden of G. mellonella infected with A. lentulus and treated with VRC compared to the untreated ones might be explained because VRC is able to eliminate or to reduce fungal growth to some extend but not enough to improve survival of the Galleria at the VRC concentration that was tested. We cannot discard that A. lentulus would respond in vivo if higher concentrations of VRC are used.
Altogether, this data indicates that G. mellonella is a simple model that can also be used to evaluate competition and in vivo fitness in mixed infections by fungi. Moreover, we were able to simultaneously detect 2 species closely related such as A. fumigatus and A. lentulus by using real time PCR, showing a promising tool for clinical detection of Aspergillus section Fumigati in single or mixed infections. Given the differences in antifungal susceptibility between both species, antifungal pressure against A. fumigatus might be relevant for selective infection by A. lentulus. In conclusion, the presence of antifungal resistant species would likely complicate the management of fungal infections, thus representing a threat for patients under long-term azole treatment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Gema del Rio for technical assistance.
Funding
EM was supported by Fondo de Investigacion Sanitaria (FIS:PI12_02376). LA-F was funded by Fondo de Investigación Sanitaria with a Miguel Servet fellowship (FIS:CP11/00026).
References
- 1. Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, Ito J, Andes DR, Baddley JW, Brown JM, et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001-2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis 2010; 50: 1091-100; PMID:20218877; http://dx.doi.org/ 10.1086/651263. [DOI] [PubMed] [Google Scholar]
- 2. Pagano L, Akova M, Dimopoulos G, Herbrecht R, Drgona L, Blijlevens N. Risk assessment and prognostic factors for mould-related diseases in immunocompromised patients. J Antimicrob Chemother 2011; 66 Suppl 1: i5-14; PMID:21177404; http://dx.doi.org/ 10.1093/jac/dkq437. [DOI] [PubMed] [Google Scholar]
- 3. Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, et al. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin Infect Dis 2010; 50: 1101-11; PMID:20218876; http://dx.doi.org/ 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- 4. Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot Cell 2005; 4: 625-32; PMID:15755924; http://dx.doi.org/ 10.1128/EC.4.3.625-632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alastruey-Izquierdo A, Mellado E, Pelaez T, Peman J, Zapico S, Alvarez M, Rodríguez-Tudela JL, Cuenca-Estrella M; FILPOP Study Group . Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrob Agents Chemother 2013; 57: 3380-7; PMID:23669377; http://dx.doi.org/ 10.1128/AAC.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based identification. Antimicrob Agents Chemother 2008; 52: 1244-51; PMID:18212093; http://dx.doi.org/ 10.1128/AAC.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zbinden A, Imhof A, Wilhelm MJ, Ruschitzka F, Wild P, Bloemberg GV, Mueller NJ. Fatal outcome after heart transplantation caused by Aspergillus lentulus. Transpl Infect Dis 2012; 14: E60-E63; PMID:22988985; http://dx.doi.org/ 10.1111/j.1399-3062.2012.00779.x. [DOI] [PubMed] [Google Scholar]
- 8. Alhambra A, Catalan M, Moragues MD, Brena S, Ponton J, Montejo JC, del Palacio A. Isolation of Aspergillus lentulus in Spain from a critically ill patient with chronic obstructive pulmonary disease. Rev Iberoam Micol 2008; 25: 246-9; PMID:19071895; http://dx.doi.org/ 10.1016/S1130-1406(08)70058-5. [DOI] [PubMed] [Google Scholar]
- 9. Montenegro G, Sanchez PS, Jewtuchowicz VM, Pinoni MV, Relloso S, Temporitti E, Iovannitti CA, Mujica MT. Phenotypic and genotypic characterization of Aspergillus lentulus and Aspergillus fumigatus isolates in a patient with probable invasive aspergillosis. J Med Microbiol 2009; 58: 391-5; PMID:19208894; http://dx.doi.org/ 10.1099/jmm.0.005942-0. [DOI] [PubMed] [Google Scholar]
- 10. Symoens F, Haase G, Pihet M, Carrere J, Beguin H, Degand N, Mely L, Bouchara JP. Unusual Aspergillus species in patients with cystic fibrosis. Med Mycol 2010; 48 Suppl 1: S10-S16; PMID:21067321; http://dx.doi.org/ 10.3109/13693786.2010.501345. [DOI] [PubMed] [Google Scholar]
- 11. Balajee SA, Weaver M, Imhof A, Gribskov J, Marr KA. Aspergillus fumigatus variant with decreased susceptibility to multiple antifungals. Antimicrob Agents Chemother 2004; 48: 1197-203; PMID:15047520; http://dx.doi.org/ 10.1128/AAC.48.4.1197-1203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mellado E, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. Role of Aspergillus lentulus 14-α sterol demethylase (Cyp51A) in azole drug susceptibility. Antimicrob Agents Chemother 2011; 55: 5459-68; PMID:21947395; http://dx.doi.org/ 10.1128/AAC.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desalermos A, Fuchs BB, Mylonakis E. Selecting an invertebrate model host for the study of fungal pathogenesis. PLoS Pathog 2012; 8: e1002451; PMID:22319439; http://dx.doi.org/ 10.1371/journal.ppat.1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuchs BB, Mylonakis E. Using non-mammalian hosts to study fungal virulence and host defense. Curr Opin Microbiol 2006; 9: 346-51; PMID:16814595; http://dx.doi.org/ 10.1016/j.mib.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 15. Jacobsen ID. Galleria mellonella as a model host to study virulence of Candida. Virulence 2014; 5: 237-9; PMID:24384470; http://dx.doi.org/ 10.4161/viru.27434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vicentefranqueira R, Moreno MA, Leal F, Calera JA. The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot Cell 2005; 4: 837-48; PMID:15879518; http://dx.doi.org/ 10.1128/EC.4.5.837-848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 2006; 5: 207-11; PMID:16400184; http://dx.doi.org/ 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang CM, Cohen J, Krausz T, Van NS, Holden DW. The alkaline protease of Aspergillus fumigatus is not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect Immun 1993; 61: 1650-6; PMID:8478053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta 1966; 113: 51-6; PMID:5940632; http://dx.doi.org/ 10.1016/S0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 20. Tang CM, Cohen J, Holden DW. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol Microbiol 1992; 6: 1663-71; PMID:1495393; http://dx.doi.org/ 10.1111/j.1365-2958.1992.tb00891.x. [DOI] [PubMed] [Google Scholar]
- 21. Tang X, Bartlett MS, Smith JW, Lee CH. A single-tube nested PCR for Pneumocystis carinii f. sp. hominis. J Clin Microbiol 1997; 35: 1597-9; PMID:9163492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mellado E, Garcia-Effron G, Buitrago MJ, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. Targeted gene disruption of the 14-α sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob Agents Chemother 2005; 49: 2536-8; PMID:15917566; http://dx.doi.org/ 10.1128/AAC.49.6.2536-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amich J, Vicentefranqueira R, Mellado E, Ruiz-Carmuega A, Leal F, Calera JA. The ZrfC alkaline zinc transporter is required for Aspergillus fumigatus virulence and its growth in the presence of the Zn/Mn-chelating protein calprotectin. Cell Microbiol 2014; 16: 548-64; PMID:24245710; http://dx.doi.org/ 10.1111/cmi.12238. [DOI] [PubMed] [Google Scholar]
- 24. Mesa-Arango AC, Forastiero A, Bernal-Martinez L, Cuenca-Estrella M, Mellado E, Zaragoza O. The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med Mycol 2013; 51: 461-72; PMID:23170962; http://dx.doi.org/ 10.3109/13693786.2012.737031. [DOI] [PubMed] [Google Scholar]
- 25. Slater JL, Gregson L, Denning DW, Warn PA. Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med Mycol 2011; 49 Suppl 1: S107-S113; PMID:20950221; http://dx.doi.org/ 10.3109/13693786.2010.523852. [DOI] [PubMed] [Google Scholar]
- 26. Rueda C, Cuenca-Estrella M, Zaragoza O. Paradoxical growth of Candida albicans in the presence of caspofungin is associated with multiple cell wall rearrangements and decreased virulence. Antimicrob Agents Chemother 2014; 58: 1071-83; PMID:24295973; http://dx.doi.org/ 10.1128/AAC.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pelaez Garcia T, Gama B, Garcia-Gil V, Alcazar-Fuoli L, Escribano P, Guinea J, Muñoz P, Mellado E, Bouza E. Incidence of mixed invasive aspergillosis in a general hospital in Madrid: An underestimated entity? In 24th European Congress of Clinical Microbiology and Infectious Diseases, Barcelona, Spain, 10–13 May 2014. Abstract P0005A. [Google Scholar]
- 28. David R.Soll. Mixed Mycotic Infections, In: Brogden KA GJ, ed. Polymicrobial Diseases. Washington: (DC: ): ASM Press, 2002. [PubMed] [Google Scholar]
- 29. Meyers JD. Fungal infections in bone marrow transplant patients. Semin Oncol 1990; 17: 10-3; PMID:2353204. [PubMed] [Google Scholar]
- 30. S J, Vipparti H. Mixed fungal lung infection with Aspergillus fumigatus and Candida albicans in a immunocomprimised patient: case report. J Clin Diagn Res 2014; 8: DD08-DD10; PMID:24959447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orzechowski XM, Pasqualotto AC, Uchoa Sales MP, Bittencourt SC, Peixoto Camargo JJ, Severo LC. Invasive pulmonary aspergillosis due to a mixed infection caused by Aspergillus flavus and Aspergillus fumigatus. Rev Iberoam Micol 2008; 25: 176-8; PMID:18785789; http://dx.doi.org/ 10.1016/S1130-1406(08)70041-X. [DOI] [PubMed] [Google Scholar]
- 32. Pelaez T, Alvarez-Perez S, Mellado E, Serrano D, Valerio M, Blanco JL, Garcia ME, Muñoz P, Cuenca-Estrella M, Bouza E. Invasive aspergillosis caused by cryptic Aspergillus species: a report of two consecutive episodes in a patient with leukaemia. J Med Microbiol 2013; 62: 474-8; PMID:23161769; http://dx.doi.org/ 10.1099/jmm.0.044867-0. [DOI] [PubMed] [Google Scholar]
- 33. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List . Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 2012; 109: 6241-6; PMID:22454494; http://dx.doi.org/ 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balajee SA, Houbraken J, Verweij PE, Hong SB, Yaghuchi T, Varga J, Samson RA. Aspergillus species identification in the clinical setting. Stud Mycol 2007; 59: 39-46; PMID:18490954; http://dx.doi.org/ 10.3114/sim.2007.59.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Serrano R, Gusmao L, Amorim A, Araujo R. Rapid identification of Aspergillus fumigatus within the section Fumigati. BMC Microbiol 2011; 11: 82; PMID:21510879; http://dx.doi.org/ 10.1186/1471-2180-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergin D, Brennan M, Kavanagh K. Fluctuations in haemocyte density and microbial load may be used as indicators of fungal pathogenicity in larvae of Galleria mellonella. Microbes Infect 2003; 5: 1389-95; PMID:14670452; http://dx.doi.org/ 10.1016/j.micinf.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 37. Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Jr., Mylonakis E. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 2009; 53: 2605-9; PMID:19332683; http://dx.doi.org/ 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garcia-Rodas R, Casadevall A, Rodriguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS One 2011; 6: e24485; PMID:21915338; http://dx.doi.org/ 10.1371/journal.pone.0024485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 2005; 73: 3842-50; PMID:15972469; http://dx.doi.org/ 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brennan M, Thomas DY, Whiteway M, Kavanagh K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Microbiol 2002; 34: 153-7; PMID:12381467; http://dx.doi.org/ 10.1111/j.1574-695X.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 41. Gago S, Garcia-Rodas R, Cuesta I, Mellado E, Alastruey-Izquierdo A. Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the non-conventional host Galleria mellonella. Virulence 2014; 5: 278-85; PMID:24193303; http://dx.doi.org/ 10.4161/viru.26973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomaz L, Garcia-Rodas R, Guimaraes AJ, Taborda CP, Zaragoza O, Nosanchuk JD. Galleria mellonella as a model host to study Paracoccidioides lutzii and Histoplasma capsulatum. Virulence 2013; 4: 139-46; PMID:23302787; http://dx.doi.org/ 10.4161/viru.23047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gomez-Lopez A, Forastiero A, Cendejas-Bueno E, Gregson L, Mellado E, Howard SJ, Livermore JL, Hope WW, Cuenca-Estrella M. An invertebrate model to evaluate virulence in Aspergillus fumigatus: the role of azole resistance. Med Mycol 2014; 52: 311-9; PMID:24577012; http://dx.doi.org/ 10.1093/mmy/myt022. [DOI] [PubMed] [Google Scholar]
- 44. Lewis RE, Liao G, Wang W, Prince RA, Kontoyiannis DP. Voriconazole pre-exposure selects for breakthrough mucormycosis in a mixed model of Aspergillus fumigatus-Rhizopus oryzae pulmonary infection. Virulence 2011; 2: 348-55; PMID:21788730; http://dx.doi.org/ 10.4161/viru.2.4.17074. [DOI] [PubMed] [Google Scholar]
- 45. Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol 2005; 3: 547-56; PMID:15953931; http://dx.doi.org/ 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- 46. Wargo AR, Huijben S, de Roode JC, Shepherd J, Read AF. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc Natl Acad Sci U S A 2007; 104: 19914-9; PMID:18056635; http://dx.doi.org/ 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]